Abstract
Background:
Our present study of the microRNA (miRNA) expression signature in castration-resistant prostate cancer (CRPC) revealed that the clustered miRNAs microRNA-221 (miR-221) and microRNA-222 (miR-222) are significantly downregulated in cancer tissues. The aim of this study was to investigate the functional roles of miR-221 and miR-222 in prostate cancer (PCa) cells.
Methods:
A CRPC miRNA signature was constructed by PCR-based array methods. Functional studies of differentially expressed miRNAs were analysed using PCa cells. The association between miRNA expression and overall survival was estimated by the Kaplan–Meier method. In silico database and genome-wide gene expression analyses were performed to identify molecular targets regulated by the miR-221/222 cluster.
Results:
miR-221 and miR-222 were significantly downregulated in PCa and CRPC specimens. Kaplan–Meier survival curves showed that low expression of miR-222 predicted a short duration of progression to CRPC. Restoration of miR-221 or miR-222 in cancer cells revealed that both miRNAs significantly inhibited cancer cell migration and invasion. Ecm29 was directly regulated by the miR-221/222 cluster in PCa cells.
Conclusions:
Loss of the tumour-suppressive miR-221/222 cluster enhanced migration and invasion in PCa cells. Our data describing targets regulated by the tumour-suppressive miR-221/222 cluster provide insights into the mechanisms of PCa and CRPC progression.
Keywords: microRNA, prostate cancer, castration-resistant prostate cancer, expression signature, miR-221, miR-222, tumour suppressor, Ecm29
Prostate cancer (PCa) is the most frequently diagnosed cancer and the second leading cause of cancer-related death among men in developed countries (Siegel et al, 2013). Androgen signalling through the androgen receptor (AR) is an important oncogenic pathway for PCa progression. Most patients initially respond to androgen-deprivation therapy (ADT), but eventually acquire resistance and progress to castration-resistant prostate cancer (CRPC). One of the most important features of PCa is the heterogeneity of PCa cells, which results in diverse outcomes in patients with PCa, even for those in the same risk group (Scher et al, 1999; Colloca, 2012). Prostate-specific antigen (PSA) does not accurately reflect the progression of CRPC. Thus, effective biomarkers for follow-up and detection of CRPC are needed. Moreover, most clinical trials for advanced PCa have shown limited benefits, eventually resulting in disease progression and metastasis (Chi et al, 2009). Therefore, understanding the molecular mechanisms of the androgen-independent and metastatic signalling pathways underlying PCa using current genomic approaches would help to improve therapies for and prevention of the disease.
MicroRNAs (miRNAs) are endogenous small noncoding RNA molecules (18–25 bases in length) that regulate protein-coding gene expression by repressing translation or cleaving RNA transcripts in a sequence-specific manner (Bartel, 2004). Aberrantly expressed miRNAs contribute to the initiation, development, and metastasis of several types of cancers, including PCa (Esquela-Kerscher and Slack, 2006). MicroRNAs are unique in their ability to regulate multiple protein-coding genes, and normal regulatory mechanisms can be disrupted by the aberrant expression of tumour-suppressive or oncogenic miRNAs in cancer cells. Therefore, identification of aberrantly expressed miRNAs is an important first step towards elucidating miRNA-mediated oncogenic targets.
In this study, we constructed the miRNA expression signature of CRPC using clinical specimens because the development of therapeutic strategies is a central theme in the advancement of PCa treatments. Using CRPC expression signature data, we investigated the specific roles of miRNAs in PCa and CRPC oncogenesis by examining differentially expressed miRNAs. Data from our previous PCa signature (Fuse et al, 2012) and present CRPC signature showed that the clustered miRNAs miR-221 and miR-222 were significantly downregulated in PCa and CRPC tissues, suggesting that these miRNAs may act as tumour suppressors. In contrast, previous studies have indicated that miR-221 and miR-222 function as oncogenes in PCa cells (Sun et al, 2014; Yang et al, 2014). Moreover, overexpression of these clustered miRNAs has been observed in breast cancer (Shah and Calin, 2011). Therefore, the functional roles of miR-221 and miR-222 in human cancers, including PCa, are still controversial.
The aim of this study was to investigate the functional significance of miR-221 and miR-222 and to identify molecular targets and pathways regulated by this miRNA cluster in PCa cells. We expect that this analysis will provide important insights into the potential molecular mechanisms of PCa oncogenesis and metastasis and will facilitate the development of novel diagnostic and therapeutic strategies for the treatment of PCa.
Materials and Methods
Patients and clinical prostate specimens
Clinical prostate specimens were obtained from patients admitted to the Teikyo University Chiba Medical Centre Hospital from 2008 to 2013. Ninety-two patients with elevated PSA levels underwent transrectal prostate needle biopsy, and three patients who died of CRPC underwent autopsies. Prostate cancer tissues (n=54), noncancerous prostate tissues (non-PCa, n=38), and CRPC tissues (n=8) were used in this study. The patients' backgrounds and clinicopathological characteristics are summarised in Supplementary Tables 1 and 2. Supplementary Figure 1 describes the clinical courses of the patients with CRPC. Samples were staged according to the UICC TNM classification (Edge et al, 2010). Written consent for tissue donation for research purposes was obtained from each patient before tissue collection. The protocol was approved by the Institutional Review Board of Teikyo University.
For pathological verification of tissue composition, a pair of needle biopsy specimens was collected from the same region as from patients in this study, and one was subjected to pathological verification; no cancerous tissue was found in non-PCa specimens. Castration-resistant prostate cancer was defined according to guidelines published by the European Association of Urology (Heidenreich et al, 2014a).
Construction of the miRNA expression signature of CRPC
miRNA expression patterns were evaluated using the TaqMan LDA Human microRNA Panel v2.0 (Applied Biosystems, Foster City, CA, USA). The assay procedure was performed as described previously (Nohata et al, 2011; Fuse et al, 2012; Fukumoto et al, 2014). A description of the real-time PCR assay and the list of human miRNAs included in the panel can be found on the manufacturer's website (http://www.appliedbiosystems.com). Analysis of relative miRNA expression data was performed using GeneSpring GX software version 7.3.1 (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's instructions. A cutoff P-value of less than 0.05 was used to narrow down the candidates after global normalisation of the raw data. After global normalisation, additional normalisation was carried out with U6.
Cell culture
Human PCa cells (RWPE-1, LNCaP, C4-2, PC3, and DU145 cells) were obtained from the American Type Culture Collection (Manassas, VA, USA). LNCaP, C4-2, PC3, and DU145 cells were maintained in RPMI-1640 medium supplemented with 10% fetal bovine serum in a humidified atmosphere of 5% CO2 and 95% air at 37 °C. RWPE-1 cells were cultured in keratinocyte serum-free medium containing 5 ng/ml epidermal growth factor and 50 μg/ml bovine pituitary extract.
RNA isolation
Total RNA was isolated using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's protocol. The quality of RNA was confirmed using an Agilent 2100 Bioanalyzer (Agilent Technologies) as described previously (Kinoshita et al, 2013; Nohata et al, 2013; Goto et al, 2014a).
Quantitative real-time reverse transcription–PCR (RT–qPCR)
The procedure for PCR quantification was carried out as previously described (Kinoshita et al, 2013; Nohata et al, 2013; Goto et al, 2014a). The expression levels of miR-221 (Assay ID: 000524) and miR-222 (Assay ID: 002276) were analysed by TaqMan RT–qPCR (TaqMan MicroRNA Assay; Applied Biosystems) and normalised to RNU48 (Assay ID: 001006). TaqMan probes and primers for Ecm29/KIAA0368 (P/N: Hs00322433_m1) and GUSB (P/N: Hs00939627_m1) as an internal control were obtained from Applied Biosystems (Assay-On-Demand Gene Expression Products).
