Abstract
Sterility is a serious problem that can affect all bionts. In teleosts, double haploids (DHs) induced by mitogynogenesis are often sterile. This sterility severely restricts the further application of DHs for production of clones, genetic analysis, and breeding. However, sterile DH individuals are good source materials for investigation of the molecular mechanisms of gonad development, especially for studies into the role of genes that are indispensable for fish reproduction. Here, we used the Illumina sequencing platform to analyze the transcriptome of sterile female DH Japanese flounder in order to identify major genes that cause sterility and to provide a molecular basis for an intensive study of gonadal development in teleosts. Through sequencing, assembly, and annotation, we obtained 52,474 contigs and found that 60.7% of these shared homologies with existing sequences. A total of 1225 differentially expressed unigenes were found, including 492 upregulated and 733 downregulated genes. Gene Ontology and KEGG analyses showed that genes showing significant upregulation, such as CYP11A1, CYP11B2, CYP17, CYP21, HSD3β, bcl2l1, and PRLR, principally correlated with sterol metabolic process, steroid biosynthetic process, and the Jak-stat signaling pathway. The significantly downregulated genes were primarily associated with immune response, antigen processing and presentation, cytokine–cytokine receptor interaction, and protein digestion and absorption. Using a co-expression network analysis, we conducted a comprehensive comparison of gene expression in the gonads of fertile and sterile female DH Japanese flounder. Identification of genes showing significantly different expression will provide further insights into DH reproductive dysfunction and oocyte maturation processes in teleosts.
Introduction
Double haploid (DH) fish are created by the process of mitogynogenesis. In mitogynogenesis, cell division is activated in haploid oocytes using irradiated sperm and diploidy is restored by temperature shock or hydrostatic pressure, which block the first cleavage to produce a diploid zygote. Offspring produced by mitogynogenesis are 100% homozygous, because a single set of chromosomes is duplicated [1]. Double haploidy is an advantageous situation for genetic mapping and genome sequencing studies [2]. As a rule, the effects of extremely high homozygosity are first noticed in fertility-related traits, especially in females [3]. In tilapia, only 10 of 77 gynogenetic DH females produced viable eggs [4]. In androgenetic common carp, the fertility rate was even lower with only 4 of 48 presumed females producing viable eggs [5]. The same fertility problem also occurs in Japanese flounder DH fish in which a few are fertile and the remainder are sterile and show varying levels of dysgenesis [6]. However, the sterile DH individuals do provide a good source material for research into the molecular mechanisms of gonad development, especially for identifying genes that are indispensable for fish reproduction. Reproductive processes in fish can be modified by environmental factors such as nutrition [7], temperature [8], and endocrine disruptors [9]. The majority of studies to date on female teleosts have investigated the effect of these factors on circulating sex hormone levels or on reproductive success in terms of spawning performance. Consequently, there are major gaps in our understanding of the molecular and cellular mechanisms of infertility.
RNA-seq has been used for quantitative gene expression analyses of biological processes in selected tissues or cells of a range of species [3, 10]. This method can be used to study genome-wide differences in gene expression through new generation high-throughput sequencing and information analysis platforms.
In the present study, we used RNA-seq to analyze the differential expression of genes in gonads of fertile and sterile female DH Japanese flounder. For some differentially expressed genes, real-time PCR was used to validate the differences. Our aim was to screen for the major genes that cause sterility in the DH flounder, and thus provide a molecular basis for an intensive study of oocyte maturation processes in teleosts.
Results
RNA-seq of sterile and fertile DH Japanese flounder
Two RNA-seq libraries were constructed from sterile and fertile fish using the Illumina Hi-Seq 2000 Genome Analyzer platform. This generated 5.1–7.2 billion clean reads sufficient for analysis of gene expression (Tables 1–3). These reads were then used in a blast analysis of target species with a cutoff of 1e-5. We identified 31,831 unigenes with homology to zebrafish transcripts, 30,147 unigenes with homology to mouse transcripts, and 30,264 unigenes with homology to human transcripts, accounting for 60.6%, 57.5%, and 57.7% of the total unigenes, respectively (Table 4). The gene ontology (GO) annotation process uses a dynamically structured control vocabulary that can be applied to describe gene function; the genes are first classified into three major categories, namely Biological Process, Molecular Function, and Cellular Component, and then into various sub-categories. Pathway Annotation is used to determine gene interactions in different metabolic pathways. We identified 29,416 unigenes assigned to biological process, 29,475 unigenes to molecular function, 29,464 unigenes to cellular component; 11,505 unigenes were assigned for further study by Pathway Annotation. Differential gene expression analysis in the control (fertile) group and case (sterile) group was performed using the DEseq Algorithm; 1225 differentially expressed unigenes were identified, of which 492 showed upregulation and 733 showed downregulation (see S1 File).
Table 1. Raw data and clean data statistics.
| Sample name | Before filter (reads) | After filter (reads) | Base filter % |
|---|---|---|---|
| F1 | 5372771647 | 5198504943 | 96.77% |
| F2 | 6049180053 | 5866858828 | 96.99% |
| F3 | 5952567280 | 5765483482 | 96.86% |
| S1 | 5198681376 | 5044969881 | 97.04% |
| S2 | 7459885244 | 7191224734 | 96.40% |
| S3 | 7037492168 | 6821752968 | 96.93% |
*Note: F1, F2, F3 and S1, S2, and S3 represent 3 fertile DH gonads and 3 sterile DH gonads, respectively; the same code is used in other Tables.
Table 3. Result of CAP3 clustering.
| Sample name | All unigenes |
|---|---|
| Number of contigs | 52474 |
| Number of characters | 75641345 |
| Maximum contig length (bp) | 17137 |
| Minimum contig length (bp) | 201 |
| Median contig length (bp) | 706 |
| Contig N50 length (bp) | 2895 |
Table 4. Unigene annotation and Blast result.
| Unigene blast | Annotated gene | Unigene annotation | Annotated gene |
|---|---|---|---|
| All unigenes | 52474 | All unigenes | 52474 |
| Blast to zebrafish | 31831 | GO-Biological Process (BP) | 29416 |
| Blast to mouse | 30147 | GO-Molecular Function (MF) | 29475 |
| Blast to human | 30264 | GO-Cellular Component (CC) | 29464 |
| Pathway | 11505 |
Table 2. Result of the Trinity transcripts assembly.
