Abstract
Pdx1 is a transcription factor of fundamental importance to pancreas formation and adult islet β-cell function. However, little is known about the positive- and negative-acting coregulators recruited to mediate transcriptional control. Here we isolated numerous Pdx1-interacting factors possessing a wide range of cellular functions linked with this protein, including, but not limited to, coregulators associated with transcriptional activation/repression, DNA damage response, and DNA replication. Because chromatin remodeling activities are essential to developmental lineage decisions and adult cell function, our analysis focused on investigating the influence of the Swi/Snf chromatin remodeler on Pdx1 action. The two mutually exclusive and indispensible Swi/Snf core ATPase subunits, Brg1 and Brm, distinctly affected target gene expression in β-cells. Furthermore, physiological and pathophysiological conditions dynamically regulated Pdx1 binding to these Swi/Snf complexes in vivo. We discuss how context-dependent recruitment of coregulatory complexes by Pdx1 could impact pancreas cell development and adult islet β-cell activity.
Graphical Abstract
Introduction
The Pdx1 protein has profound transcriptional regulatory properties and is an essential driver of normal pancreas development and mature β-cell function. For example, embryonic formation of ductal, acinar and islet cells that compose the pancreas are derived from a common Pdx1-expressing progenitor pool (Jonsson et al., 1994; Offield et al., 1996), with pancreas agenesis manifested in mice and humans unable to produce the functional protein (Jonsson et al., 1994; Offield et al., 1996; Stoffers et al., 1997). Developing mouse β-(Gannon et al., 2008) and/or acinar-cell (Hale et al., 2005) formation is also severely blunted upon Pdx1 removal from the progenitors of these cell populations. Moreover, forced and persistent Pdx1 expression in embryonic Neurogenin 3 (Ngn3)+ pan-endocrine cell progenitors causes islet glucagon hormone+ α-cells to become insulin+ β-cells postnatally (Yang et al., 2011). Conversely, specific deletion of Pdx1 from adult islet β-cells, which are the principal adult pancreatic cell type producing this protein, results in their rapid acquisition of an α-like cell identity (Gao et al., 2014).
Like all transcription factors, Pdx1 binds to specific cis-acting sequences within target gene control regions, often referred to as enhancer regions (Pasquali et al., 2014). Transcriptional modulation is then imposed through dynamic changes in chromatin structure that causes the underlying DNA to become more or less accessible to RNA polymerase II. However, even powerful transcription factors like Pdx1 lack inherent chromatin-modifying properties and alone cannot modulate gene transcription effectively. Instead, cell-signaling events drive context-dependent protein-protein interactions between these DNA-binding factors and coregulators, which possess transcriptional modifying functions. For example, acute exposure of β-cell lines to glucose, the most important physiological effector of cell function in vivo, causes a rapid activator/repressor switch in Pdx1 activity on Insulin gene transcription. This involves the differential recruitment of histone acetyltransferases (HAT) coactivator at high, stimulating glucose concentrations, and histone deacetylase (HDAC) corepressors at low, inhibitory glucose levels (Mosley and Ozcan, 2004; Mosley et al., 2004).
Coregulators that alter chromatin structure can do so by both enzymatic and non-enzymatic means. Non-enzymatic coregulators such as Mediator harbor protein-protein and protein-DNA/RNA interaction surfaces that influence transcription by altering epigenetic patterns, chromatin compaction, as well as recruitment of distinct cofactors and RNA Polymerase II (Poss et al., 2013). The coregulators that function enzymatically can be divided into two main mechanistically distinct classes. First are those that alter chromatin through covalent modifications to DNA (e.g. methylation) and DNA binding proteins (e.g. histones, transcription factors and coregulators by acetylation, methylation, phosphorylation, ubiquitination, sumoylation, and/or glycosylation) (Bhaumik et al., 2007; Chen and Li, 2004; Flotho and Melchior, 2013; Wells et al., 2003). Second are those that use the energy of ATP hydrolysis to destabilize nucleosomes and alter accessibility of DNA to the transcriptional machinery (Sudarsanam and Winston, 2000). While there are over 250 transcriptional coregulators in mammalian cells, relatively few have been ascribed to Pdx1 specifically (Pcif1, p300, HDAC1/2, Set7/9, and Bridge1 (Francis, 2005; Liu et al., 2004; Mosley and Ozcan, 2004; Qiu et al., 2002; Stanojevic et al., 2005)) or other islet-enriched transcription factors (Nkx2.2 (Grg3, HDAC1, DNMT3a (Papizan et al., 2011)), Isl1 (Ldb1/2 (Hunter et al., 2013)), HNF1β (PCAF/CBP (Barbacci, 2004)), NeuroD1 (Bridge1, p300/CBP (Qiu et al., 1998; Thomas et al., 1999)). Significantly, essentially all of these transcription factor associations were made in studies using a small subset of candidate coregulators.
Here we have used an unbiased chemical cross-linking, antibody precipitation, and mass spectrometry strategy to identify endogenous Pdx1-binding proteins in β-cells. Although many new and interesting coregulatory factors were found using this in-cell cross-linking approach, we chose to specifically focus on investigating whether Swi/Snf chromatin remodeling complex recruitment was linked to the positive- and/or negative-acting control properties of Pdx1. Our results strongly suggest that Pdx1 interacts with functionally distinct Swi/Snf complexes in a highly dynamic manner in islet β-cells. Hence, Swi/Snf complexes containing the core Brg1 ATPase subunit were demonstrated to be involved in Pdx1-mediated activation, while the Brm ATPase subunit containing complexes imposed transcriptional repression. Evidence is also presented indicating that physiological and pathophysiological conditions influence Pdx1 binding to these distinct complexes in β-cells in vivo, and we propose that the antagonistic actions of Brg1-Swi/Snf and Brm-Swi/Snf have significant implications to glucose homeostasis. Our findings have identified many different possible coregulators of Pdx1 and shed light on how their recruitment and consequently transcription factor function is impacted normally and in the context of dysfunctional T2DM β-cells.
Results
Pdx1 interacts with many functionally distinct coregulatory proteins in β-cells
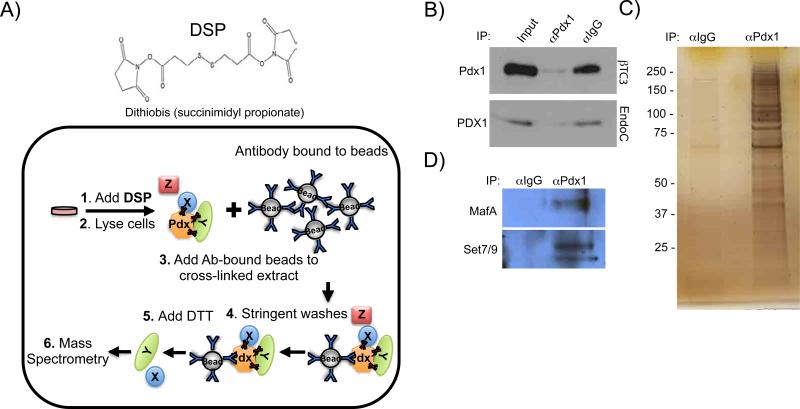
An unbiased strategy utilizing reversible cross-linking, co-immunoprecipitation (ReCLIP (Smith et al., 2011)) and mass spectrometry was used to identify Pdx1-interacting proteins in βTC3 cells, a murine β-cell line (Figure 1A). Employment of the cell permeable, lysine reactive, thiol cleavable, dithiobis(succinimidyl propionate) (DSP) cross-linking reagent allowed for stringent detergent-based washes, which led to a very clear difference in the proteins obtained from the Pdx1 antibody and control precipitates (Figure 1B, C). As anticipated, established Pdx1-interacting factors such as the MafA transcription factor (Zhao et al., 2005) and the Set7/9 coregulator (Francis, 2005) were found by immunoblot analysis after Pdx1 precipitation (Figure 1D). Additionally, we identified a number of novel candidate Pdx1 binding factors by mass spectrometry (Table 1, Table S1). Many of the candidates are likely linked with Pdx1 transcriptional regulation, while others may be associated with context-dependent recruitment in, for example, DNA repair, proliferation, and apoptosis (Babu et al., 2007; Lebrun et al., 2005).
Figure 1. ReCLIP enriches for Pdx1 interacting proteins.
A) Diagram of ReCLIP showing the lysine-reactive cross-linking reagent, DSP, that is first incubated with β-cells from which nuclear extract is generated for Pdx1 antibody or IgG immunoprecipitation. The control and α-Pdx1 bound complexes were washed with RIPA buffer and then RIPA+DTT to elute bound cross-linked proteins. B) Nearly all of Pdx1 was removed from the co-IP supernatants with Pdx1 antibody. The eluted proteins were C) SDS-PAGE separated and submitted for MS analysis or D) immunoblotted with MafA or Set7/9 antibodies (Table 1, Table S1). N≥3.
Table 1.