Transfection with miRNA mimic and small interfering RNA (siRNA)
The following miRNA mimic species were used in this study: Ambion Pre-miR miRNA precursor for hsa-miR-221-3p (A) (product ID: PM10337; Austin, TX, USA) and miRIDIAN miRNA Human hsa-miR-221-3p-Mimic for hsa-miR-221(T) (product ID: C-300578-05; Thermo Fisher Scientific, Waltham, MA, USA); Ambion Pre-miR miRNA precursor for hsa-miR-222-3p (A) (product ID: PM11376) and miRIDIAN miRNA Human hsa-miR-222-3p-Mimic for hsa-miR-222(T) (product ID: C-300579-07; Thermo Fisher Scientific). The following siRNAs were used: Stealth Select RNAi siRNA; si-KIAA0368 (Ecm29) (cat no. HSS146372; Invitrogen); and negative control miRNA/siRNA (P/N: AM17111, Applied Biosystems). RNAs were incubated with OPTI-MEM (Invitrogen) and Lipofectamine RNAiMAX reagent (Invitrogen). The transfection procedures and transfection efficiencies of miRNA in PC3 and DU145 cells were reported previously (Kinoshita et al, 2013; Nohata et al, 2013; Goto et al, 2014a).
Cell proliferation, migration, and invasion assays
To investigate the functional significance of the miR-221/222 cluster or gene silencing by siRNA knockdown, we performed cell proliferation, migration, and invasion assays using PC3 and DU145 cells, as previously described (Kinoshita et al, 2013; Nohata et al, 2013; Goto et al, 2014a).
Genome-wide gene expression and in silico analyses for the identification of genes regulated by the miR-221/222 cluster
We performed a combination of in silico and genome-wide gene expression analyses. First, genes regulated by the miR-221/222 cluster were listed using the TargetScan database, as described previously (Kinoshita et al, 2013; Nohata et al, 2013; Goto et al, 2014a). Next, to identify upregulated genes in PCa, we analysed a publicly available gene expression data set in GEO (accession number: GSE29079). Finally, we performed genome-wide gene expression analysis using miR-221- and miR-222-transfected PC3 and DU145 cells. A SurePrint G3 Human GE 60K Microarray (Agilent Technologies) was used for expression profiling of each miRNA transfectant in comparison with negative control miRNA transfectants. Finally, downregulated mRNAs containing miR-221/222 target sites were listed as putative target genes of these miRNAs.
Western blotting
Immunoblotting was performed with rabbit anti-KIAA0368 (Ecm29) antibodies (1 : 1000, PA5-29467; Pierce Antibodies, Thermo Scientific, Fremont, CA, USA), and anti-GAPDH antibodies (1 : 1000, ab8245; Abcam, Cambridge, UK) were used as an internal loading control. Membranes were washed and incubated with anti-rabbit IgG horseradish peroxidase-linked antibodies (7074; Cell Signaling Technology, Danvers, MA, USA). Complexes were visualised with Clarity Western ECL Substrate (Bio-Rad, Hercules, CA, USA). The experimental procedures were performed as described in our previous studies (Kinoshita et al, 2013; Nohata et al, 2013; Goto et al, 2014a).
Plasmid construction and dual-luciferase reporter assay
Partial wild-type sequences of the KIAA0368 (Ecm29) 3′ untranslated region (UTR) or those with deleted miR-221/222 target sites (position 97–103 of the KIAA0368 3′ UTR) were inserted between the XhoI–PmeI restriction sites in the 3′ UTR of the hRluc gene in the psiCHECK-2 vector (C8021; Promega, Madison, WI, USA). The protocol for vector construction was described previously (Kinoshita et al, 2013; Nohata et al, 2013; Goto et al, 2014a).
Immunohistochemistry
A total of eight CRPC specimens were used (Supplementary Table 2). Tissue specimens were immunostained following the manufacturer's protocol with the Ultra-Vision Detection System (Thermo Scientific). Primary rabbit polyclonal antibodies against AR (1 : 50, ab9474; Abcam), antibodies against PSA (1 : 500, HPA000764; Sigma-Aldrich), and antibodies against Ecm29 (1 : 500, PA5-29467; Pierce Antibodies, Thermo Scientific) were used for immunochemistry. The slides were treated with biotinylated goat antibodies (Histofine SAB-PO kit; Nichirei, Tokyo, Japan).
Statistical analysis
The relationships between two groups and the numerical values obtained by RT–qPCR were analysed using Mann–Whitney U-tests. Spearman's rank test was used to evaluate the correlations between the expression of miR-221 and miR-222. The relationships among more than three variables and numerical values were analysed using the Bonferroni-adjusted Mann–Whitney U-test. Survival analysis was evaluated by the Kaplan–Meier method and log-rank test. A multivariate Cox proportional hazards model was used to establish independent factors for survival. The Kaplan–Meier method and log-rank test were performed using Stat Mate software (version 4.01, ATMS Co., Tokyo, Japan); all other analyses were performed using Expert StatView (version 5, SAS Institute Inc., Cary, NC, USA).
Results
Identification of downregulated miRNAs in CRPC specimens by miRNA expression signatures
We evaluated the expression levels of mature miRNAs in CRPC specimens by PCR-based array analysis. Of the eight CRPC specimens used in this study, we used six specimens (Nos. 3–8, Supplementary Table 2) for array analysis. Expression signatures revealed that 42 miRNAs were significantly downregulated in CRPC specimens compared with our previous miRNA signature of normal prostate tissue (Table 1). In addition, we compared miRNA expressions in CRPC with our previous miRNA signature of hormone naïve PCa specimens (Supplementary Table 3). We focused on downregulated miRNAs in CRPC compared with normal prostate tissue in this study. Three miRNA clusters, that is, miR-221/222, miR-23b/27b/24, and miR-106a/20b, were found in this signature. We focused on the miR-221/222 cluster for further studies.
Table 1. Downregulated miRNAs in CRPC.