| Sample name | F1 | F2 | F3 | S1 | S2 | S3 |
|---|---|---|---|---|---|---|
| Raw reads | 53728010 | 60492160 | 59526022 | 51987136 | 74599184 | 70375224 |
| Clean reads | 52205350 | 58855834 | 57841646 | 50585486 | 72116290 | 68417302 |
| Number of contigs | 36499 | 37287 | 37645 | 36953 | 38528 | 39647 |
| Number of characters | 49304573 | 50626218 | 51400958 | 48397183 | 53368664 | 51946657 |
| Average contig length (bp) | 1350.85 | 1357.74 | 1365.41 | 1309.70 | 1385.19 | 1310.23 |
| Median contig length (bp) | 770 | 782 | 779 | 737 | 790 | 734 |
| Contig N50 length (bp) | 2493 | 2498 | 2515 | 2394 | 2544 | 2414 |
| Reads mapping to all unigenes (%) | 99.30 | 99.13 | 99.44 | 99.37 | 99.59 | 99.28 |
Functional enrichment analysis
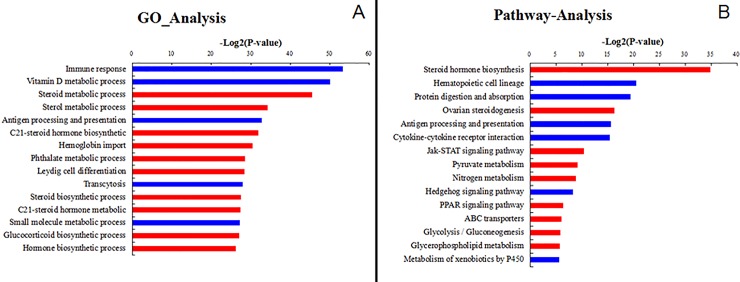
Based on the results of the unigene annotation, a GO analysis of expressed unigenes was applied and significant p-values were confirmed by Fisher’s exact tests. Of 1225 differentially expressed unigenes, 573 unigenes were annotated to 351 significant biological process go(p < 0.01): 175 were enriched by over-expressed genes in sterile gonads, 278 were enriched by under-expressed genes in sterile gonads. Among these biological processes, the genes highly expressed in the sterile gonads were mainly associated with immune response, vitamin D metabolic process, transcytosis, and small molecular metabolic process; the highest expressed genes in the fertile gonads were closely related to sterol metabolic process, steroid metabolic process, and C21-steroid hormone biosynthetic process (Fig 1A). CYP11A1, CYP11B2, CYP17A1, CYP21A1, HSD3β, and PRLR were the six genes showing the highest expression in fertile compared to sterile gonads (see S2 File).
Fig 1. Gene ontology analysis and pathway analysis of all differentially expressed unigenes.
(A) Gene ontology analysis of differentially expressed unigenes. The blue bar indicates up-regulated genes in sterile gonads compared with fertile gonads; the red bar indicates down-regulated genes in sterile gonads compared with sterile gonads. (B) Pathway analysis of all differentially expressed unigenes. The blue bar indicates up-regulated genes in sterile gonads compared with fertile gonads; the red bar indicates down-regulated genes in sterile gonads compared with sterile gonads.
A pathway analysis using the KEGG database was used to identify the significant pathways involving differentially expressed gene sets. Out of 1225 differentially expressed unigenes, 301 were successfully annotated. The significance level was calculated by Fisher’s exact test. In total, 46 pathway categories were found to be significantly enriched, including 26 pathways significantly enriched by under-expressed genes in the sterile gonads and 36 by over-expressed genes. Steroid hormone biosynthesis was the most significant pathway (Fig 1B) with six differentially expressed genes (CYP11A1, CYP11B2, CYP17A1, CYP21A1, HSD3B6, and AKR1D1) that showed lower expression in sterile gonads. The pathways showing greatest downregulation were involved in immune response, protein digestion and absorption, and cytokine–cytokine receptor interaction. The genes CTSS, CD74, B2M, C1QB, C1QC, SLC3A2, DPP4, CPA1, CTRB1, TGFB3, CSF3R, and TNFSF10 were involved in these pathways (see S3 File).
Differential expression: keyword analysis
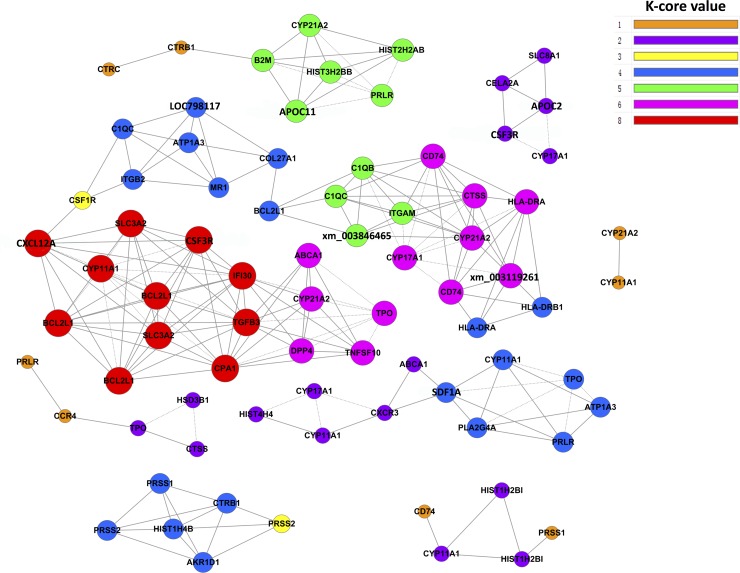
Gene co-expression networks were constructed to identify pivotal gene or genes. In the network, cycle nodes represent genes, and edges between two nodes represent interactions between genes, which are quantified by degrees. Degrees within the network describe the number of single genes that regulate other genes and represent the size of the cycle node; the higher the degree, the more centrally the gene occurs within the network [11]. The network was constructed using 72 differentially expressed genes in sterile gonads (Fig 2). These genes were attributed to the pathways steroid hormone biosynthesis, Staphylococcus aureus infection, hematopoietic cell lineage, protein digestion and absorption, antigen processing and presentation, cytokine–cytokine receptor interaction, proximal tubule bicarbonate reclamation, and Jak-stat signaling. Three of these pathways were upregulated and five were downregulated. Core regulatory genes involved in the eight pathways were determined using k-core differences between the sterile and fertile groups. As shown in Table 5, TGFB3 and SLC3A2 had the biggest k-core differences, followed by CYP11A1 and DPP4. Among these four genes, CYP11A1 was upregulated and more highly expressed in the fertile group and at a lower level in the sterile group; it directly regulates 10 neighboring genes that interact according to their degrees. The other three genes are related to protein digestion and absorption and cytokine–cytokine interaction, and showed downregulated expression. Other upregulated genes with large k-core differences were related to steroid hormone biosynthesis and the Jak-stat signaling pathway, including CYP21A1, CYP17A1, HSD3B6, CYP11B2, and PRLR.
Fig 2. Co-expression network of differentially expressed genes in sterile gonads.
