Abstract
Nuclear hormone receptors (NHRs) are transcription factors regulated by small molecules. The functions of NHRs range from development of primary and secondary lymphoid organs, to regulation of differentiation and function of DCs, macrophages and T cells. The human genome has 48 classic (hormone and vitamin receptors) and non-classic (all others) NHRs; 17 non-classic receptors are orphans, meaning that the endogenous ligand is unknown. Understanding the function of orphan NHRs requires the identification of their natural ligands. The mevalonate pathway, including its sterol and non-sterol intermediates and derivatives, is a source of ligands for many classic and non-classic NHRs. For example, cholesterol biosynthetic intermediates (CBIs) are natural ligands for RORγ/γt. CBIs are universal endogenous metabolites in mammalian cells, and to study NHRs that bind CBIs requires ligand-free reporters system in sterol auxotroph cells. Furthermore, RORγ/γt shows broad specificity to sterol lipids, suggesting that RORγ/γt is either a general sterol sensor or specificity is defined by an abundant endogenous ligand. Unlike other NHRs, which regulate specific metabolic pathways, there is no connection between the genetic programs induced by RORγ/γt and ligand biosynthesis. In this review we summarize the roles of non-classic NHRs and their potential ligands in the immune system.
Keywords: immunology, nuclear hormone receptors, cholesterol biosynthesis, lymphoid tissues, RORγt, sterols
Introduction
Nuclear hormone receptors (NHRs) are a superfamily of transcription factors that contain a DNA binding domain and a ligand binding domain (LBD) that binds small molecules, such as lipids, hormones, vitamins and metabolites of amino acids (reviewed in [1, 2]). NHRs can be grouped into 6 groups based on sequence homology [1] (summarized in Table 1). The presence or absence of the ligand in the LBD modulates the structural conformation of NHRs. The conformational change in the NHR induced by binding of an agonist ligand promotes the recruitment of nuclear receptor coactivators (Ncoas) and subsequently drives the transcription of NHR target genes [2]. In contrast, the conformation of ligand-free NHRs promotes the recruitment of nuclear receptor corepressors (Ncor) that inhibit the transcription of NHR target genes [2].
Table 1.
NHR Classification, distribution and function in immune cells.a)
| Official Name | Gene Symbolb | Common Name | Ligands | Expressionc | Function |
|---|---|---|---|---|---|
| NR1A1 | THRA | Thyroid hormone receptor α | Thyroid hormones | preT, ILC3, S | T/B dev [97] |
| NR1A2 | THRB | Thyroid hormone receptor β | Thyroid hormones | DC,M,S | ND |
| NR1B1 | RARA | Retinoic acid receptor α | Retinoic acid (RA) | M, Ne | Reg B, DC, T, Lti [48, 98] |
| NR1B2 | RARB | Retinoic acid receptor β | RA | DC, M | Reg B, DC, T, Lti [48, 98] |
| NR1B3 | RARG | Retinoic acid receptor γ | RA | γδT cells | Reg B, DC, T, Lti [48, 98] |
| NR1C1 | PPARA | Pparα | Fatty acids, phospholipid [99] | no signal | DC, M diff and function [100] |
| NR1C2 | PPARD | Pparδ | Fatty acids | ILCs, Ne, S | ND |
| NR1C3 | PPARG | Pparγ | Fatty acids, PGJ2 | αβT, ILC2, M, S | ND |
| NR1D1 | NR1D1 | Rev-erbα | Heme [101] | ILC2/3, γδT, S | TLR IECs [58] |
| NR1D2 | NR1D2 | Rev-erbβ | Heme [101] | uncertain | ND |
| NR1F1 | RORA | RAR-related orphan receptor α | CBIs [42], cholesterol, CS | αβT, ILCs | TLR IECs [58], T cells dev. |
| NR1F2 | RORB | RAR-related orphan receptor β | retinoic acid | no signal | ND |
| NR1F3 | RORC | RAR-related orphan receptor γ | CBIs, oxysterols [42, 67] | αβT, γδT, ILC3 | Lymphoid, Th17[55, 56, 93] |