| miRNA | Log2 ratio (CRPC/non-PCa) | non-PCa | CRPC | P-value |
|---|---|---|---|---|
| hsa-miR-205 | −11.34 | 0.00318 | 1.228E-06 | 0.0401 |
| hsa-miR-378 | −11.15 | 0.00336 | 1.479E-06 | 0.0485 |
| hsa-miR-222 | −8.40 | 0.11068 | 0.0003286 | 0.0161 |
| hsa-miR-143 | −8.34 | 0.05562 | 0.0001715 | 0.0465 |
| hsa-miR-133a | −7.70 | 0.01215 | 5.847E-05 | 0.0203 |
| hsa-miR-23b | −6.50 | 0.00103 | 1.138E-05 | 0.0276 |
| hsa-miR-345 | −6.45 | 0.00266 | 3.035E-05 | 0.0264 |
| hsa-miR-150 | −6.25 | 0.00945 | 0.0001244 | 0.0084 |
| hsa-miR-139-5p | −6.22 | 0.00299 | 4.018E-05 | 0.0075 |
| hsa-miR-221 | −6.21 | 0.00208 | 2.822E-05 | 0.0355 |
| hsa-miR-484 | −5.68 | 0.01537 | 0.0003003 | 0.0144 |
| hsa-miR-29c | −5.55 | 0.01459 | 0.0003123 | 0.0447 |
| hsa-miR-28-3p | −5.44 | 0.00672 | 0.0001547 | 0.0262 |
| hsa-miR-27b | −5.28 | 0.00134 | 3.444E-05 | 0.0362 |
| hsa-miR-320 | −5.26 | 0.01826 | 0.0004779 | 0.0382 |
| hsa-miR-30a* | −5.21 | 0.00730 | 0.000197 | 0.0048 |
| hsa-miR-532-3p | −5.21 | 0.00113 | 3.05E-05 | 0.0415 |
| hsa-miR-574-3p | −5.15 | 0.02286 | 0.0006437 | 0.0048 |
| hsa-miR-30e* | −5.13 | 0.00553 | 0.0001576 | 0.0105 |
| hsa-miR-24 | −5.06 | 0.14362 | 0.0043124 | 0.0215 |
| hsa-miR-196b | −5.01 | 0.00183 | 5.666E-05 | 0.0023 |
| hsa-miR-199a-3p | −4.50 | 0.01030 | 0.0004562 | 0.0434 |
| hsa-miR-218 | −4.43 | 0.00137 | 6.354E-05 | 0.0233 |
| hsa-miR-27a | −4.31 | 0.00235 | 0.0001182 | 0.0426 |
| hsa-miR-152 | −4.25 | 0.00175 | 9.14E-05 | 0.0341 |
| hsa-miR-660 | −3.96 | 0.00165 | 0.0001059 | 0.0396 |
| hsa-miR-126* | −3.61 | 0.00169 | 0.0001386 | 0.0191 |
| hsa-miR-146a | −3.55 | 0.00718 | 0.0006132 | 0.0099 |
| hsa-miR-20b | −3.52 | 0.00430 | 0.0003738 | 0.0333 |
| hsa-miR-193b | −3.38 | 0.00966 | 0.0009259 | 0.0213 |
| hsa-miR-106a | −2.93 | 0.03526 | 0.00462 | 0.0359 |
| hsa-miR-125a-5p | −2.86 | 0.00179 | 0.0002467 | 0.0062 |
| hsa-miR-149 | −2.81 | 0.00538 | 0.000767 | 0.0211 |
| hsa-miR-17 | −2.77 | 0.02746 | 0.0040348 | 0.0460 |
| hsa-miR-223 | −2.66 | 0.01701 | 0.0026952 | 0.0189 |
| hsa-miR-454 | −2.46 | 0.00136 | 0.0002463 | 0.0077 |
| hsa-miR-186 | −2.46 | 0.00464 | 0.0008433 | 0.0093 |
| hsa-miR-151-3p | −2.34 | 0.00238 | 0.0004702 | 0.0353 |
| hsa-miR-16 | −2.07 | 0.05451 | 0.0129807 | 0.0193 |
| hsa-miR-342-3p | −2.04 | 0.00623 | 0.0015125 | 0.0207 |
| hsa-miR-200c | −1.58 | 0.03969 | 0.0132931 | 0.0324 |
| hsa-miR-126 | −1.22 | 0.04305 | 0.0184953 | 0.0434 |
Abbreviations: CRPC=castration-resistant prostate cancer; miRNA=microRNA; PCa=prostate cancer.
Expression levels of miR-221 and miR-222 in PCa specimens and cell lines
The chromosomal location of miR-221/222 in the human genome is shown in Supplementary Figure 2. Database analysis demonstrated that these miRNAs were closely located on human chromosome Xp11.3 within 800 base pairs.
We evaluated the expression levels of the clustered miRNAs (miR-221/222) in non-PCa (n=38), PCa (n=54), and CRPC (n=8) tissues. In patients from whom non-PCa tissues were collected, the median PSA level was 7.315 ng ml−1 (range=4.3–35.5 ng ml−1). In contrast, in patients from whom PCa tissues were collected, PSA levels were quite high, with a median of 212 ng ml−1 (range=3.45–3750 ng ml−1). Forty-four patients with PCa had progressive disease classified as stage IV according to TNM classification (Supplementary Table 1). The results of H&E staining and immunohistochemical staining of AR in CRPC specimens are shown in Supplementary Figure 3.
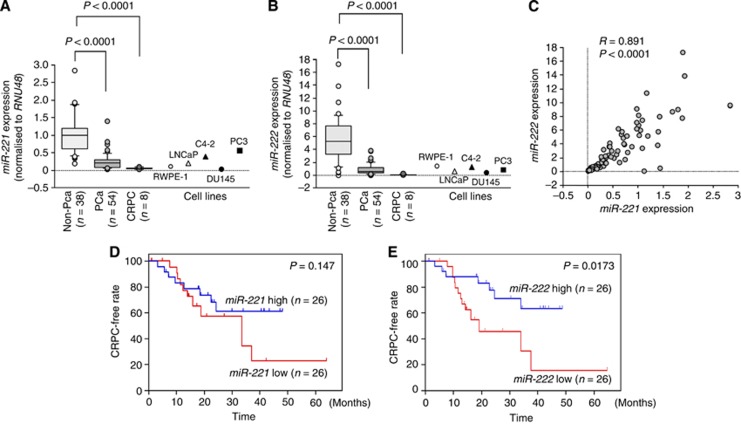
The expression levels of miR-221 and miR-222 were significantly downregulated (P<0.0001) in cancer tissues compared with noncancerous tissues (Figures 1A and B). Furthermore, the levels of these miRNAs were significantly downregulated (P<0.0001) in CRPC tissues compared with noncancerous tissues (Figures 1A and B). Spearman's rank test showed positive correlations between the expression of miR-221 and miR-222 (R=0.891 and P<0.0001; Figure 1C).
Figure 1.
Expression levels of miR-221/222 in prostate specimens and associations between miR-221/222 expression and CRPC-free interval. Expression levels of miR-221 (A) and miR-222 (B) in clinical prostate specimens. RNU48 was used for normalisation. (C) Correlations among the relative expression levels of miR-221/miR-222. Kaplan–Meier survival curves for CRPC progression-free survival based on (D) miR-221 and (E) miR-222 expression in PCa patients. P-values were calculated using the log-rank test.
PC-3 cells had slightly higher miR-221 expression level than LNCaP or C4–2 cell lines. DU145 cells exhibited lowest miR-221 expression levels among these cell lines. As for miR-222, the expression levels were not consistent with aggressiveness of cell lines.
Associations between the expression levels of miR-221 and miR-222 and clinicopathological features in PCa specimens
Among 54 patients with PCa, 52 underwent ADT with luteinising hormone-releasing hormone agonist and anti-androgens (Supplementary Table 1). A total of 20 ADT-treated patients progressed to CRPC over a median follow-up of 17.2 months. The risk of progression to CRPC was then analysed in patients with high vs low miR-221/222 expression. Low expression of miR-222 was associated with shorter progression-free interval (P=0.0173; Figure 1E). However, miR-221 did not predict the time to CRPC in these PCa patients (P=0.147; Figure 1D). Similarly, various clinicopathological parameters and CRPC progression-free intervals were evaluated. cT4 or cM1 was associated with shorter CRPC progression-free interval (P=0.0403 and P=0.0309; Supplementary Figures 4A and C). However, no significant differences in cN stage or PSA levels were observed between the two groups in this cohort (Supplementary Figures 4B and D).
Univariate and multivariate Cox proportional hazards models were used to assess independent predictors of time to progression to CRPC, including Gleason score, cT stage, cN stage, cM stage, PSA, age, and miR-222 expression. miR-222 expression was a prognostic factor of patient outcomes for PCa patients treated with ADT both in the univariate and multivariate analyses (univariate: hazard ratio=0.352, 95% confidence interval=0.135–0.917, P=0.0326; multivariate: hazard ratio=0.206, 95% confidence interval=0.067–0.637, P=0.0060; Table 2).
