Abstract
Background
Galectin-3, a β-galactoside binding lectin, has been described as a mediator of cardiac fibrosis in experimental studies and as a risk factor associated with cardiovascular events in subjects with heart failure. Previous studies have evaluated the genetic susceptibility to Chagas disease in humans, including the polymorphisms of cytokine genes, demonstrating correlations between the genetic polymorphism and cardiomyopathy development in the chronic phase. However, the relationship between the galectin-3 single nucleotide polymorphism (SNP) and phenotypic variations in Chagas disease has not been evaluated.
Objective
The present study aimed to determine whether genetic polymorphisms of galectin-3 may predispose to the development of cardiac forms of Chagas disease.
Methods
Fifty-five subjects with Chagas disease were enrolled in this observational study. Real-time polymerase chain reaction (PCR) was used for genotyping the variants rs4644 and rs4652 of the galectin-3 gene.
Results
For the SNP rs4644, the relative risk for the cardiac form was not associated with the genotypes AA (OR = 0.79, p = 0.759), AC (OR = 4.38, p = 0.058), or CC (OR = 0.39, p = 0.127). Similarly, for the SNP rs4652, no association was found between the genotypes AA (OR = 0.64, p = 0.571), AC (OR = 2.85, p = 0.105), or CC (OR = 0.49, p = 0.227) and the cardiac form of the disease.
Conclusion
Our results showed no association between the different genotypes for both SNPs of the galectin-3 gene and the cardiac form of Chagas disease.
Keywords: Chagas Disease; Galectin 3; Heart Failure; Polymorphism, Genetic; Chagas Cardiomyopathy
Introduction
Chagas disease (CD), caused by the intracellular parasite Trypanosoma cruzi, is a major public health problem in Latin America and affects millions of people worldwide1. About 30% of the patients develop chronic cardiomyopathy, which is the most severe form of CD and associated with worse prognosis in heart failure2. Since there is no effective treatment in this stage of the disease, it is crucial to identify biomarkers that might be used in the early detection of cardiac disease and prognostic stratification.
Previous studies have evaluated the genetic susceptibility to CD in humans. Polymorphisms of molecules involved in host damage, induced by the parasite, such as IL-4, IL-6, IL-10, IL-12, TNF-α, and IFN-γ have been determined in subjects with the disease, demonstrating the association between genetic polymorphism and the development of cardiomyopathy in the chronic phase of CD3-8.
Recently, our group has demonstrated that the galectin-3 gene is overexpressed in the heart of animals chronically infected with Trypanosoma cruzi9,10. Galectin-3 is a member of the galectin family that binds β-galactosides and is produced by activated macrophages and fibroblasts11. It has been shown to be a mediator of cardiac fibrosis in experimental studies11,12 and to be related to cardiovascular events and heart failure severity13,14.
Previous studies have shown that genetic variants at two galectin-3 gene single nucleotide polymorphism (SNP) sites (rs4644 and rs4652, corresponding to LGALS3 +191 A>C and LGALS3 +292 A>C, respectively) are able to change the protein levels15. Galectin-3 plays an important role in inflammation and fibrosis process, and the relationship between the SNPs in the galectin-3 gene and the phenotypic variations in CD have not been evaluated. We hypothesized that galectin-3 SNPs can influence the development of cardiac forms of Chagas disease. Therefore, the present study aimed to evaluate the correlation of genetic polymorphisms of galectin-3 with severity of the cardiac form of Chagas disease.
Methods
Study population
A retrospective observational study was performed between January 2011 and December 2013. A convenience sample of 55 subjects attending the CD outpatient clinic at Hospital São Rafael was studied.
Inclusion criteria were: CD diagnosis confirmed microbiologically by two serological tests (indirect hemagglutination and indirect immunofluorescence), and age from 18 to 70 years. Exclusion criteria were: previous myocardial infarction or history of coronary artery disease, primary valve disease, dialysis treatment for end-stage renal disease, active liver disease, hematologic, neoplastic or bone diseases, and contraindications to magnetic resonance imaging (MRI).
