Abstract
A central goal of biology is to uncover the genetic basis for the origin of new phenotypes. A particularly effective approach is to examine the genomic architecture of species that have secondarily lost a phenotype with respect to their close relatives. In the eusocial Hymenoptera, queens and workers have divergent phenotypes that may be produced via either expression of alternative sets of caste-specific genes and pathways or differences in expression patterns of a shared set of multifunctional genes. To distinguish between these two hypotheses, we investigated how secondary loss of the worker phenotype in workerless ant social parasites impacted genome evolution across two independent origins of social parasitism in the ant genera Pogonomyrmex and Vollenhovia. We sequenced the genomes of three social parasites and their most-closely related eusocial host species and compared gene losses in social parasites with gene expression differences between host queens and workers. Virtually all annotated genes were expressed to some degree in both castes of the host, with most shifting in queen-worker bias across developmental stages. As a result, despite >1 My of divergence from the last common ancestor that had workers, the social parasites showed strikingly little evidence of gene loss, damaging mutations, or shifts in selection regime resulting from loss of the worker caste. This suggests that regulatory changes within a multifunctional genome, rather than sequence differences, have played a predominant role in the evolution of social parasitism, and perhaps also in the many gains and losses of phenotypes in the social insects.
Keywords: social parasite, ant, caste, phenotypic plasticity, genome
Introduction
A goal of the genomic revolution is to understand the genetic architecture underlying phenotypic diversity. Genomic studies have been instrumental in helping to integrate the contributions of distinct mechanisms for generating alternative phenotypes, including coding sequence variation, regulatory changes, and gene by environment interactions (Anholt and Mackay 2004; Jones et al. 2012). In addition to answering mechanistic questions for individual species, comparative genomic approaches made possible by advances in high-throughput sequencing and bioinformatics have added a novel perspective on the evolutionary processes that generate both diversification and convergence of organismal phenotypes.
One area that has benefited immensely from genomic approaches is the study of sociality, particularly in the social insects (Robinson et al. 2005; Page and Amdam 2007; Smith et al. 2008; Gadau et al. 2012). In the ants, the evolution of reproductive division of labor between queens and workers was accompanied by pronounced phenotypic shifts in workers from the ancestral hymenopteran female body plan. There are two primary hypotheses to explain the origin of worker-specific traits. First, queen-worker divergence may result from disruptive selection on ancestral gene regulation patterns, co-opting, and exaggerating preexisting regulatory changes across life history stages (West-Eberhard 1987; Gadagkar 1997; Amdam et al. 2006; Toth et al. 2007). More recently, however, studies of gene expression have consistently identified caste-specific “social” genes that are overexpressed in either workers or reproductive individuals (Grozinger et al. 2003; Toth et al. 2007; Bonasio et al. 2010; Wurm et al. 2011; Feldmeyer et al. 2014; Berens et al. 2015; Mikheyev and Linksvayer 2015; Schrader et al. 2015), and comparative genomics studies have revealed that these include an overabundance of evolutionarily novel genes (i.e., “orphan” genes without apparent homology in related organisms) (Bonasio et al. 2010; Johnson and Tsutsui 2011; Feldmeyer et al. 2014). This work has led to an alternate hypothesis that the phenotypic distinctiveness of queens and workers extends to the genetic level (Hunt et al. 2011; Sumner 2014), which releases genes from universal evolutionary constraints and facilitates worker specialization.
Studies investigating the genetic architecture of the worker phenotype generally focus on comparing gene expression of queens and workers within species. However, a potentially more powerful approach is to investigate the genomic impacts of evolutionary losses of the worker phenotype using phylogenetically informed comparisons (Lahti et al. 2009; Cini et al. 2015). Although the vast majority of ant species produce both reproductive and worker castes, there have been multiple independent secondary losses of the worker caste during the transition to a socially parasitic life style, particularly in the ant subfamily Myrmicinae (Hölldobler and Wilson 1990). Workerless social parasites (inquilines) obligately invade host colonies where the parasitic queen cohabits the nest with the resident host queen, and produces exclusively reproductive winged male and female progeny who are reared by the host workers (Buschinger 1986; Buschinger 2009). Like other evolutionary transitions to a parasitic lifestyle, the obligate dependence on a host species to provide all or many vital functions is expected to relax selection on portions of the parasite genome associated with those functions. This may be detectable as genome reduction (Tsai et al. 2013), structural or functional gene loss, or shifts in evolutionary signatures, e.g., from purifying to relaxed selection.
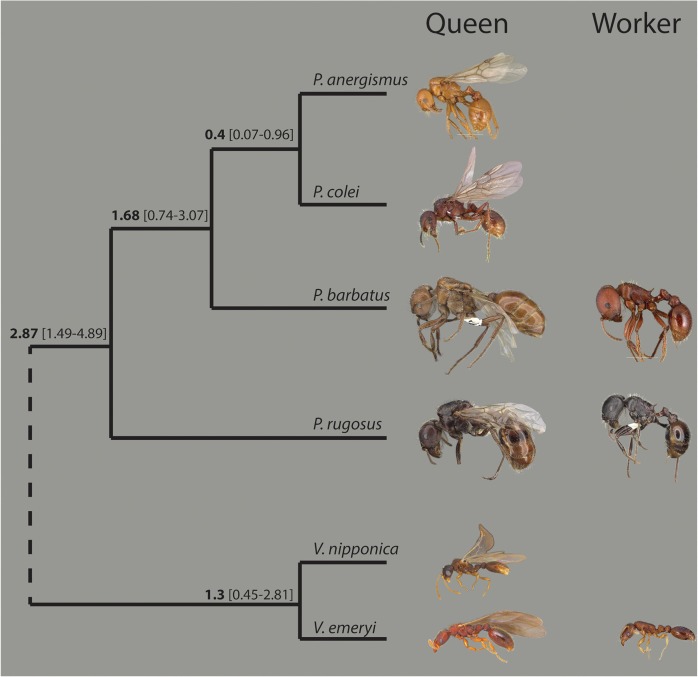
In this study, we examined the genomic effects of the loss of the worker caste in two independent transitions to social parasitism in the myrmicine genera Pogonomyrmex and Vollenhovia. In the seed harvester ant genus Pogonomyrmex, social parasitism has evolved at least once, giving rise to a clade of two parasitic species, Pogonomyrmex anergismus and Pogonomyrmex colei (Johnson et al. 1996; Parker and Rissing 2002), which form a monophyletic group with their two host species, Pogonomyrmex rugosus and Pogonomyrmex barbatus (fig. 1). In the genus Vollenhovia, the workerless species Vollenhovia nipponica parasitizes the closely related species, Vollenhovia emeryi (Kinomura and Yamauchi 1992) (fig. 1). Fossil-calibrated phylogenetic dating suggests that the Pogonomyrmex social parasites and hosts separated ∼1.7 Ma, while the Vollenhovia host-parasite pair diverged ∼1.3 Ma (fig. 1, supplementary fig. S1, Supplementary Material online). We coupled genome sequencing of the three social parasites (P. anergismus, P. colei, and V. nipponica) and their hosts (P. barbatus, P. rugosus, and V. emeryi) with gene expression data from different developmental stages for both queens and workers of two of the host species (P. barbatus and V. emeryi), to determine the genomic changes that are associated with the loss of the worker phenotype. We predicted that, if the queen and worker castes were produced by differentiated sets of worker and queen-specific genes, we should detect signatures of relaxed selection (including accelerated evolutionary rate, accumulation of damaging mutations, or gene loss) in the parasite genomes. In particular, we expected a strong effect in genes homologous to those in their host species that are exclusively expressed, or have biased expression, in the worker caste.
Fig. 1.
The evolutionary relationships, and divergence dates (with 95% CI), for the two monophyletic groups of host and parasites in the subgenus Myrmicinae which are included in this study. The images represent the known castes present in each species; social parasites are those lacking an image for the worker caste. Images from antweb.org.