Table 2. Univariate and multivariate Cox proportional analysis for the prediction of CRPC progression-free survival.
|
Univariate analysis |
Multivariate analysis |
|||||
|---|---|---|---|---|---|---|
| Covariant | HR | 95% CI | P-value | HR | 95% CI | P-value |
| miR-222 | 0.352 | 0.135–0.917 | 0.0326 | 0.206 | 0.067–0.637 | 0.006 |
| cT stage | 2.497 | 1.009–6.18 | 0.0477 | 1.311 | 0.4448–3.833 | 0.621 |
| cN stage | 3.768 | 0.873–16.269 | 0.0755 | 4.196 | 0.792–22.23 | 0.0918 |
| cM stage | 3.54 | 1.033–12.135 | 0.0443 | 2.618 | 0.607–11.299 | 0.1971 |
| PSA at diagnosis | 1 | 1–1.001 | 0.1674 | 1 | 1.000–1.001 | 0.3442 |
| Gleason score | 1.663 | 0.756–3.659 | 0.2061 | 1.433 | 0.637–3.223 | 0.3846 |
| Age | 1.014 | 0.951–1.081 | 0.6657 | 1.129 | 1.019–1.25 | 0.0198 |
Abbreviations: CI=confidence interval; CRPC=castration-resistant prostate cancer; HR=hazard ratio; PSA=prostate-specific antigen. P<0.05 is shown in bold characters.
Effects of restoring miR-221 or miR-222 expression on cell proliferation, migration, and invasion in PC3 and DU145 cells
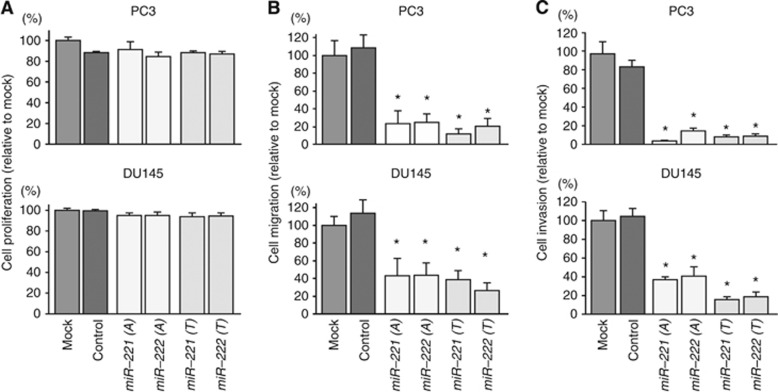
To investigate the functional roles of the miR-221/222 cluster, we performed gain-of-function studies using miRNA transfection in PC3 and DU145 cells. We used two sources of miR-221 and miR-222 mimic (miR-221-A and miR-222-A: Ambion; miR-221-T and miR-222-T: Thermo Scientific Dharmacon, Waltham, MA, USA) to ensure the reproducibility of the data.
As observed using XTT assays, cell proliferation was not inhibited in miR-221 and miR-222 transfectants in comparison with mock- or miR-control-transfected cells (Figure 2A). However, miR-221 and miR-222 transfection significantly inhibited cell migration as compared with mock- or miR-control-transfected cells (P<0.0001; Figure 2B). Similarly, Matrigel invasion assays demonstrated that cell invasion activity was significantly inhibited in miR-221 and miR-222 transfectants in comparison with mock or miR-control transfectants (P<0.0001; Figure 2C).
Figure 2.
Effects of miR-221/222 transfection on cell proliferation, migration, and invasion in PC3 and DU145 cells. (A) Cell proliferation was determined 72 h after transfection with miR-221/222 using XTT assays. (B) Cell migration activity was determined 48 h after transfection with miR-221/222 using migration assays. (C) Effects of miR-221/222 transfection on cell invasion in PC3 and DU145 cells. Cell invasion activity was determined 48 h after transfection with miR-221/222 using Matrigel invasion assays. miR-221-A and miR-222-A: Ambion, miR-221-T and miR-222-T: Thermo Scientific Dharmacon. *P<0.0001. Experiments were performed triplicate. The bars mean s.d.
In addition, we investigated cell cycle and apoptosis assay by using miR-221 or miR-222 transfectant in PC3 cells. No positive data were observed in both assays (Supplementary Figures 6 and 7).
Identification of target genes regulated by the miR-221/222 cluster in PCa
To identify target genes of miR-221/222, we performed in silico analysis and oligomicroarray analysis. The TargetScan programme showed that 2275 genes had putative target sites for miR-221/222 in their 3′ UTRs. To gain further insights into which genes were affected by the tumour-suppressive miR-221/222 cluster in PCa, we investigated their expression status in PCa clinical specimens and examined gene expression profiles in the GEO database (accession numbers GSE29079) to evaluate upregulated genes in PCa specimens. Among the 2275 putative target genes of the miR-221/222 cluster, 135 genes were significantly upregulated in PCa specimens compared with non-PCa tissues (log2 ratio >0.5). Finally, we performed genome-wide gene expression analysis using PC3 and DU145 cells (GEO accession number GSE56243). Genes downregulated (log2 ratio <−0.1) by transfection with miR-221 and miR-222 were selected as putative target genes. A total of 17 putative candidate genes for miR-221/222 regulation were identified. In this study, we sorted these candidate genes in order of GEO expression data (Table 3), because we considered that high expression genes in cancer tissues functioned as an oncogene. As a result, Ecm29 was the most upregulated gene in the putative candidate genes. Moreover, Ecm29 has a conserved binding site for miR-221/222. We focused on Ecm29 for further studies. Our strategy for selection of miR-221/222 cluster-targeted genes is shown in Supplementary Figure 5.
Table 3. Downregulated genes in miR-221/222 transfectants and upregulated genes in the GEO database.