Pdxl bound chromatin remodeling complexes identified by ReCLIP/MS.
| Coregulator | Protein | % Coverage | Peptides |
|
|---|---|---|---|---|
| IgG control | Pdx1 IP | |||
| NuRD complex | Rbbp4 | 9 | 1 | 10 |
| Hdac1/2 | 4 | 0 | 5 | |
| Mi-2β | 3 | 0 | 7 | |
| Swi/Snf complex | Baf47 | 11 | 0 | 1 |
| Baf177 | 3 | 0 | 3 | |
| Baf57 | 9 | 0 | 2 | |
| Baf170 | 3 | 0 | 3 | |
| Brg1 | 2 | 0 | 5 | |
| Tif1β | Tif1β | 14 | 0 | 6 |
The coregulator complex is shown as well as the MS results illustrating percent protein subunit coverage and the number of peptides found by IgG or Pdx1 antibody precipitation. These results are representative of data averaged from at least three independently performed IgG and Pdx1 antibody treatments.
Several of the transcriptional coregulators were of particular interest because of their ability to potentially activate and/or repress Pdx1 trans-regulation (e.g. Tif1β, Swi/Snf and NuRD) (Flowers et al., 2009; Iyengar et al., 2011; Miccio et al., 2010). The binding of Tif1β, Swi/Snf and NuRD complex proteins to Pdx1 was independently confirmed in co-IP experiments performed with both mouse and human β-cell lines (Figure 2A). The authenticity of the interactions was further verified by the ability to precipitate Pdx1 with specific coregulator antibodies (Figure 2B). In addition, the roughly 42 kDa Pdx1 protein was found to migrate as a high molecular weight complex after sucrose gradient sedimentation of βTC3 nuclear extracts (Figure 2C), and to bind to Swi/Snf complex subunits by co-IP analysis (Figure 2E). Moreover, Pdx1-Swi/Snf was shown to retain specific cis-control element binding properties in electrophoretic mobility shift assays (Figure 2D). Notably, few of the published Pdx1 coregulators and none of the interacting islet-enriched transcription factors were detected by mass spectrometry (Table 1, Table S1). We presume that these represent relatively rare complexes observed by more sensitive analytical methods (e.g. immune-based analysis). Significantly, these experiments identified a number of new, interesting interacting candidate proteins involved in Pdx1 action.
Figure 2. Proteins in NuRD, Tif1β and Swi/Snf immunoprecipitated with Pdx1 in mouse βTC3 and human EndoC-βH1 cells.
A) Proteins of the Swi/Snf (Brg1, Brm), NuRD (Rbbp4, Mi2) complexes, and Tif1β were enriched in Pdx1 antibody ReCLIP precipitations of βTC3 (left) and EndoC-βH1 (right) extracts. B) Reverse co-IPs illustrate Pdx1 enrichment in βTC3 nuclear extracts immunoprecipitated with Tif1β, Swi/Snf and NuRD coregulator subunit antibodies (Figure S1). C,D,E) Sucrose gradient sedimentation demonstrates that the Pdx1 C) migrating in the highest molecular weight fraction retains the ability to D) bind Pdx1 regulated control element DNA and E) coprecipitate with SWI/SNF complex subunits. N≥3 for each co-IP.
Pdx1 interacts in the developing and adult pancreas with Brg1 and Brm, the catalytic ATPase subunits of the Swi/Snf complex
The contribution of Swi/Snf to Pdx1 activity was further investigated because of the prominent actions of this coregulator in developing neuron, cardiomyocyte and lymphocyte cell proliferation and lineage decisions (Chi et al., 2003; Lessard et al., 2007; Lickert et al., 2004). Immunofluorescence analysis revealed that the ATPase subunits of Swi/Snf, Brg1 and Brm, were broadly expressed in the embryonic and adult pancreas, with no observable difference in protein level between cell types within each developmental stage (Figure S2A,B). Because of the unique and dynamic expression pattern of Pdx1 in the pancreas, this transcription factor would be coexpressed with Brm- and Brg1-Swi/Snf within multipotent pancreatic progenitor cells during early embryogenesis (i.e. embryonic day E12.5, (Offield et al., 1996)), and then principally in the insulin+ cells of developing (E15.5, E18.5) and adult islet β-cells that express relatively high levels of Pdx1 (termed Pdx1High (Boyer et al., 2006)), and not acinar, ductal or other islet cell types (i.e. α, ε, δ and PP) (Guz et al., 1995; Ohlsson et al., 1993).
The proximity ligation assay (PLA) was next used to evaluate Pdx1 binding to Brg1 and Brm in the forming and adult pancreas. In this assay, a fluorescent signal is generated if Pdx1 is within 30-40 nm of these Swi/Snf proteins, a physical distance only producing a signal for neighboring proteins. However, the signals are not a reflection of the absolute quantity of Pdx1:Brg1-Swi/Snf or Pdx1:Brm1-Swi/Snf complexes in a cell due to antibody quality and/or other vagaries of the assay. Significantly, Pdx1:Brg1 and Pdx1:Brm signals were detected in nearly all Pdx1+ pancreatic progenitors of the E12.5 pancreatic epithelium (Figure S3A,B). In contrast, no Brg1 or Brm PLA binding was observed using antibodies specific for Sox9 (Seymour et al., 2007) or Ptf1a (Krapp et al., 1998), other key transcription factors enriched in this multipotent progenitor population (data not shown). Notably, Pdx1:Brg1 and Pdx1:Brm signals became confined to insulin+ cells later in development and in islets (Figure S3A,B). These experimental results demonstrate that Pdx1 interacts with the core catalytic subunits of Swi/Snf in the developing and adult mouse pancreas, which we conclude represents greater Swi/Snf complex recruitment. Furthermore, the progressive restriction of binding to insulin+ cells supports the specific nature of these interactions, and implies functional relevance upon consideration of the many established roles of Swi/Snf in other cell types (Chi et al., 2003; Lessard et al., 2007; Lickert et al., 2004).
Blood glucose concentration dynamically regulates Pdx1:Brg1 levels in mouse islet β-cells
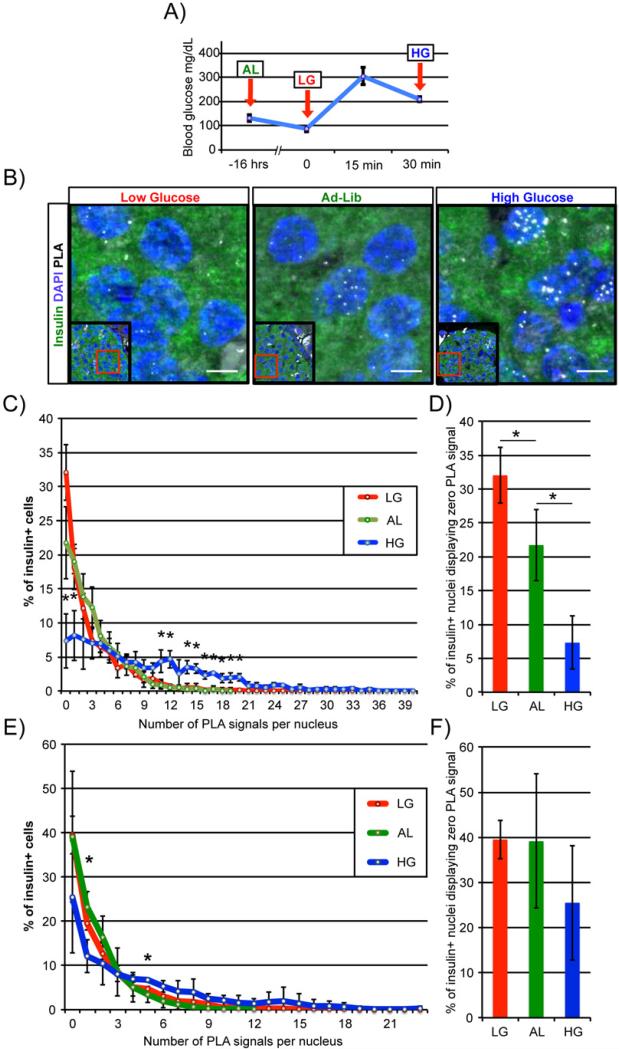
We next asked whether Pdx1:Swi/Snf complex formation was regulated in a glucose-dependent manner in vivo by comparing PLA signal numbers in pancreata prepared from fasted mice with low blood glucose levels to those fasted then given an intraperitoneal injection of a high glucose solution (Figure 3A). Strikingly, the number of Pdx1:Brg1 complexes was significantly increased compared to fasted and ad-lib fed controls 30 minutes after glucose treatment, as shown quantitatively by the specific increase per β-cell nucleus after high glucose treatment (Figure 3B, C). Additionally, ad-lib fed and fasted mouse islets had roughly three-and five-fold more β-cells displaying zero detectible PLA signals than glucose-injected animals (Figure 3D). These data illustrate a strong, positive relationship between high glucose conditions that stimulate Pdx1 β-cell activity and Pdx1:Brg1 binding (Figure 3C). Supporting the specific nature of these interactions, no PLA signals were detected under these conditions between Pdx1 and Isl1 transcription factor coregulator, Ldb1 (Hunter et al., 2013) (Figure S4). Interestingly, although the Pdx1:Brm signals changed some under these conditions, relatively few of the data points were significantly different between low, ad-lib and high glucose conditions (Figure 3E). In addition, the number of β-cells harboring zero detectible Pdx1:Brm PLA signals was not altered (Figure 3F). These results indicate that blood glucose signaling dynamically regulates Pdx1 and Brg1-Swi/Snf association.