Results and Discussion
Genome Sequencing
We sequenced the genomes of P. anergismus, P. colei, and V. nipponica, representing two independent origins of parasitism, and the genomes of two host species, P. rugosus and V. emeryi. A third host species, P. barbatus, had already been sequenced and annotated (Smith et al. 2011), and served as the mapping reference for P. rugosus, P. anergismus, and P. colei. Vollenhovia nipponica was mapped to the assembled genome of V. emeryi. Our analyses only include genes with at least 90% of the coding region covered by mapped reads. The mapped genomes covered 98–99% of annotated genes in P. barbatus and 93% in V. emeryi (supplementary table S1, Supplementary Material online). Therefore, at least at the level of the protein-coding genome, there is no evidence of major genomic loss in social parasites as is the case in some non-social parasites (e.g., tapeworms) relative to their free-living ancestors (Tsai et al. 2013).
By using paired-end sequencing reads that generally mapped uniquely within a genome, we established explicit orthologous relationships between genes in the host and parasite species and limited accumulation of assembly and annotation errors to those present in the reference genomes. This approach may, however, have had less power to detect major structural variation relative to comparing assembled genomes, though independent genome assemblies would each have errors, which would then amplify error rates in genome comparisons.
In Search of “Worker Genes”
To test the prediction that genes that primarily function in the worker caste would degrade in the social parasites due to relaxed selection (Linksvayer and Wade 2009), we performed RNA sequencing (RNAseq) on the two host species, P. barbatus and V. emeryi, to establish the degree of queen-worker gene expression bias. For Pogonomyrmex, we used the genetic caste determining J-lineages of P. barbatus, which allowed us to identify the caste fate of larvae prior to morphological differentiation (Anderson et al. 2006; Schwander et al. 2007a; Smith et al. 2012). We generated RNAseq from four different developmental stages with two biological replicates for each caste (queen and worker) per developmental stage (supplementary table S2, Supplementary Material online): adults, pupae, and two sizes of larvae, small (workers and queens of equal mass and equal head capsule width) and large (queens much more massive than workers, but equal in head capsule width). Only adult workers and gynes were sampled in V. emeryi (supplementary table S3, Supplementary Material online).
We found that gene expression differences between worker and queen castes are rarely absolute. To filter out spurious caste-specificity resulting from low expression, we looked for genes with at least 100 reads in one caste, but completely lacking in expression in the other caste; 100 reads is near the median number of reads (44th to 65th percentile) per sample across data sets. Using the RNAseq data from P. barbatus and V. emeryi, we found only three genes with caste-exclusive expression, one uniquely expressed in workers of P. barbatus and the other two in queens of V. emeryi. The worker-exclusive gene (PB25546, likely a histidine-rich glycoprotein) is expressed in all developmental stages of P. barbatus workers but primarily in pupae. Both V. emeryi queen-exclusive genes had no homology in other ants and may be non-coding RNA (Supplement). To examine whether our finding of few caste-exclusive genes was anomalous, we did the same search using the published RNAseq data sets of Camponotus floridanus and Harpegnathos saltator (Bonasio et al. 2010). We found three genes exclusively expressed in queens of C. floridanus, though none with a clear function with regard to caste-specific development or function (Supplement). Thus, genes expressed in only one caste appear to be largely absent in ant genomes, roughly only one in ten thousand genes.
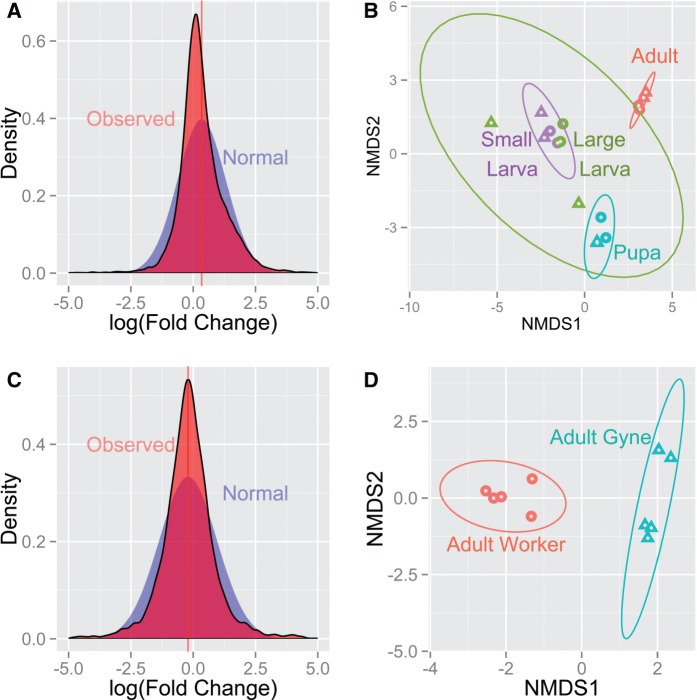
Even at a quantitative level, the transcriptomic data from both P. barbatus and V. emeryi showed an excess of genes with little expression difference between castes relative to what would be expected if gene expression bias was normally distributed (Kolmogorov–Smirnov: D = 0.112 and 0.0704, P < 0.001, for P. barbatus and V. emeryi, respectively) (fig. 2A and C). Fewer than 1% (30 genes) were differentially expressed between P. barbatus queens and workers when all developmental stages were analyzed together (supplementary table S4, Supplementary Material online). For V. emeryi, roughly 58% (5,909) of genes showed statistically significant caste bias toward either adult queens or workers (supplementary table S5, Supplementary Material online). However, even some genes with only a small difference in expression between castes were statistically significant in V. emeryi; the average log fold change for genes with false discovery rate (FDR) < 0.05 was 1.2.
Fig. 2.
Gene expression data for Pogonomyrmex barbatus (top, A–B) and Vollenhovia emeryi (bottom, C–D). On the left (A, C) are the observed and expected normal distributions of logFC (fold change) between queens and workers; negative values represent queen bias, and positive worker bias. Both species show a slightly positive mean (horizontal line) with reads near zero and reads with a more extreme worker-bias being overrepresented relative to the expected normal (a small number of extreme values are excluded from this graph for the purpose of visualization). The right panels (B, D) show NMDS plots coded by caste (triangles are queens and circles are workers) and/or developmental stage (color). Only adults were sequenced in V. emeryi but multiple developmental stages of queens and workers were sequenced for P. barbatus (adult, pupa, large, clearly differentiated, larvae, and small, undifferentiated, larvae).
