Abstract
Background:
Genetic association studies have traditionally focused on associations between individual single nucleotide polymorphisms (SNPs) and disease. Standard analysis ignores interactions between multiple SNPs and environmental exposures explaining a small portion of disease heritability: the often-cited issue of “missing heritability.”
Methods:
We present a novel three-step analytic framework for modeling gene-environment interactions (GEIs) between an angiogenesis candidate-gene pathway and three lifestyle exposures (dietary protein, smoking, and alcohol consumption) on colon cancer risk and survival. Logic regression was used to summarize the gene-pathway effects, and GEIs were modeled using logistic regression and Cox proportional hazards models. We analyzed data from 1541 colon cancer case patients and 1934 control subjects in the Diet, Activity and Lifestyle as a Risk Factor for Colon Cancer Study.
Results:
We identified five statistically significant GEIs for colon cancer risk. For risk interaction, odds ratios (ORINT) and 95% confidence intervals (CIs) were FLT1(rs678714) and BMP4(rs17563) and smoking (ORINT = 1.64, 95% CI = 1.11 to 2.41 and ORINT = 1.60, 95% CI = 1.10 to 2.32, respectively); FLT1(rs2387632 OR rs9513070) and protein intake (ORINT = 1.69, 95% CI = 1.03 to 2.77); KDR(rs6838752) and TLR2(rs3804099) and alcohol (ORINT = 1.53, 95% CI = 1.10 to 2.13 and ORINT = 1.59, 95% CI = 1.05 to 2.38, respectively). Three GEIs between TNF, BMP1, and BMPR2 genes and the three exposures were statistically significant at the 5% level in relation to colon cancer survival but not after multiple-testing adjustment.
Conclusions:
Adopting a comprehensive biologically informed candidate-pathway approach identified GEI effects on colon cancer. Findings may have important implications for public health and personalized medicine targeting prevention and therapeutic strategies. Findings from this study need to be validated in other studies.
Colon cancer is a multifactorial disease with well-documented genetic and nongenetic risk factors (1). Several lines of evidence indicate a prominent role of dietary and lifestyle factors in colon cancer etiology including wide geographical variations in incidence across countries (2) and migrant populations, especially of Asian descent, moving from low-risk to high-risk countries acquiring the host country’s high levels of risk (3,4). Additional evidence comes from Japan, a country with historically one of the lowest incidence rates of colon cancer becoming one of the highest incidence rates in the world over several decades (1,5,6). Although evidence on lifestyle/environmental exposures’ effects on colon cancer survival is limited, some evidence suggests pre- and/or postdiagnostic dietary patterns, smoking, and alcohol consumption may have an impact on colon cancer mortality (7).
Considerable efforts have been made to identify highly and moderately penetrant rare variants in association with colon cancer and, more recently, common low-penetrance risk alleles through genome-wide association studies (GWAS), with only modest success (8). This has reinforced the hypothesis that the large unexplained hereditary component of colon cancer risk referred to as “missing heritability” (by which is meant, more correctly, “missing explanations for familial aggregation”) may be partially explained by epistatic and/or gene-environment interactions (GEIs) (9). The standard “marginal” analysis approach analyzes single nucleotide polymorphisms (SNPs) one at a time. This does not take into account the possibility of interaction between individual genetic variants and will, thus, either fail to observe or detect only weak associations. Such an approach ignores the inherent coordination among genes or their proteins, which is better captured by a pathway structure comprised of multiple genes with related biologic functions jointly contributing to risk in different environmental contexts (10).
It is essential to focus on a biologic pathway relevant to the disease and environmental exposures relevant to the pathway. One of the genetic pathways of particular interest in colon cancer outcomes is the angiogenesis pathway, which mediates the process of growing of blood vessels from existing ones to support tumor growth and progression. A tumor microenvironment with poor oxygen and nutrient supply is an important trigger of the angiogenesis process (11). Expression of several proteins is involved in tumor angiogenesis including the vascular endothelial growth factor (VEGF), which acts as one of the most potent angiogenic factors (12,13), and hypoxia-inducible factor 1 (HIF-1) (14). Activation of HIF-1α signaling pathway under glucose deprivation has recently been shown to lead to colon cancer cells acquiring anti-apoptosis functions (15). We selected three environmental exposures with evidence of associations with colon cancer and relevant to the angiogenesis pathway: dietary protein intake, cigarette smoking, and alcohol consumption (16–18). We hypothesized that these three environmental exposures stimulate tumor angiogenesis under conditions of hypoxia and hypoglycemia (19–21).
In this study we examined GEIs between the angiogenesis-gene pathway and the three environmental factors in association with colon cancer risk and survival. We applied a hypothesis-driven candidate-pathway approach that considered a gene pathway rather than individual SNPs.