Figure 3. Pdx1:Brg1 complex formation is acutely regulated by blood glucose concentration in vivo.
A) Diagram showing mouse blood glucose concentration and time of Ad-Lib (AL), Low glucose (16hr fast (LG)) and High glucose (16hr fast + glucose injection (HG)) conditions; arrows represent time of sacrifice and pancreas fixation. B) Pdx1:Brg1 PLA/immunofluorescence analysis of islet β-cell nuclei reveals that Pdx1:Brg1 signals become more abundant with increasing blood glucose concentration. Representative images are shown with PLA signals appearing as white spots; lower magnification insets are provided to orient each image. Scale bars indicate 5μM. C) The distribution of insulin+ PLA signals under high glucose conditions were significantly greater than in low or Ad-Lib β-cells, although the E) Pdx1:Brm population was essentially unchanged. Islets from high glucose conditions possessed significantly fewer β-cells displaying zero detectible D) Pdx1:Brg1 signals than low or Ad-Lib conditions. This pattern was not observed for F) Pdx1:Brm between glucose conditions. N≥3, *P<0.05
Brg1 and Brm play opposing roles in the regulation of β-cell specific genes
Brg1 or Brm are incorporated into Swi/Snf complexes in a mutually exclusive manner (Wang et al., 1996). These two ATPases share ~74% amino acid sequence identity and have similar in vitro biochemical activity (Khavari et al., 1993; Phelan et al., 1999). Brg1 and Brm have the capacity to compensate for one another in heterozygous ATPase subunit mutant mice during development, although their combined gene dosage is critical to function (Bultman et al., 2000; Smith-Roe and Bultman, 2013; Willis et al., 2012). Null mutants of nearly ubiquitously expressed Brg1 and Brm manifest different phenotypes in vivo. Thus, Brg1−/− mice die at the preimplantation stage of development (Bultman et al., 2000), while Brm−/− mice survive to adulthood with only a roughly 15% increase in body mass, which is attributed to greater bone density (Reyes et al., 1998). Intriguingly, these ATPases have distinct and antagonistic roles in human osteoblast formation, with depletion of BRG1 hindering differentiation and BRM accelerating (Flowers et al., 2009). BRM-SWI/SNF represses transcription of differentiation genes such as osteocalcin in osteoblast progenitors, at least in part through recruitment of the HDAC1 corepressor. In contrast, BRM and HDAC1 dissociate from the osteocalcin promoter during differentiation, allowing activating BRG1-specific complexes to initiate transcription. These experiments reveal how recruitment of BRG1 or BRM containing SWI/SNF complexes can produce different transcriptional outcomes.
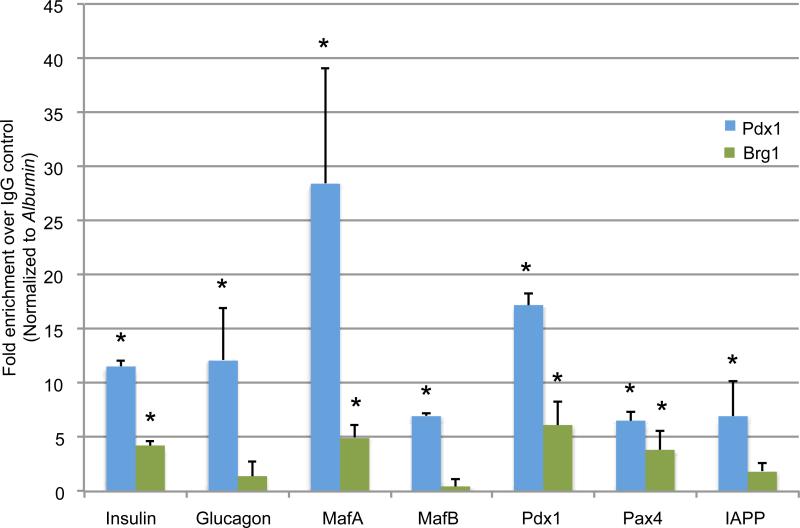
Pdx1 both activates and represses gene transcription in islet β-cells (Gao et al., 2014). Quantitative ChIP-qPCR assays were performed in mouse Min6 β-cells with Brg1 and Brm-specific antibodies to examine their pattern of recruitment to genes that Pdx1 either directly stimulates (i.e. β-cell-enriched Insulin, MafA andPdx1) or inhibits (i.e. islet α-cell-enriched MafB and Glucagon (Artner et al., 2006; Gao et al., 2014)). Brg1 was recruited to genes stimulated by Pdx1 in β-cells (Figure 4). However, antibody reagent limitations precluded our ability to detect Brm control region binding, as illustrated by our inability to even detect control, bona fide Brm binding sites in this assay (Figure S5A).
Figure 4. Brg1 binds to Pdx1 activated genes in Min6 β-cells.
ChIP assays illustrating Pdx1 and Brg1 genomic binding, with Brg1 enriched at activated (Insulin, MafA R3, Pdx1 AI/II, Pax4) Pdx1 target genes in Min6 cells. Notably, Brg1 did not bind to these control regions in Brg1+, PDX1− HeLa cells (Figure S5B). N=4, *P<0.05
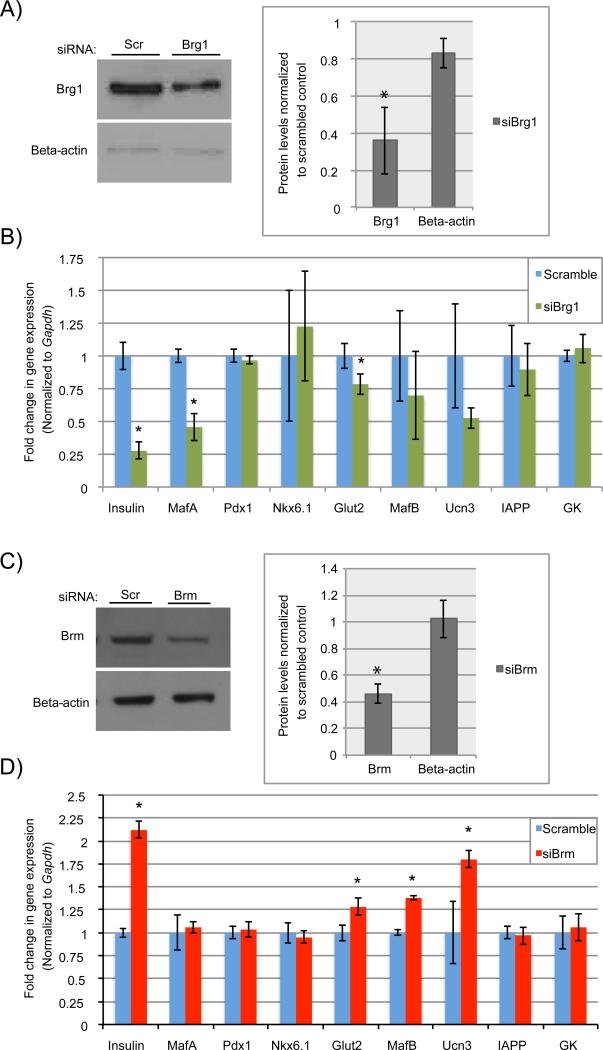
Depletion of Pdx1 from either islet β-cells in vivo or β-cell lines causes a rapid loss in cell identity marker expression (e.g. MafA, Glut2, insulin), and activation of islet α-cell-specific genes (e.g. MafB, glucagon) (Gao et al., 2014). The induction of islet α-cell-enriched MafB transcription factor expression was a driving force in glucagon transcription. These studies clearly illustrate the critical role of Pdx1 in maintaining β-cell identity. We therefore sought to investigate how depletion of Brg1 or Brm in rat INS-1 β-cells influenced Pdx1-mediated transcriptional control. Brg1 and Brm siRNA targeting decreased protein levels by roughly 65% (Figure 5A) and 55% (Figure 5C), respectively. As expected from the ChIP results, Brg1 knockdown resulted in a significant reduction in Insulin, MafA and Glut2 transcript levels (Figure 5B). Interestingly, Brm depletion led to increased Insulin, Glut2, Ucn3, and MafB transcripts (Figure 5D). Our data strongly indicate that Brg1-Swi/Snf serves as a coactivator of Pdx1-mediated gene expression in islet β-cells, whereas Brm-Swi/Snf acts in a corepressive manner.