| NR1H3 | LXRA | Liver X receptor α | CBIs [68, 70] oxysterols | M,,S | M [68], T,B, prolfieration [102] |
| NR1H2 | LXRB | Liver X receptor β | CBIs [68, 70] oxysterols | no signal | M [68], T,B, prolfieration [102] |
| NR1H4 | FXR | Farnesoid x receptor | Bile acids | no signal | ND |
| NR1H5 | FXRB | Farnesoid x receptor β | Lanosterol | no signal | ND |
| NR1I1 | VDR | Vitamin D receptor | Vitamin D | Aβ/γδT, DC,M,S | Suppress B,DC,T cells [48] |
| NR1I2 | PXR | Pregnane X receptor | xenobiotics | no signal | IEC TLR expression [59] |
| NR1I3 | CAR | Constitutive androstane receptor | xenobiotics | no signal | ND |
| NR2A1 | HNF4A | Hepatocyte nuclear factor 4α | fatty acid [103] | SC, preT | ND |
| NR2A2 | HNF4G | Hepatocyte nuclear factor 4γ | orphan | S | ND |
| NR2B1 | RXRA | Retinoic X receptor α | 9-cis RA [104, 105] | γδT, M, | Pleiotropic |
| NR2B2 | RXRB | Retinoic X receptor β | 9-cis RA [104, 105] | no signal | Pleiotropic |
| NR2B3 | RXRG | Retinoic X receptor γ | 9-cis RA [104, 105] | ILC2 | Pleiotropic |
| NR2C1 | TR2 | Testicular receptor 2 | orphan | no signal | ND |
| NR2C2 | TR4 | Testicular receptor 4 | orphan | no signal | granulomas [66] |
| NR2E1 | TLX | Tailless | orphan | no signal | ND |
| NR2E3 | PNR | Photoreceptor nuclear receptor | orphan | ILCs? | ND |
| NR2F1 | COUP-TFI | COUP transcription factor 1 | orphan | S | ND |
| NR2F2 | COUP-TFII | COUP transcription factor 2 | orphan | S | Lymphatic vessel dev [62] |
| NR2F6 | EAR2 | ErbA-related protein 2 | orphan | weak signal | Th17 cells [49] |
| NR3A1 | ESR1 | Estrogen receptor 1 | estrogens | proB, preT, SC, S | Pleiotropic |
| NR3A2 | ESR2 | Estrogen receptor 2 | estrogens | weak signal | Pleiotropic |
| NR3B1 | ESRRA | Estrogen related receptor α | orphan | weak signal | Teff and Tmem [50] |
| NR3B2 | ESRRB | Estrogen related receptor β | orphan | weak signal | ND |
| NR3B3 | ESRRG | Estrogen related receptor γ | orphan | S | Fe load mac [57] |
| NR3C1 | GR | Glucocoticoid receptor | Glucocorticoid | broad | T cell dev [47] |
| NR3C2 | MLR | Mineralocorticoid receptor | Mineralocorticoid | B, DC, S | ND |
| NR3C3 | PGR | Progesterone receptor | Progesterone | B cells? | ND |
| NR3C4 | AR | Androgen receptor | androgens | γδT, ILC2, S | ND |
| NR4A1 | NGFIB | Nerve growth factor IB | orphan | DC,ILCs,M, S, T | T cells, M dev [51] [52, 53] |
| NR4A2 | NURR1 | Nuclear receptor related 1 | orphan | DC, ILCs, M, T | T cells dev [51] |
| NR4A3 | NOR1 | Neuron-derived orphan receptor 1 | orphan | DC, ILCs, M, T | T cells dev [51] |
| NR5A1 | SF1 | Steroidogenic factor 1 | Phosphatidyl inositols [63] | weak signal | Spleen dev [64, 65] |
| NR5A2 | LRH-1 | Liver receptor homolog-1 | Phosphatidyl inositols [63] | S | ND |
| NR6A1 | GCNF | Germ cell nuclear factor | orphan | weak signal | ND |
| NR0B1 | DAX-1 | DAX-1** | orphan | weak signal | ND |
| NR0B2 | SHP | small Heterodimer partner | orphan | SC, M | TLR signaling mac [54] |
Human NHR classification from [1]. References added for natural ligands unknown in 2006. Classic NHRs (yellow boxes). Receptors with known accessory iLBPs (grey boxes). Receptors with broad specificity for ligands (brown boxes). Biological function of ligand unknown (red text). Examples of functions discussed in this review (blue boxes). Abreviations: M=myeloid cells, Ne=neutrophil, S=stroma, SC=stem cells, ND=no data, Reg=positive and negative modulation of function, dev=development, Teff=T effector cells, Tmem=T memory cells, mac=macrophage. FXRB is a pseudogene in the human lineage and a lanosterol receptor in mice [69].