| Entrez gene ID | Symbol | Gene name | Location | PC3 miR-221 transfectant | PC3 miR-222 transfectant | DU145 miR-221 transfectant | DU145 miR-222 transfectant | Average | GEO fold change | No. of conserved sites | No. of poorly conserved sites |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 23392 | KIAA0368 | Ecm29 (KIAA0368) | 9q31.3 | −1.28 | −1.05 | −0.82 | −0.62 | −0.94 | 0.999 | 1 | 0 |
| 159195 | USP54 | Ubiquitin-specific peptidase 54 | 10q22.2 | −0.73 | −0.65 | −0.19 | −0.15 | −0.43 | 0.869 | 0 | 1 |
| 51809 | GALNT7 | UDP-N-acetyl-alpha-D-galactosamine:polypeptide N-acetylgalactosaminyltransferase 7 (GalNAc-T7) | 4q34.1 | −0.75 | −0.67 | −0.85 | −0.94 | −0.80 | 0.833 | 0 | 1 |
| 10238 | DCAF7 | DDB1- and CUL4-associated factor 7 | 17q23.3 | −0.78 | −0.87 | −0.58 | −0.47 | −0.67 | 0.767 | 1 | 1 |
| 55920 | RCC2 | Regulator of chromosome condensation 2 | 1p36.13 | −0.48 | −0.53 | −0.12 | −0.18 | −0.33 | 0.734 | 0 | 1 |
| 23097 | CDK19 | Cyclin-dependent kinase 19 | 6q21 | −2.77 | −2.18 | −0.56 | −0.18 | −1.42 | 0.732 | 1 | 2 |
| 58508 | MLL3 | Myeloid/lymphoid or mixed-lineage leukaemia 3 | 7q36.1 | −1.44 | −1.40 | −0.42 | −0.28 | −0.88 | 0.697 | 0 | 1 |
| 9870 | KIAA0317 | KIAA0317 | 14q24.3 | −0.67 | −0.47 | −0.36 | −0.41 | −0.48 | 0.621 | 0 | 1 |
| 54874 | FNBP1L | Formin-binding protein 1-like | 1p22.1 | −1.20 | −1.25 | −0.22 | −0.21 | −0.72 | 0.613 | 0 | 1 |
| 55884 | WSB2 | WD repeat and SOCS box containing 2 | 12q24.23 | −2.64 | −2.71 | −1.58 | −1.21 | −2.03 | 0.569 | 1 | 1 |
| 6319 | SCD | stearoyl-CoA desaturase (delta-9-desaturase) | 10q24.31 | −1.78 | −1.81 | −0.49 | −0.15 | −1.06 | 0.545 | 0 | 1 |
| 2017 | CTTN | Cortactin | 11q13.3 | −1.12 | −1.01 | −0.32 | −0.14 | −0.65 | 0.542 | 0 | 1 |
| 5451 | POU2F1 | POU class 2 homeobox 1 | 1q24.2 | −0.46 | −0.43 | −0.21 | −0.26 | −0.34 | 0.524 | 0 | 1 |
| 204851 | HIPK1 | Homeodomain interacting protein kinase 1 | 1p13.2 | −2.00 | −1.70 | −0.81 | −0.56 | −1.27 | 0.519 | 1 | 0 |
| 9265 | CYTH3 | Cytohesin 3 | 7p22.1 | −0.53 | −0.43 | −0.58 | −0.39 | −0.48 | 0.518 | 0 | 1 |
| 23126 | POGZ | Pogo transposable element with ZNF domain | 1q21.3 | −0.64 | −0.80 | −0.26 | −0.16 | −0.46 | 0.515 | 1 | 0 |
| 2799 | GNS | Glucosamine (N-acetyl)-6-sulfatase | 12q14.3 | −0.65 | −1.16 | −0.16 | −0.29 | −0.57 | 0.513 | 0 | 1 |
Ecm29 was a direct target of miR-221/222 in PCa cells
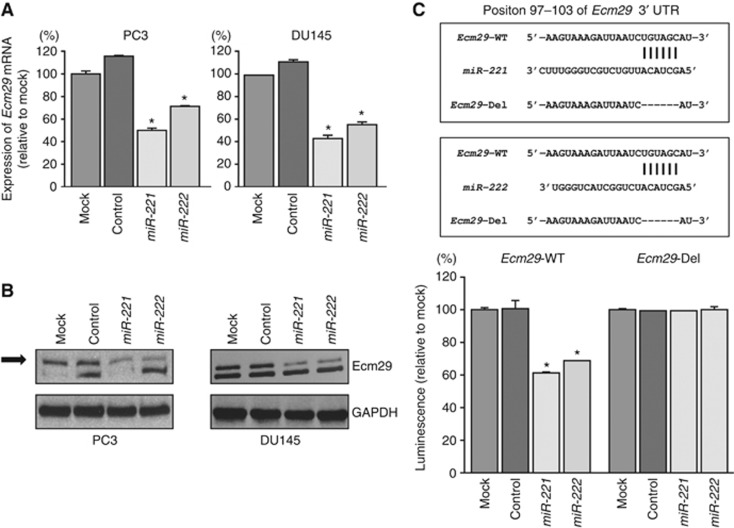
We performed real-time RT–qPCR and western blotting in PC3 and DU145 cells to investigate whether restoration of miR-221 and miR-222 altered the expression of the Ecm29 gene and Ecm29 protein. The mRNA and protein expression levels of Ecm29/Ecm29 were significantly repressed in miR-221/222 transfectants as compared with mock- or miR-control-transfected cells (P<0.0001; Figures 3A and B and Supplementary Figure 8).
Figure 3.
Downregulation of Ecm29 expression by miR-221/222 in PC3 and DU145 cells. (A) Ecm29 mRNA expression 72 h after transfection with miR-221/222. GUSB was used as an internal control. *P<0.0001. (B) Ecm29 protein expression 48 h after transfection with miR-221/222. GAPDH was used as a loading control. (C) miR-221/222-binding sites in Ecm29 mRNA. Luciferase reporter assays were carried out using a vector encoding the putative miR-221/222 target site in the Ecm29 3'-UTR (position 97–103) for wild-type and deletion constructs. *P<0.0001. Experiments were performed triplicate. The bars mean s.d.
Therefore, we next performed luciferase reporter assays in PC3 cells to determine whether Ecm29 mRNA had target sites for miR-221 and miR-222. The TargetScan database predicted that miR-221 and miR-222 bound at position 97–103 in the 3′ UTR of Ecm29. We used vectors encoding a partial wild-type sequence of the 3′ UTR of Ecm29 mRNA, including the predicted miR-221 and miR-222 target site, or a vector lacking the miR-221 and miR-222 target site. We found that the luminescence intensity was significantly reduced by cotransfection with miR-221 or miR-222 and the vector carrying the wild-type 3′ UTR of Ecm29. On the other hand, the luminescence intensity was not decreased when the seed sequence of the target site was deleted from the vectors (P<0.0001; Figure 3C).
Effects of silencing Ecm29 on cell proliferation, migration, and invasion in PCa cell lines
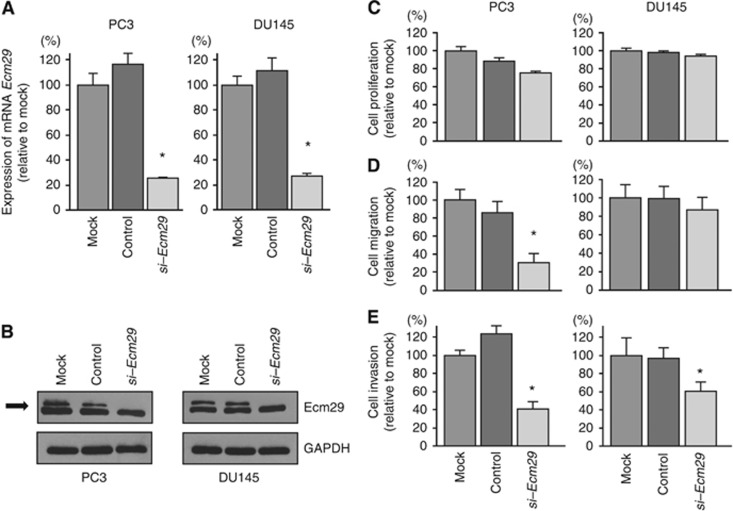
To investigate the functional role of Ecm29, we performed loss-of-function studies using si-Ecm29 transfectants. First, we evaluated the knockdown efficiency of si-Ecm29 transfection in PC3 and DU145 cells. RT–qPCR and western blotting indicated that si-Ecm29 transfection effectively downregulated Ecm29 mRNA and protein expression in PC3 and DU145 cells (Figures 4A and B).
Figure 4.
Effects of silencing Ecm29 mRNA and protein expression by si-Ecm29 transfection in PCa cells on cell proliferation, migration, and invasion in PCa cell lines. (A) Ecm29 mRNA expression was determined at 72 h after transfection with si-Ecm29. GUSB was used as an internal control. (B) Ecm29 protein expression was evaluated by western blotting at 48 h after transfection with si-Ecm29. GAPDH was used as a loading control. *P<0.0001. (C) Cell proliferation was determined by XTT assays. (D) Cell migration activity was determined by wound healing assays. (E) Cell invasion activity was determined by Matrigel invasion assays. *P<0.0001. mock: untransfected cells, control: control siRNA-transfected cells. Experiments were performed triplicate. The bars mean s.d.
In functional assays, cell proliferation and migration were not inhibited by transfection with si-Ecm29 in comparison with mock- or si-control-transfected DU145 cells (Figures 4C and D). However, cell invasion (Figure 4E) assays demonstrated that cancer cell invasion activity was significantly inhibited in si-Ecm29 transfectants in comparison with mock- or si-control-transfected PC3 and DU145 cells.