Methods
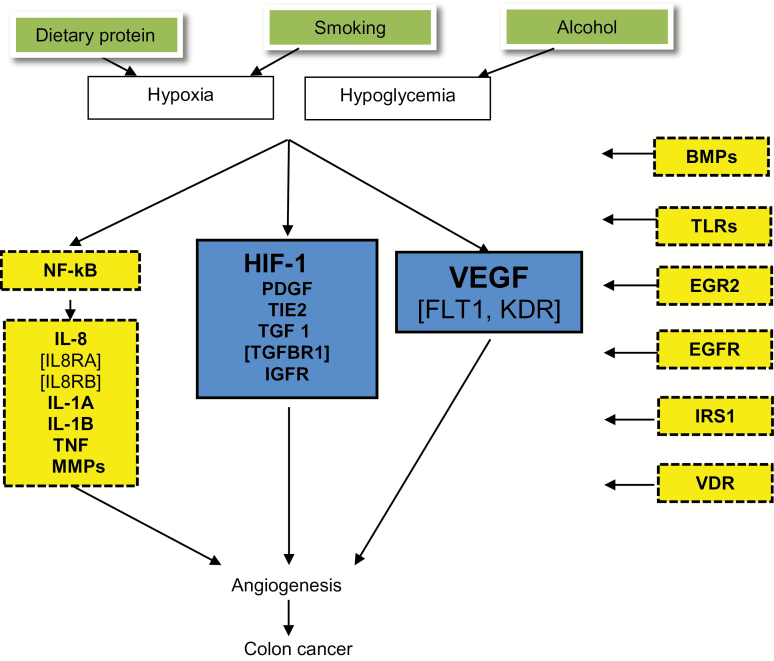
The study used data from the population-based Diet, Activity and Lifestyle as a Risk Factor for Colon Cancer study (22). Subjects completed two in-person interview questionnaires: 1) the health and lifestyle questionnaire (eg, demographic characteristics, medical history, lifestyle habits) and 2) a diet history questionnaire adapted from the validated CARDIA diet history (23,24). Information on stage at diagnosis, months of survival after diagnosis, and vital status was obtained from local tumor registries. Follow-up information was available for at least five years for all subjects from date of diagnosis up to date of last follow-up or death. All participants provided written informed consent. The original and current studies were approved by ethics committees at their respective study locations. The current analysis included data only from participants who agreed to use of their information for further studies (roughly 99%). Genetic markers were genotyped using a multiplexed bead-array assay format based on Golden Gate chemistry (Illumina Human Hap550k, San Diego, CA) and TaqMan assay from Applied Biosystems (Foster City, CA). The candidate angiogenesis gene-pathway was constructed through extracted information from three recognized web-based resources: The BioCarta Pathways: “VEGF, Hypoxia, and Angiogenesis Pathway”; Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway database: “VEGF Signaling Pathway”; Cell Signaling Technologies Pathways: the “Angiogenesis Signaling Pathway” (Supplementary Figures 1–3, available online). We searched online gene databases and PubMed for evidence on the biologic activity and function of the candidate genes and in relation to tumor angiogenesis. Figure 1 shows the working pathway figure used as a guide to the analysis.
Figure 1.
Working figure of the angiogenesis pathway genes. BMP = bone morphogenetic proteins; EGFR = epidermal growth factor receptor; EGR2 = early growth response 2; FLT1 = vascular endothelial growth factor receptor 1; HIF-1 = hypoxia-inducible factor 1; IGFR = insulin-like growth factor receptor; IL-1A = interleukin-1, alpha; IL-1B = interleukin-1, beta; IL-8 = interleukin-8; IL8RA = interleukin-8 receptor, alpha; IL8RB = interleukin-8 receptor, beta; IRS1 = insulin receptor substrate 1; KDR = vascular endothelial growth factor receptor 2; MMP = matrix metallopeptidases; NF-kB = nuclear factor of kappa light polypeptide gene enhancer in B-cells; PDGF = platelet-derived growth factor; TIE2 = tyrosine-protein kinase receptor; TLR = toll-like receptors; TNF = tumor necrosis factor; VEGF = vascular endothelial growth factor.
For environmental exposure we considered long-term prolonged exposure patterns. Smoking status was based on regular cigarette smoking and subjects were categorized as having 20 or more pack-years, less than 20 pack-years, or having never smoked. Long-term exposure to alcohol, based on consumption of any type of alcoholic beverage 10 and 20 years prior to the referent year, was categorized in two levels (none to moderate and high alcohol consumption, cutoff was 20 gms/week for men and 10 gms/week for women). For dietary protein we calculated an animal/vegetable protein intake ratio and used a cutoff corresponding to the median of animal protein proportion of total protein intake equivalent to 60% of total protein intake being animal based. This resulted into two categories (low and high animal/vegetable protein intake ratio).