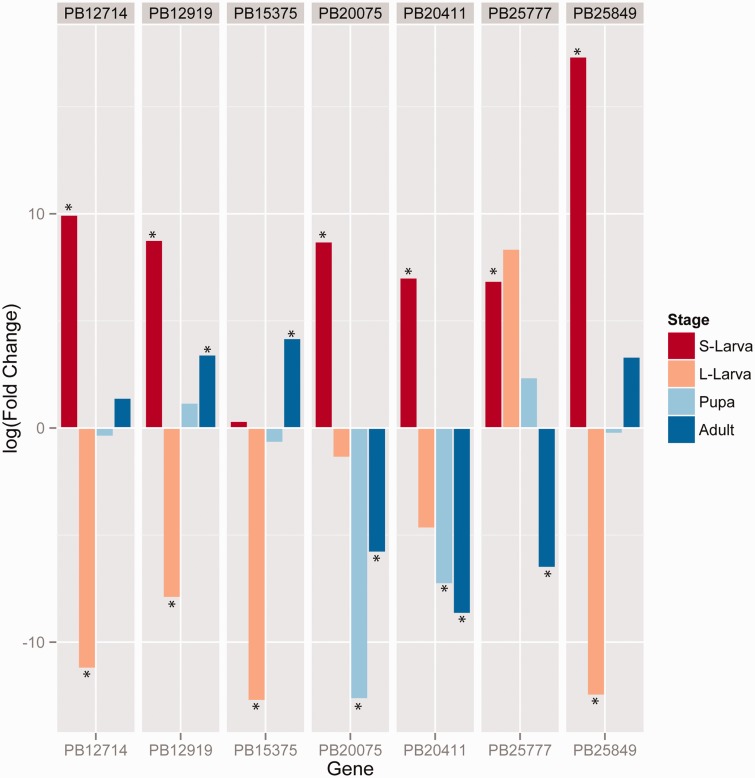
The low level of caste specialization observed across all samples in P. barbatus may reflect the comprehensive developmental comparison afforded by genetic caste determination, which uniquely allows the caste fate of developing larvae to be identified genetically prior to the development of morphological differentiation (Smith et al. 2012). Despite the much lower biological replication, there were more genes in P. barbatus identified as differentially expressed between castes at particular developmental stages, such as adults (109 genes), pupae (174 genes), large larvae (200 genes), or small larvae (568 genes) (supplementary table S4, Supplementary Material online, fig. 2, supplementary fig. S2–9, Supplementary Material online). Comparisons between developmental stages, however, revealed up to 45-times more genes with expression differences compared with comparisons between castes (supplementary table S4, Supplementary Material online, supplementary fig. S9, Supplementary Material online). There were few genes (18%, 177 of 972) with caste-biased expression that were not also different among developmental stages (supplementary fig. S9, Supplementary Material online). Inclusion of multiple, distinct, developmental stages revealed considerable shifts in both the extent and direction of caste bias across development, such that genes that appeared to be strongly biased at one particular stage were not biased when considered across all stages (fig. 3). For example, across caste comparisons within each developmental stage, there were 27 genes that were significantly (FDR < 0.05) upregulated in both workers and queens, but in different developmental stages. Figure 3 shows the seven most variable such genes. Furthermore, 89% (11,151 of 12,460) of genes changed their direction of expression (fold change) between queen and worker across development. This finding is congruent with recent work in evolutionary developmental biology showing that individual genes and gene regulatory networks are reused in different contexts during different developmental stages and tissues. This result also strongly supports the more classic notion of co-option of preexisting genes and networks to produce novel phenotypes (True and Carroll 2002; Monteiro 2012).
Fig. 3.
Seven of the 27 genes with significant differential expression between castes in multiple developmental stages. Each color represents a different developmental stage (S-Larva and L-Larva refer to small and large larvae, respectively). The y-axis is log fold change where positive values represent upregulation in workers and negative values upregulation in queens. Asterisks denote significant differential expression with FDR < 0.05.
Although genes with worker-biased expression represent a small proportion of the overall transcriptome (supplementary table S4, Supplementary Material online), worker traits may still be disproportionately produced by recruitment of novel, taxonomically restricted genes (Simola et al. 2013; Feldmeyer et al. 2014; Jasper et al. 2015; Sumner 2014). From previous phylogenomic analyses on sequenced arthropod genomes, a list of 2,036 taxonomically restricted “orphan” genes was available for P. barbatus (Wissler et al. 2013). Orphan genes are genes that lack homologs in other species (with sequenced genomes). At least in the ants it was found that most orphan genes arise de novo from intergenic regions (Wissler et al. 2013). In our analyses, orphan genes in P. barbatus were not enriched among those genes with higher expression in workers, or in genes with any caste bias (P > 0.13 in all cases, supplementary table S6 and S7, Supplementary Material online). However, orphan genes were disproportionately underrepresented in our RNAseq data compared with genes without RNAseq coverage (orphan genes represented 7% of expressed genes, but 25% of unexpressed genes, χ2 = 977, df, = 1, P < 0.0001), and overall expression levels were lower in orphan compared with non-orphan genes (3.36 vs. 4.57 log[copies per million]: t934 = 14.3, P < 0.0001). This suggests that many orphan genes may be non-functional or were improperly annotated, and that those that are expressed may be differentially regulated compared with more conserved genes. In agreement with previous analyses (Domazet-Loso and Tautz 2003; Wissler et al. 2013), P. barbatus-specific orphan genes are evolving at a significantly faster rate than other genes in P. barbatus (F3,9594 = 16.83, P < 0.0001, P. barbatus orphan genes evolved faster than all other groups of genes, Tukey’s HSD P < 0.05 in all comparisons, supplementary fig. S10, Supplementary Material online). Genes unique to ants, and shared among ant lineages (ant-specific orphan genes), evolved at a rate similar to non-orphan genes (supplementary fig. S5, Supplementary Material online, Tukey HSD P > 0.75 in all cases), suggesting that the rapid evolution of P. barbatus-specific orphan genes is the result of their recent evolutionary history and possibly relaxed selection (assuming that all described orphan genes are true genes). Pogonomyrmex barbatus orphan genes are not evolving at different rates in the social parasites compared with the hosts (t = −1.2, df = 1170, P = 0.3, supplementary fig. S11, Supplementary Material online), suggesting that they are not playing a major role in the evolution of workerless social parasites.
Genes expressed primarily in the worker caste are also expected to evolve more rapidly, even when they are not novel, because selection on them is indirect (Linksvayer and Wade 2009). Additionally, some studies have found that genes that are differentially expressed in polyphenisms, in solitary and social taxa alike, tend to have higher evolutionary rates, even prior to the evolution of novel phenotypes (Hunt et al. 2011; Leichty et al. 2012; Purandare et al. 2014). In Pogonomyrmex, genes with worker-biased expression, queen-biased expression, or unbiased expression (FDR < 0.05 for worker-queen bias in any developmental stage comparison) did not significantly differ in their evolutionary rate (ANOVA: F2,9678 = 2.1, P = 0.12, supplementary fig. S12, Supplementary Material online). When evolutionary rates are examined between caste-biased and non-biased genes across different developmental stages, there is no clear pattern (supplementary fig. S13, Supplementary Material online). For example, worker-biased genes do show an accelerated evolutionary rate in adults (F2,9762 = 7.3, P < 0.001), but in pupal and larval stages queen-biased genes evolved faster (F2,9762 = 6.5 and 7.2, P < 0.005 and P < 0.001, respectively; supplementary fig. S13, Supplementary Material online). That our data do not show a clear pattern suggests that the relationship between evolutionary rate and caste-bias may be much more complicated than previously theorized (Helanterä and Uller 2014), and that the inclusion of multiple developmental stages may be necessary to capture the overall role of genes in polyphenic development. Discerning morph-bias from a single stage may in fact yield incorrect conclusions about evolutionary pattern.
Parasite Genes and Domain Loss
Although the gene expression results suggest that the majority of genes across the genome are likely to be under selection in both castes, RNAseq and differential expression analysis capture only a subset of the predicted transcriptome (72% in P. barbatus and 38% in V. emeryi after filtering), and even genes with universal expression may only be functionally important for one caste, while in the other the gene is expressed as a correlated response. To address these limitations, we also looked for the expected effects of worker loss on the evolution of worker-specific genes due to relaxed selection. To test whether we had sufficient statistical power to detect genomic effects of worker loss for each host-parasite pair, we used coalescent simulations to simulate two alternative scenarios representing the two extremes of a continuum of caste-specificity: purifying selection, where all genes are universal and remain under the same selection regime even when the worker caste is lost, and relaxed selection, where genes are worker-specific and become effectively non-functional with the loss of the worker caste, such that the parasite lineage evolves without codon bias and at an accelerated rate characteristic of introns. The in silico results suggest that there is sufficient power to detect an effect of relaxed selection on genes specialized for worker function. For Pogonomyrmex, when selection was relaxed there was a high likelihood (67.8% of simulation runs) that a frameshift or stop-codon mutation within the gene would become fixed given the divergence time estimate for the P. colei/P. anergismus clade from P. barbatus. Simulation results were similar in Vollenhovia, though with a lower probability (24% of simulation runs; 95% CI: 0–50) that damaging mutations become fixed in V. nipponica; the lesser likelihood compared with Pogonomyrmex probably reflects the more recent divergence between the Vollenhovia host and parasite (fig. 1).