Expression of Ecm29 in clinical PCa specimens
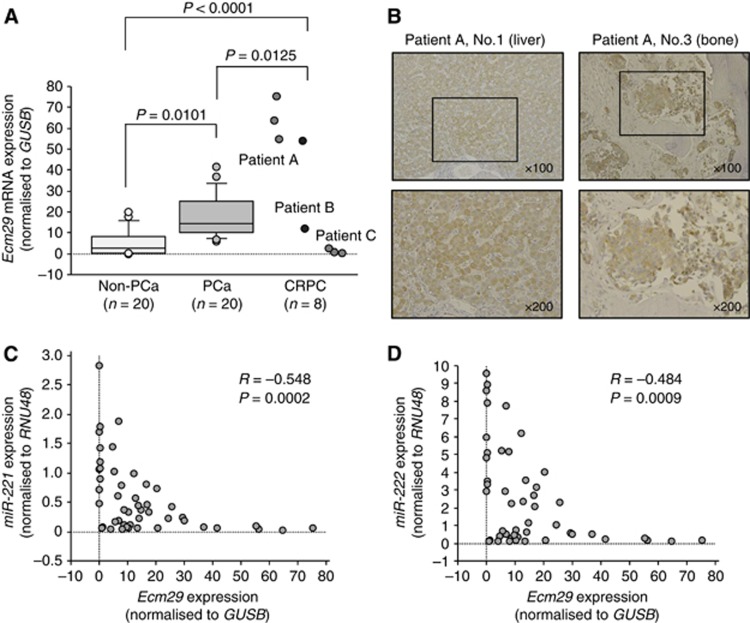
A total of 20 PCa samples (Supplementary Table 1, Nos. 1–20), 20 non-PCa samples (Supplementary Table 1, Nos. 55–74), and 8 CRPC samples were used to analyse Ecm29 mRNA expression in this study. RT-qPCR analysis showed that the expression of Ecm29 mRNA was significantly higher in clinical PCa specimens than in non-PCa specimens (P=0.0101; Figure 5A). Furthermore, expression of Ecm29 mRNA was significantly higher in CRPC specimens than in PCa and non-PCa specimens (P=0.0125 and P<0.0001, respectively; Figure 5A). In these 48 clinical specimens, Spearman's rank test showed that there was a negative correlation between the expression level of miR-221 and that of Ecm29 (R=−0.548 and P=0.0002; Figure 5C). Similarly, a negative correlation was found between the expression level of miR-222 and that of Ecm29 (R=−0.484 and P=0.0009; Figure 5D). Immunohistochemical staining of Ecm29 in CRPC specimens demonstrated high expression of Ecm29 in the cytoplasm of CRPC cells and low expression in the nucleus (Figure 5B).
Figure 5.
Expression of Ecm29 in clinical PCa specimens. (A) Ecm29 mRNA expression levels were determined by qRT–PCR analysis in PCa, non-PCa, and CRPC specimens. GUSB was used as an internal control. (B) Ecm29 expression in CRPC specimens. High expression of Ecm29 was found in the cytoplasm of CRPC cells, whereas the expression was low in the nucleus. (C) Inverse correlation between Ecm29 mRNA and miR-221 expression. (D) Inverse correlation between Ecm29 mRNA and miR-222 expression.
Discussion
A growing body of evidence has shown that miRNAs are involved in several biological processes and are tightly correlated with human oncogenesis and metastasis (Nelson and Weiss, 2008). Recent studies from our laboratory have identified a variety of novel molecular targets and pathways regulated by tumour-suppressive miRNAs in PCa based on PCa miRNA signatures (Goto et al, 2014b; Nishikawa et al, 2014). Moreover, we have begun to analyse clustered miRNAs and have reported that several miRNA clusters, including miR-1/133, miR-23b/27b/24-1, and miR-143/145, act as tumour suppressors and contribute substantially to PCa oncogenesis and metastasis (Kojima et al, 2012, 2014; Goto et al, 2014a). Although elucidation of the molecular networks in CRPC specimens is needed to improve therapies for and prevention of the disease, advancement of genomic analysis is difficult because of the challenges with obtaining clinical CRPC specimens.
Evaluation of miRNA expression signatures using CRPC specimens is an indispensable tool for cancer research. In this study, we have constructed a new CRPC miRNA signature and identified several downregulated miRNAs in CRPC tissues compared with PCa and normal prostate tissues. According to our previous PCa signatures and other studies, the clustered miRNAs miR-221 and miR-222 were significantly downregulated in PCa specimens (Porkka et al, 2007; Ambs et al, 2008; Schaefer et al, 2010; Szczyrba et al, 2010; Carlsson et al, 2011; Fuse et al, 2012; Wach et al, 2012). Furthermore, our present CRPC signature revealed that both miR-221 and miR-222 were also significantly reduced in CRPC specimens. These findings suggested that downregulation of the molecular networks regulated by the miR-221/222 cluster contributes substantially to PCa and CRPC progression.
Currently, a variety of diverse treatment options are available for patients with PCa, particularly those with localised PCa (Heidenreich et al, 2014b). However, it is not clear which treatment is suitable for achieving an optimal prognosis in patients with high-risk PCa. Prediction of the time to progression to CRPC under ADT using the expression status of miRNAs from needle biopsies during initial diagnosis will improve the accuracy of treatment decisions for patients with high-risk PCa. We found that miR-222 expression status was a good prognostic marker for time to progression to CRPC using hormone-naïve prostate biopsy specimens. A recent study demonstrated that miR-221 expression is progressively reduced in aggressive PCa and predicts recurrence-free survival after radical prostatectomy in patients with high-risk PCa (Kneitz et al, 2014). These results strongly suggest that the miR-221/222 cluster may be useful in PCa diagnosis. A large-scale cohort study will be necessary to determine whether miR-221 and miR-222 are effective markers for CRPC-free interval.
In this study, we also investigated the functional significance of miR-221 and miR-222 using two different types of miR-221 and miR-222 mimic in gain-of-function studies. We found that restoration of miR-221 and miR-222 using these four constructs significantly inhibited cancer cell migration and invasion. These results strongly suggested that miR-221 and miR-222 functioned as tumour suppressors in PCa cells. Our expression data of miR-221/222 transfectants in PC3 cells (GEO accession number GSE56243) demonstrated that several downregulated genes were categorised to ‘ECM-receptor interaction', ‘Cell adhesion molecules', and ‘focal adhesion pathways'. These differentially expressed genes were deeply contributed to cancer cell migration and invasion in PCa. Moreover, a recent study showed that miR-221 regulates cell growth, invasion, and apoptosis by targeting the oncogenes SOCS3 and IRF2, suggesting that miR-221 has a tumour-suppressive role in PCa cells (Kneitz et al, 2014). The tumour-suppressive functions of the miR-221/222 cluster have been reported in other types of cancers, such as tongue squamous cell carcinomas and gastric cancers (Garofalo et al, 2012). These findings strongly support that the miR-221/222 cluster functions as a tumour suppressor in PCa cells.
In contrast to tumour-suppressive function of miR-221/222, previous studies have shown that miR-221 and miR-222 act as oncogenes. Upregulation of miR-221 and miR-222 were observed in CRPC cell lines and these miRNAs promote cancer cell proliferation (Sun et al, 2014). Furthermore, in vivo mouse model study demonstrated that inhibition of miR-221/222 in PC3 cells with LNA antisense oligonucleotides reduced tumour growth (Mercatelli et al, 2008). Several studies of breast cancer have demonstrated that the miR-221/222 cluster acts as an onco-miR, targeting tumour-suppressive genes, such as p27Kip1 and oestrogen receptor alpha (Shah and Calin, 2011). The oncogenic functions of miR-221 and miR-222 have also been reported in glioblastoma and melanoma (Garofalo et al, 2012). It is unclear why the expression statuses of these miRNAs vary in different types of cancer cells. Detailed molecular studies of the expression control of these miRNAs by the cancer types is necessary.