Statistical Analysis
Three-Step Approach of Candidate Pathway–Based Gene-Environment Interaction Analysis
Our approach to examining gene-environment interactions at the pathway level, adjusting for gene and gene-gene interaction effects, attempted to integrate biologic and logical reasoning using a three-step analysis approach. Analysis was conducted for colon cancer risk and colon cancer survival separately using the same three-step procedures. Each step provided a “product” to be used in the following steps. Step 1: For each gene in the pathway, we summarized SNP-set interactions within the gene. Specifically, we developed gene-specific trees (GSTs) that captured SNP-set interactions in the gene using logic regression (25). Step 2: Epistatic interactions of genes on the pathway (gene-gene interactions) were modeled using the GSTs from Step 1 to develop pathway tree(s). Pathway trees represented interactions of the genes without considering the environmental exposures and were used as adjustment variable(s) in the GEI models of the third step. Step 3: we modeled pathway GEIs between the GSTs and the three environmental exposures. The pathway genes were grouped into nine mutually exclusive subpathways of closely related genes (eg, a gene for a protein and its receptor genes) as illustrated in Figure 1. The GEI models were built using backward selection; GEIs statistically significant at the 5% significance level from the subpathway summary models were jointly tested in the final GEI model for the entire pathway. A P value of less than .05 was considered statistically significant, and all statistical tests were two-sided. A summary of the three steps in the analysis approach is shown in Table 1 for colon cancer risk and survival, respectively (see Supplementary Materials, available online, for details). To further assess the statistical significance of GEIs adjusting for multiple testing we permuted the 0/1 values of each of the GSTs and the three environmental exposures and repeated Step 3 of our analysis to test for GEIs: This is analogous to selecting random genes and exposures with the same respective prevalence levels as the original genes/exposures. Note that the preceding steps (Steps 1 and 2) are independent of the GEI tests in Step 3, and therefore permuting at Step 3 only is appropriate (26).
Table 1.
Summary of the three-step candidate-pathway gene-environment interaction approach*
| Analysis step | Interaction of interest | Variable of interest | Model | Specific procedures | Product |
|---|---|---|---|---|---|
| Colon cancer risk analysis steps | |||||
| Step 1: Summarize gene effects | SNP-set interaction within gene | SNPs on each gene separately | Logic regression with logit link | Cross-validation to determine optimal model size | Gene-specific trees |
| Step 2: Summarize pathway effects | Gene-gene interaction within pathway | All GSTs on the pathway | Logic regression with logit link | Cross-validation to determine optimal model size | Pathway trees |
| Step 3: Test gene- environment interaction | Gene-environment interaction within pathway | a. Subpathway- specific GSTxEb. Full pathway GSTxE | Logistic regression model† | Statistical significance testing | Pathway GEIs |
| Colon cancer survival analysis steps | |||||
| Step 1: Summarize gene effects | SNP-set interaction within gene | SNPs on each gene separately | Logic regression fitting exponential survival models | Cross-validation to determine optimal model size | Gene-specific trees |
| Step 2: Summarize pathway effects | Gene-gene interaction within pathway | All GSTs on the pathway | Logic regression fitting exponential survival models | Cross-validation to determine optimal model size | Pathway trees |
| Step 3: Test gene- environment interaction | Gene-environment interaction within pathway | a. Subpathway- specific GSTxEb. Full pathway GSTxE | Cox proportional hazards model‡ | Statistical significance testing | Pathway GEIs |
* GEI = gene-environment interaction; GST = gene-specific tree; GSTxE = gene-specific tree–environment interaction; SNP = single nucleotide polymorphism.
† Models adjusted for age, sex, race, study center, and pathway tree.
‡ Models adjusted for age, sex, race, study center, and pathway trees, stratified by cancer stage.
Results
The study included data on 1541 colon cancer case patients and 1934 control subjects. Follow-up data and vital status for use in the survival analysis were available for only 1408 of the 1541 case patients. Overall five-year survival probability was 68.3%. Case patients with missing follow-up belonged mainly to the Northern California and Minnesota study centers; patients may have moved out of state or were not able to be tracked by their respective local tumor registry. They, however, did not differ from case patients with follow-up information with regards to baseline variables (age, sex, race, or cancer stage).
The angiogenesis candidate-gene pathway included a total of 257 SNPs in 34 genes (Table 2). Results of the first two steps of the analysis are shown in Supplementary Results (available online). Results of the third step of the analysis modeling interactions between GSTs and the three environmental exposures are displayed in Tables 3 and 4. Overall, the magnitude of the main effects of the statistically significant GSTs increased with increasing levels of the three exposures.
Table 2.
Angiogenesis pathway gene list
| Genes | Name |
|---|---|
| Major drivers of angiogenesis | |
| VEGFA | Vascular endothelial growth factor A |
| FLT1 | Vascular endothelial growth factor receptor 1 |
| KDR | Vascular endothelial growth factor receptor 2 |
| HIF-1α | Hypoxia-inducible factor 1, alpha |
| PDGF | Platelet-derived growth factor |
| TIE2 | Tyrosine-protein kinase receptor |
| TGFβ | Transforming growth factor, beta |
| TGFβR | Transforming growth factor, beta receptor |
| IGF-IR | Insulin-like growth factor-I receptor |
| Interacting inflammatory genes | |
| NFKB1 | Nuclear factor of kappa light polypeptide gene enhancer in B-cells 1 |
| IL8 | Interleukin-8 |
| IL8RA | Interleukin-8 receptor, alpha |
| IL8RB | Interleukin-8 receptor, beta |
| IL1A | Interleukin-1, alpha |
| IL1B | Interleukin-1, beta |
| TNF | Tumor necrosis factor |
| MMPs | Matrix metallopeptidases (MMP1, MMP3, MMP7, MMP9) |
| BMPs | Bone morphogenetic proteins (BMP1, BMP2, BMP4, BMPR1A, BMPR1B, BMPR2, GDF10) |
| TLRs | Toll-like receptors (TLR2, TLR3, TLR4) |
| EGR2 | Early growth response 2 |
| EGFR | Epidermal growth factor receptor |
| IRS1 | Insulin receptor substrate 1 |
| VDR | Vitamin D Receptor |
Table 3.