Figure 5. The Pdx1 recruited Brg1- and Brm-containing Swi/Snf complexes appear to play opposing regulatory roles in β-cells.
Treatment with A) siBrg1 or C) siBrm effectively reduced protein levels over scrambled RNAs in INS-1 cells as demonstrated by immunoblot and densitometry analysis. B) Expression of Pdx1 activated Insulin, MafA and Glut2 were significantly compromised following Brg1 knockdown, while D) Brm knockdown had the opposite effect, causing up-regulation of Insulin, Glut2 and non-Brg1 targets (MafB, Ucn3). The siRNA transfections were performed in triplicate on three separate occasions. N=3, *P<0.05
PDX1 recruitment of BRG1-SWI/SNF is compromised in human T2DM islet β-cells
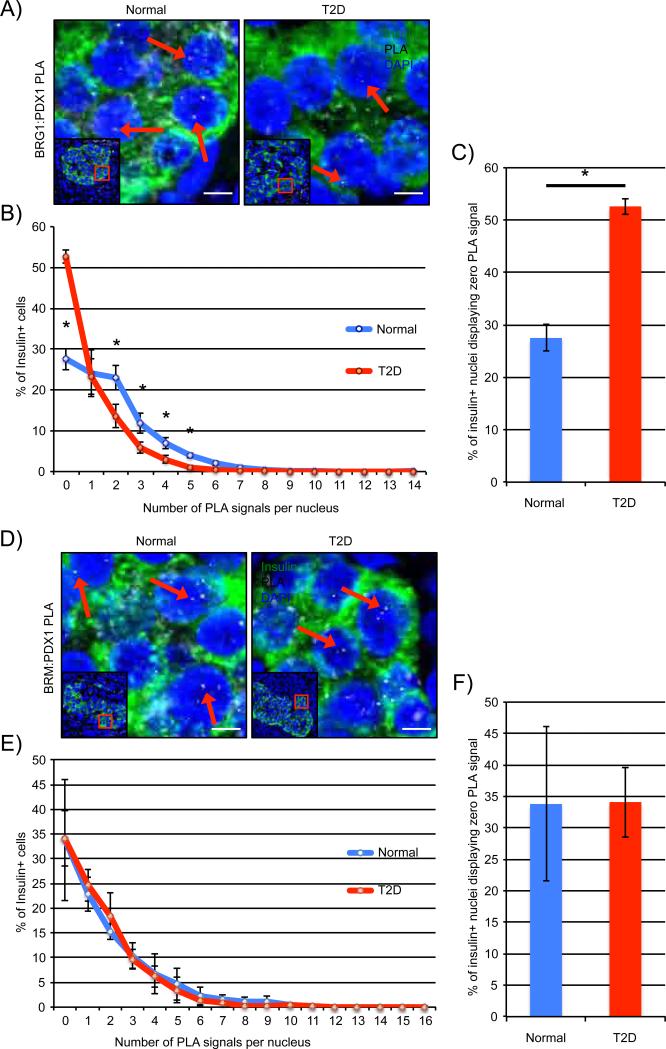
Reduced levels of a small subset of islet-enriched transcription factors contribute to T2DM β-cell dysfunction, with PDX1 among the specifically affected factors (Guo et al., 2013). We therefore sought to investigate whether alterations in PDX1:SWI/SNF complex formation could be contributing to diminished β-cell activity. PDX1 binding to BRG1 (Figure 6A) and BRM (Figure 6D) was readily detected in normal human islet β-cell nuclei. However, only insulin+ PDX1:BRG1 (Figure 6B) signal numbers were significantly diminished in age-, sex- and BMI-matched T2DM samples, and not PDX1:BRM (Figure 6E). The change in PDX1:BRG1 levels paralleled that of PDX1 in T2DM islet β-cells (Guo et al., 2013), while there is no apparent change in BRG1 or BRM levels compared to normal tissue (Figure 3C,D). In addition, there was a roughly two-fold increase in the number of β-cells with zero PLA signals in the PDX1:BRG1 T2DM samples (Figure 6C), even though PDX1:BRM levels were unaffected (Figure 6F). This suggests that the quantitative change in the amount of PDX1 protein affects BRG1-SWI/SNF coactivator binding in T2DM β-cells, and not the BRMSWI/SNF corepressor. Collectively, these results strongly indicate that association of PDX1 to the SWI/SNF chromatin remodeler is dynamically regulated under both physiological and pathophysiological conditions in islet β-cells.
Figure 6. PDX1:BRG1-SWI/SNF activator levels are compromised in T2DM islet β-cells.
Representative PLA signals for PDX1 and A) BRG1 or D) BRM complexes in age-, sex- and BMI-matched normal and T2DM human pancreas sections; arrows indicate PLA signals and lower magnification insets are provided to orient each image. Scale bars indicate 5μM. B) PDX1:BRG1, but not E) PDX1:BRM signal levels were significantly attenuated in T2DM β-cell nuclei. C) T2DM islets contain two-fold more β-cells displaying no detectible PDX1:BRG1 signal than normal controls. No significant difference in PDX1:BRM PLA signal E) distribution per β-cell or F) number of β-cells presenting zero was observed between T2DM and normal human islets. Paraffin sections from three individual T2DM and normal donors were independently analyzed and quantitated. N=3 for each donor of each type, *P<0.05
Discussion
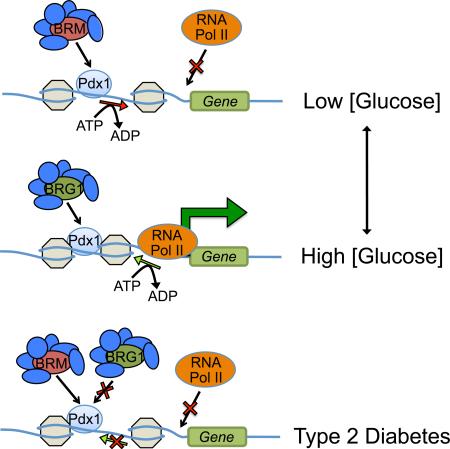
Modulation of target gene expression by transcription factors is contingent upon their ability to recruit coregulator proteins, such as those with a capacity to modify chromatin structure. Surprisingly, despite the fundamental importance of transcription factors such as Pdx1, MafB, Nkx2.2, and Nkx6.1 to islet β-cell development and function, there is a glaring deficiency in our knowledge of how coregulators influence these, and other islet enriched transcription factors. Thus, the goal here was to generate an impartial and comprehensive list of coregulators binding to Pdx1 in β-cells. Our ReCLIP/mass spectrometry screen returned a substantial number of candidate coregulators with a vast array of cellular functions, including several capable of mediating the positive and negative transcriptional actions linked to Pdx1 (e.g. TIF1β (Iyengar et al., 2011), DNA methyltransferase 1 (Dnmt1 (Dhawan et al., 2011)), NuRD (Miccio et al., 2010), and Swi/Snf (Flowers et al., 2009)) (Figure 2, Table 1, Table S1). We focused our analysis on determining the significance of the Pdx1:Swi/Snf complex in modulating β-cell activity. Our results strongly suggest that recruitment of Swi/Snf complexes harboring the Brg1 ATPase is essential for Pdx1 transactivation, while the Brm-Swi/Snf complexes mediate repression (Figure S6).
Brg1 and Brm were widely produced throughout the pancreatic epithelium and in the surrounding mesenchyme of the E12.5 pancreas (Figure S2A, B). PLA analyses revealed Pdx1 interacts with both ATPases in nearly all cells of the multipotent Pdx1+ epithelium, but not in the surrounding mesenchyme where Pdx1 is not produced (Figure S3A, B). Notably, a hypoplastic pancreas phenotype was found in mice upon removal of floxed Brg1 early in pancreas development by Ptf1a-Cre (von Figura et al., 2014). Interestingly, all of the mature pancreatic cell types are formed and Brg1 mutant animals are otherwise healthy, albeit final pancreas size was reduced by 50%. Because the number MPCs limits pancreas size (Stanger et al., 2007), we propose that reduced Brg1 decreases Pdx1+/Ptf1a+/Sox9+ progenitor numbers by affecting their proliferative capacity and/or health. Moreover, we further suggest that this is principally due to the actions of Pdx1 recruitment of Swi/Snf to MPC target genes, and not Ptf1a or Sox9, as neither associated with Swi/Snf in this cell population. It will be interesting to determine if Brm-Swi/Snf compensates for the loss of Brg1 in this context and how this coregulator influences Pdx1 action in adult islet β-cells.