Gene symbol shared by human and mouse NHR.
Expression of NHRs in immune cells are from (http://www.immgen.org/index_content.html).
The history of NHRs is fascinating and could be divided into 4 eras. The first one, from 1840–1926 physiologists postulated the existence of hormones [3] and vitamins [4] – special classes of signaling molecules produced by glands (hormones) or obtained from the diet (vitamins) and transported by the circulatory system to target distant organs to regulate physiology and behavior. The search for these substances led to the isolation of the thyroid hormone [5] and the discoveries of vitamins A and D [6, 7], all compounds with unknown structures or receptors at that time. The determination of the chemical structure and synthesis of the thyroid hormone [8] gave way to the “orphan ligand” or second era (1927–1984), in which most known hormones and vitamins were identified, structurally characterized and synthesized, but still with no receptors identified. The cloning of the receptors for glucocorticoids [9] and vitamin A [10, 11] ended the era of “orphan ligands” as these orphan ligands were “adopted” by their respective, discovered receptors (1985–1997 – third era). For sake of simplicity we will call these receptors, which recognize the hormones and vitamins identified during the first era: Classic NHRs. The sequence homologies of classic NHRs suggested that hormone and vitamin receptors are members of a large superfamily of nuclear receptors [11–13]. Furthermore, the cloning and sequencing of classic receptors revealed the existence of related proteins for which there were no known ligands [12]. However, the extend of the diversity of non-classic NHRs could not be accessed. The sequencing of the C. elegans genome in 1998 revealed the full diversity of the NHR family and marks the beginning of the fourth era of “non-classic NHRs”: in the C. elegans genome 1.5% of all predicted protein sequences are NHRs and the majority of those are non-classic and orphans for their ligands [14]. The human genome project identified 48 NHRs, of which only 12 belong to the classic receptor group, while 17 of the non-classic receptors are still orphans for their ligands [1].
Non-classic NHRs are broadly expressed in tissues and have been shown to be important in the regulation of metabolism and the development in many organ systems, including the immune system. This review focuses on the roles of non-classic NHRs in immunity with particular attention paid to those receptors that are orphans (see Table 1). We will not discuss in detail the peroxisome proliferator activated receptors (PPARs) and the retinoid X receptors, as there is extensive literature about them and they would deserve each a separate review.
The mevalonate pathway: Source of CBIs and NHR ligands
The mevalonate pathway is divided into a pre-sterol and sterol phase [15] (Fig. 1). Pre-sterol intermediates of the mevalonate pathway are cholesterol biosynthetic intermediates (CBIs) but they are not exclusive for the cholesterol pathway and can be used as intermediates for other biosynthetic pathways and cellular functions like the synthesis of Heme A and O, Isopentenyl-adenine (tRNA), ubiquinone, dolichol and protein prenylation [15] (Fig. 1). These intermediates have important functions in cell biology and immunity. Isopentenyl-PP is a pre-sterol metabolite that is recognized by human γδT cells in tumors and in cells infected “in vitro” with the bacteria Escherichia coli and Staphylococcus aureus [16, 17]. Another pre-sterol is Farnesyl-PP that is required for post-translational modification (farnesylation) of proteins, like Ras, which is essential for the differentiation of Th1 cells [18]. Furthermore, a mutation in the enzyme mevalonate kinase (MVK), occurring in the pre-sterol part of the mevalonate pathway, has been linked to hyper-IgD syndrome [19, 20]. The transition between the pre-sterol and sterol phases of the pathway is marked by the cyclization of squalene and related metabolites into sterols which are compounds containing 4 structural rings: A, B, C, D (Fig. 1). Cholesterol itself is processed further into oxysterols, bile acids, estrogens, androgens, corticoid and mineralocorticoid hormones and vitamin D in what we could call post-cholesterol products of the pathway (Fig.1).