Effects of gene-environment interactions statistically significant at 5% level between colon cancer gene-specific trees and environmental factors on colon cancer risk
| Gene-specific tree | Gene | Chr | Case patients No. (%) | Control subjects No. (%) | Gene OR (95%CI) | Env factor | Category | No. (%) | Gene OR by env factor (95% CI) | ORINT (95% CI) | P INT* |
|---|---|---|---|---|---|---|---|---|---|---|---|
| rs678714 (TA or AA) | FLT1 | 13q12 | 276 (18.1) | 417 (21.5) | 0.82 (0.69 to 0.97) | Smoking | Non | 1563 (44.6) | 0.72 (0.55 to 0.94) | Ref | |
| <20 PY | 668 (19.1) | 0.58 (0.40 to 0.86) | 0.80 (0.50 to 1.28) | .35 | |||||||
| ≥20 PY | 1272 (36.3) | 1.16 (0.88 to 1.54) | 1.64 (1.11 to 2.41) | .01 | |||||||
| rs2387632 (CC or CT) | FLT1 | 13q12 | 1333 (85.6) | 1738 (88.8) | 0.64 (0.51 to 0.80) | Animal/ vegetable protein ratio | Low | 1010 (28.7) | 0.40 (0.26 to 0.61) | Ref | |
| OR | |||||||||||
| rs9513070 (GG) | 238 (15.3) | 335 (17.11) | High | 2506 (71.3) | 0.76 (0.59 to 0.99) | 1.69 (1.03 to 2.77) | .04 | ||||
| rs6838752 (TT) | KDR | 4q11-q12 | 925 (59.9) | 1098 (56.7) | 1.11 (0.97 to 1.27) | Alcohol | Non/ moderate | 2744 (77.0) | 1.01 (0.86 to 1.18) | Ref | |
| Heavy | 819 (23.0) | 1.53 (1.14 to 2.04) | 1.53 (1.10 to 2.13) | .01 | |||||||
| rs17563 (CC or CT) | BMP4 | 14q22-q23 | 1193 (76.6) | 1568 (80.2) | 0.84 (0.71 to 0.99) | Smoking | Non | 1562 (44.6) | 0.70 (0.54 to 0.89) | Ref | |
| <20 PY | 668 (19.1) | 0.74 (0.51 to 1.09) | 1.05 (0.67 to 1.65) | .82 | |||||||
| ≥20 PY | 1271 (36.3) | 1.12 (0.85 to 1.47) | 1.60 (1.10 to 2.32) | .01 | |||||||
| rs3804099 (TT or TC) | TLR2 | 4q32 | 1257 (80.7) | 1531 (78.2) | 1.20 (1.02 to 1.42) | Alcohol | Non/ moderate | 2714 (77.3) | 1.08 (0.89 to 1.31) | Ref | |
| Heavy | 798 (22.7) | 1.72 (1.21 to 2.45) | 1.59 (1.05 to 2.38) | .03 |
* Two-sided P values were calculated using logistic regression models adjusted for age, sex, race, study center, and pathway tree. Chr = chromosome; CI = confidence interval; env = environmental; INT = interaction; OR = odds ratio; PY = pack-years.
Table 4.
Effects of gene-environment interactions statistically significant at 5% level between colon gene-specific trees and environmental factors on colon cancer survival
| Gene- specific tree | Gene | Chr | Case patients No. (%) | Gene HR (95% CI) | Env Factor | Category | No. (%) | Gene HR by Env Factor (95% CI) | HRINT (95% CI) | P INT* |
|---|---|---|---|---|---|---|---|---|---|---|
| rs1800630 (CA or AA) | TNF | 6p21.3 | 466 (31.5) | 0.93 (0.77 to 1.14) | Animal/ vegetable protein ratio | Low | 399 (27.0) | 0.62 (0.41 to 0.93) | Ref | |
| High | 1079 (73.0) | 1.07 (0.86 to 1.35) | 1.74 (1.09 to 2.76) | .02 | ||||||
| rs13257482 (GG) | BMP1 | 8p21 | 850 (58.3) | 1.45 (1.13 to 1.87) | Smoking | Non | 614 (41.7) | 1.12 (0.76 to 1.65) | Ref | |
| OR | ||||||||||
| rs4075478 (TC or CC) | 902 (61.8) | <20 PY | 279 (19.0) | 1.39 (0.68 to 2.84) | 1.51 (0.68 to 3.36) | .32 | ||||
| ≥20 PY | 578 (39.3) | 2.04 (1.37 to 3.04) | 1.79 (1.03 to 3.10) | .04 | ||||||
| rs12477602 (GG or GA) | BMPR2 | 2q33-q34 | 1389 (98.7) | 1.09 (0.55 to 2.18) | Alcohol | Non/moderate | 1060 (74.9) | 0.60 (0.28 to 1.29) | Ref | |
| Heavy | 356 (25.1) | 2.64 (0.57 to 11.33) | 7.91 (1.57 to 39.74) | .01 |
* Two-sided P values were calculated using Cox proportional hazards models adjusted for age, sex, race, study center, and pathway trees, baseline hazard stratified by cancer stage. Chr = chromosome; CI = confidence interval; env = environmental; HR = hazard ratio; INT = interaction; OR = odds ratio; PY = pack-years.