Genes from pathways showing high expression differences were analyzed and identified using a gene co-expression network with a k-core algorithm. Cycle nodes represent genes, and the size of the node represents the power of the interrelationships among nodes; the edges between two nodes represent interactions between genes, and the greater the number of edges associated with a gene, the more it is connected to other genes and the more central is its role within the network.
Table 5. Eighteen genes identified by gene co-expression network with k-core algorithm.
| Gene symbol | Fertile | Sterile | Dif-k-core | Expression in sterile gonads relative to fertile gonads | ||
|---|---|---|---|---|---|---|
| Degree | k-core | Degree | k-core | |||
| TGFB3 | 1 | 1 | 14 | 8 | -7 | up |
| SLC3A2 | 1 | 1 | 9 | 8 | -7 | up |
| CYP11A1 | 3 | 2 | 10 | 8 | -6 | down |
| DPP4 | 0 | 0 | 7 | 6 | -6 | up |
| CPA1 | 3 | 3 | 13 | 8 | -5 | up |
| CTSS | 2 | 1 | 10 | 6 | -5 | up |
| BCL2L1 | 3 | 3 | 10 | 8 | -5 | down |
| CYP21A | 1 | 1 | 7 | 6 | -5 | down |
| ABCA1 | 1 | 1 | 6 | 6 | -5 | up |
| CXCR3 | 10 | 8 | 15 | 9 | -5 | up |
| APOC1I | 1 | 1 | 5 | 5 | -4 | up |
| CSF3R | 4 | 4 | 10 | 8 | -4 | up |
| CYP17A1 | 3 | 3 | 8 | 6 | -3 | down |
| TNFSF10 | 3 | 3 | 7 | 6 | -3 | up |
| HSD3β | 5 | 4 | 2 | 2 | 2 | down |
| CXCL12A | 11 | 11 | 9 | 8 | 3 | up |
| CYP11B2 | 6 | 5 | 3 | 2 | 3 | down |
| PRLR | 8 | 5 | 1 | 1 | 4 | down |
qRT-PCR validation of gene expression
To validate the results obtained by RNA-seq, the 10 candidate genes with the highest k-core differences were selected from the co-expression network analysis for confirmation by qRT-PCR. Eight candidate genes (SLC3A2, DPP4, CYP21A, CXCR3, CSF3R, CYP17A1, CXCL12A, and APOC1I) showed k-core differences that were too low to allow proper quantification in both groups (sterile gonads and fertile gonads). The qRT-PCR analysis confirmed the outcome of the RNA-seq analysis regarding changes in expression of the 10 candidate genes (Table 6).
Table 6. Verification of gene expression changes by qRT-PCR.
| Gene symbol | Expression in sterile gonads | Sequence of primers used in q-PCR | Expression of fold change (log2) | |
|---|---|---|---|---|
| RNA-seq | qPCR | |||
| TGFB3 | over | CAAGATTGGTCTGTCGGTA (F) | -1.25 | -2.34 |
| AGCCAAACAGCGTTACAT (R) | ||||
| CYP11A1 | under | ACGCCTGTTTGACCTCTG (F) | 4.69 | 3.81 |
| TGAGCATTTCTGTTGGGAG (R) | ||||
| CPA1 | over | GGTGTAAGCGTAGCCGTCAG (F) | -4 | -3.85 |
| TCCAGGGAATGGGTGTCG (R) | ||||
| CTSS | over | CAAACAAGCGAGTTATGAT (F) | -1.36 | -1.6 |
| AACCTCTTTCGGAGACAA (R) | ||||
| BCL2L1 | under | GTGATGGACGAGGTGTTC (F) | 1.22 | 0.19 |
| ATCCTGTCCACCAGCGAA (R) | ||||
| ABCA1 | over | GCCACAGAGTGCCGTTAT (F) | -2 | -3.1 |
| TTCCTTCGGGTGATGAGT (R) | ||||
| TNFSF10 | over | CCTCGGTTCAAAGATGGAT (F) | -1.22 | -2.18 |
| CGTTGGCAAAGCAAGGAA (R) | ||||
| HSD3β | under | TCGTATCCCGTCATCCAT (F) | 1.84 | 0.73 |
| TGAGGTTTTCCTACAGCAAG (R) | ||||
| CYP11B2 | under | TATTCTGCGTCCCAAACA (F) | 4.67 | 3.78 |
| TTCGTCCAGAGCAGTATCA (R) | ||||
| PRLR | under | CAATAATGAGCCAATAACG (F) | 5.12 | 4.06 |
| GTGGGTCTGTGGATGTTA (R) | ||||
Discussion
In general, the effects of inbreeding are first noticed in fertility related traits, especially in females [3]. However, in fish, only a few detailed studies have been performed on the fertility of DH individuals; these studies largely concentrated on delayed natural spawning time, decreased ovulation responses to hormonal induction, and reduced egg size and quality [12]. In this study, we used transcriptome sequencing to investigate the biological mechanisms of homozygotic sterility in female DH Japanese flounder.
Genes showing high levels of expression in fertile ovaries were primarily associated with steroid biosynthesis, while genes expressed highly in sterile gonads were mainly associated with other metabolic processes, such as immune response, cytokine–cytokine receptor interaction, and protein digestion and absorption. In the transcriptomes of sterile fish, seven genes showed the greatest upregulation and played important roles in steroid biosynthesis and in the Jak-stat signaling pathway: CYP11A1, CYP11B2, CYP17, CYP21, HSD3B, BCL2L1, and PRLR. Other researchers have confirmed that the variation in the expression of genes for steroid biosynthesis can affect hormone levels and cause metabolic disorders. For example, polycystic ovary syndrome, a common endocrine disease in women of reproductive age, is mainly caused by abnormal expression of steroidogenic enzymes, which also can induce abnormal follicular maturation [13]. CYP11A1 is the first and rate-limiting step of steroid biosynthesis, and catalyzes the conversion of cholesterol to pregnenolone, the gene controls the rate of synthesis of all steroid hormones. Thus, CYP11A1 is critical for steroidogenesis at the gonad development stage [10]. Additionally, CYP11A1 is important for placental progesterone synthesis, which is essential for the maintenance of pregnancy in mammals [14]. In humans, mutation of CYP11A1 can cause congenital lipoid adrenal hyperplasia, with a range of symptoms such as male sex reversal, high plasma adrenocorticotropic hormone levels, and increased plasma concentrations of gonadotrophins [15]. Similarly to humans, Cyp11a1-null mice have exceedingly high levels of adrenocorticotropic hormone but very little corticosterone and aldosterone in the plasma [16]. CYP11A1 is also regarded as a critical regulator for gonadal development in fish. In zebrafish, CYP11A1 is synthesized as a maternal transcript, and targeted knockdown of the gene leads to a shortened embryonic axis and epiboly defects, presumably because of the lack of adequate maternal steroid supply [17]. CYP11A1 is also indispensable for gonadal development and maturation in catfish [18]. Here, CYP11A1 was found to be expressed at extremely low levels in sterile ovaries, and at much higher levels in fertile ovaries. We suggest that CYP11A1 has a significant effect on ovary development in the Japanese flounder and that lower levels of expression impair oogenesis.