Regulation mechanisms of miR-221/222 cluster are reported in some articles. As for colorectal cancer, miR-221/222 is upregulated in cancer tissue and positively regulates NF-kB-STAT3 pathway and vice versa, forming feedback loop, which has critical role for colorectal cancer growth (Di Leva et al, 2010). Similarly, in breast cancer, miR-221/222 directly regulates ERα and ERα represses miR-221/222 expression recruiting co-repressor proteins (Liu et al, 2014). The mechanisms of downregulation of miR-221/222 in PCa are yet to be clear. Thus, elucidation of the epigenetic mechanisms controlling the expression of clustered miRNAs in different types of cancer cells is an important theme in cancer research.
Normal regulatory mechanisms can be disrupted by the aberrant expression of tumour-suppressive or oncogenic miRNAs in cancer cells. Therefore, one of the next problems to address is identification of oncogenic genes that are regulated by the miR-221/222 cluster in PCa cells. We selected 17 putative candidate genes using a combination of in silico and genome-wide gene expression analyses in the present study. Among them, we focused on the Ecm29 gene as a responsible oncogene in PCa oncogenesis and metastasis because the functional significance of Ecm29 is not well understood in PCa cells. Ecm29 is a conserved 210-kDa protein composed almost entirely of HEAT-like repeats (Gorbea et al, 2004). It is a scaffold protein that links the 26S proteasome to motor proteins, resulting in inhibition of proteasome activity and functioning as a proteasome quality control protein (Gorbea et al, 2010; Lehmann et al, 2010; De La Mota-Peynado et al, 2013). However, the precise mechanisms of Ecm29 in cancer cells remain unknown. Our data demonstrated that Ecm29 is significantly upregulated in PCa clinical specimens. Also, overexpressed Ecm29 was observed in some CRPC specimens. Large number of PCa and CRPC specimens are necessary to investigate the clinical significance of Ecm29 expression. Gene expression database search showed that upregulation of Ecm29 was observed in other types of cancer, such as muscle invasive bladder cancer and pancreatic cancer (GSE15471 and GSE3167). These expression results support our present data of PCa (Supplementary Figure 9). Knockdown of Ecm29 by siRNAs significantly inhibited migration of PC3 cells (not DU145 cells) and inhibited invasion of both PC3 and DU145 cells. Our data are the first report demonstrating that Ecm29 contributed to cancer cell migration and invasion in PCa cell. We showed that Ecm29 was under direct regulation of miR-221/222 in PCa cells, however, many targets of miR-221/222 regulation are still unclear. Elucidation of other candidate targets of the tumour-suppressive miR-221/222 cluster regulation presented in this study may improve our understanding of PCa progression and metastasis.
Conclusions
Downregulation of the miR-221/222 cluster was identified based on the miRNA expression signature of CRPC in this study. Moreover, Kaplan–Meier survival curves showed that low expression of miR-222 predicted a short duration of progression to CRPC. miR-221 and miR-222 were shown to function as tumour suppressors in PCa cells. To the best of our knowledge, this is the first report demonstrating that the tumour-suppressive miR-221/222 cluster directly regulated Ecm29 in PCa cells. The identification of novel molecular pathways and targets regulated by the miR-221/222 cluster based on CRPC signatures may lead to a better understanding of PCa and the development of new therapeutic strategies to treat this disease.
Acknowledgments
This study was supported by the KAKENHI 24592590 (C), 26462430 (C), and 25293333 (B).
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on British Journal of Cancer website (http://www.nature.com/bjc)
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 4.0 Unported License.
Supplementary Material
References
- Ambs S, Prueitt RL, Yi M, Hudson RS, Howe TM, Petrocca F, Wallace TA, Liu CG, Volinia S, Calin GA, Yfantis HG, Stephens RM, Croce CM (2008) Genomic profiling of microRNA and messenger RNA reveals deregulated microRNA expression in prostate cancer. Cancer Res 68: 6162–6170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116: 281–297. [DOI] [PubMed] [Google Scholar]
- Carlsson J, Davidsson S, Helenius G, Karlsson M, Lubovac Z, Andren O, Olsson B, Klinga-Levan K (2011) A miRNA expression signature that separates between normal and malignant prostate tissues. Cancer Cell Int 11: 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi KN, Bjartell A, Dearnaley D, Saad F, Schroder FH, Sternberg C, Tombal B, Visakorpi T (2009) Castration-resistant prostate cancer: from new pathophysiology to new treatment targets. Eur Urol 56: 594–605. [DOI] [PubMed] [Google Scholar]
- Colloca G (2012) Prostate-specific antigen kinetics as a surrogate endpoint in clinical trials of metastatic castration-resistant prostate cancer: a review. Cancer Treat Rev 38: 1020–1026. [DOI] [PubMed] [Google Scholar]
- De La Mota-Peynado A, Lee SY, Pierce BM, Wani P, Singh CR, Roelofs J (2013) The proteasome-associated protein Ecm29 inhibits proteasomal ATPase activity and in vivo protein degradation by the proteasome. J Biol Chem 288: 29467–29481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, Iorio MV, Li M, Volinia S, Alder H, Nakamura T, Nuovo G, Liu Y, Nephew KP, Croce CM (2010) MicroRNA cluster 221-222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst 102: 706–721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edge S, Byrd DR, Compton CC, Fritz AG, Greene FL, Trotti A (2010) American Joint Committee on Cancer Staging Manual 7th edn. Springer: New York. [Google Scholar]
- Esquela-Kerscher A, Slack FJ (2006) Oncomirs–microRNAs with a role in cancer. Nat Rev Cancer 6: 259–269. [DOI] [PubMed] [Google Scholar]
- Fukumoto I, Kinoshita T, Hanazawa T, Kikkawa N, Chiyomaru T, Enokida H, Yamamoto N, Goto Y, Nishikawa R, Nakagawa M, Okamoto Y, Seki N (2014) Identification of tumour suppressive microRNA-451a in hypopharyngeal squamous cell carcinoma based on microRNA expression signature. Br J Cancer 111: 386–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuse M, Kojima S, Enokida H, Chiyomaru T, Yoshino H, Nohata N, Kinoshita T, Sakamoto S, Naya Y, Nakagawa M, Ichikawa T, Seki N (2012) Tumor suppressive microRNAs (miR-222 and miR-31) regulate molecular pathways based on microRNA expression signature in prostate cancer. J Hum Genet 57: 691–699. [DOI] [PubMed] [Google Scholar]
- Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G (2012) miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med 12: 27–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbea C, Goellner GM, Teter K, Holmes RK, Rechsteiner M (2004) Characterization of mammalian Ecm29, a 26 S proteasome-associated protein that localizes to the nucleus and membrane vesicles. J Biol Chem 279: 54849–54861. [DOI] [PubMed] [Google Scholar]