For colon cancer risk, variants on FLT1 interacted with smoking (interaction odds ratios [ORINT] =1.64 and 95% confidence intervals [CIs] =1.11 to 2.41, P = .01) and animal protein intake (ORINT = 1.69, 95% CI = 1.03 to 2.77, P = .04); and KDR gene with alcohol (ORINT = 1.53, 95% CI = 1.10 to 2.13, P = .01). Interactions between BMP4 gene smoking (ORINT = 1.60, 95% CI = 1.10 to 2.32, P = .01) and TLR2 gene and alcohol (ORINT = 1.59, 95% CI = 1.05 to 2.38, P = .03) were statistically significant. These interactions remained statistically significant after allowing for the multiple-testing adjustment: the results from the 1000 permutation runs showed that only 32 runs resulted in five or more statistically significant GEIs. Because we have found five statistically significant GEIs in our original data analysis, our GEI findings on colon cancer risk are unlikely to be because of chance alone.
Three GEIs of the inflammatory genes had statistically significant nominal P values in association with colon cancer survival, each with one of the three exposures: TNF gene and animal protein intake (P = .02), BMP1 gene and smoking (P = .04), and BMPR2 gene and alcohol (P = .01). The permutation-based multiple testing adjustment for the colon cancer survival, however, showed that in the 1000 permutation runs 204 runs resulted in three or more statistically significant GEIs. Therefore, although being biologically plausible and hypothesized a priori, the three GEIs we identified in the original data analysis in association with colon cancer survival did not remain statistically significant following the multiple-testing adjustment and need to be interpreted with caution.
Discussion
Prior to the advent of GWAS, candidate-gene studies specified genes to be investigated a priori based on their functional significance to the disease. An approach to investigating entire pathways systematically, however, has been lacking, and seldom has the biologic reasoning used for the selection of candidate genes been carried through to the analysis (27). We developed a novel candidate-pathway framework to assess GEIs and illustrated its use for colon cancer risk and survival. Our framework emphasized the biologically informed hypothesis throughout the process, starting from the selection of the candidate genes and the specific lifestyle exposures, and carried the logic to the three steps of the analysis. We started by developing GSTs that captured biologically plausible forms of SNP-set interactions within each gene; hence, our building blocks of gene-gene and gene-environment analysis represented the genes rather than individual SNPs. Our next step provided a summary of the full pathway’s genetic effects. Because the same environmental exposure could be interacting with different genes in the same pathway, whether through similar or different mechanisms (28), we dissected the pathway into mutually exclusive subpathways involving groups of genes sharing similar or closely related functions, guided by the working pathway figure. This grouping allowed for genes in the subpathways to interact with all three exposures and precluded missing potentially important GEIs. Indeed, we observed interactions between the same environmental exposure and different components of the angiogenesis-gene pathway.
Interest in identifying GEIs in colon cancer has been on the rise. In genome-wide settings, GEI has been examined through genome-wide scans and/or a candidate approach focusing on previously identified GWAS loci and known colon cancer risk/survival factors. One GWAS that used three methods to test GEI (a traditional case-control test, a case-only test and a two-step method proposed by Murcray and colleagues that involves a screening test followed by a traditional case-control test of GEI) did not identify any genome-wide statistically significant GEIs, yet using a candidate approach of analyzing previously reported colon cancer GWAS susceptibility loci they identified seven nominally statistically significant GEIs, one of which was between alcohol and an SNP in CHD1 (chromosome 16q22.1) (29). Another study that examined 10 published colon cancer GWAS loci, and 12 environmental risk factors identified a single interaction with vegetable consumption and an SNP on chromosome 8q23.3 (30). A third study was able to identify an interaction with overweight and an SNP on chromosome 11q23.3 but none from the candidate approach (31). In contrast to focusing on previous empirical GWAS findings as candidate genes for GEI testing, our approach to selecting candidate genes and a pathway was based on biologic relevance and a prespecified hypothesis. Despite analyzing genes in only one pathway, we were able to identify a number of statistically significant interactions with all three exposures on both cancer risk and survival with the magnitude of the interaction odds ratio ranging between 1.53 and 1.69 for risk and 1.80 and 7.78 for survival. We believe our findings could be extended by focusing on more colon cancer–related pathways and their relevant environmental exposures.