CYP11B2, aldosterone synthase gene, encodes a cytochrome P450 membrane-bound heme-containing enzyme that accepts electrons from NADPH via accessory proteins; it participates in hydroxylation and other oxidative conversions of target molecules. CYP11B2 is normally expressed only the in zona glomerulosa where it catalyzes three sequential reactions: 11β-hydroxylation, 18-hydroxylation, and 18-oxidation to form aldosterone [19–20]. Mutation of CYP11B2 can cause aldosterone synthase deficiency in humans [21]. Fish CYP11B also catalyzes the 11β-hydroxylation step that produces 11β-hydroxytestosterone in the gonad [21]. Expression of CYP11B in fish shows a sexually dimorphic pattern in the gonad, as it only expressed in testis and not in the ovary of either developing or adult fish [19, 22–24]. In contrast, CYP11B2 was expressed in DH ovaries with a higher level of expression in gonads of fertile compared to sterile flounder. We speculate that gonadal development in DH flounder may differ from the standard pattern.
Germ cell development in sterile DH ovaries was inhibited at an early stage and vitellogenesis was not completed to enable final oocyte maturation. It is known that sex steroids regulate liver metabolism to produce vitellogenin (VTG), the egg yolk precursor protein. Additionally, 17α,20β-dihydroxy-4-pregnen-3-one is an essential hormone for final oocyte maturation in fish [25–26] and CYP17 has a vital role in its production in gonadal tissues and also for production of sex steroids [27]. The expression of CYP17 in the ovaries of trout and eel increases during development and maturation of the follicles [28]. It was reported that the level of CYP17 expression increases continually during gonad development in Japanese flounder [29]. Mutation of the coding region of CYP17 or changes to its methylation status might influence expression and, consequently, reproductive endocrine levels [30]. In common carp, CYP17 deficiency leads to inter-renal hyperplasia [31]. Our study also indicated that low expression of CYP17 may affect the production of sex steroids and further restrict gonadal development.
In the transcriptome from sterile ovaries, the least expressed genes were HSD3β and CYP21. HSD3β is responsible for the second step of steroidogenesis, namely, the conversion of P5 into progesterone. CYP21 catalyzes the final steps in the production of cortisol and functions in sex steroid production [32]. CYP21 deficiency in humans leads to congenital adrenal hyperplasia, an autosomal recessive disorder associated with deficiency in adrenocortical enzymes necessary for cortisol biosynthesis [33]. Deficient expression of the CYP21 gene results in the accumulation of 17-α-hydroxy progesterone and its conversion to androgens. Excessive adrenal androgen production can induce clinical hyperandrogenism, anovulatory cycles, and infertility [34]. In addition to the five genes described above, PRLR is also involved in steroid biosynthesis. PRLR is a member of the class 1 cytokine receptor superfamily and forms a transmembrane chain that is embedded in the cell membrane; it functions in combination with prolactin [35]. The role of PRLR in reproduction in mice was demonstrated through analysis of a germ-line null mutation. Female PRLR-/- mice exhibit total sterility because of the regression of the corpora lutea, the absence of sufficient progesterone to support implantation and the subsequent development and maintenance of the placenta [36]. PRLR mRNA and protein have been found in the gonads of some fish, such as Mozambique tilapia [37], Nile tilapia [38], and seabream [39]. Furthermore, PRLR levels often change during the breeding cycle. The highest PRLR level in female Nile tilapia plasma occurs after spawning and during vitellogenesis [40], suggesting that PRLR may be involved in vitellogenesis and/or ovulation. These observations are consistent with our experimental results that PRLR is expressed at low levels in sterile DH fish that show yolk accumulation deficiency. This implies that PRLR has an important role in ovarian development in Japanese flounder.
VTG is synthesized mainly in the liver under the regulation of E2. It is transferred to the oocyte via thecal capillaries to the granulosa layer, and passes to the oocyte surface through pore canals in the zona radiata. Subsequently, the VTG enters the cell by receptor-mediated endocytosis [41]. In the present study, we found upregulation of the VTG gene in sterile DH fish. VTG is involved in vitamin metabolic process, transcytosis, lipoprotein transport, proteolysis, and receptor-mediated endocytosis. All of these processes are related to yolk deposition (vitellogenesis). Thus, although the DH fish were sterile, they still showed higher expression of the gene for yolk protein accumulation. The numerous oocytes in the sterile gonad presumably still continue vitellogenesis. We speculate that steroid hormone disorder caused by key enzyme genes for steroid biosynthesis (as described above) is associated with the high level of VTG expression.
The vitellogenic period is characterized by high production of RNAs, proteins, lipids, vitamins, and hormones [42]. In the sterile ovaries, we found higher expression of genes related to cytokine–cytokine receptor interaction, and protein digestion and absorption, namely, TGFB3, CSF3R, CXCR3, SLC3A2, and DPP4. Of these, TGFB3, CSF3R, and CXCR3 have been reported to be expressed in the preovulatory ovary of fish [43]. SLC3A2 is expressed in a wide variety of tissues in mammals, specifically in the yolk syncytial layer in the embryo [44]. DPP4 plays an important role in enzymatic degradation of incretin peptides [45]. We speculate that these genes are correlated to vitellogenesis in the sterile ovaries.
Follicular atresia is a widespread phenomenon in fish ovaries under both natural and experimental conditions; in this process, a number of ovarian follicles recruited into the vitellogenesis pool fail to complete maturation and ovulation [46]. Atresia mainly occurs during the post-spawning period, but can also be observed in other stages of the reproductive cycle [47]. In this study, although the appearance of the sterile DH gonads differed from that of follicular atresia in fish described in other studies [48–49], some genes related to atresia were more highly expressed in the sterile DH gonads. APOC1-I encodes a protein component of chylomicrons, high-density lipoproteins (HDLs) involved in lipid transport in the bloodstream [50]. ABCA1 mediates the transport of cellular cholesterol and phospholipids to APOA-1 to generate nascent HDL particles [51]. The expression of ABCA1 and APOC1-I is required for the clearance of excess cholesterol and phospholipids from hepatocytes and for the reduction in hepatic lipid accumulation [52]. During follicular atresia in rainbow trout, there is massive transfer of the oocyte yolk proteins and possibly lipids into the bloodstream combined with HDLs [53] because of the ingestion and digestion of the yolk by the follicular cells. The study of Senegalese sole has shown the importance of lipid-metabolic process during follicular atresia in fish [54]. In humans, serum APOC1-I has been proposed to be an early marker for metabolic abnormalities in women with polycystic ovary syndrome [55]. Similarly, APOC1-I may be a useful marker to identify factors involved in premature ovarian regression in cultured fish [54]. Therefore, lipid-metabolic processes during follicular atresia in fish may have evolved to facilitate the redistribution of energy-rich yolk materials from oocytes that failed to develop properly [56]. In the sterile DH females, gonadal development arrested at the vitellogenic stage, presumably stimulating the redistribution of energy from the oocytes. The finding of high APOC1-I and ABCA1 transcriptional levels in sterile ovaries of Japanese flounder provides additional evidence for this inference.