The study complied with the Declaration of Helsinki and was approved by the Ethics Committee. All subjects signed a written informed consent before their inclusion in the study. All subjects underwent a structured medical history and physical examination, blood analysis, 12-lead Electrocardiography (ECG), chest X-Ray, 24-h Holter monitoring, conventional Doppler echocardiogram, and cardiac magnetic resonance.
Genotyping of galectin-3 gene SNPs
Samples of genomic DNA were obtained from peripheral blood mononuclear cells. DNA extraction was performed using the commercial kit QIAamp DNA Blood Mini Kit (Qiagen GmbH, Hilden, Germany), according to the manufacturer’s recommendations. The samples were stored in a freezer at -20°C. Genotyping of the SNPs rs4644 and rs4652 of the galectin-3 gene was performed by real-time polymerase chain reaction (PCR) using TaqMan® (Life Technologies, Carlsbad, USA), and the SNPs were amplified using the TaqMan® Universal PCR protocol (Life Technologies). Genotypes of each SNP were determined using the Sequence Detection System software version 1.3.1 (Life Technologies). The probes and primers for the selected SNPs were supplied by the same company.
Doppler echocardiogram
Standard transthoracic echocardiographic examination was performed using the Vivid 7 digital ultrasound system (GE Vingmed Ultrasound AS, Horten, Norway). Three cardiac cycles were stored in cineloop format for offline analysis. Left ventricle (LV) and left atrial dimensions were measured according to the American Society of Echocardiography’s recommendations16. The LV ejection fraction (LVEF) was measured using the biplane Simpson’s method.
Cardiac magnetic resonance imaging
Cardiac magnetic resonance imaging was performed using a Sigma HDx 1.5-T system (General Electric; Fairfield, CT, USA). For assessment of the LV function, electrocardiography-gated, breath-hold long-axis, short-axis, and four-chamber views were acquired in the same location in different sequences. Acquisition parameters used for the dynamic sequence included a repetition time (RT) of 3.5 ms, an echo time (ET) of 1.5 ms, flip angle of 60°, a receiver bandwidth of 125 kHz, a 35 x 35 cm field of view (FOV), a matrix of 256x148, a temporal resolution (TR) of 35 ms, and a slice thickness of 8.0 mm without gap. Delayed enhancement images were acquired every heartbeat, 10 to 20 min after the administration of a gadolinium-based contrast (0.1 mmol/kg) using RT of 7.1 ms, ET of 3.1 ms, flip angle of 20°, first cardiac phase, views per segment 16/32, matrix size of 256 x 192, slice thickness of 8.0 mm, gap between slices of 2 mm, field of view 32 to 38 cm, inversion time 150 to 300 ms, receiver bandwidth of 31.25 kHz, number of excitations of 2. The myocardial delayed enhancement (MDE) technique was used to investigate myocardial fibrosis, which was estimated by a quantitative visual method.
Statistical analysis
Categorical data were expressed as numbers (percentages, 95% confidence interval), and continuous data were expressed as mean ± SD or median (interquartile range). The genotype distribution of the SNPs rs4644 and rs4652 (AA, AC and CC) in subjects with indeterminate form of CD was compared with that in subjects with the cardiac form of CD using the Chi-square test or the Fisher’s exact test. The percentage of myocardial fibrosis was compared between the groups with the 3 different genotypes Kruskal-Wallis test. Mann-Whitney test was used to assess differences in the median of myocardial fibrosis between subjects with and without the SNPs. The association between the cardiac form of CD and each of the genotypes (AA, AC and CC) was estimated by Odds Ratio (OR) and 95% confidence interval (CI). Cases with missing data were dropped from the analysis. Analyses were performed using SPSS version 20.0 (IBM), and p < 0.05 (two-tailed) was considered statistically significant.