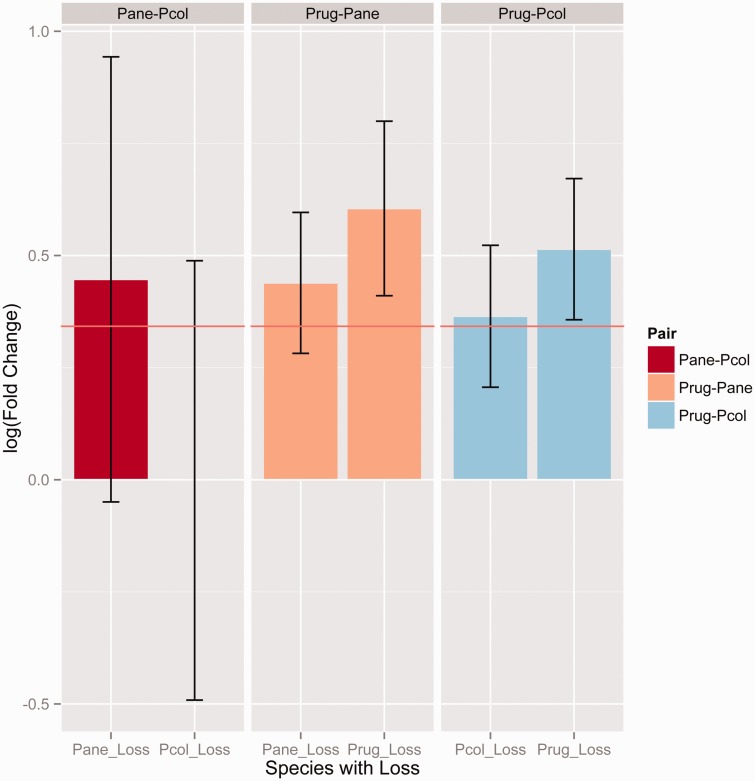
In the Pogonomyrmex system, we were able to compare observed patterns of gene loss in the parasites with those in the second host P. rugosus, thus not confounding true gene loss with potential artifacts of mapping to the genome of P. barbatus. This also allowed us to estimate the amount of “normal” gene loss between closely related eusocial species (no worker loss), although the divergence time between P. barbatus and P. rugosus is slightly greater than that between the social parasites and their hosts (fig. 1). Contrary to our expectation, the social parasites did not have an elevated rate of damaging mutations relative to their hosts. Genes with damaging mutations were marginally worker-biased in all Pogonomyrmex species (ANOVA: F2,8561 = 4.4, P < 0.05) but the Pogonomyrmex parasites did not differ in the extent of this bias from their host P. rugosus (Tukey HSD: P = 0.45, supplementary fig. S14, Supplementary Material online). Similarly, there was no difference between V. emeryi and V. nipponica in queen-worker gene expression bias between genes with damaging mutations and those without (t21 = 0.91, P = 0.37, supplementary fig. S14, Supplementary Material online), though the number of genes with differential loss between these species was very small (n = 22). In addition to examining damaging mutations across the entire gene length, we also conducted an analysis restricted to functional protein domains. Again, genes in Pogonomyrmex with lost domains were worker-biased, in both social parasites and P. rugosus (fig. 4, t142 > 3.8, P < 0.0005 in all cases). No domain differences were detected between V. emeryi and V. nipponica. Taken together, these results indicate that worker-biased genes, or protein domains, are more likely to be lost, regardless of species and life-history strategy. Indirect selection may cause more losses of genes with worker-biased expression even when such losses are weakly maladaptive. High turnover of genes with caste-biased expression, especially those biased toward the worker caste, is consistent with recent comparative transcriptomic data suggesting that organisms that evolved sociality independently have relatively few genes with common responses during caste determination, despite the fact that a similar set of broader functional processes are consistently implicated in caste differences (Berens et al. 2015). Developmental regulatory networks are expected to contain considerable functional redundancies to maintain developmental homeostasis; different elements of which may be modified to produce and shape polyphenisms (Ohno 1970).
Fig. 4.
Gene expression (logFC, fold change—positive is worker bias, negative is queen bias) in genes that have lost protein domains in each of three lineage pairs, Pcol-Pane (social parasites), Pcol-Prug (parasite-host), and Pane-Prug (parasite-host), where Pcol = Pogonomyrmex colei, Pane = Pogonomyrmex anergismus, and Prug = Pogonomyrmex rugosus. Genes with lost domains have a greater than average worker-biased expression in parasite-host comparisons (t > 2, df > 75, P < 0.05 in all cases), regardless of whether the loss was in the host or parasite. The horizontal line is the mean logFC value for all genes; bars and whiskers are means ± 95% CI.
Evidence for Selection
In addition to losing worker-specific functions (e.g., foraging, nest maintenance, defense, etc.), social parasites require a set of novel traits that may also be reflected at the genomic level (Cini et al. 2015). Social parasite queens are smaller than their respective hosts, have reduced mating flights and/or intranidal mating, and have to evolve novel behaviors to infiltrate host colonies and evade host defenses (Buschinger 2009). We therefore expected to see signatures of positive selection in genes that are responsible for changes in life history traits in the social parasites. We used a branch-site model to detect genes under positive selection in the social parasites compared with their hosts. There were only three genes with a signature of positive selection in the harvester ant social parasites, and none in Vollenhovia (Supplement). Although this would seem a surprisingly small number of genes under selection in the parasites, this result is broadly in agreement with the simulation data in suggesting that the majority of the genome is under purifying selection. Despite the simulations predicting that genes under relaxed selection would increase in evolutionary rate (ANOVA, F1,198 = 59.22, P < 0.0001; fig. 5A), in actuality, genes in the social parasites show no significant acceleration in evolutionary rate relative to their hosts, even for genes with worker-biased expression (One-sided t-tests: Overall, t9738 = 2.56, P = 0.99; Worker-biased only, t106 = 1.54, P = 0.94; fig. 5B). Genes with queen-biased expression did show evidence of accelerated evolution in the hosts relative to both unbiased and worker-biased genes (ANOVA: F2,9736 = 5.22, P < 0.01; Tukey’s HSD: queen-biased vs. unbiased, P < 0.005, queen-biased vs. worker-biased, P < 0.01; fig. 5B). This result may reflect the loss of worker-associated trade-offs in the social parasites that normally act on such genes and would enhance the effect of purifying selection for queen function (Hall et al. 2013).
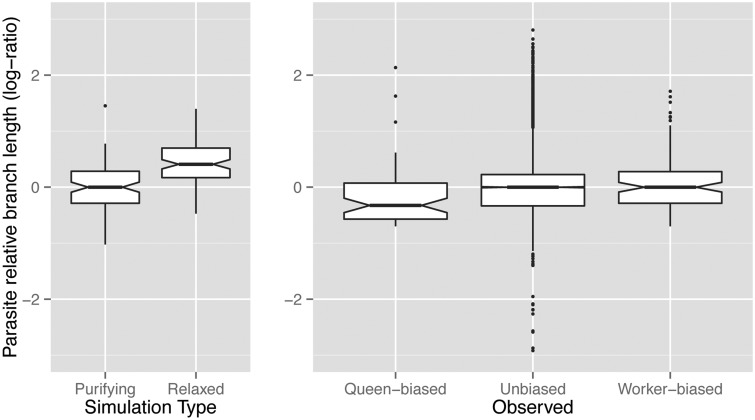
Fig. 5.
Simulated and observed evolutionary rates (host-parasite branch length) of genes in Pogonomyrmex barbatus. Simulations (left) demonstrate that genes under relaxed selection are expected to evolve rapidly (and accrue deleterious mutations). Observed results in P. barbatus (right) suggest that genes, regardless of whether they show a caste bias in gene expression, are evolving consistently under purifying selection.