In the transcriptome of sterile ovaries, the genes showing the highest upregulation were related to immune response. During follicular atresia in fish, immune cells may act synergistically with follicular cells in the resorption of the oocyte through release of lytic enzymes [57]. Granulocytes appear in the atretic follicles of some fish species [54, 58], and there is evidence of a relationship between follicular regression and immune cells [48]. In this study, a series of chemokines (CCR4, CMKLR1, CXCL12A, and CXCR3) in the immune response process drew our attention. In the mammalian ovary, chemokines play an important role in follicular atresia and energy reassignment [57]. In atretic follicles of Senegalese sole, the chemokine LECT2 shows high expression levels [54]. However, the molecular pathways through which chemokines act in atretic follicles are largely unknown. Ovarian expression of CCR4, CMKLR1, CXCL12A, and CXCR3 have not yet been reported in fish and, therefore, the structural and functional relationships of these 4 chemokines genes in Japanese flounder requires further investigation. Atresia is considered as an apoptotic process in many organisms, and the proteolytic degradation of the oocyte yolk proteins, mediated by the differential activation of lysosomal cathepsins, has been proposed as the initial event leading to oocyte cell death [56]. Two key genes, TNFSF10 and CTSS, involved in the regulation of cell apoptosis also appeared in the immune response process category. TNFSF10 induces apoptotic cell death in cancer by binding to its functional death receptors [59]. CTSS is an important cathepsin that inhibits apoptosis; null mutation of the gene results in decreased DNA injury and apoptosis [60]. We speculate that non-functional oocytes in sterile DH gonads will become apoptotic and there will be a redistribution of cellular energy as a result of follicular atresia.
Conclusions
In this study, we conducted a comprehensive comparison of gene expression in fertile and sterile gonads of DH Japanese flounder. We identified many genes responsible for gonad development through transcriptome sequencing. In combination with the sequencing results, we speculated that genes involved in steroid biosynthesis that showed reduced levels of expression might affect the development of the ovary and possibly cause sterility in DH Japanese flounder. We also found that genes related to apoptosis and to digestion of the yolk for energy reallocation showed higher expression in sterile DH gonads. These results indicated that the arrested oocytes might become apoptotic and that their energy might be redistributed. These significant changes in gene expression patterns provide further insights not only into DH reproductive dysfunction, but also into oogenesis in Japanese flounder and the molecular mechanisms of piscine oocyte maturation.
Materials and Methods
Ethics Statement
Experimental treatment of the Japanese flounder in this study was performed strictly in accordance with the Guide for Care and Use of Laboratory Animals of the Chinese Association for Laboratory Animal Sciences (No. 2011–2); all the experiments were approved by the animal care and use committee of Beidaihe Central Experiment Station.
Screening of sterile DH Japanese flounder
The DH Japanese flounder were produced in April 2009, from a single wild female that captured from the Qinghuangdao area of The Bohai Sea and reared at Beidaihe Central Experiment Station, Qinghuangdao, following the standard protocol for mitogynogenesis [16].
From 2012, the DH females were manually checked for egg production every year. Eggs were extracted from fertile individuals, and the progenies survived. Approximately, 70% of DHs had a flat abdomen and appeared sterile. In May 2014, three sterile DHs that could not produce eggs were euthanized using 300 mg/L tricaine methanesulfonate (MS222), along with three fertile DHs who produced viable progeny. The gonads were dissected from each female, immediately frozen in liquid nitrogen, and stored at -80°C until use.
RNA preparation, cDNA synthesis, sequencing, and de novo assembly
Total RNA for each sample was extracted with an RNeasy Mini Kit and digested with DNase I following the manufacturer’s instructions (Qiagen, Hilden, Germany). Integrity and size distribution of the RNA samples were verified using an Agilent 2200 Bioanalyser (Agilent Technologies, Germany). Samples with an RNA Integrity Number ≥ 8.0 were used for cDNA library preparation. The concentration of RNA in each extracted sample was measured using a Qubit 1.0 Fluorometer (Invitrogen, Carlsbad, USA). The cDNA libraries were constructed for each pooled RNA sample using the TruSeqTM RNA sample preparation kit (Illumina, Inc.) according to the manufacturer’s instructions. The tagged cDNA libraries were pooled in equal ratio and used for 101 bp paired-ends sequencing with an Illumina HiSeqTM 2000.
Clean reads were obtained from the raw reads by removing the adaptor sequences, reads with >5% ambiguous bases (noted as N), and low-quality reads containing more than 30 percent of bases with qualities of <20. Contig assembly was carried out using Trinity software [17] and the contig of the 6 different samples was clustered using CAP3 software [18] to achieve the final unigenes result. Gene Annotation was performed using tBlastx (National Center for Biotechnology Information; http://www.ncbi.nlm.nih.gov/) on zebrafish, mouse, and human transcripts and by filtering using the criterion “E-Value < 1e-5”. Coding sequence prediction was obtained using EST scan software [61]. BWA [62] software was used for mapping the reads to the unigene assembly and the counts were calculated with Picard software. We applied the DEseq algorithm to filter the differentially expressed unigenes, after the analyses of significance and false discovery rate (FDR), with the following criteria: i) Fold Change >2 or <0.5; and ii) FDR < 0.05 [63].