Results
Clinical and imaging characteristics
The study comprised 55 subjects, 42% were men, with mean age of 58 ± 9 years. The prevalence of hypertension, diabetes, hypercholesterolemia and smoking were 71, 14, 42 and 27% respectively. Regarding the clinical forms, the subjects were distributed as follows: 16 (29%) with the indeterminate form (subjects with no evidence of cardiac involvement or heart failure), 16 (29%) with the cardiac form without ventricular dysfunction and 23 (42%) with the cardiac form with ventricular dysfunction. Seventeen subjects were in NYHA (New York Heart Association) functional class III-IV (31%). The mean LVEF, measured using the biplane Simpson’s method, was 54 ± 15%, and the median percentage of myocardial fibrosis was 9.4% (2.2-17.3). Clinical and demographic characteristics of the subjects are described in Table 1.
Table 1.
Subjects' clinical and demographic characteristics
| Subjects (n = 55) | |
|---|---|
| Male gender | 23 (41.8) |
| Age (years) | 58 ± 9 |
| Indeterminate form | 16 (29) |
| Cardiac form without ventricular dysfunction | 16 (29) |
| Cardiac form with ventricular dysfunction | 23 (42) |
| NYHA III ou IV | 17 (30.9) |
| Hypertension | 39 (70.9) |
| Diabetes | 8 (14.5) |
| Current smoking | 15 (27.3) |
| Hypercholesterolemia | 23 (41.8) |
| RBBB | 27 (49.1) |
| Hemoglobin (g/dl) | 14.0 ± 0.97 |
| Creatinin (mg/dl) | 0.87 ± 0.16 |
| LVEF (%) | 53.7 ± 15.4 |
| LVESV (ml) | 79.1 ± 65.4 |
| LVEDV (ml) | 177.3 ± 80.6 |
| LVmass (g) | 172.8 ± 57.3 |
| Myocardial fibrosis (%) | 9.4 (2.2-17.3)* |
Data are expressed as number (percentage) for categorical variables, and as mean ± SD or median (interquartile interval) for continuous variables; NYHA: New York Heart Association CMR: RBBB: Right bundle-branch block; LVEF: Left ventricular ejection fraction; LVESV: Left ventricular end-systolic volume; LVEDV: Left ventricular end-diastolic volume; LV: Left ventricular mass.
n = 40 (delayed enhancement cardiac magnetic resonance).
Genotyping of LGALS3 SNPs
The SNPs genotype distribution is described in Table 2. There was no significant association between LGALS3 genotypes (AA, AC, CC) of the SNPs rs4644 and rs4652 and the presence of the cardiac form of CD. For the SNP rs4644, the relative risk of the cardiac form was not associated with the genotype AA (OR = 0.79, 95%CI = 0.17 to 3.63, p = 0.759), AC (OR = 4.38, 95%CI = 0.87 to 22.02, p = 0.058), or CC (OR = 0.39, 95%CI = 0.11 to 1.33, p = 0.127). Similarly, for the SNP rs4652, the genotypes AA (OR = 0.64, 95%CI = 0.13 to 3.06, p = 0.571), AC (OR = 2.85, 95%CI = 0.78 to 10.40, p = 0.105), and CC (OR = 0.49, 95%CI = 0.15 to 1.58, p = 0.227) were not associated with the cardiac form of CD.
Table 2.
Genotype and allele frequency of LGALS3 + 191 and LGALS3+292 in 16 subjects with the indeterminate form and 39 subjects with the cardiac form of Chagas disease
| LGALS3 polymorphism | Indeterminate form (n=16) | Cardiac form (n = 39) | OR (95% CI) | p value |
|---|---|---|---|---|
| LGALS3 +191 | ||||
| AA | 3 (19, 7-43) | 6 (15, 7-30) | 0.79 (0.17-3.63) | 0.759 |
| AC | 2 (12, 4-36) | 15 (38, 25-54) | 4.38 (0.87-22.02) | 0.058 |
| CC | 11 (69, 44-86) | 18 (46, 32-61) | 0.39 (0.11-1.33) | 0.127 |
| LGALS3 +292 | ||||
| AA | 3 (19, 7-43) | 5 (13, 6-27) | 0.64 (0.13-3.06) | 0.571 |
| AC | 4 (25, 10-50) | 19 (49, 34-64) | 2.85 (0.78-10.40) | 0.105 |
| CC | 9 (56, 33-77) | 15 (38, 25-54) | 0.49 (0.15-1.58) | 0.227 |
Data are expressed as number (percentage, 95% confidence interval).