In contrast to the extensive interactions of Pdx1:Swi/Snf within the pancreas MPC population, complex formation became restricted to the insulin+ cells produced later in development and in islet β-cells (Figure S3A,B). However, as islet somatostatin hormone+ δ-cells and acinar cells produce low levels of Pdx1, these results indicate some change within Pdx1 (potentially a post-translational modification) or simply the higher levels in β-cells allows specific recruitment of widely distributed Brg1-Swi/Snf and Brm-Swi/Snf. Strikingly, knockdown of Brg1 and Brm in β-cell lines revealed that Brg1 was important for coactivation of Pdx1 target genes (Insulin, MafA, Glut2), and Brm corepression (Insulin, Glut2, MafB, Ucn3) (Figure 5A-D). The acute conditions of our analysis presumably enabled detection of this regulatory pattern despite an only a 50% reduction in ATPase subunit levels, and not compensation by the unaffected coregulator that has been observed in vivo (Bultman et al., 2000; Smith-Roe and Bultman, 2013). Furthermore, Brg1 was found to directly bind within Pdx1 binding control element containing regions of these activated genes in ChIP assays (Figure 4) (Bultman et al., 2000; Smith-Roe and Bultman, 2013; Willis et al., 2012).
We next examined if Pdx1 binding to Brg1-Swi/Snf and Brm-Swi/Snf was acutely regulated by changes in blood glucose levels in vivo, specifically examining low (fasting) to high (fed) conditions that influence Pdx1 activity in β-cell lines. For example, Insulin gene transcription is enhanced by recruitment of the p300 coactivator, which catalyzes histone H4 hyperacetylation within the proximal promoter region that correlates with gene activation (Mosley et al., 2004). Conversely, Hdac1/2 recruitment by Pdx1 at non-stimulating, low glucose levels inhibits transcription (Mosley and Ozcan, 2004). Similarly, Pdx1:Brg1-activator complex formation was rapidly and significantly increased in islet β-cells by elevated blood glucose levels, while Pdx1:Brm-repressor binding was unaffected (Figure 3A-F). This was observed under circumstances where nuclear Pdx1, Brg1 and Brm levels were unaffected by changing glucose levels (Figure S7). We conclude that high glucose concentrations amplified Pdx1:Brg1-Swi/Snf activator complex formation over Pdx1:Brm-Swi/Snf, causing transcriptional activation by directly influencing nucleosome occupancy and phasing in an ATP-dependent manner (Figure S6A,B). This represents a distinct mechanism from Pdx1 recruited p300 gene activation described above. Collectively, these results highlight how context-dependent recruitment of physiologically regulated and compositionally distinct Swi/Snf complexes dictate Pdx1 action and β-cell function.
Peripheral insulin resistance and islet β-cell dysfunction are the fundamental causes of T2DM, and symptoms often present slowly, after many years of progressive loss of β-cell function and metabolic control. The initial response to peripheral insulin resistance is augmentation of Islet β-cell mass and function (Talchai et al., 2009). However, the increasing metabolic demand imposed on β-cells is strongly linked to their accumulation of destructive stress molecules, such as reactive oxygen species. The resulting oxidative stress conditions induce deleterious posttranslational modifications on proteins, affecting activity, stability and subcellular localization. Indeed, a selective decline in transcription factors central to glucose sensing and insulin secretion is observed in T2DM islet β-cells (i.e. MAFA, MAFB, PDX1, and NKX6.1) (Guo et al., 2013). Notably, MAFA and MAFB are highly susceptible to oxidative stress and their rapid loss is proposed to dictate first phase insulin secretion defects early in disease progression. Persistent insults to PDX1 and NKX6.1, however, cause a more prolonged decline (years to decades) in activity, ultimately resulting in overt diabetes manifesting from severe β-cell dysfunction.
Although it remains unclear how these T2DM associated transcription factors are rendered inoperative in vivo, we speculate that pathophysiological conditions affect coregulator binding, and consequentially, transcription factor function. Significantly, coregulator catalyzed changes in epigenetic DNA methylation and histone modifications are found in T2DM islets, which influences promoter and enhancer structures driving islet-specific gene expression (Dayeh et al., 2014; Parker et al., 2013). In addition, our PLA results illustrated that PDX1:BRG1-activator binding was diminished in T2DM β-cells, while PDX1:BRM-repressor complex formation was unaffected (Figure 6A-F). Presumably, decreased PDX1:BRG1-activator formation simply reflects the diminished levels of PDX1 protein in T2DM β-cells (Guo et al., 2013), while how BRM binding to PDX1 is selectively retained is unclear. Posttranslational modifications of PDX1 and/or SWI/SNF subunits likely influence their interactions, since, for example, Pdx1 transcriptional activity (i.e. Hdac1/2 and p300 recruitment (Mosley and Ozcan, 2004; Mosley et al., 2004)) and protein destruction (Pcif1-Cul3 (Claiborn et al., 2010)) are impacted by such events. We propose that PDX1:BRM-SWI/SNF activity contributes to the loss in T2DM β-cell function (Figure S6). Moreover, conditions affecting PDX1 coactivator and corepressor recruitment occur relatively soon after exposure to insulin resistance in the context of T2DM disease process, and well before overt transcription factor loss.
Overall, our results clearly demonstrate the prominent role of Swi/Snf in regulating Pdx1 activity in β-cells. Furthermore, we have uncovered that Pdx1 recruitment of Brg1-Swi/Snf and Brm-Swi/Snf is influenced by physiological and pathophysiological settings, which are hypothesized to have significant implications on transcription factor activity and β-cell function. In addition, this study illustrates how a ReCLIP/mass spectrometry strategy can be used to identify coregulators of other islet-enriched transcription factors. Such knowledge will provide valuable mechanistic insight into how these transcription factors regulate islet cell formation, function, and survival.
Methods
ReCLIP/MS
Mouse βTC-3 and human EndoC-βH1 cells were grown as defined earlier (Nagamatsu and Steiner, 1992; Ravassard et al., 2011). The ReCLIP procedure was performed as described (Smith et al., 2011) with minor modifications. Briefly, fourteen 15cm culture plates of βTC-3 cells at 70% confluency (~108 cells) were exposed to freshly made DSP (stock 20mM in DMSO) diluted to a final concentration of 1mM in phosphate-buffered saline (PBS) at pH 7.4 for 45 minutes at 37°C. Nuclear extract was prepared as previously (Schreiber et al., 1989), except that DTT was withheld from the extraction buffer. The nuclear extract was incubated with either goat α-Pdx1 (generated by Dr. Chris Wright (Vanderbilt University)) antibody or goat IgG (control) bound Protein G Dynabeads for 3 hours at 4°C, and then washed with RIPA buffer (100mM NaCl, 1% Nonidet P40, 0.5% deoxycholic acid, 0.1% SDS, 50mM Tris/HCl, pH 8.0, and 1mM EDTA). Pdx1 binding partners were eluted with RIPA buffer supplemented with 200mM DTT. Eluted proteins were analyzed via MudPIT (Multidimensional Protein Identification Technology) as previously described (Martinez et al., 2012) in the Vanderbilt University Medical Center Proteomics Core. MS analysis was performed on at least 3 independent ReCLIP preparations, with each showing similar enrichment of specific peptides.
Coimmunoprecipitations and immunoblotting
βTC-3 nuclear extracts were incubated with either goat α-Pdx1 (generated by Dr. Chris Wright (Vanderbilt University)), rabbit α-Brg1 (Santa-Cruz H88-X), rabbit α-Tif1β (Abcam ab10483), rabbit α-Mi2β (Abcam, ab72418) antibody or species matched IgG (control) bound Protein G Dynabeads for 3 hours at 4°C, as describe previously (Hunter et al., 2013). The immunoprecipitates were fractionated by SDS/PAGE (NuPAGE 10% Bis-Tris gels), transferred on to PVDF membrane, and probed with the following antibodies: goat α-Pdx1 (1:10,000); rabbit α-Brg1 (Santa-Cruz H88-X, 1:5,000); rabbit α-Brm (Cell Signaling 6889S, 1:2,000); rabbit α-Tif1β (Abcam ab10483, 1:2,000); rabbit α-RBBP4 (Bethyl Laboratories A301-206A, 1:2,000); rabbit α-Mi2β (Abcam, ab72418, 1:2,000); rabbit α-Beta-Actin (Cell Signaling 4967S, 1:2,000); HRP-conjugated α-rabbit IgG (Promega 1:2,000), HRP-conjugated α-goat IgG (Santa-Cruz, 1:2,000). The experiments were performed at least 3 times and quantitated using NIH ImageJ software.
Sucrose gradient ultracentrifugation and Electrophoretic Mobility Shift Assay (EMSA)
Sucrose gradients were performed as previously described (Guo et al., 2010). Briefly, βTC-3 nuclear extract (1.5-2.5mg) were separated over a 10-35% sucrose gradient (4.5 mL total volume). Fractions (300 μL each, excluding the first 500 μL) were analyzed by immunoblotting, mouse MafA Region 3 element binding in electrophoretic mobility shift assays (Guo et al., 2010), and by immunoprecipitation.