Figure 1.
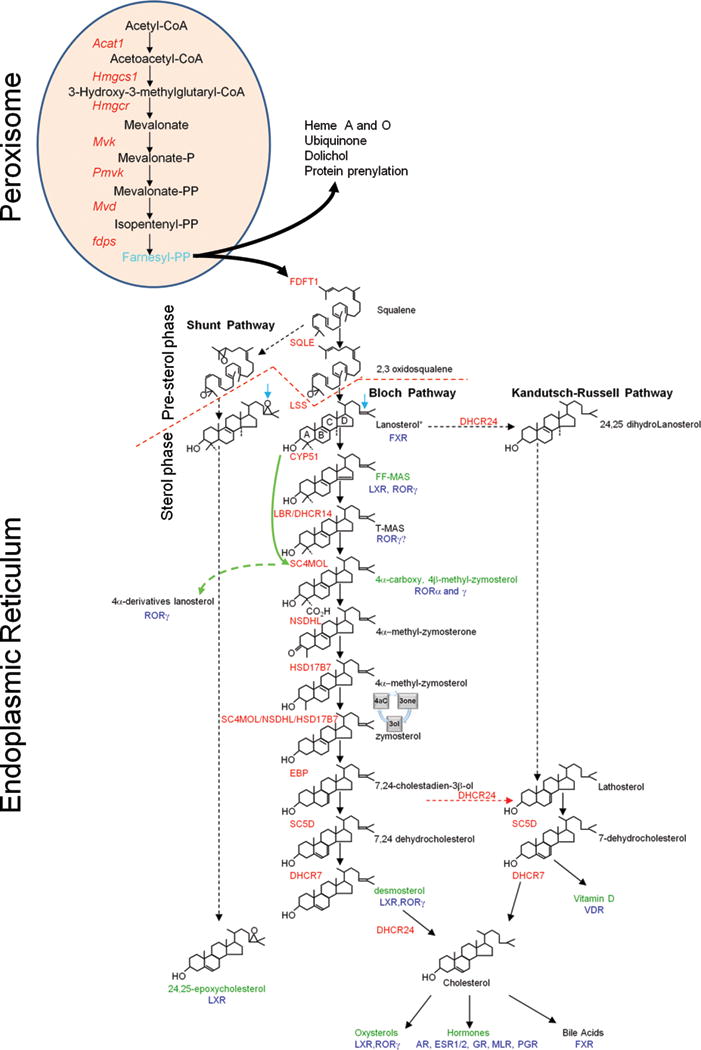
The mevalonate pathway, NHRs and immunity. The mevalonate pathway is divided into pre-sterol and sterol stages (red segmented line). Compound structures and names without immune functions are in black and enzymes of the cholesterol pathway are named in (red). The pre-sterol intermediates are mainly produced in peroxisomes. Peroxisomes transform Acetyl-CoA into farnesyl-diphosphate (light blue) which is also a precursor for Heme A and O, Ubiquinone, dolichol, protein prenylation and sterol biosynthesis. The sterol-stage occurs in the endoplasmic reticulum. It starts when squalene or its derivatives, which are linear precursors are cyclicized by the enzyme LSS giving origin to sterol lipids which are molecules with 4 structural rings (A,B,C,D) shown here in the molecule lanosterol. For sake of simplicity only the Bloch pathway is shown in its totality. Substrates have similar structures for each pathway but the shunt pathway has a epoxy group in the side-chain (blue arrow) while the enzyme DHCR24 removes the Δ24 double bond of any CBI (blue arrow in lanosterol) to produce the intermediates of the Kandutsch-Russell pathway. The preferred substrate of DHCR24 is 7,24-cholestadien-3β-ol (red segmented arrow).The removal of methyl groups at C4 from T-MAS requires two cycles of processing by the enzymes SC4MOL, NSDHL and HSD17B7. Shown is only the first cycle, the second is represented by a circle (4aC to 3one to 3ol). The CBIs from lanosterol to 4α-methyl-zymosterol are also called methylsterols or C4-methylated sterols. Non-canonical CBIs are produced when one intermediate in the pathway is processed “out of order” like lanosterol (green arrow) producing 4α-derivatives of Lanosterol. The CBIs that were identified as NHR ligands with immune functions are named in (olive green) and NHR name is in (blue); for NHR names see Table 1. Many CBIs bind RORγ/γt but the immune function of these compounds has not been demonstrated. For a complete list of sterols that bind RORγ/γt and promote reporter activity in ligand-free systems see [42].