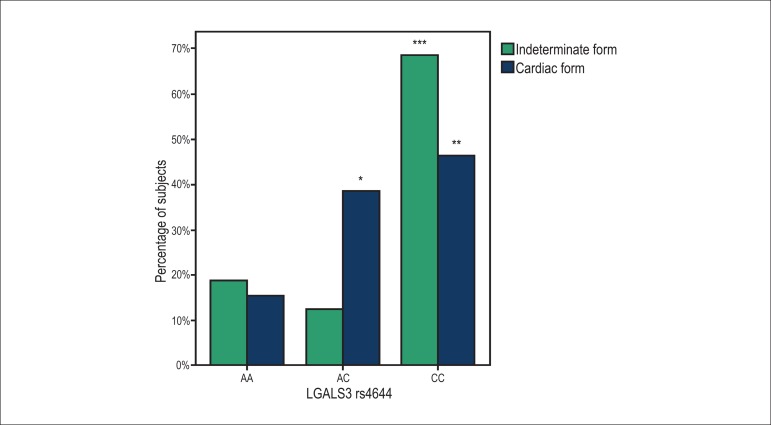
The SNP rs4644 genotype frequencies were not statistically different between the subjects with the indeterminate form and with the cardiac form of CD. (AA [19% vs 15%, p = 0.710], AC [12% vs 38%, p = 0.106] and CC [69% vs 46%, p = 0.149]) (Figure 1). In subjects with the indeterminate form, the most frequent genotype was the CC genotype, with a significantly higher prevalence than the genotypes AA and AC (p = 0.011 and p = 0.003, respectively). In subjects with the cardiac form, two genotypes were found to be more prevalent, AC and CC, as compared with the AA genotype (p = 0.04 and p = 0.006, respectively), with no difference between the prevalence of AC and CC (p = 0.647).
Figure 1.
Prevalence of the genotypes AA, AC and CC of the LGALS3 rs4644 in the indeterminate form and cardiac form of Chagas disease. The p-values using two-sided Fisher’s exact test were p = 0.710, p = 0.106 and p = 0.149, respectively. * p = 0.04, compared to AA genotype; ** p = 0.006, compared to AA genotype AA; *** p = 0.011, compared to AA genotype, and p = 0.003, compared to AC genotype
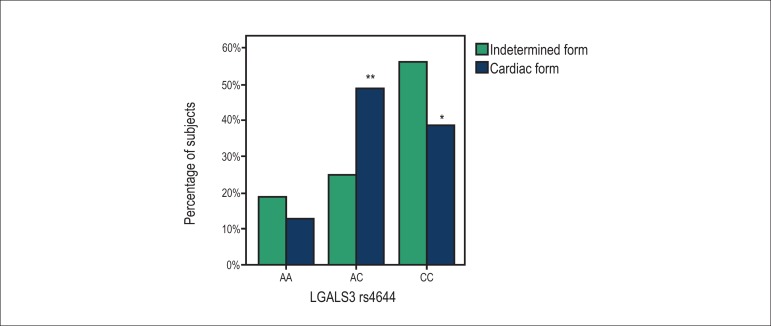
Similar results were observed for the SNP rs4652 genotype distribution between the groups (Figure 2). In subjects with the indeterminate form and subjects with the cardiac form of CD, respectively, the prevalence of the AA genotype was 19% vs 13% (p = 0.678), the prevalence of AC was 25% vs 49% (p = 0.138), and the prevalence of CC was 56% vs 38% (p = 0.249). In subjects with the indeterminate form, there was no significant difference between the three genotypes (AA vs CC, p = 0.066; AA vs AC, p = 1.00; and AC vs AA, p = 0.149). Similarly to the SNP rs4644, in subjects with the cardiac form, genotypes AC and CC were more prevalent, as compared with genotype AA (p = 0.001 and p = 0.018, respectively), with no difference between the prevalence of AC and CC (p = 0.494).