Analysis of GO category, pathway, gene-act-network, and gene co-expression
Gene ontology (GO) analysis was performed to elucidate the biological implications of unique genes in the significant or representative profiles of differentially expressed genes in the experiment [64]. We downloaded the GO annotations from NCBI (http://www.ncbi.nlm.nih.gov/), UniProt (http://www.uniprot.org/), and Gene Ontology (http://www.geneontology.org/). Fisher’s exact test was applied to identify significant GO categories and FDR was used to correct the p-values. Pathway analysis was used to identify significant pathways associated with the differentially expressed genes according to the KEGG database. We also used Fisher’s exact test to select significant pathways, and the threshold of significance was defined by p-value and FDR [65]. The KEGG database was used to build a network of genes according to the relationships among genes, proteins, and compounds in the database. Gene Ontology is structured as a directed acyclic graph, and each term has defined relationships to one or more other terms. We built gene co-expression networks to show the relationships among genes [66]. Gene co-expression networks were based on normalized expression values of genes selected from those genes in significant GO terms and pathway terms. For each pair of genes, we calculated the Pearson correlation and chose the pairs with a significant correlation (FDR < 0.05) to construct the network [67]. In this network analysis, degree centrality was the most simple and important measure to determine the relative importance of a gene to the network. Degree centrality was defined as the number of links of one node to others [68]. The properties of the networks were also analyzed using k-cores, derived from graph theory as a method to simplify graph topology analysis. The k-core of a network is a sub-network in which all nodes are connected to at least k other genes. The k-core of a protein–protein interaction network usually contains cohesive groups of proteins [69].
Real-time qPCR assays
Real-time qPCR was performed to validate gene expression data from the RNA-Seq analysis. Total RNA was reverse transcribed using a Superscript III kit (Invitrogen) according to the manufacturer’s instructions. Real-time qPCR was performed using a 7500 Fast Real Time PCR system (Applied Biosystems, Foster City, USA) with SYBR green I nucleic acid kit (Invitrogen) in 20 μL reactions. Each reaction consisted of 2 ng of total RNA and 0.5 μL of each primer. PCR was performed in a thermocycler under the following conditions: 2 min at 95°C, then 40 cycles of 10 s at 95°C, 30 s at 60°C, and 45 s at 72°C, followed by 5 min at 72°C. To verify the presence of a specific product, a melting curve analysis of amplification products was performed at the end of each PCR. Primer pairs for qPCR amplification were designed using Netprimer (http://www.premierbiosoft.com/netprimer/). The comparative CT method of quantification was used to quantify the relative expression of specific genes. All samples were run in triplicate, and the relative expression levels of target genes were calculated with the 2-∆∆Ct method, with the β-actin gene used for normalization of the data.
Supporting Information
(XLS)
(XLS)
(XLS)
Acknowledgments
We thank Mr. Dai Chen (Novel Bioinformatics Ltd., Co, Shanghai, China) for his technical assistance in bioinformatics analysis. We aso thank Dr. Yuanshuai Fu (Shanghai Ocean University) for his technical assisstance in real-time qPCR assays.
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
The work was funded by National High Technology Research and Development Program in China (2012BAD26B01) and Special Scientific Research Funds for Central Non-profit Institutes Chinese Academy of Fishery Sciences (2014A03XK02).
References
- 1. Streisinger G, Walker C, Dower N, Knauber D, Singer F. Production of clones of homozygous diploid zebra fish (Brachydanio rerio). Nature. 1981;291(5813):293–6. [DOI] [PubMed] [Google Scholar]
- 2. Komen H, Thorgaard GH. Androgenesis, gynogenesis and the production of clones in fishes: A review. Aquaculture. 2007;269(1–4):150–173. [Google Scholar]
- 3.Keller L. Inbreeding effects in wild populations. In; 2002. p. 230–241.
- 4. Müller-Belecke A, Horstgen-Schwark G. Sex determination in tilapia (Oreochromis niloticus) sex ratios in homozygous gynogenetic progeny and their offspring. Aquaculture. 1995;137(1–4):57–65. [Google Scholar]
- 5.Bongers ABJ, Zandieh-Doulabi B. Viable androgenetic YY genotypes of common carp (Cyprinus carpio L.). In; 1999.
- 6. Zhang X, HJWG. Primary study on sterile mitogynogenetic Paralichthys olivaceus: Histological observation and microsatellite screening. Marine fishery. 2014;36:503–510. [Google Scholar]
- 7. Izquierdo MS, Fernández-Palacios H, Tacon AGJ. Effect of broodstock nutrition on reproductive performance of fish In: Donaldson CLM, editor Reproductive Biotechnology in Finfish Aquaculture. Amsterdam: Elsevier; 2001. p. 25–42. [Google Scholar]
- 8.de Vlaming VL. Environmental control of teleost reproductive cycles: a brief review. In; 1972. p. 131–140.
- 9. Arukwe A. Cellular and molecular responses to endocrine-modulators and the impact on fish reproduction. Mar Pollut Bull. 2001;42(8):643–55. [DOI] [PubMed] [Google Scholar]
- 10. Huang W, Khatib H. Comparison of transcriptomic landscapes of bovine embryos using RNA-Seq. BMC Genomics. 2010;11(1):711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chen F, Zhu HH, Zhou LF, Li J, Zhao LY, Wu SS, et al. Genes related to the very early stage of ConA-induced fulminant hepatitis: a gene-chip-based study in a mouse model. BMC Genomics. 2010;11:240 10.1186/1471-2164-11-240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kj Rsvik E, Mangor-Jensen A, Holmefjord I. Egg Quality in Fishes In: HSBA J., editor Advances in Marine Biology: Academic Press; 1990. p. 71–113. [Google Scholar]
- 13. Gunawan A, Sahadevan S, Neuhoff C, Grosse-Brinkhaus C, Gad A, Frieden L, et al. RNA deep sequencing reveals novel candidate genes and polymorphisms in boar testis and liver tissues with divergent androstenone levels. PLoS One. 2013;8(5):e63259 10.1371/journal.pone.0063259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cerdà J, Bobe J, Babin PJ, Admon A, Lubzens E. Functional genomics and proteomic approaches for the study of gamete formation and viability in farmed finfish. Rev Fish Sci. 2008;16:54–70. [Google Scholar]
- 15. Hayashi M, Sato M, Iwasaki Y, Terasawa M, Tashiro M, Yokoyama S, et al. Combining next-generation sequencing with microarray for transcriptome analysis in rainbow trout gonads. Mol Reprod Dev. 2012;79(12):870–8. 10.1002/mrd.22127 [DOI] [PubMed] [Google Scholar]
- 16. Yamamoto E. Studies on sex-manipulation and production of cloned populations in hirame, Paralichthys olivaceus (Temminck et Schlegel). Aquaculture. 1999;173(1–4):235–246. [Google Scholar]