Conclusions
The evolution of complex societies is considered one of the major transitions of life (Maynard Smith and Szathmáry 1995). In stark contrast to the prediction that novel eusocial phenotypes are generated by caste-specific gene sets, our results suggest that few, if any, genes involved in generating the worker phenotype have been lost or released from purifying selection in workerless social parasites. Instead, eusocial insect genomes appear to be largely multifunctional: genes serve multiple roles that differ between the castes and among developmental stages, suggesting that differential regulation of genes, rather than coding sequence change, is the major driver of caste evolution. Subtle changes, such as responsiveness to nutrition or hormones, may be sufficient to alter the expression of genes within regulatory networks and lead to drastically different phenotypes. The vast majority of genes are expressed in both castes and shift in their direction of expression bias between castes over development. Given this, greater caution should be given to inferring evolutionary patterns from expression studies of single developmental stages, and more emphasis should be given to the generation of gene expression studies that span the life history of each species and are comparable between species.
A major implication of regulatory control is that once evolved, phenotypic plasticity may be easily lost via a change in the environment which fails to trigger an alternative pathway, or a change in the regulatory response to the environment, which might explain why social parasitism has evolved many times independently. There are similar examples of mating incompatibilities between genotypes or genetic lineages producing significant caste bias or complete loss of one phenotype, such as in species with genetic caste determination (Anderson et al. 2008; Smith et al. 2008; Schwander et al. 2010; Libbrecht et al. 2011). It has been speculated that both genetic caste determination and social parasitism are related phenomena that could arise from dysregulation as a result of genomic mixing during interspecific hybridization (Helms Cahan and Keller 2003) or the formation of caste-biasing supergenes (Linksvayer et al. 2013). Notably, along with social parasitism, examples of genetic caste determination occur in both Pogonomyrmex and Vollenhovia (Helms Cahan and Keller 2003; Ohkawara et al. 2006; Anderson et al. 2008).
Given that regulatory changes are not protein-coding, phenotypes may also be recovered because the underlying genetic architecture is not lost with the phenotype (Rajakumar et al. 2012), and therefore, some genomes may harbor many unexpressed, cryptic, phenotypes that were favored under bygone environments but are currently inaccessible due to changes in either regulatory machinery or the absence of necessary environmental cues. It is likely that the ability to produce workers is not lost in Pogonomyrmex and Vollenhovia social parasites, but instead that developmental plasticity is cryptic, such as has been demonstrated previously for the super-soldier caste in some Pheidole ants (Rajakumar et al. 2012). Studies have documented a nearly complete loss of phenotypic plasticity in genetic caste-determining Pogonomyrmex ants (Helms Cahan et al. 2004); however, interlineage queens (Schwander et al. 2007; Anderson et al. 2009) and intralineage workers (Helms Cahan et al. 2004) have all been detected, though rare. Similarly, sexually produced queens are known from Vollenhovia (Okamoto et al. 2015). With such great diversity of worker phenotypes in the social insects, it is not beyond reason that even species with monomorphic workers harbor the capacity to produce multiple worker phenotypes if the selective environment changes (Rajakumar et al. 2012).
Materials and Methods
Phylogenetic Analysis
We inferred phylogeny and divergence dates with BEAST v2.1.2 (Drummond et al. 2012) under an uncorrelated lognormal relaxed clock model (Drummond et al. 2006). As a framework for this analysis, we used molecular data from a phylogenetic study of Myrmicinae (Ward et al. 2015) that included five Pogonomyrmex species and one Hylomyrma species (which is nested within Pogonomyrmex). We assembled sequences from the nine protein-coding genes generated by Ward et al. (2015) and added the homologous loci from the four Pogonomyrmex and two Vollenhovia genomes, resulting in 5,490 bp of aligned nucleotides. The nine genes used were abdominal-A (Abd-A), elongation factor 1-alpha F1 copy (EF1aF1), elongation factor 1-alpha F2 copy (EF1aF2), long wavelength rhodopsin (LW Rh), arginine kinase (ArgK), topoisomerase 1 (Top1), ultrabithorax (Ubx), wingless (Wg), and rudimentary (CAD). Data partitioning (by gene and codon position) and substitution models were determined using PartitionFinder v1.1.1 (Lanfear et al. 2012). Substitution models were unlinked and clock and tree models were linked among partitions. The tree prior was a Yule process. We used two a priori age distributions to calibrate our analysis. The first prior calibrated the stem-group node of the most recent common ancestor (MRCA) of Pogonomyrmex (striatinodis + subdentatus) based on Florissant Formation fossils (Carpenter 1930), using a lognormal distribution with an offset (minimum bound) = 34 Ma, median = 50 Ma, and 95% quantile = 73 Ma (equating to a log-transformed mean = 2.77 and a log-transformed standard deviation = 0.54). The second prior calibrated the MRCA of Pogonomyrmex and Vollenhovia using a normal distribution with a mean = 95.4 Ma and 95% confidence interval of 85.2–106.0 Ma, based on the results of Ward et al. (2015). We conducted multiple BEAST runs, each for 500 million generations with a burn-in of 100 million generations. Stationarity and burn-in were determined by observing high ESS values and the consistency of likelihood values and other results among independent runs. We combined results from five independent runs (totaling 2 billion post–burn-in generations) and visualized the resulting topology and divergence times using FigTreev1.4.0 (http://tree.bio.ed.ac.uk/software/figtree/, last accessed August 9, 2015).
Genome Sequencing, Assembly, Annotation, and Mapping
Pogonomyrmex Genome Sequencing
Shotgun libraries for the three Pogonomyrmex genomes were prepared from single-male samples using TruSeq library preparation kits (Illumina), according to the manufacturer’s instructions, and sequenced on the IlluminaHiSeq 2000. The reads were then mapped to the P. barbatus reference genome using bowtie2 (Langmead and Salzberg 2012). Variants were called simultaneously on all three samples using UnifiedGenotyper in GATK (v. 2.2-16) (McKenna et al. 2010) in haploid mode. We followed the GATK best practices recommendations, including realignment of reads around indels, variant, and base quality recalibration (Van der Auwera et al. 2002; DePristo et al. 2011). The coordinates of the P. barbatus annotation were then adjusted to accommodate insertions and deletions introduced by the variant calling pipeline. We filtered any gene models from the P. barbatus reference genome where all of the other species were predicted to have a stop codon or a frame shift, and were suggestive of incorrect assembly or annotation of the reference genome. Many of these sites were in homopolymer regions, and were consistent with the error profile of Roche 454 sequencing technology, which was used for the P. barbatus genome (Smith et al. 2011).
RNA Extraction
For P. barbatus, DNA and RNA were extracted from 16 individuals, from both queen and worker castes, and representing four developmental stages (supplementary table S2, Supplementary Material online) of J-lineage P. barbatus. All pupae and adults were sampled at the same time from a lab colony reared from a single mated queen (∼5 years old); the colony was collected north of Lordsburg, NM (N 32.44250, W108.66605). Larvae were collected from two field colonies (label numbers 41 and 44 in supplementary table S2, Supplementary Material online) near Portal, AZ (N 31.31.93180, W 109.20538). Larval mass and head size in P. barbatus (Smith, unpublished data) suggest that all of these larvae were in the last (fourth) instar despite their major difference in wet mass (supplementary table S2, Supplementary Material online). DNA and RNA were extracted using TRIzol (Life Technologies). The manufacturer’s protocols were used with the following modifications, 1) 100 μl was used as the starting volume of TRIzol and all subsequent reagent amounts were adjusted according to the protocol and 2) two sodium citrate washes were used to clean the DNA. The extracted DNA was used for microsatellite genotyping of larvae (adults and pupae were not genotyped because caste can be morphologically determined) using the following loci: Myrt3, Pb8, Pr1, and L18 (Evans 1993; Foitzik et al. 1997; Volny and Gordon 2002; Gadau et al. 2003). The former three loci have no allele overlap and the protocols for PCR and polyacrylamide gel electrophoresis were as in (Smith et al. 2012). The conditions used for Pr1 were as in (Schwander et al. 2007b), though we used modified primers to shorten the locus length (Pr1aF: AAGTGGGTCACGAGAACGAG, Pr1aR: ATGCCAAGTGAAAGGATTCG). For V. emeryi, RNA was extracted using RNAqueous-Micro Total RNA Isolation Kit (Ambion) according to the manufacturer’s specifications; five specimens were pooled per sample. For V. emeryi, RNA was extracted from pools of 5–15 individuals from populations collected in Tokyo, Ishikawa, and Toyama areas in Japan, using RNeasy Micro Kits (QIAGEN).