Figure 2.
Prevalence of the genotypes AA, AC and CC of the LGALS3 rs4652 in the indeterminate form and cardiac form of Chagas disease. The p-values using twosided Fisher’s exact test were p = 0. 678, p = 0.138 and p = 0.249, respectively; * p = 0.018, compared to the AA genotype; ** p = 0.001, compared to the AA genotype
Myocardial fibrosis
There was no statistically significant difference in the median percentage of myocardial fibrosis between subjects with and without the presence of any of the SNPs rs4644 and rs4652 genotypes (Table 3).
Table 3.
Median percentage of myocardial fibrosis in subjects with and without galectin-3 SNPs by genotype
| Without the SNP | With the SNP | p value | |
|---|---|---|---|
| rs4644 AA | 4.3 (0.0-14.4) | 1.6 (0.0-4.7) | 0.261 |
| rs4644 AC | 4.4 (0.0-9.8) | 1.6 (0.0-19.2) | 0.969 |
| rs4644 CC | 1.6 (0.0-15.0) | 5.2 (0.2-18.5) | 0.441 |
| rs4652 AA | 4.3 (0.0-14.0) | 2.0 (0.3-10.6) | 0.743 |
| rs4652 AC | 4.4 (0.0-10.0) | 1.7 (0.0-15.1) | 0.862 |
| rs4652 CC | 1.9 (0.0-14.1) | 4.8 (0.0-10.0) | 0.681 |
SNP: Single nucleotide polymorphism; Data are expressed as median (interquartile interval); p-values of Mann-Whitney U test.
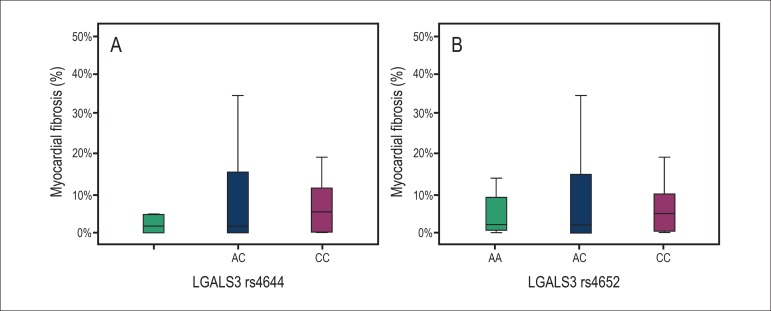
The median percentage of myocardial fibrosis was not statistically different between the AA, AC and CC genotypes (p = 0.508), or between SNP rs4644 and SNP rs4652 (p = 0.903 (Figure 3).
Figure 3.
Percentage of myocardial fibrosis in subjects with the AA, AC and CC genotypes of the LGALS3 rs4644 (A) and rs4652 (B), p = 0.508 and p = 0.903, respectively.
Discussion
This study evaluated the association between genetic variants of the LGALS3 gene, encoding for galectin-3, and disease severity in subjects with CD. Since galectin-3 has been previously found to be highly correlated with fibrosis, we also evaluated the association of the three genotypes of the SNPs rs4644 and rs4652 with myocardial fibrosis. We found no significant difference in the prevalence of any of the genotypes between the subjects with the indeterminate form and the cardiac form of the disease. Also, there was no difference in the median of myocardial fibrosis between subjects with the AA, AC and CC genotypes of the SNPs in question.
The genetic predisposition to galectin-3 polymorphism has been previously associated with rheumatoid arthritis, another inflammatory disease. Hu et al15 described an association between this rheumatic disease and the LGALS3 +292C allele. Although we have shown that the LGALS3 +191C and LGALS3 + 292C allele carriage was relatively high in subjects with the cardiac form of CD, the CC genotype, at least for the SNP rs4644, was relatively more frequent among subjects with the indeterminate form. Thus, we could not confirm the possible role of the presence of CC genotype or AA genotype for both SNPs as a protective or a risk factor for developing the cardiac form of CD. Similarly, no relationship between the genetic polymorphisms in the gene encoding galectin-3 and the median of myocardial fibrosis was observed.