- Gorbea C, Pratt G, Ustrell V, Bell R, Sahasrabudhe S, Hughes RE, Rechsteiner M (2010) A protein interaction network for Ecm29 links the 26 S proteasome to molecular motors and endosomal components. J Biol Chem 285: 31616–31633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Kojima S, Nishikawa R, Enokida H, Chiyomaru T, Kinoshita T, Nakagawa M, Naya Y, Ichikawa T, Seki N (2014. a) The microRNA-23b/27b/24-1 cluster is a disease progression marker and tumor suppressor in prostate cancer. Oncotarget 5: 7748–7759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goto Y, Nishikawa R, Kojima S, Chiyomaru T, Enokida H, Inoguchi S, Kinoshita T, Fuse M, Sakamoto S, Nakagawa M, Naya Y, Ichikawa T, Seki N (2014. b) Tumour-suppressive microRNA-224 inhibits cancer cell migration and invasion via targeting oncogenic TPD52 in prostate cancer. FEBS Lett 588: 1973–1982. [DOI] [PubMed] [Google Scholar]
- Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F, Mottet N (2014. a) EAU Guidelines on Prostate Cancer. Part II: Treatment of Advanced, Relapsing, and Castration-Resistant Prostate Cancer. Eur Urol 65: 467–479. [DOI] [PubMed] [Google Scholar]
- Heidenreich A, Bastian PJ, Bellmunt J, Bolla M, Joniau S, van der Kwast T, Mason M, Matveev V, Wiegel T, Zattoni F, Mottet N (2014. b) EAU guidelines on prostate cancer. part 1: screening, diagnosis, and local treatment with curative intent-update 2013. Eur Urol 65: 124–137. [DOI] [PubMed] [Google Scholar]
- Kinoshita T, Nohata N, Hanazawa T, Kikkawa N, Yamamoto N, Yoshino H, Itesako T, Enokida H, Nakagawa M, Okamoto Y, Seki N (2013) Tumour-suppressive microRNA-29s inhibit cancer cell migration and invasion by targeting laminin-integrin signalling in head and neck squamous cell carcinoma. Br J Cancer 109: 2636–2645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kneitz B, Krebs M, Kalogirou C, Schubert M, Joniau S, van Poppel H, Lerut E, Kneitz S, Scholz CJ, Strobel P, Gessler M, Riedmiller H, Spahn M (2014) Survival in patients with high-risk prostate cancer is predicted by miR-221, which regulates proliferation, apoptosis, and invasion of prostate cancer cells by inhibiting IRF2 and SOCS3. Cancer Res 74: 2591–2603. [DOI] [PubMed] [Google Scholar]
- Kojima S, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nohata N, Fuse M, Ichikawa T, Naya Y, Nakagawa M, Seki N (2012) Tumour suppressors miR-1 and miR-133a target the oncogenic function of purine nucleoside phosphorylase (PNP) in prostate cancer. Br J Cancer 106: 405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima S, Enokida H, Yoshino H, Itesako T, Chiyomaru T, Kinoshita T, Fuse M, Nishikawa R, Goto Y, Naya Y, Nakagawa M, Seki N (2014) The tumor-suppressive microRNA-143/145 cluster inhibits cell migration and invasion by targeting GOLM1 in prostate cancer. J Hum Genet 59: 78–87. [DOI] [PubMed] [Google Scholar]
- Lehmann A, Niewienda A, Jechow K, Janek K, Enenkel C (2010) Ecm29 fulfils quality control functions in proteasome assembly. Mol Cell 38: 879–888. [DOI] [PubMed] [Google Scholar]
- Liu S, Sun X, Wang M, Hou Y, Zhan Y, Jiang Y, Liu Z, Cao X, Chen P, Chen X, Tao Y, Xu C, Mao J, Cheng C, Li C, Hu Y, Wang L, Chin YE, Shi Y, Siebenlist U, Zhang X (2014) A microRNA 221- and 222-mediated feedback loop maintains constitutive activation of NFkappaB and STAT3 in colorectal cancer cells. Gastroenterology 147: 847–859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercatelli N, Coppola V, Bonci D, Miele F, Costantini A, Guadagnoli M, Bonanno E, Muto G, Frajese GV, De Maria R, Spagnoli LG, Farace MG, Ciafre SA (2008) The inhibition of the highly expressed miR-221 and miR-222 impairs the growth of prostate carcinoma xenografts in mice. PLoS One 3: e4029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson KM, Weiss GJ (2008) MicroRNAs and cancer: past, present, and potential future. Mol Cancer Ther 7: 3655–3660. [DOI] [PubMed] [Google Scholar]
- Nishikawa R, Goto Y, Sakamoto S, Chiyomaru T, Enokida H, Kojima S, Kinoshita T, Yamamoto N, Nakagawa M, Naya Y, Ichikawa T, Seki N (2014) Tumor-suppressive microRNA-218 inhibits cancer cell migration and invasion via targeting of LASP1 in prostate cancer. Cancer Sci 105: 802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kikkawa N, Sakurai D, Fujimura L, Chiyomaru T, Kawakami K, Yoshino H, Enokida H, Nakagawa M, Katayama A, Harabuchi Y, Okamoto Y, Seki N (2011) Tumour suppressive microRNA-874 regulates novel cancer networks in maxillary sinus squamous cell carcinoma. Br J Cancer 105: 833–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nohata N, Hanazawa T, Kinoshita T, Inamine A, Kikkawa N, Itesako T, Yoshino H, Enokida H, Nakagawa M, Okamoto Y, Seki N (2013) Tumour-suppressive microRNA-874 contributes to cell proliferation through targeting of histone deacetylase 1 in head and neck squamous cell carcinoma. Br J Cancer 108: 1648–1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porkka KP, Pfeiffer MJ, Waltering KK, Vessella RL, Tammela TL, Visakorpi T (2007) MicroRNA expression profiling in prostate cancer. Cancer Res 67: 6130–6135. [DOI] [PubMed] [Google Scholar]
- Schaefer A, Jung M, Mollenkopf HJ, Wagner I, Stephan C, Jentzmik F, Miller K, Lein M, Kristiansen G, Jung K (2010) Diagnostic and prognostic implications of microRNA profiling in prostate carcinoma. Int J Cancer 126: 1166–1176. [DOI] [PubMed] [Google Scholar]
- Scher HI, Kelly WM, Zhang ZF, Ouyang P, Sun M, Schwartz M, Ding C, Wang W, Horak ID, Kremer AB (1999) Post-therapy serum prostate-specific antigen level and survival in patients with androgen-independent prostate cancer. J Natl Cancer Inst 91: 244–251. [DOI] [PubMed] [Google Scholar]
- Shah MY, Calin GA (2011) MicroRNAs miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med 3: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel R, Naishadham D, Jemal A (2013) Cancer statistics, 2013. CA Cancer J Clin 63: 11–30. [DOI] [PubMed] [Google Scholar]
- Sun T, Wang X, He HH, Sweeney CJ, Liu SX, Brown M, Balk S, Lee GS, Kantoff PW (2014) MiR-221 promotes the development of androgen independence in prostate cancer cells via downregulation of HECTD2 and RAB1A. Oncogene 33: 2790–2800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szczyrba J, Loprich E, Wach S, Jung V, Unteregger G, Barth S, Grobholz R, Wieland W, Stohr R, Hartmann A, Wullich B, Grasser F (2010) The microRNA profile of prostate carcinoma obtained by deep sequencing. Mol Cancer Res 8: 529–538. [DOI] [PubMed] [Google Scholar]
- Wach S, Nolte E, Szczyrba J, Stohr R, Hartmann A, Orntoft T, Dyrskjot L, Eltze E, Wieland W, Keck B, Ekici AB, Grasser F, Wullich B (2012) MicroRNA profiles of prostate carcinoma detected by multiplatform microRNA screening. Int J Cancer 130: 611–621. [DOI] [PubMed] [Google Scholar]
- Yang X, Yang Y, Gan R, Zhao L, Li W, Zhou H, Wang X, Lu J, Meng QH (2014) Down-regulation of mir-221 and mir-222 restrain prostate cancer cell proliferation and migration that is partly mediated by activation of SIRT1. PLoS One 9: e98833. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.