RNA-Seq Library Preparation and Sequencing
Extracted RNA was used for cDNA generation using the RACE protocol from Aird et al. (2013), except that we added ERCC92 spike-in controls (Invitrogen) alternately to each library, according to the manufacturer’s specification. Four hundred nanograms of RNA and 13 cycles of PCR were used for each P. barbatus sample. For V. emeryi, five replicate libraries were sequenced for workers and queens using pools of five individuals for each. One hundred nanograms of total RNA was used for each library, and 12 cycles of PCR. The samples were sequenced on the IlluminaHiSeq 2000 in paired end 200-cycle configuration.
Vollenhovia Genome Sequencing
V. emeryi has an unusual reproductive system, where males exist as a single clonal lineage. The V. emeryi reference genome was sequenced from a single male clone, using a shotgun library for Roche 454 FLX+ (454 Life Sciences), prepared according to the manufacturer’s instruction. These were assembled using the Newbler assembler using large genome mode (v. 2.6) (454 Life Sciences). The genome was annotated using MAKER (v.2.1) (Cantarel et al. 2008; Holt and Yandell 2011), using protein predictions from other ants, and RNA-seq data from V. emeryi that were mapped to the reference genome using TopHat (Trapnell et al. 2009), and extracted using Cufflinks (Trapnell et al. 2012). This resulted in a higher number of predicted genes than found in other ants. Because Vollenhovia is only distantly related to other sequenced ant species (Ward et al. 2015), we kept these predictions to increase the number of possible targets. The V. nipponica genome was sequenced using Illumina technology. The library preparation, sequencing, and variant calling were as described above for the Pogonomyrmex samples.
Reference-Guided Gene Expression Analysis
Read counts were obtained by mapping the reads to the predicted transcript sequences using RSEM (Li and Dewey 2011), which were then used to find significantly differentially expressed genes using edgeR (Robinson et al. 2010). Prior to differential expression analysis, we used the svseq function (Leek 2014) of the sva package (Leek et al. 2015) to identify, estimate, and account for unknown sources of variation. Data were filtered in two ways and the results compared. With conservative filtering, genes without at least 100 reads in at least two of the samples were filtered out. With liberal filtering, genes without at least two reads in at least four of the samples were filtered out. The results were similar and the total number of differentially expressed genes from each filter are highly correlated (r = 0.97, P < 0.001) with a slope of 0.34 (i.e., the conservative filter detects one third the number of differentially expressed genes as compared with the liberal filter) (supplementary table S4 and S5, Supplementary Material online). The reported results are from the liberal filter. Libraries were normalized using the trimmed mean of M-values (TMM) method (Robinson and Oshlack 2010). Differential expression was compared between castes and developmental stages using linear models with contrasts (and the surrogate variables from sva, above, as a factor). Reported statistical significance is after correction for FDR using a 0.05 threshold because this is the most common practice, and thus the results we present will be comparable with other studies.
Damaging Mutations and Domain Losses
Data Generation
Reciprocal best hit BLAST was used to establish ortholog relationships between the P. barbatus sequences and six other sequenced ant species (H. saltator, Linepithema humile, C. floridanus, Acromyrmex echinatior, and Atta cephalotes). Pogonomyrmex barbatus genes were projected onto the genome alignments. The CDSs were then extracted for each gene and translated into amino acids. On encountering a stop codon the translation was stopped. Therefore, the extracted genes of P. rugosus, P. colei, and P. anergismus can be shorter but not longer than the reference genes of P. barbatus. Domains for all genes were predicted using the Pfam database (v27) (Finn et al. 2014). Two domain annotations were generated using pfam_scan.pl, one with standard threshold and one with relaxed thresholds (e-value threshold = 10).
Domain Counting
The considered species were divided into two sets: hosts (P. barbatus/P. rugosus) and parasites (P. colei/P. anergismus). A gene was then considered to be changed if a change has been occurring in all species of a group, meaning that the minimum number a domain occurs in one set is higher than the maximum number in the other. To be more specific:
Given 2 sets of species (H and P) and the counts of a specific domain of a specific gene in a given species x using standard threshold () and the relaxed threshold (), the domain is considered to have a group specific change if:
or
To avoid counting exchanges between very similar domains (e.g., →), domains that belong to the same clan were treated as being the same.
Loss Analysis
Wagner parsimony was used to analyze the domain family changes in the hosts and parasites in case of repeat number changes. For complete loss of domain types in one of the groups Dollo parsimony was used. For this the losses after the split of host and parasite species were calculated. This analysis was performed for all genes for which a 1-1 ortholog relationship could be determined, including at least one species inside the six outgroup species. The same procedure was repeated for the Vollenhovia species.
Coalescent Simulations of Damaging Mutations in Social Parasites
To estimate the expected probability of loss-of-function mutations (stop codons and frameshifts) for worker-specific genes in the social parasite clade, as well as to test the power to detect relaxed selection associated with loss of worker function, we conducted coalescent simulations of gene evolution with Indel-Seq-Gen, a flexible DNA simulator that can model lineage-specific shifts in selection for both point mutations and insertion/deletions (Rambaut and Grassly 1997; Strope et al. 2007). Mutations evolve along a user-provided guide tree, with evolutionary rates defined by the branch lengths and distributed according to specified codon usage rates, insertion/deletion frequencies, and length distributions. Two scenarios were simulated, representing the two extreme ends of a continuum of gene caste-specificity: in the first, genes were fully functional in both castes and thus retained their function even when the worker phenotype was lost. In the second, genes lost their functional expression in the clade leading to the two social parasites and thus became effectively neutral. This was reflected in the simulations for the clade leading to the social parasite(s) by a shift of substitutions across codon positions from a third-position bias to a uniform rate, retention of indels of up to 6 bp in length at one-tenth the rate of substitutions drawn from a Chang & Benner length distribution, and an overall accelerated substitution rate for novel mutations. The expected substitution rate of social parasite genes relative to host genes under relaxed selection was estimated by comparing the mean substitution rate of the introns and exons of each gene across the annotated transcriptome of P. barbatus (=2X the rate of exons). To generate a guide tree for the host and parasite taxa, we used the majority-rules scaffold topology, with branch lengths averaged from those scaffolds that matched that topology. One hundred replicates of each scenario were conducted for each species with simulated gene sequences equaling the median gene length across the transcriptome (P. barbatus = 699 bp, V. emeryi = 535 bp) (Smith et al. 2011). Confidence limits around the resulting estimates were generated by scaling branch lengths to the relative rates of the bottom and top 5% of genes. Simulated sequences were aligned and assessed for the presence of stop codons and frameshift indels.
Supplementary Material
Supplementary figures S1–S14 and tables S1–S7 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the OIST sequencing center for preparing the V. emeryi genomic libraries and for carrying out the sequencing. At the University of Illinois, they thank Hugh Robertson and Kim Walden for preparing the P. anergismus genomic libriaries and the Keck Center for carrying out the sequencing. They are grateful to M.M.Y. Tin and M. Grau-Lopez for assistance with laboratory work and bioinformatics, respectively. They thank the OIST workshop committee for helping to organize the Genomics of Social Parasite Evolution workshop. Funding for this project was provided by OIST and JSPS Kakenhi grants to ASM (25221206 and 24770034), and by grants from NSF (DEB 1020979 to AVS, DUE 1139893 to CRS, and DEB 0919052 to SIHC). They thank “Uncle” Bob Johnson for P. anergismus specimens as well as E. Cash and K. E. Anderson for lab colonies, and B. Smith and N. Hamada Fernside for larva collection.