Conclusion
Studies at the levels of genes and proteins contribute to the understanding of the role of genetic variants in the etiology of diseases, as well as to elucidate their potential as therapeutic targets, risk predictors and prognostic markers. Our findings did not lead to a significant conclusion on the role of galectin-3 polymorphism in the development of the cardiac form of CD. In addition, further studies with long-term follow up are necessary to evaluate whether other genetic variants play a role in inflammation and fibrosis in the indeterminate form of CD, to identify possible variants associated with disease progression in CD.
Acknowledgments
The authors thank Dr. Kyan James Allahdadi, PhD, for carefully reviewing the manuscript.
Footnotes
Author contributions
Conception and design of the research: Cruz GS, Angelo ALD, Souza BSF, Santos RR, Soares MBP; Acquisition of data: Cruz GS, Angelo ALD, Larocca TF, Macedo CT, Noya- Rabelo M, Torreão JÁ; Analysis and interpretation of the data: Cruz GS, Angelo ALD, Larocca TF, Macedo CT, Noya- Rabelo M, Correia LCL, Torreão JA, Souza BSF, Soares MBP; Statistical analysis: Cruz GS, Angelo ALD, Larocca TF, Noya- Rabelo M, Correia LCL; Obtaining financing: Santos RR, Soares MBP; Writing of the manuscript: Cruz GS, Larocca TF, Souza BSF; Critical revision of the manuscript for intellectual content: Cruz GS, Angelo ALD, Larocca TF, Macedo CT, Noya- Rabelo M, Correia LCL, Souza BSF, Santos RR, Soares MBP.
Potential Conflict of Interest
No potential conflict of interest relevant to this article was reported.
Sources of Funding
This study was funded by FAPESB.
Study Association
This study is not associated with any thesis or dissertation work.
References
- 1.World Health Organization . Chagas disease: control and elimination. Geneva: 2010. [Google Scholar]
- 2.Silva CP, Del Carlo CH, Oliveira MT, Junior, Scipioni A, Strunz-Cassaro C, Ramirez JA, et al. Why do patients with chagasic cardiomyopathy have worse outcomes than those with non-chagasic cardiomyopathy? Arq Bras Cardiol. 2008;91(6):358–362. doi: 10.1590/s0066-782x2008001800006. [DOI] [PubMed] [Google Scholar]
- 3.Beraún Y, Nieto A, Collado MD, González A, Martín J. Polymorphisms at tumor necrosis factor (TNF) loci are not associated with Chagas' disease. Tissue Antigens. 1998;52(1):81–83. doi: 10.1111/j.1399-0039.1998.tb03028.x. [DOI] [PubMed] [Google Scholar]
- 4.Costa GC, da Costa Rocha MO, Moreira PR, Menezes CA, Silva MR, Gollob KJ, et al. Functional IL-10 gene polymorphism is associated with Chagas disease cardiomyopathy. J Infect Dis. 2009;199(3):451–454. doi: 10.1086/596061. [DOI] [PubMed] [Google Scholar]
- 5.Drigo SA, Cunha-Neto E, Ianni B, Mady C, Faé KC, Buck P, et al. Lack of association of tumor necrosis factor-alpha polymorphisms with Chagas disease in Brazilian patients. Immunol Lett. 2007;108(1):109–111. doi: 10.1016/j.imlet.2006.10.008. [DOI] [PubMed] [Google Scholar]
- 6.Flórez O, Martín J, González CI. Interleukin 4, interleukin 4 receptor-a and interleukin 10 gene polymorphisms in Chagas disease. Parasite Immunol. 2011;33(9):506–511. doi: 10.1111/j.1365-3024.2011.01314.x. [DOI] [PubMed] [Google Scholar]