Although some evidence exists linking VEGFA gene common polymorphisms with colon cancer (32), reported associations with VEGF receptor genes (FLT1 and KDR) have been limited to date (33,34). KDR is considered as the principal mediator of VEGF angiogenesis signaling involving stimulation of endothelial cell migration, proliferation, and survival, whereas FLT1 is believed to have a protective role against cancer through modulating binding of KDR to VEGF. Evidence from our analysis on statistically significant GEIs between KDR and FLT1 and our three exposures highlights the important role of these receptor genes in solid tumors, including colon cancer. Despite evidence of associations between toll-like receptor (TLR) expression levels and colon cancer prognosis, variants on TLR genes have not been identified from GWAS and have not been targeted for investigation in association with colon cancer. We have observed previously from a single-SNP analysis associations between TLR genes and colon cancer risk and survival using pACT to adjust for multiple comparisons (35). In our interaction analysis for colon cancer risk, we identified GEI between TLR2 and alcohol consumption. Biological plausibility of interaction between TLR2 and alcohol is supported by an alcohol induced inflammatory response together with the mediator roles TLRs play in gut inflammation.
BMPs (bone morphogenetic proteins) are multifunctional growth factors, part of the TGFβ superfamily (36) recently shown to have tumor suppressor properties (37,38). Loci on BMP4 gene were previously identified in association with colon cancer from a GWAS meta-analysis (39) and a fine mapping study of the BMP pathway (40). We have previously published associations between SNPs on BMP-signaling pathway genes and colon cancer risk, including BMP2, BMP4, and BMPR2 (41). In this study, we identified GEIs with BMP genes in association with colon cancer risk and suggestive evidence with colon cancer survival. Our study is among the few examining GEI in relation to colon cancer survival. In our results, we detected three nominally statistically significant GEIs associated with colon cancer survival, but these did not survive the stringent permutation-based multiple-testing adjustment.
A GWAS employs an agnostic data-driven approach where prior knowledge of SNP function is not required and most GWASs have not investigated GEI, primarily because of lack of data on environmental exposures (42). On the other hand, a candidate pathway in a candidate-gene study is based on hypotheses derived from existing knowledge of the pathway as well as genes and SNPs defining that pathway. Approaches based on informed candidate-gene selection may be more suited to examining GEI effects than GWAS loci. The low-penetrance GWAS loci are harder to identify, have smaller effect sizes, and the likelihood of them being functional variants is low: the discovered SNPs are markers of an underlying haplotype that includes the functional variants (42). In contrast, our candidate-pathway approach has several advantages over an empirical data-driven approach: 1) candidate genes and exposures were selected based on biologic relevance; 2) it allows for the interaction of multiple SNPs within each gene, thus potentially capturing the full gene effect; and 3) a multiple testing adjustment for testing many nonhypothesized associations in GWAS may not be required because the associations were specified a priori (40). Furthermore, GWAS analyses that focus only on SNPs with statistically significant marginal effects will miss interactions with variants with weak or no marginal effects.
A few limitations of our study are related to the design of the case-control study, which suffers from inherent forms of bias including recall bias. In this study, this was minimized by: using a rapid-reporting system to identify case patients, conducting the majority of interviews within four months of diagnosis, and limiting the referent period of the study questionnaires to two to three years prior to diagnosis. We obtained long-term alcohol consumption and cigarette smoking history and collected extensive diet history to capture more detail compared with self-administered questionnaires. We considered all colon cancer case patients and did not stratify by distal and proximal site. Because of the large size of the GEI models, we limited the adjustment variables to select colon cancer risk and survival predictors. Despite the advantage of our approach that considers SNP-set interactions at the gene level and gene-gene interactions at the pathway level, interactions between the candidate angiogenesis pathway and other candidate pathways were not considered. We did, however, include both angiogenesis and angiogenesis-related inflammatory genes in our candidate-gene pathway.
The GEIs detected from our analysis in association with colon cancer outcomes emphasize the need to employ an approach based on hypotheses when examining GEIs. The low-penetrance markers and the environmental exposures are common in the population, and the magnitude of the interaction is often larger than their individual effects, thus potentially explaining a large proportion of the risk variation in the population (43). Furthermore, knowledge of personalized risk profiles based on individuals’ genetic and environmental exposure patterns can have a larger impact on behavioral modification. Despite difficulties in making lifestyle and health behavior changes beneficial for colon cancer prevention, evidence suggests that colon cancer–related morbidity and mortality can be reduced through modifications of the lifestyle risk factors and/or adhering to screening guidelines (44). An effective discovery approach that identifies relationships between genes and lifestyle/environmental factors may present important public health and personalized medicine applications. The findings could guide the development and testing of innovative interventions for the primary prevention and screening of colon cancer. Specifically, genotyping and profiling an individual’s risk based on the gene-environment interactions could inform the design of regular self-monitoring tools of modifiable lifestyle/health-behavioral risk factors specific to the individual.
It is important to note that our analysis is exploratory in nature and findings, despite being biologically plausible, need to be validated using other studies. This would require the initial step of harmonizing environmental data across datasets. We encourage other researchers in the field to utilize our approach to attempt replication of the findings. We also plan to pursue this approach ourselves.