References
- Aird SD, Watanabe Y, Villar-Briones A, Roy MC, Terada K, Mikheyev AS. 2013. Quantitative high-throughput profiling of snake venom gland transcriptomes and proteomes (Ovophis okinavensis and Protobothrops flavoviridis). BMC Genomics 14:790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amdam GV, Csondes A, Fondrk MK, Page RE. 2006. Complex social behaviour derived from maternal reproductive traits. Nature 439:76–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson KE, Gadau J, Mott BM, Johnson RA, Altamirano A, Strehl C, Fewell JH. 2006. Distribution and evolution of genetic caste determination in Pogonomyrmex seed-harvester ants. Ecology 87:2171–2184. [DOI] [PubMed] [Google Scholar]
- Anderson KE, Linksvayer TA, Smith CR. 2008. The causes and consequences of genetic caste determination in ants (Hymenoptera: Formicidae). Myrmecol News 11:119–132. [Google Scholar]
- Anderson KE, Smith CR, Linksvayer TA, Mott BM, Gadau J, Fewell JH. 2009. Modeling the maintenance of a dependent lineage system: the influence of positive frequency-dependent selection on sex ratio. Evolution 63:2142–2152. [DOI] [PubMed] [Google Scholar]
- Anholt RR, Mackay TF. 2004. Quantitative genetic analyses of complex behaviours in Drosophila. Nat Rev Genet. 5:838–849. [DOI] [PubMed] [Google Scholar]
- Berens AJ, Hunt JH, Toth AL. 2015. Comparative transcriptomics of convergent evolution: different genes but conserved pathways underlie caste phenotypes across lineages of eusocial insects. Mol Biol Evol. 32:690–703. [DOI] [PubMed] [Google Scholar]
- Bonasio R, Zhang G, Ye C, Mutti NS, Fang X, Qin N, Donahue G, Yang P, Li Q, Li C. 2010. Genomic comparison of the ants Camponotus floridanus and Harpegnathos saltator. Science 329:1068–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschinger A. 1986. Evolution of social parasitism in ants. Trends Ecol Evol. 1:155–160. [DOI] [PubMed] [Google Scholar]
- Buschinger A. 2009. Social parasitism among ants: a review (Hymenoptera: Formicidae). Myrmecol News 12:219–235. [Google Scholar]
- Cantarel BL, Korf I, Robb SMC, Parra G, Ross E, Moore B, Holt C, Sánchez Alvarado A, Yandell M. 2008. MAKER: An easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18:188–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter FM. 1930. The fossil ants of North America. Bull Mus Comp Zool. 70:1–66. [Google Scholar]
- Cini A, Patalano S, Segonds-Pichon A, Busby GBJ, Cervo R, Sumner S. 2015. Social parasitism and the molecular basis of phenotypic evolution. Front Genet. 6:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 43:491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D. 2003. An evolutionary analysis of orphan genes in Drosophila. Genome Res. 13:2213–2219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Ho SY, Phillips MJ, Rambaut A. 2006. Relaxed phylogenetics and dating with confidence. PLoS Biol. 4:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond AJ, Suchard MA, Xie D, Rambaut A. 2012. Bayesian phylogenetics with BEAUti and the BEAST 1.7. Mol Biol Evol. 29:1969–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldmeyer B, Elsner D, Foitzik S. 2014. Gene expression patterns associated with caste and reproductive status in ants: worker-specific genes are more derived than queen-specific ones. Mol Ecol. 23:151–161. [DOI] [PubMed] [Google Scholar]
- Evans JD. 1993. Parentage analyses in ant colonies using simple sequence repeat loci. Mol Ecol. 2:393–397. [DOI] [PubMed] [Google Scholar]
- Finn RD, Bateman A, Clements J, Coggill P, Eberhardt RY, Eddy SR, Heger A, Hetherington K, Holm L, Mistry J, et al. 2014. Pfam: the protein families database. Nucleic Acids Res. 42:D222–D230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foitzik S, Haberl M, Gadau J, Heinze J. 1997. Mating frequency of Leptothorax nylanderi ant queens determined by microsatellite analysis. Insect Soc. 44: 219–227. [Google Scholar]
- Gadagkar R. 1997. The evolution of caste polymorphism in social insects: genetic release followed by diversifying evolution. J Genet. 76:167–179. [Google Scholar]
- Gadau J, Helmkampf M, Nygaard S, Roux J, Simola DF, Smith CR, Suen G, Wurm Y, Smith CD. 2012. The genomic impact of 100 million years of social evolution in seven ant species. Trends Genet. 28:14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadau J, Strehl CP, Oettler J, Hölldobler B. 2003. Determinants of intracolonial relatedness in Pogonomyrmex rugosus (Hymenoptera: Formicidae): mating frequency and brood raids. Mol Ecol. 12:1931–1938. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Sharabash NM, Whitfield CW, Robinson GE. 2003. Pheromone-mediated gene expression in the honey bee brain. Proc Natl Acad Sci. 100:14519–14525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall DW, Soojin VY, Goodisman MA. 2013. Kin selection, genomics and caste-antagonistic pleiotropy. Biol Lett. 9:20130309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helanterä H, Uller T. 2014. Neutral and adaptive explanations for an association between caste-biased gene expression and rate of sequence evolution. Front Genet. 5: 297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helms Cahan SH, Julian GE, Rissing SW, Schwander T, Parker JD, Keller L. 2004. Loss of phenotypic plasticity generates genotype-caste association in harvester ants. Curr Biol. 14:2277–2282. [DOI] [PubMed] [Google Scholar]
- Helms Cahan SH, Keller L. 2003. Complex hybrid origin of genetic caste determination in harvester ants. Nature 424:306–309. [DOI] [PubMed] [Google Scholar]
- Hoülldobler B, Wilson EO. 1990. The ants. Cambridge (MA): Belknap Press of Harvard University Press. [Google Scholar]
- Holt C, Yandell M. 2011. MAKER2: an annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12:491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt BG, Ometto L, Wurm Y, Shoemaker D, Soojin VY, Keller L, Goodisman MA. 2011. Relaxed selection is a precursor to the evolution of phenotypic plasticity. Proc Natl Acad Sci. 108:15936–15941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasper WC, Linksvayer TA, Atallah J, Friedman D, Chiu JC, Johnson BR. 2015. Large scale coding sequence change underlies the evolution of post-developmental novelty in honey bees. Mol Biol Evol. 32:334–346. [DOI] [PubMed] [Google Scholar]
- Johnson BR, Tsutsui ND. 2011. Taxonomically restricted genes are associated with the evolution of sociality in the honey bee. BMC Genomics 12:164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RA, Parker JD, Rissing SW. 1996. Rediscovery of the workerless inquiline ant Pogonomyrmex colei and additional notes on natural history (Hymenoptera: Formicidae). Insectes Sociaux 43:69–76. [Google Scholar]
- Jones FC, Grabherr MG, Chan YF, Russell P, Mauceli E, Johnson J, Swofford R, Pirun M, Zody MC, White S. 2012. The genomic basis of adaptive evolution in threespine sticklebacks. Nature 484:55–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinomura K, Yamauchi K. 1992. A new workerless socially parasitic species of the genus Vollenhovia (Hymenoptera: Formicidae) from Japan. Jpn J Entomol. 60:203–206. [Google Scholar]
- Lahti DC, Johnson NA, Ajie BC, Otto SP, Hendry AP, Blumstein DT, Coss RG, Donohue K, Foster SA. 2009. Relaxed selection in the wild. Trends Ecol Evol. 24:487–496. [DOI] [PubMed] [Google Scholar]
- Lanfear R, Calcott B, Ho SY, Guindon S. 2012. PartitionFinder: combined selection of partitioning schemes and substitution models for phylogenetic analyses. Mol Biol Evol. 29:1695–1701. [DOI] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. 2012. Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT. 2014. svaseq: removing batch effects and other unwanted noise from sequencing data. Nucleic Acids Res. 42:e161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Johnson WE, Parker HS, Fertig EJ, Jaffe AE, Storey JD. 2015. sva: Surrogate Variable Analysis. R package version 3.14.0. Available from: http://www.bioconductor.org/packages/release/ bioc/html/sva.htm.l. [Google Scholar]