- 7.Torres OA, Calzada JE, Beraún Y, Morillo CA, González A, González CI, et al. Lack of association between IL-6-174G/C gene polymorphism and Chagas disease. Tissue Antigens. 2010;76(2):131–134. doi: 10.1111/j.1399-0039.2010.01478.x. [DOI] [PubMed] [Google Scholar]
- 8.Zafra G, Morillo C, Martín J, González A, González CI. Polymorphism in the 3' UTR of the IL12B gene is associated with Chagas' disease cardiomyopathy. Microbes Infect. 2007;9(9):1049–1052. doi: 10.1016/j.micinf.2007.04.010. [DOI] [PubMed] [Google Scholar]
- 9.Soares MB, Lima RS, Souza BS, Vasconcelos JF, Rocha LL, Dos Santos RR, et al. Reversion of gene expression alterations in hearts of mice with chronic chagasic cardiomyopathy after transplantation of bone marrow cells. Cell Cycle. 2011;10(9):1448–1455. doi: 10.4161/cc.10.9.15487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vasconcelos JF, Souza BS, Lins TF, Garcia LM, Kaneto CM, Sampaio GP, et al. Administration of granulocyte colony-stimulating factor induces immunomodulation, recruitment of T regulatory cells, reduction of myocarditis and decrease of parasite load in a mouse model of chronic Chagas disease cardiomyopathy. FASEB J. 2013;27(12):4691–4702. doi: 10.1096/fj.13-229351. [DOI] [PubMed] [Google Scholar]
- 11.Sharma UC, Pokharel S, van Brakel TJ, van Berlo JH, Cleutjens JP, Schroen B, et al. Galectin-3 marks activated macrophages in failure-prone hypertrophied hearts and contributes to cardiac dysfunction. Circulation. 2004;110(19):3121–3128. doi: 10.1161/01.CIR.0000147181.65298.4D. [DOI] [PubMed] [Google Scholar]
- 12.Liu YH, D'Ambrosio M, Liao TD, Peng H, Rhaleb NE, Sharma U, et al. N-acetyl-seryl-aspartyl-lysyl-proline prevents cardiac remodeling and dysfunction induced by galectin-3, a mammalian adhesion/growth-regulatory lectin. Am J Physiol Heart Circ Physiol. 2009;296(2):H404–H412. doi: 10.1152/ajpheart.00747.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Felker GM, Fiuzat M, Shaw LK, Clare R, Whellan DJ, Bettari L, et al. Galectin-3 in ambulatory patients with heart failure: results from the HF-ACTION study. Circ Heart Fail. 2012;5(1):72–78. doi: 10.1161/CIRCHEARTFAILURE.111.963637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fermann GJ, Lindsell CJ, Storrow AB, Hart K, Sperling M, Roll S, et al. Galectin 3 complements BNP in risk stratification in acute heart failure. Biomarkers. 2012;17(8):706–713. doi: 10.3109/1354750X.2012.719037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hu CY, Chang SK, Wu CS, Tsai WI, Hsu PN. Galectin-3 gene (LGALS3) +292C allele is a genetic predisposition factor for rheumatoid arthritis in Taiwan. Clin Rheumatol. 2011;30(9):1227–1233. doi: 10.1007/s10067-011-1741-2. [DOI] [PubMed] [Google Scholar]
- 16.Lang RM, Bierig M, Devereux RB, Flachskampf FA, Foster E, Pellikka PA, et al. Chamber Quantification Writing Group. American Society of Echocardiography's Guidelines and Standards Committee. European Association of Echocardiography Recommendations for chamber quantification: a report from the American Society of Echocardiography's Guidelines and Standards Committee and the Chamber Quantification Writing Group, developed in conjunction with the European Association of Echocardiography, a branch of the European Society of Cardiology. J Am Soc Echocardiogr. 2005;18(12):1440–1463. doi: 10.1016/j.echo.2005.10.005. [DOI] [PubMed] [Google Scholar]