Since the pre-sterol metabolites of the pathway can be used as intermediates in other metabolic pathways, the term CBIs is here limited to the sterol compounds produced by the cyclization of squalene and its derivatives. The sterol phase of the mevalonate pathway is branched into two main routes for the production of cholesterol, the Bloch pathway [21] and the Kandutsch-Russell pathway [22] (Fig. 1). Besides the production of cholesterol, a shunt in the mevalonate pathway leads to the production of 24,25 epoxycholesterol [23], a potent LXR ligand [24, 25]. Most studies have been done with CBIs derived from the Bloch pathway, because most of these are commercially available. Therefore, it is possible that metabolites from the Kandutsch-Russell and shunt pathways produce ligands for NHRs that are as potent as the products of the Bloch pathway tested so far. The Bloch and Kandutsch-Russell routes of cholesterol biosynthesis differ by the timing of removal of a double bond at Δ24 by the enzyme DHCR24 (Fig. 1). This can happen very early with the conversion of lanosterol into 24,25-dihydrolanosterol (Fig. 1). However, while any CBI can be processed by DHCR24, the preferential substrate for DHCR24 is 24-dihydrolathosterol (Fig.1) and that most likely in living cells, cholesterol biosynthesis branches preferentially from the Bloch into the Kandutsch-Russell pathway downstream of zymosterol [26] (Fig. 1). In the Kandutsch-Russell pathway the immediate precursor of cholesterol is 7-dehydrocholesterol (7-DHC) which is converted into cholesterol by the enzyme DHCR7 (Fig.1). In the Bloch pathway, the immediate precursor of cholesterol is desmosterol (24-dehydrocholesterol) whose double bound at Δ24 is removed by DHCR24 (Fig.1). The physiological significance of the branching in cholesterol biosynthesis is not fully understood. However, genetic polymorphisms of DHCR7 which is the last enzyme in the Kandutsch-Russell pathway are associated with susceptibility to autoimmune diseases like multiple sclerosis [27] and type 1 diabetes [28]. This could be related to the fact that 7-DHC, the last CBI in the Kandutsch-Russell pathway, is the main intermediate for vitamin D3 [29], a very potent mediator of immune function as we will discuss in detail below. It is also possible that the different CBIs produced by the two alternative pathways have different functions in cells and that the use of one branch or another will depend on cell type and expression of DHCR24. Nevertheless, the increased production of cholesterol by overexpression of Dhcr24 has a protective role against oxidative stress in mammalian cells [30]. Maybe, the presence of the Δ24 double bond protects desmosterol from side-chain oxidations that result in the production of oxysterols like 25-hydroxycholesterol [31]. Thus, it is tempting to speculate that by regulating the conversion of desmosterol into cholesterol, the Bloch pathway limits the concentration of precursor cholesterol available for the enzymatic and non-enzymatic production of oxysterols which act as signaling components on several aspects of the biology of dendritic cells, macrophages and T cells (reviewed in [32]).