- Leichty AR, Pfennig DW, Jones CD, Pfennig KS. 2012. Relaxed genetic constraint is ancestral to the evolution of phenotypic plasticity. Int Comp Biol. 52:16–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. 2011. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libbrecht R, Schwander T, Keller L. 2011. Genetic components to caste allocation in a multiple-queen ant species. Evolution 65:2907–2915. [DOI] [PubMed] [Google Scholar]
- Linksvayer TA, Busch JW, Smith CR. 2013. Social supergenes of superorganisms: do supergenes play important roles in social evolution? BioEssays News Rev Mol Cell Dev Biol. 35:683–689. [DOI] [PubMed] [Google Scholar]
- Linksvayer TA, Wade MJ. 2009. Genes with social effects are expected to harbor more sequence variation within and between species. Evolution 63:1685–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maynard Smith J, Szathmáry E. 1995. The major transitions in evolution. Nature 374:227–232. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikheyev AS, Linksvayer TA. 2015. Genes associated with ant social behavior show distinct transcriptional and evolutionary patterns. eLife 4:e04775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro A. 2012. Gene regulatory networks reused to build novel traits. BioEssays 34:181–186. [DOI] [PubMed] [Google Scholar]
- Ohkawara K, Nakayama M, Satoh A, Trindl A, Heinze J. 2006. Clonal reproduction and genetic caste differences in a queen-polymorphic ant, Vollenhovia emeryi. Biol Lett. 2:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohno S. 1970. Evolution by gene duplication. New York: Springer Science and Business Media. [Google Scholar]
- Okamoto M, Kobayashi K, Hasegawa E, Ohkawara K. 2015. Sexual and asexual reproduction of queens in a myrmicine ant, Vollenhovia emeryi (Hymenoptera: Formicidae). Myrmecol News 21:13–17. [Google Scholar]
- Page RE, Amdam GV. 2007. The making of a social insect: developmental architectures of social design. Bioessays 29:334–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JD, Rissing SW. 2002. Molecular evidence for the origin of workerless social parasites in the ant genus Pogonomyrmex. Evolution 56:2017–2028. [DOI] [PubMed] [Google Scholar]
- Purandare SR, Bickel RD, Jaquiery J, Rispe C, Brisson JA. 2014. Accelerated evolution of morph-biased genes in pea aphids. Mol Biol Evol. 31:2073–2083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajakumar R, San Mauro D, Dijkstra MB, Huang MH, Wheeler DE, Hiou-Tim F, Khila A, Cournoyea M, Abouheif E. 2012. Ancestral developmental potential facilitates parallel evolution in ants. Science 335:79–82. [DOI] [PubMed] [Google Scholar]
- Rambaut A, Grassly NC. 1997. Seq-Gen: an application for the Monte Carlo simulation of DNA sequence evolution along phylogenetic trees. Comput Appl Biosci. 13:235–238. [DOI] [PubMed] [Google Scholar]
- Robinson GE, Grozinger CM, Whitfield CW. 2005. Sociogenomics: social life in molecular terms. Nat Rev Genet. 6:257–270. [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, Oshlack A. 2010. A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader L, Simola DF, Heinze J, Oettler J. 2015. Sphingolipids, transcription factors, and conserved toolkit genes: developmental plasticity in the ant Cardiocondyla obscurior. Mol Biol Evol. 32:1474–1486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwander T, Helms Cahan S, Keller L. 2007a. Characterization and distribution of Pogonomyrmex harvester ant lineages with genetic caste determination. Mol Ecol. 16:367–387. [DOI] [PubMed] [Google Scholar]
- Schwander T, Keller L, Helms Cahan S. 2007b. Two alternate mechanisms contribute to the persistence of interdependent lineages in Pogonomyrmex harvester ants. Mol Ecol. 16:3533–3543. [DOI] [PubMed] [Google Scholar]
- Schwander T, Lo N, Beekman M, Oldroyd BP, Keller L. 2010. Nature versus nurture in social insect caste differentiation. Trends Ecol Evol. 25:275–282. [DOI] [PubMed] [Google Scholar]
- Simola DF, Wissler L, Donahue G, Waterhouse RM, Helmkampf M, Roux J, Nygaard S, Glastad KM, Hagen DE, Viljakainen L. 2013. Social insect genomes exhibit dramatic evolution in gene composition and regulation while preserving regulatory features linked to sociality. Genome Res. 23:1235–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Mutti NS, Jasper WC, Naidu A, Smith CD, Gadau J. 2012. Patterns of DNA methylation in development, division of labor and hybridization in an ant with genetic caste determination. PloS One 7:e42433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Smith CD, Robertson HM, Helmkampf M, Zimin A, Yandell M, Holt C, Hu H, Abouheif E, Benton R. 2011. Draft genome of the red harvester ant Pogonomyrmex barbatus. Proc Natl Acad Sci. 108:5667–5672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith CR, Toth AL, Suarez AV, Robinson GE. 2008. Genetic and genomic analyses of the division of labour in insect societies. Nat Rev Genet. 9:735–748. [DOI] [PubMed] [Google Scholar]
- Strope CL, Scott SD, Moriyama EN. 2007. indel-Seq-Gen: a new protein family simulator incorporating domains, motifs, and indels. Mol Biol Evol. 24:640–649. [DOI] [PubMed] [Google Scholar]
- Sumner S. 2014. The importance of genomic novelty in social evolution. Mol Ecol. 23:26–28. [DOI] [PubMed] [Google Scholar]
- Toth AL, Varala K, Newman TC, Miguez FE, Hutchison SK, Willoughby DA, Simons JF, Egholm M, Hunt JH, Hudson ME. 2007. Wasp gene expression supports an evolutionary link between maternal behavior and eusociality. Science 318:441–444. [DOI] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc. 7:562–578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- True JR, Carroll SB. 2002. Gene co-option in physiological and morphological evolution. Annu Rev Cell Dev Biol. 18:53–80. [DOI] [PubMed] [Google Scholar]
- Tsai IJ, Zarowiecki M, Holroyd N, Garciarrubio A, Sanchez-Flores A, Brooks KL, Tracey A, Bobes RJ, Fragoso G, Sciutto E. 2013. The genomes of four tapeworm species reveal adaptations to parasitism. Nature 496:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, et al. 2002. From FastQ data to high-confidence variant calls: the genome analysis toolkit best practices pipeline. Current Protocols in Bioinformatics. 11:11.10:11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volny VP, Gordon DM. 2002. Characterization of polymorphic microsatellite loci in the red harvester ant, Pogonomyrmex barbatus. Mol Ecol Notes 2:302–303. [Google Scholar]
- Ward PS, Brady SG, Fisher BL, Schultz TR. 2015. The evolution of myrmicine ants: phylogeny and biogeography of a hyperdiverse ant clade (Hymenoptera: Formicidae). Syst Entomol. 40:61–81. [Google Scholar]
- West-Eberhard MJ. 1987. Flexible strategy and social evolution. In: Ito Y, Brown JL, Kikkawa J, editors. Animal societies: theories and facts. Tokyo: Japan Scientific Societies Press. p. 35–51. [Google Scholar]
- Wissler L, Gadau J, Simola DF, Helmkampf M, Bornberg-Bauer E. 2013. Mechanisms and dynamics of orphan gene emergence in insect genomes. Genome Biol Evol. 5:439–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wurm Y, Wang J, Riba-Grognuz O, Corona M, Nygaard S, Hunt BG, Ingram KK, Falquet L, Nipitwattanaphon M, Gotzek D. 2011. The genome of the fire ant Solenopsis invicta. Proc Natl Acad Sci. 108:5679–5684 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.