- 17. Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–52. 10.1038/nbt.1883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Huang X, Madan A. CAP3: A DNA sequence assembly program. Genome Res. 1999;9(9):868–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. von Schalburg KR. A Comprehensive Survey of the Genes Involved in Maturation and Development of the Rainbow Trout Ovary. Biology of Reproduction. 2005;72(3):687–699. [DOI] [PubMed] [Google Scholar]
- 20. Bureik M, Lisurek M, Bernhardt R. The Human Steroid Hydroxylases CYP11B1 and CYP11B2. Biol Chem. 2002;383(10):1537–1551. [DOI] [PubMed] [Google Scholar]
- 21. Lisurek M, Bernhardt R. Modulation of aldosterone and cortisol synthesis on the molecular level. Mol Cell Endocrinol. 2004;215(1–2):149–59. [DOI] [PubMed] [Google Scholar]
- 22. Jiang JQ, Young G, Kobayashi T, Nagahama Y. Eel (Anguilla japonica) testis 11beta-hydroxylase gene is expressed in interrenal tissue and its product lacks aldosterone synthesizing activity. Mol Cell Endocrinol. 1998;146(1–2):207–11. [DOI] [PubMed] [Google Scholar]
- 23. Liu S, Govoroun M, D'Cotta H, Ricordel MJ, Lareyre JJ, McMeel OM, et al. Expression of cytochrome P450(11beta) (11beta-hydroxylase) gene during gonadal sex differentiation and spermatogenesis in rainbow trout, Oncorhynchus mykiss. J Steroid Biochem Mol Biol. 2000;75(4–5):291–8. [DOI] [PubMed] [Google Scholar]
- 24. Kusakabe M, Kobayashi T, Todo T, Mark LP, Nagahama Y, Young G. Molecular cloning and expression during spermatogenesis of a cDNA encoding testicular 11beta-hydroxylase (P45011beta) in rainbow trout (Oncorhynchus mykiss). Mol Reprod Dev. 2002;62(4):456–69. [DOI] [PubMed] [Google Scholar]
- 25. Kagawa H, Young G, Nagahama Y. Relationship between seasonal plasma estradiol-17 beta and testosterone levels and in vitro production by ovarian follicles of amago salmon (Oncorhynchus rhodurus). Biol Reprod. 1983;29(2):301–9. [DOI] [PubMed] [Google Scholar]
- 26. Nagahama Y, Adachi S. Identification of maturation-inducing steroid in a teleost, the amago salmon (Oncorhynchus rhodurus). Dev Biol. 1985;109(2):428–35. [DOI] [PubMed] [Google Scholar]
- 27. Zhou LY, Wang DS, Shibata Y, Paul-Prasanth B, Suzuki A, Nagahama Y. Characterization, expression and transcriptional regulation of P450c17-I and -II in the medaka, Oryzias latipes. Biochem Biophys Res Commun. 2007;362(3):619–25. [DOI] [PubMed] [Google Scholar]
- 28. Kazeto Y, Ijiri S, Todo T, Adachi S, Yamauchi K. Molecular Cloning and Characterization of Japanese Eel Ovarian P450c17 (CYP17) cDNA. General and Comparative Endocrinology. 2000;118(1):123–133. [DOI] [PubMed] [Google Scholar]
- 29. Ding Y, He F, Wen H, Li J, Ni M, Chi M, et al. DNA methylation status of cyp17-II gene correlated with its expression pattern and reproductive endocrinology during ovarian development stages of Japanese flounder (Paralichthys olivaceus). Gene. 2013;527(1):82–88. 10.1016/j.gene.2013.05.037 [DOI] [PubMed] [Google Scholar]
- 30. Ding Y, He F, Wen H, Li J, Qian K, Chi M, et al. Polymorphism in exons CpG rich regions of the cyp17-II gene affecting its mRNA expression and reproductive endocrine levels in female Japanese flounder (Paralichthys olivaceus). General and Comparative Endocrinology. 2012;179(1):107–114. 10.1016/j.ygcen.2012.08.003 [DOI] [PubMed] [Google Scholar]
- 31. Nematollahi MA, van Pelt-Heerschap H, Komen H. High corticosterone and sex reversal in common carp (Cyprinus carpio L.) with adrenal hyperplasia caused by P450c17a2 deficiency. Aquaculture. 2014;418–419:165–170. [Google Scholar]
- 32. Hsu H, Lin J, Chung B. Zebrafish cyp11a1 and hsd3b genes: Structure, expression and steroidogenic development during embryogenesis. Molecular and Cellular Endocrinology. 2009;312(1–2):31–34. 10.1016/j.mce.2009.07.030 [DOI] [PubMed] [Google Scholar]
- 33. Reisch N. Substitution therapy in adult patients with congenital adrenal hyperplasia. Best Practice & Research Clinical Endocrinology & Metabolism. 2015;29(1):33–45. [DOI] [PubMed] [Google Scholar]
- 34. Bannister LA, Pezza RJ, Donaldson JR, de Rooij DG, Schimenti KJ, Camerini-Otero RD, et al. A dominant, recombination-defective allele of Dmc1 causing male-specific sterility. PLoS Biol. 2007;5(5):e105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bazan JF. A novel family of growth factor receptors: a common binding domain in the growth hormone, prolactin, erythropoietin and IL-6 receptors, and the p75 IL-2 receptor beta-chain. Biochem Biophys Res Commun. 1989;164(2):788–95. [DOI] [PubMed] [Google Scholar]
- 36. Bole-Feysot C, Goffin V, Edery M, Binart N, Kelly PA. Prolactin (PRL) and its receptor: actions, signal transduction pathways and phenotypes observed in PRL receptor knockout mice. Endocr Rev. 1998;19(3):225–68. [DOI] [PubMed] [Google Scholar]
- 37. Edery M, Young G, Bern H, Steiny S. Prolactin receptors in tilapia (Sarotherodon mossambicus) tissues: binding studies using I-125 labeled ovine prolactin. Gen Comp Endocrinol. 1984;56:19–23. [DOI] [PubMed] [Google Scholar]
- 38. Sandra O, Le Rouzic P, Cauty C, Edery M, Prunet P. Expression of the prolactin receptor (tiPRL-R) gene in tilapia Oreochromis niloticus: tissue distribution and cellular localization in osmoregulatory organs. J Mol Endocrinol. 2000;24(2):215–24. [DOI] [PubMed] [Google Scholar]
- 39.Cavaco JEB, Santos CRA, Ingleton P, Canario AVM, Power DM. Quantification of prolactin (PRL) and PRL receptor messenger RNA in gilthead Seabream (Sparus aurata) after treatment with Estradiol-17 beta. In; 2003. p. 588–594. [DOI] [PubMed]
- 40. Tacon P, Baroiller JF, Le Bail PY, Prunet P, Jalabert B. Effect of egg deprivation on sex steroids, gonadotropin, prolactin, and growth hormone profiles during the reproductive cycle of the mouthbrooding cichlid fish Oreochromis niloticus. Gen Comp Endocrinol. 2000;117(1):54–65. [DOI] [PubMed] [Google Scholar]
- 41. Wallace RA, Selman K. Ultrastructural aspects of oogenesis and oocyte growth in fish and amphibians. J Electron Microsc Tech. 1990;16(3):175–201. [DOI] [PubMed] [Google Scholar]
- 42. SJ TC. Oocyte growth and development in teleosts. rev fish biol fish. 1996;6:287–318. [Google Scholar]
- 43. Bobe J, Nguyen T, Fostier A. Ovarian function of the trout preovulatory ovary: New insights from recent gene expression studies. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2009;153(1):63–68. [DOI] [PubMed] [Google Scholar]
- 44. Takesono A, Moger J, Farooq S, Cartwright E, Dawid IB, Wilson SW, et al. Solute carrier family 3 member 2 (Slc3a2) controls yolk syncytial layer (YSL) formation by regulating microtubule networks in the zebrafish embryo. Proceedings of the National Academy of Sciences. 2012;109(9):3371–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhong J, Rao X, Deiuliis J, Braunstein Z, Narula V, Hazey J, et al. A Potential Role for Dendritic Cell/Macrophage-Expressing DPP4 in Obesity-Induced Visceral Inflammation. Diabetes. 2012;62(1):149–157. 10.2337/db12-0230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Saidapur SK. Follicular atresia in the ovaries of nonmammalian vertebrates. Int Rev Cytol. 1978;54:225–44. [DOI] [PubMed] [Google Scholar]
- 47.Guraya SS. Gonadal development and production of gametes in fish. In; 1994. p. 15–32.