Immunofluorescence
Tissue fixation, embedding, and immunofluorescence labeling were performed as previously described (Matsuoka et al., 2003). The primary antibodies used were goat α-Pdx1 (1:15,000), rabbit α-Brg1 (1:300), rabbit α-Brm (Abcam ab72418, 1:300), and guinea pig α-insulin (Dako A0546, 1:500). The secondary Cy2-, Cy3-, or Cy5-conjugated donkey α-rabbit, α-guinea pig, and α-goat IgGs were obtained from Jackson ImmunoResearch Laboratories. Nuclear counterstaining was performed using DAPI (Invitrogen). Immunofluorescence images were acquired by fluorescence microscopy using a Zeiss Axioimager M2 and images processed by NIH ImageJ software.
Proximity Ligation Assay (PLA)
Paraffin embedded human pancreas, mouse embryonic and adult pancreas samples were analyzed by procedures described by the PLA kit manufacturer (OLink Bioscience), with minor modifications. Slides were de-waxed/rehydrated as previously described (Matsuoka et al., 2003). Heat antigen retrieval in 1X TEG (25mM Tris HCl pH 8, 10mM EDTA, 50mM glucose) was followed by three 10-minute 1XPBS washes. A 1% BSA/PBS blocking solution supplemented with 5% normal donkey serum was applied for 2 hours at room temperature. The following primary antibodies were incubated in a humidity chamber overnight at 4°C: goat α-Pdx1 (1:15,000), rabbit α-Brg1 (1:300), rabbit α-Brm (Abcam ab72418, 1:300), mouse α-glucagon (Sigma g2654-.5mL, 1:4,000), and guinea pig α-insulin (1:500). Immunofluorescence images were acquired on a Zeiss Axioimager M2 (Zeiss) and processed for counting analysis using ImageJ (National Institutes of Health) software. The discrete fluorescent nuclear PLA spots were counted from at least three pancreas sections per experimental animal or human donor. The Gift of Hope Organ Procurement Organization in Chicago generously provided human pancreata, which were obtained from 3 normal and 3 T2DM de-identified cadaver donors: normal, 2 male and 1 female (59.3 ± 8.5 years [range: 51 to 68], BMI 22.6 ± 2.4 [range: 21.1 to 25.4]); T2DM, 2 male, 1 female, (57.0 ± 5.3 years [range: 51 to 61], BMI 25.9 ± 7.1 [range: 21.2 to 34].
Chromatin immunoprecipitation assays
Mouse Min6 β-cells (~4 × 106 cells) were 1% formaldehyde cross-linked, and the sonicated protein-DNA complexes isolated under conditions described previously (Gerrish et al., 2001). The fragmented chromatin was incubated with α-Pdx1, α-Brg1, α-Brm or species-matched IgG (Bethyl Laboratories), and the complexes isolated with BSA- or herring sperm ssDNA (Abcam ab46666)-blocked protein-A Dynabeads (Invitrogen). Quantitative real-time PCR was performed using SYBR Green master mix in a Roche LightCycler 480II. The PCR primers were as follows: MafB -742 to -938 (site1): Forward: 5’-TTAGCGCAGACAGAGCTACCGAAA-3’, Reverse: 5’-ATACTCTTTACACTCCCACCCTCG-3’; Glucagon +70 to -148 (contains G1 element): Forward: 5’-CGTAAAAAGCAGATGAGCAAAGTG-3’, Reverse: 5’-GAACAGGTGTAGACAGAGGGAGTCC-3’; Pax4 -1841 to -1966 (contains β-cell-specific enhancer (Stein 2010 MCB)) Forward: 5’-CCAACGATCCAGGCTCTACATC-3’, Reverse: 5’-CGGGTTTGGGGCTAATTGTCC-3’; Pdx1 -2471 to -2598 (contains Area I): Forward: 5’-TGGCTCGGGAAGGCTCTTG-3’, Reverse: 5’-CCATCAGGTGGCTAAATCCATTATG-3’; IAPP -97 to -190: Forward: 5’-TCACCCACACAAAGGCACTCAG-3’ Reverse: 5’-GGTTTCATTGGCAGATGGAGC-3’; MafA R3: Forward: 5’-CTGGAAGATCACCGCACA-3’, Reverse: 5’-ATTTACCAAGCCCCAAACG-3’; Insulin (proximal promoter): Forward: 5’-GCCATCTGCTGACCTACCC-3’, Reverse: 5’-CCCCTGGACTTTGCTGTTT-3’; Albumin -3164 to -3342 (distal TAAT-containing region): Forward: 5’-TGGGAAAACTGGGAAAACCATC-3’, Reverse: 5’-CACTCTCACACATACACTCCTGCTG-3’. Experiments were performed with at least three independently isolated chromatin preparations.
siRNA knockdown and RNA analysis
Rat INS-1 cells (7.5×105/well) were seeded into 6-well plates and targeting Brg1, Brm or scrambled control siRNAs (Dharmacon) introduced using Lipofectamine 2000 (Invitrogen). RNA and immunoblot protein studies were performed 72 hours following treatment. Cellular RNA was isolated using the QIAGEN RNeasy Mini Kit. cDNA was prepared from RNA using the iScript cDNA synthesis kit (Bio-Rad) and quantitative real-time PCR performed using SYBR Green master mix and a Roche LightCycler 480II. The experimental data were normalized to glyceraldehyde-3-phosphate dehydrogenase (Gapdh) mRNA levels and relative changes calculated by the comparative DCt method (Chakrabarti et al., 2002).
Glucose treatment conditions
C57BL/6 mice (Charles River Laboratories) were fasted from 18:00 to 10:00 hours, and then blood glucose measurements were taken from the tail tip using an Aviva glucometer (Accu Chek). Glucose was administered to the ‘high’ group by intraperitoneal injection of a 20% glucose solution to achieve a final dose of 2g/kg body weight. Glucose measurements were taken at 0, 15 and 30 minutes post-injection. Pancreata were harvested from each group and tissue embedded as previously described (Matsuoka et al., 2003).
Statistics
Data are expressed as the means ± SEM. P values were calculated with a Student's 2-tailed test. Results were considered significant at P < 0.05.
Study approval
The IRB at the University of Chicago and Vanderbilt University approved the use of human tissues in these studies. The Vanderbilt University IACUC approved all studies involving animals.
Supplementary Material
Acknowledgments
We thank Dr. Chris Wright at Vanderbilt for generously providing goat raised Pdx1 antibody. This work was supported by NIH grants (DK050203 to R.S.; Vanderbilt Molecular Endocrinology Training Program grant 5T32 DK07563 to B.M.; DK-020595 to the University of Chicago Diabetes Research and Training Center (Animal Models Core), DK-072473, AG-042151 to M.H.; and CA55724 and CA111947 to A.B.R.) M.H. was also supported by a gift from the Kovler Family Foundation and R.S. from the Mark Collie Endowment fund). Imaging was performed with NIH support at the Vanderbilt University Medical Center's Cell Imaging Shared Resource (CA68485, DK20593, DK58404, HD15052, DK59637, and EY08126).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: The authors have declared that no conflict of interest exists.