Funding
This work was supported by an operating grant from the Canadian Institutes of Health Research to YY. The study was funded by National Cancer Institute grants CA48998 and CA615757. This research also was supported by the Utah Cancer Registry, which is funded by Contract #N01-PC-67000 from the National Cancer Institute, with additional support from the State of Utah Department of Health, the Northern California Cancer Registry, and the Sacramento Tumor Registry. YY is also supported by the Canada Research Chair Program, and YY and NS are supported by Alberta Innovates-Health Solutions. NS is supported by Alberta Cancer Foundation and the University of Alberta Dissertation Fellowship.
Supplementary Material
The study funders had no role in design of the study, the collection, analysis, or interpretation of the data, the writing of the manuscript, nor the decision to submit the manuscript for publication.
The contents of this manuscript are solely the responsibility of the authors and do not necessarily represent the official view of the National Cancer Institute.
We would like to acknowledge the contributions of Sandra Edwards, Roger Edwards, Leslie Palmer, Donna Schaffer, Dr. Kristin Anderson, and Judy Morse for data management and collection.
References
- 1. Potter JD. Colorectal cancer: molecules and populations. J Natl Cancer Inst. 1999;91(11):916–932. [DOI] [PubMed] [Google Scholar]
- 2. Boyle P, Levin B. World cancer report 2008. Lyon: International Agency for Research on Cancer; 2008. http://www.iarc.fr/en/publications/pdfs-online/wcr/2008/wcr_2008.pdf. (Accessed May 28, 2014). [Google Scholar]
- 3. Marchand LL. Combined influence of genetic and dietary factors on colorectal cancer incidence in Japanese Americans. J Natl Cancer Inst Monogr. 1999;(26):101–105. [DOI] [PubMed]
- 4. Flood DM, Weiss NS, Cook LS, et al. Colorectal cancer incidence in Asian migrants to the United States and their descendants. Cancer Causes Control. 2000;11(5):403–411. [DOI] [PubMed] [Google Scholar]
- 5. Oba S, Shimizu N, Nagata C, et al. The relationship between the consumption of meat, fat, and coffee and the risk of colon cancer: a prospective study in Japan. Cancer Lett. 2006;244(2):260–267. [DOI] [PubMed] [Google Scholar]
- 6. Takachi R, Tsubono Y, Baba K, et al. Red meat intake may increase the risk of colon cancer in Japanese, a population with relatively low red meat consumption. Asia Pac J Clin Nutr. 2011;20(4):603–612. [PubMed] [Google Scholar]
- 7. Pelser C, Arem H, Pfeiffer RM, et al. Prediagnostic lifestyle factors and survival after colon and rectal cancer diagnosis in the National Institutes of Health (NIH)-AARP Diet and Health Study. Cancer. 2014;120(10):1540–1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Whiffin N, Houlston RS. Architecture of inherited susceptibility to colorectal cancer: a voyage of discovery. Genes (Basel). 2014;5(2):270–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Manolio TA, Collins FS, Cox NJ, et al. Finding the missing heritability of complex diseases. Nature. 2009;461(7265):747–753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Kraft P, Raychaudhuri S. Complex diseases, complex genes: keeping pathways on the right track. Epidemiology. 2009;20(4):508–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Folkman J, Shing Y. Angiogenesis. J Biol Chem. 1992;267(16):10931–10934. [PubMed] [Google Scholar]
- 12. Ferrara N. Molecular and biological properties of vascular endothelial growth factor. J Mol Med (Berl). 1999;77(7):527–543. [DOI] [PubMed] [Google Scholar]
- 13. Lohela M, Bry M, Tammela T, et al. VEGFs and receptors involved in angiogenesis versus lymphangiogenesis. Curr Opin Cell Biol. 2009;21(2):154–165. [DOI] [PubMed] [Google Scholar]
- 14. Semenza GL. Defining the role of hypoxia-inducible factor 1 in cancer biology and therapeutics. Oncogene. 2010;29(5):625–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Nishimoto A, Kugimiya N, Hosoyama T, et al. HIF-1alpha activation under glucose deprivation plays a central role in the acquisition of anti-apoptosis in human colon cancer cells. Int J Oncol. 2014;44(6):2077–2084. [DOI] [PubMed] [Google Scholar]
- 16. Gonzalez CA, Riboli E. Diet and cancer prevention: Contributions from the European Prospective Investigation into Cancer and Nutrition (EPIC) study. Eur J Cancer. 2010;46(14):2555–2562. [DOI] [PubMed] [Google Scholar]
- 17. Poynter JN, Haile RW, Siegmund KD, et al. Associations between smoking, alcohol consumption, and colorectal cancer, overall and by tumor microsatellite instability status. Cancer Epidemiol Biomarkers Prev. 2009;18(10):2745–2750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cheng J, Chen Y, Wang X, et al. Meta-analysis of prospective cohort studies of cigarette smoking and the incidence of colon and rectal cancers. Eur J Cancer Prev. 2015;24(1):6–15. [DOI] [PubMed] [Google Scholar]
- 19. Vigne P, Frelin C. A low protein diet increases the hypoxic tolerance in Drosophila. PLoS One. 2006;1:e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Wong HP, Yu L, Lam EK, et al. Nicotine promotes colon tumor growth and angiogenesis through beta-adrenergic activation. Toxicol Sci. 2007;97(2):279–287. [DOI] [PubMed] [Google Scholar]