- 48. Miranda AC, Bazzoli N, Rizzo E, Sato Y. Ovarian follicular atresia in two teleost species: a histological and ultrastructural study. Tissue Cell. 1999;31(5):480–8. 10.1054/tice.1999.0045 [DOI] [PubMed] [Google Scholar]
- 49. Lang I. Electron microscopic and histochemical investigations of the atretic oocyte of Perca fluviatilis L. (Teleostei). Cell Tissue Res. 1981;220(1):201–12. [DOI] [PubMed] [Google Scholar]
- 50. Jong MC, Hofker MH, Havekes LM. Role of ApoCs in lipoprotein metabolism: functional differences between ApoC1, ApoC2, and ApoC3. Arterioscler Thromb Vasc Biol. 1999;19(3):472–84. [DOI] [PubMed] [Google Scholar]
- 51. Oram JF, Heinecke JW. ATP-binding cassette transporter A1: a cell cholesterol exporter that protects against cardiovascular disease. Physiol Rev. 2005;85(4):1343–72. [DOI] [PubMed] [Google Scholar]
- 52. Ma D, Liu W, Wang Y. ApoA-I or ABCA1 expression suppresses fatty acid synthesis by reducing 27-hydroxycholesterol levels. Biochimie. 2014;103:101–108. 10.1016/j.biochi.2014.04.010 [DOI] [PubMed] [Google Scholar]
- 53. Babin PJ. Apolipoproteins and the association of egg yolk proteins with plasma high density lipoproteins after ovulation and follicular atresia in the rainbow trout (Salmo gairdneri). J Biol Chem. 1987;262(9):4290–6. [PubMed] [Google Scholar]
- 54. Tingaud-Sequeira A, Chauvigne F, Lozano J, Agulleiro MJ, Asensio E, Cerda J. New insights into molecular pathways associated with flatfish ovarian development and atresia revealed by transcriptional analysis. BMC Genomics. 2009;10:434 10.1186/1471-2164-10-434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Huang S, Qiao J, Li R, Wang L, Li M. Can serum apolipoprotein C-I demonstrate metabolic abnormality early in women with polycystic ovary syndrome? Fertil Steril. 2010;94(1):205–10. 10.1016/j.fertnstert.2009.03.005 [DOI] [PubMed] [Google Scholar]
- 56. Wood AW, Van der Kraak G. Yolk proteolysis in rainbow trout oocytes after serum-free culture: evidence for a novel biochemical mechanism of atresia in oviparous vertebrates. Mol Reprod Dev. 2003;65(2):219–227. [DOI] [PubMed] [Google Scholar]
- 57. Besseau L, Faliex E. Resorption of unemitted gametes in Lithognathus mormyrus (Sparidae, Teleostei): a possible synergic action of somatic and immune cells. Cell Tissue Res. 1994;276(1):123–32. [DOI] [PubMed] [Google Scholar]
- 58. Santos HB, Rizzo E, Bazzoli N, Sato Y, Moro L. Ovarian regression and apoptosis in the South American teleost Leporinus taeniatus Lütken (Characiformes, Anostomidae) from the Sao Francisco Basin. Journal of Fish Biology. 2005;67(5):1446–1459. [Google Scholar]
- 59. Kuribayashi K, Krigsfeld G, Wang W, Xu J, Mayes PA, Dicker DT, et al. TNFSF10 (TRAIL), a p53 target gene that mediates p53-dependent cell death. Cancer Biol Ther. 2008;7(12):2034–8. [DOI] [PubMed] [Google Scholar]
- 60. Carnevali O, Cionna C, Tosti L, Lubzens E, Maradonna F. Role of cathepsins in ovarian follicle growth and maturation. General and Comparative Endocrinology. 2006;146(3):195–203. [DOI] [PubMed] [Google Scholar]
- 61.Iseli C, Jongeneel CV, Bucher P. ESTScan: a program for detecting, evaluating, and reconstructing potential coding regions in EST sequences. In; 1999. p. 138–48. [PubMed]
- 62. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. 10.1093/bioinformatics/btp324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W, et al. Count-based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc. 2013;8(9):1765–86. 10.1038/nprot.2013.099 [DOI] [PubMed] [Google Scholar]
- 64. Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25(1):25–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Draghici S, Khatri P, Tarca AL, Amin K. A systems biology approach for pathway level analysis. In; 2007. p. 1537–1545. [DOI] [PMC free article] [PubMed]
- 66. Pujana MA, Han JD, Starita LM, Stevens KN, Tewari M, Ahn JS, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat Genet. 2007;39(11):1338–49. [DOI] [PubMed] [Google Scholar]
- 67. Prieto C, Risueno A, Fontanillo C, De Las RJ. Human gene coexpression landscape: confident network derived from tissue transcriptomic profiles. PLoS One. 2008;3(12):e3911 10.1371/journal.pone.0003911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Barabasi AL, Oltvai ZN. Network biology: understanding the cell's functional organization. Nature Reviews Genetics. 2004;5(2):101–113. [DOI] [PubMed] [Google Scholar]
- 69. Ravasz E, Somera AL, Mongru DA, Oltvai ZN, Barabasi AL. Hierarchical organization of modularity in metabolic networks. Science. 2002;297(5586):1551–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLS)
(XLS)
(XLS)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.