References
- Artner I, Le Lay J, Hang Y, Elghazi L, Schisler JC, Henderson E, Sosa-Pineda B, Stein R. MafB An Activator of the Glucagon Gene Expressed in Developing Islet α-and β-Cells. Diabetes. 2006;55:297–304. doi: 10.2337/diabetes.55.02.06.db05-0946. [DOI] [PubMed] [Google Scholar]
- Babu DA, Deering TG, Mirmira RG. A feat of metabolic proportions: Pdx1 orchestrates islet development and function in the maintenance of glucose homeostasis. Mol. Genet. Metab. 2007;92:43–55. doi: 10.1016/j.ymgme.2007.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbacci E. HNF1 /TCF2 mutations impair transactivation potential through altered co-regulator recruitment. Hum. Mol. Genet. 2004;13:3139–3149. doi: 10.1093/hmg/ddh338. [DOI] [PubMed] [Google Scholar]
- Bhaumik SR, Smith E, Shilatifard A. Covalent modifications of histones during development and disease pathogenesis. Nat. Struct. 38 Mol. Biol. 2007;14:1008–1016. doi: 10.1038/nsmb1337. [DOI] [PubMed] [Google Scholar]
- Boyer DF, Fujitani Y, Gannon M, Powers AC, Stein RW, Wright CVE. Complementation rescue of Pdx1 null phenotype demonstrates distinct roles of proximal and distal cis-regulatory sequences in pancreatic and duodenal expression. Dev. Biol. 2006;298:616–631. doi: 10.1016/j.ydbio.2006.07.020. [DOI] [PubMed] [Google Scholar]
- Bultman S, Gebuhr T, Yee D, La Mantia C, Nicholson J, Gilliam A, Randazzo F, Metzger D, Chambon P, Crabtree G, et al. A<i> Brg1</i> Null Mutation in the Mouse Reveals Functional Differences among Mammalian SWI/SNF Complexes. Mol. Cell. 2000;6:1287–1295. doi: 10.1016/s1097-2765(00)00127-1. [DOI] [PubMed] [Google Scholar]
- Chakrabarti SK, James JC, Mirmira RG. Quantitative Assessment of Gene Targeting in Vitroand in Vivo by the Pancreatic Transcription Factor, Pdx1: IMPORTANCE OF CHROMATIN STRUCTURE IN DIRECTING PROMOTER BINDING. J. Biol. Chem. 2002;277:13286–13293. doi: 10.1074/jbc.M111857200. [DOI] [PubMed] [Google Scholar]
- Chen T, Li E. Structure and function of eukaryotic DNA methyltransferases. Curr. Top. Dev. Biol. 2004;60:55–89. doi: 10.1016/S0070-2153(04)60003-2. [DOI] [PubMed] [Google Scholar]
- Chi TH, Wan M, Lee PP, Akashi K, Metzger D, Chambon P, Wilson CB, Crabtree GR. Sequential roles of Brg, the ATPase subunit of BAF chromatin remodeling complexes, in thymocyte development. Immunity. 2003;19:169–182. doi: 10.1016/s1074-7613(03)00199-7. [DOI] [PubMed] [Google Scholar]
- Claiborn KC, Sachdeva MM, Cannon CE, Groff DN, Singer JD, Stoffers DA. Pcif1 modulates Pdx1 protein stability and pancreatic β cell function and survival in mice. J. Clin. Invest. 2010;120:3713–3721. doi: 10.1172/JCI40440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dayeh T, Volkov P, Salö S, Hall E, Nilsson E, Olsson AH, Kirkpatrick CL, Wollheim CB, Eliasson L, Rönn T, et al. Genome-Wide DNA Methylation Analysis of Human Pancreatic Islets from Type 2 Diabetic and Non-Diabetic Donors Identifies Candidate Genes That Influence Insulin Secretion. PLoS Genet. 2014;10:e1004160. doi: 10.1371/journal.pgen.1004160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhawan S, Georgia S, Tschen S, Fan G, Bhushan A. Pancreatic β Cell Identity Is Maintained by DNA Methylation-Mediated Repression of Arx. Dev. Cell. 2011;20:419–429. doi: 10.1016/j.devcel.2011.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Figura G, Fukuda A, Roy N, Liku ME, Morris JP, IV, Kim GE, Russ HA, Firpo MA, Mulvihill SJ, Dawson DW, et al. The chromatin regulator Brg1 suppresses formation of intraductal papillary mucinous neoplasm and pancreatic ductal adenocarcinoma. Nat. Cell Biol. 2014;16:255–267. doi: 10.1038/ncb2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flotho A, Melchior F. Sumoylation: A Regulatory Protein Modification in Health and Disease. Annu. Rev. Biochem. 2013;82:357–385. doi: 10.1146/annurev-biochem-061909-093311. [DOI] [PubMed] [Google Scholar]
- Flowers S, Nagl NG, Beck GR, Moran E. Antagonistic Roles for BRM and BRG1 SWI/SNF Complexes in Differentiation. J. Biol. Chem. 2009;284:10067–10075. doi: 10.1074/jbc.M808782200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francis J. Pdx-1 Links Histone H3-Lys-4 Methylation to RNA Polymerase II Elongation during Activation of Insulin Transcription. J. Biol. Chem. 2005;280:36244–36253. doi: 10.1074/jbc.M505741200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gannon M, Tweedie Ables E, Crawford L, Lowe D, Offield MF, Magnuson MA, Wright CVE. pdx-1 function is specifically required in embryonic β cells to generate appropriate numbers of endocrine cell types and maintain glucose homeostasis. Dev. Biol. 2008;314:406–417. doi: 10.1016/j.ydbio.2007.10.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao T, McKenna B, Li C, Reichert M, Nguyen J, Singh T, Yang C, Pannikar A, Doliba N, Zhang T, et al. Pdx1 Maintains β Cell Identity and Function by Repressing an α Cell Program. Cell Metab. 2014;19:259–271. doi: 10.1016/j.cmet.2013.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerrish KE, Cissell MA, Stein R. The role of hepatic nuclear factor 1a and PDX-1 in transcriptional regulation of the pdx-1 gene. J. Biol. Chem. 2001 doi: 10.1074/jbc.M109244200. [DOI] [PubMed] [Google Scholar]
- Guo S, Vanderford NL, Stein R. Phosphorylation within the MafA N Terminus Regulates C-terminal Dimerization and DNA Binding. J. Biol. Chem. 2010;285:12655–12661. doi: 10.1074/jbc.M110.105759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Dai C, Guo M, Taylor B, Harmon JS, Sander M, Robertson RP, Powers AC, Stein R. Inactivation of specific β cell transcription factors in type 2 diabetes. J. Clin. Invest. 2013;123:3305–3316. doi: 10.1172/JCI65390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guz Y, Montminy MR, Stein R, Leonard J, Gamer LW, Wright CV, Teitelman G. Expression of murine STF-1, a putative insulin gene transcription factor, in beta cells of pancreas, duodenal epithelium and pancreatic exocrine and endocrine progenitors during ontogeny. Development. 1995;121:11–18. doi: 10.1242/dev.121.1.11. [DOI] [PubMed] [Google Scholar]
- Hale MA, Kagami H, Shi L, Holland AM, Elsässer H-P, Hammer RE, MacDonald RJ. The homeodomain protein PDX1 is required at mid-pancreatic development for the formation of the exocrine pancreas. Dev. Biol. 2005;286:225–237. doi: 10.1016/j.ydbio.2005.07.026. [DOI] [PubMed] [Google Scholar]
- Hunter CS, Dixit S, Cohen T, Ediger B, Wilcox C, Ferreira M, Westphal H, Stein R, May CL. Islet -, -, and -Cell Development Is Controlled by the Ldb1 Coregulator. Acting Primarily With the Islet-1 Transcription Factor. Diabetes. 2013;62:875–886. doi: 10.2337/db12-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyengar S, Ivanov AV, Jin VX, Rauscher FJ, Farnham PJ. Functional Analysis of KAP1 Genomic Recruitment. Mol. Cell. Biol. 2011;31:1833–1847. doi: 10.1128/MCB.01331-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jonsson J, Carlsson L, Edlund T, Edlund H. Insulin-promoter-factor 1 is required for pancreas development in mice. Nature. 1994;371:606–609. doi: 10.1038/371606a0. [DOI] [PubMed] [Google Scholar]
- Khavari PA, Peterson CL, Tamkun JW, Mendel DB, Crabtree GR. BRG1 contains a conserved domain of the SWI2/SNF2 family necessary for normal mitotic growth and transcription. Nature. 1993;366:170–174. doi: 10.1038/366170a0. [DOI] [PubMed] [Google Scholar]
- Krapp A, Knofler M, Ledermann B, Burki K, Berney C, Zoerkler N, Hagenbuchle O, Wellauer PK. The bHLH protein PTF1-p48 is essential for the formation of the exocrine and the correct spatial organization of the endocrine pancreas. Genes Dev. 1998;12:3752–3763. doi: 10.1101/gad.12.23.3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebrun P, Montminy MR, Van Obberghen E. Regulation of the Pancreatic Duodenal Homeobox-1 Protein by DNA-dependent Protein Kinase. J. Biol. Chem. 2005;280:38203–38210. doi: 10.1074/jbc.M504842200. [DOI] [PubMed] [Google Scholar]
- Lessard J, Wu JI, Ranish JA, Wan M, Winslow MM, Staahl BT, Wu H, Aebersold R, Graef IA, Crabtree GR. An Essential Switch in Subunit Composition of a Chromatin Remodeling Complex during Neural Development. Neuron. 2007;55:201–215. doi: 10.1016/j.neuron.2007.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lickert H, Takeuchi JK, von Both I, Walls JR, McAuliffe F, Adamson SL, Henkelman RM, Wrana JL, Rossant J, Bruneau BG. Baf60c is essential for function of BAF chromatin remodelling complexes in heart development. Nature. 2004;432:107–112. doi: 10.1038/nature03071. [DOI] [PubMed] [Google Scholar]