- 21. Gu JW, Elam J, Sartin A, et al. Moderate levels of ethanol induce expression of vascular endothelial growth factor and stimulate angiogenesis. Am J Physiol Regul Integr Comp Physiol. 2001;281(1):R365-R372. [DOI] [PubMed] [Google Scholar]
- 22. Slattery ML, Potter J, Caan B, et al. Energy balance and colon cancer--beyond physical activity. Cancer Res. 1997;57(1):75–80. [PubMed] [Google Scholar]
- 23. Slattery ML, Caan BJ, Duncan D, et al. A computerized diet history questionnaire for epidemiologic studies. J Am Diet Assoc. 1994;94(7):761–766. [DOI] [PubMed] [Google Scholar]
- 24. Liu K, Slattery M, Jacobs D, Jr, et al. A study of the reliability and comparative validity of the cardia dietary history. Ethn Dis. 1994;4(1):15–27. [PubMed] [Google Scholar]
- 25. Ruczinski I, Kooperberg C, LeBlanc M. Logic regression. J. Comp Graph Stat. 2003;12. [Google Scholar]
- 26. Dai JY, Kooperberg C, Leblanc M, et al. Two-stage testing procedures with independent filtering for genome-wide gene-environment interaction. Biometrika. 2012;99(4):929–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Thomas DC, Conti DV, Baurley J, et al. Use of pathway information in molecular epidemiology. Hum Genomics. 2009;4(1):21–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10(6):392–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Figueiredo JC, Lewinger JP, Song C, et al. Genotype-environment interactions in microsatellite stable/microsatellite instability-low colorectal cancer: results from a genome-wide association study. Cancer Epidemiol Biomarkers Prev. 2011;20(5):758–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hutter CM, Chang-Claude J, Slattery ML, et al. Characterization of gene-environment interactions for colorectal cancer susceptibility loci. Cancer Res. 2012;72(8):2036–2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Siegert S, Hampe J, Schafmayer C, et al. Genome-wide investigation of gene-environment interactions in colorectal cancer. Hum Genet. 2013;132(2):219–231. [DOI] [PubMed] [Google Scholar]
- 32. Zhao Z, Ba C, Wang W, et al. Vascular endothelial growth factor (VEGF) gene polymorphisms and colorectal cancer: a meta-analysis of epidemiologic studies. Genet Test Mol Biomarkers. 2012;16(12):1390–1394. [DOI] [PubMed] [Google Scholar]
- 33. Jang MJ, Jeon YJ, Kim JW, et al. Association of VEGF and KDR single nucleotide polymorphisms with colorectal cancer susceptibility in Koreans. Mol Carcinog. 2013;52(Suppl 1):E60-E69. [DOI] [PubMed] [Google Scholar]
- 34. Slattery ML, Lundgreen A, Wolff RK. VEGFA, FLT1, KDR and colorectal cancer: assessment of disease risk, tumor molecular phenotype, and survival. Mol Carcinog. 2014;53(Suppl 1):E140-E150. [DOI] [PubMed] [Google Scholar]
- 35. Slattery ML, Herrick JS, Bondurant KL, et al. Toll-like receptor genes and their association with colon and rectal cancer development and prognosis. Int J Cancer. 2012;130(12):2974–2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen D, Zhao M, Mundy GR. Bone morphogenetic proteins. Growth Factors. 2004;22(4):233–241. [DOI] [PubMed] [Google Scholar]
- 37. Beck SE, Jung BH, Fiorino A, et al. Bone morphogenetic protein signaling and growth suppression in colon cancer. Am J Physiol Gastrointest Liver Physiol. 2006;291(1):G135-G145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishanian TG, Kim JS, Foxworth A, et al. Suppression of tumorigenesis and activation of Wnt signaling by bone morphogenetic protein 4 in human cancer cells. Cancer Biol Ther. 2004;3(7):667–675. [DOI] [PubMed] [Google Scholar]
- 39. Study C, Houlston RS, Webb E, et al. Meta-analysis of genome-wide association data identifies four new susceptibility loci for colorectal cancer. Nat Genet. 2008;40(12):1426–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tomlinson IP, Carvajal-Carmona LG, Dobbins SE, et al. Multiple common susceptibility variants near BMP pathway loci GREM1, BMP4, and BMP2 explain part of the missing heritability of colorectal cancer. PLoS Genet. 2011;7(6):e1002105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Slattery ML, Lundgreen A, Herrick JS, et al. Genetic variation in bone morphogenetic protein and colon and rectal cancer. Int J Cancer. 2012;130(3):653–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Stranger BE, Stahl EA, Raj T. Progress and promise of genome-wide association studies for human complex trait genetics. Genetics. 2011;187(2):367–383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Le Marchand L, Wilkens LR. Design considerations for genomic association studies: importance of gene-environment interactions. Cancer Epidemiol Biomarkers Prev. 2008;17(2):263–267. [DOI] [PubMed] [Google Scholar]
- 44. Binefa G, Rodriguez-Moranta F, Teule A, et al. Colorectal cancer: from prevention to personalized medicine. World J Gastroenterol. 2014;20(22):6786–6808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.