- Liu A, Desai BM, Stoffers DA. Identification of PCIF1, a POZ Domain Protein That Inhibits PDX-1 (MODY4) Transcriptional Activity. Mol. Cell. Biol. 2004;24:4372–4383. doi: 10.1128/MCB.24.10.4372-4383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez MN, Emfinger CH, Overton M, Hill S, Ramaswamy TS, Cappel DA, Wu K, Fazio S, McDonald WH, Hachey DL, et al. Obesity and altered glucose metabolism impact HDL composition in CETP transgenic mice: a role for ovarian hormones. J. Lipid Res. 2012;53:379–389. doi: 10.1194/jlr.M019752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka T. -a., Zhao L, Artner I, Jarrett HW, Friedman D, Means A, Stein R. Members of the Large Maf Transcription Family Regulate Insulin Gene Transcription in Islet Cells. Mol. Cell. Biol. 2003;23:6049–6062. doi: 10.1128/MCB.23.17.6049-6062.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miccio A, Wang Y, Hong W, Gregory GD, Wang H, Yu X, Choi JK, Shelat S, Tong W, Poncz M, et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010;29:442–456. doi: 10.1038/emboj.2009.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosley AL, Ozcan S. The Pancreatic Duodenal Homeobox-1 Protein (Pdx-1) Interacts with Histone Deacetylases Hdac-1 and Hdac-2 on Low Levels of Glucose. J. Biol. Chem. 2004;279:54241–54247. doi: 10.1074/jbc.M410379200. [DOI] [PubMed] [Google Scholar]
- Mosley AL, Corbett JA, Özcan S. Glucose Regulation of Insulin Gene Expression Requires the Recruitment of p300 by the β-Cell-Specific Transcription Factor Pdx-1. Mol. Endocrinol. 2004;18:2279–2290. doi: 10.1210/me.2003-0463. [DOI] [PubMed] [Google Scholar]
- Nagamatsu S, Steiner DF. Altered glucose regulation of insulin biosynthesis in insulinoma cells: mouse beta TC3 cells secrete insulin-related peptides predominantly via a constitutive pathway. Endocrinology. 1992;130:748–754. doi: 10.1210/endo.130.2.1733723. [DOI] [PubMed] [Google Scholar]
- Offield MF, Jetton TL, Labosky PA, Ray M, Stein RW, Magnuson MA, Hogan BL, Wright CV. PDX-1 is required for pancreatic outgrowth and differentiation of the rostral duodenum. Development. 1996;122:983–995. doi: 10.1242/dev.122.3.983. [DOI] [PubMed] [Google Scholar]
- Ohlsson H, Karlsson K, Edlund T. IPF1, a homeodomain-containing transactivator of the insulin gene. EMBO J. 1993;12:4251. doi: 10.1002/j.1460-2075.1993.tb06109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papizan JB, Singer RA, Tschen S-I, Dhawan S, Friel JM, Hipkens SB, Magnuson MA, Bhushan A, Sussel L. Nkx2.2 repressor complex regulates islet -cell specification and prevents -to- -cell reprogramming. Genes Dev. 2011;25:2291–2305. doi: 10.1101/gad.173039.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker SCJ, Stitzel ML, Taylor DL, Orozco JM, Erdos MR, Akiyama JA, van Bueren KL, Chines PS, Narisu N, NISC Comparative Sequencing Program et al. Chromatin stretch enhancer states drive cell-specific gene regulation and harbor human disease risk variants. Proc. Natl. Acad. Sci. 2013;110:17921–17926. doi: 10.1073/pnas.1317023110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquali L, Gaulton KJ, Rodríguez-Seguí SA, Mularoni L, Miguel-Escalada I, Akerman İ, Tena JJ, Morán I, Gómez-Marín C, van de Bunt M, et al. Pancreatic islet enhancer clusters enriched in type 2 diabetes risk-associated variants. Nat. Genet. 2014;46:136–143. doi: 10.1038/ng.2870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan ML, Sif S, Narlikar GJ, Kingston RE. Reconstitution of a core chromatin remodeling complex from SWI/SNF subunits. Mol. Cell. 1999;3:247–253. doi: 10.1016/s1097-2765(00)80315-9. [DOI] [PubMed] [Google Scholar]
- Poss ZC, Ebmeier CC, Taatjes DJ. The Mediator complex and transcription regulation. Crit. Rev. Biochem. Mol. Biol. 2013;48:575–608. doi: 10.3109/10409238.2013.840259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Sharma A, Stein R. p300 mediates transcriptional stimulation by the basic helix-loop-helix activators of the insulin gene. Mol. Cell. Biol. 1998;18:2957–2964. doi: 10.1128/mcb.18.5.2957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu Y, Guo M, Huang S, Stein R. Insulin Gene Transcription Is Mediated by Interactions between the p300 Coactivator and PDX-1, BETA2, and E47. Mol. Cell. Biol. 2002;22:412–420. doi: 10.1128/MCB.22.2.412-420.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravassard P, Hazhouz Y, Pechberty S, Bricout-Neveu E, Armanet M, Czernichow P, Scharfmann R. A genetically engineered human pancreatic β cell line exhibiting glucose-inducible insulin secretion. J. Clin. Invest. 2011;121:3589–3597. doi: 10.1172/JCI58447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes JC, Barra J, Muchardt C, Camus A, Babinet C, Yaniv M. Altered control of cellular proliferation in the absence of mammalian brahma (SNF2α). EMBO J. 1998;17:6979–6991. doi: 10.1093/emboj/17.23.6979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber E, Matthias P, Müller MM, Schaffner W. Rapid detection of octamer binding proteins with “mini extracts”, prepared from a small number of cells. Nucleic Acids Res. 1989;17:6419–6419. doi: 10.1093/nar/17.15.6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seymour PA, Freude KK, Tran MN, Mayes EE, Jensen J, Kist R, Scherer G, Sander M. SOX9 is required for maintenance of the pancreatic progenitor cell pool. Proc. Natl. Acad. Sci. 2007;104:1865–1870. doi: 10.1073/pnas.0609217104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith AL, Friedman DB, Yu H, Carnahan RH, Reynolds AB. ReCLIP (Reversible Cross-Link Immuno-Precipitation): An Efficient Method for Interrogation of Labile Protein Complexes. PLoS ONE. 2011;6:e16206. doi: 10.1371/journal.pone.0016206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith-Roe SL, Bultman SJ. Combined gene dosage requirement for SWI/SNF catalytic subunits during early mammalian development. Mamm. Genome. 2013;24:21–29. doi: 10.1007/s00335-012-9433-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanger BZ, Tanaka AJ, Melton DA. Organ size is limited by the number of embryonic progenitor cells in the pancreas but not the liver. Nature. 2007;445:886–891. doi: 10.1038/nature05537. [DOI] [PubMed] [Google Scholar]
- Stanojevic V, Yao K-M, Thomas MK. The coactivator Bridge-1 increases transcriptional activation by pancreas duodenum homeobox-1 (PDX-1). Mol. Cell. Endocrinol. 2005;237:67–74. doi: 10.1016/j.mce.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Stoffers DA, Thomas MK, Habener JF. Homeodomain Protein IDX-1: A Master Regulator of Pancreas Development and Insulin Gene Expression. Trends Endocrinol. Metab. 1997;8:145–151. doi: 10.1016/s1043-2760(97)00008-8. [DOI] [PubMed] [Google Scholar]
- Sudarsanam P, Winston F. The Swi/Snf family: nucleosome-remodeling complexes and transcriptional control. TRENDS Genet. 2000;16:345–351. doi: 10.1016/s0168-9525(00)02060-6. [DOI] [PubMed] [Google Scholar]
- Talchai C, Lin HV, Kitamura T, Accili D. Genetic and biochemical pathways of β-cell failure in type 2 diabetes. Diabetes Obes. Metab. 2009;11:38–45. doi: 10.1111/j.1463-1326.2009.01115.x. [DOI] [PubMed] [Google Scholar]
- Thomas MK, Yao K-M, Tenser MS, Wong GG, Habener JF. Bridge-1, a novel PDZ-domain coactivator of E2A-mediated regulation of insulin gene transcription. Mol. Cell. Biol. 1999;19:8492–8504. doi: 10.1128/mcb.19.12.8492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Cote J, Xue Y, Zhou S, Khavari PA, Biggar SR, Muchardt C, Kalpana GV, Goff SP, Yaniv M, et al. Purification and biochemical heterogeneity of the mammalian SWI-SNF complex. EMBO J. 1996;15:5370. [PMC free article] [PubMed] [Google Scholar]
- Wells L, Whelan SA, Hart GW. O-GlcNAc: a regulatory post-translational modification. Biochem. Biophys. Res. Commun. 2003;302:435–441. doi: 10.1016/s0006-291x(03)00175-x. [DOI] [PubMed] [Google Scholar]
- Willis MS, Homeister JW, Rosson GB, Annayev Y, Holley D, Holly SP, Madden VJ, Godfrey V, Parise LV, Bultman SJ. Functional redundancy of SWI/SNF catalytic subunits in maintaining vascular endothelial cells in the adult heart. Circ. Res. 2012;111:e111–e122. doi: 10.1161/CIRCRESAHA.112.265587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y-P, Thorel F, Boyer DF, Herrera PL, Wright CVE. Context-specific -to- -cell reprogramming by forced Pdx1 expression. Genes Dev. 2011;25:1680–1685. doi: 10.1101/gad.16875711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao L, Guo M, Matsuoka T. -a., Hagman DK, Parazzoli SD, Poitout V, Stein R. The Islet Cell-enriched MafA Activator Is a Key Regulator of Insulin Gene Transcription. J. Biol. Chem. 2005;280:11887–11894. doi: 10.1074/jbc.M409475200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.