The synthesis of the pre-sterol components is distributed over the mitochondria, cytosol, peroxisomes and endoplasmatic reticulum [33] (Fig.1). However, the production of the first linear committed precursor squalene, the sterol biosynthetic enzymes and their sterol products are all located in the endoplasmatic reticulum [33, 34] (Fig.1). One exception is the lamin B receptor (LBR), which is a nuclear protein with catalytic Δ14-reductase activity which removes the Δ14 double bond of FF-MAS, converting it into T-MAS [35] (Fig. 1). The function of the nuclear enzymatic activity is not well understood but both LBR and the ER resident enzyme DHCR14 can provide substantial redundancy in the Δ14-reductase activity required for cholesterol biosynthesis, and one single Lbr allele can rescue cholesterol biosynthesis in Dhcr14−/− animals [36]. However, in both humans [37] and mice [38] viable mutations of the LBR/lbr gene have resulted in abnormal development of the neutrophil nucleus, which instead of the normal multi-lobulated pattern, show a bilobed or ovoid shape. This defect in the nuclear shape of neutrophils is called the Pelger-Huet anomaly [39]. An in vitro study of granulocyte differentiation using cells derived from mice with the ichthyosis mutation (icj), a Lbr mutant that contains a CC insertion at the Δ14-reductase domain, suggests that the phagocytosis of (icj) neutrophils is normal while chemotaxis and oxidative burst are deficient [40]. The same group has shown that it is the Δ14-reductase activity of LBR and not of the redundant enzyme DHCR14 that is necessary to partially rescue the maturation defects of (icj) neutrophils in mouse cells [41]. It is tempting to speculate that the enzymatic activity of Lbr may convert nuclear CBIs into NHR ligands. In this context, lanosterol, which is a direct precursor of RORγ/γt ligands [42], is increased in the nucleus of Lipid A-stimulated mouse macrophages [43], suggesting that signaling pathways such as TLR4 can increase the permeability of the nucleus for some C4-methylated CBIs. It would be important to examine whether the concentration of lanosterol and other CBIs is increased in the nucleus of Th17 cells or RORγt+ ILCs. Furthermore, it would be essential to define whether CBIs present in the cell nucleus can be processed further into NHR ligands by nuclear enzymes like LBR.
A line that has been little explored is the possibility that the canonical CBIs of the Bloch and Kandutsch-Russell pathway can be metabolized into alternative non-canonical metabolites of CBIs. For example, the CBIs lanosterol can be prematurely processed by the enzyme SC4MOL, leading to the formation of 4α-carboxy lanosterol [44, 45] (Fig. 1). Lanosterol can also be metabolized by enzymes outside the cholesterol biosynthetic pathway giving rise to CBI based oxysterols like lanosterol-26-diol that promotes the negative regulation of cholesterol biosynthesis in mammalian cells[46]. Since these compounds are oxysterols they could play a role in the function of oxysterol receptors LXR and RORγt in the immune system. Sterols with exact mass corresponding to non-canonical intermediates of lanosterol have been found in the thymus and are suspected to be RORγ/γt ligands [42]. The function of non-canonical CBIs and metabolites of CBIs is in its infancy and much research is still needed to determine whether they are present at physiological levels in normal tissues and whether they show specific agonist activity to NHRs and other immune receptors.
NHRs in the development and function of the immune system
Classic NHRs, such as hormone and vitamin receptors, have been shown to be important regulators of the immune system. Induction of transcription by the glucocorticoid receptor has been shown to control apoptosis in CD4+CD8+ double thymocytes and to mediate immunosuppression of the mature T-cell response in the periphery (reviewed in [47]). Signaling via the vitamin A and D receptors controls B-cell and T-cell development and innate immunity (reviewed in [48]). Vitamin A has functions that are context dependent acting either as enhancers or suppressors of B cell proliferation, antigen presentation by dendritic cells or T cell function [48]. The best example of vitamin A function in the immune system is its effect on the differentiation of Th17 cells under non-inflammatory conditions, while in the presence of TGF-β it promotes the differentiation of regulatory T (Treg) cells [48]. In contrast, vitamin D has mainly suppressive effects on B cells, dendritic cells and T cells [48]
Orphan NHRs have also been shown to be involved in immunity (Table 1). The largest group of orphan NHRs known to have an influence on immunity regulates the development and function of hematopoietic cells in mice and potentially in humans. The majority of non-classic NHRs (Table 1) expressed in hematopoietic cells have immune phenotypes restricted to specific cell lineages. Some examples include NR2F6 (EAR2) which acts as a suppressor of Th17-cell function in mice [49], and NR3B1 (ESRRA) which is a modulator of aerobic glycolysis, a process which is required for the differentiation of activated cells and other cell lineages that have precursors that are dependent on a high proliferation rate, such as effector and memory T cells in mice [50]. The members of the NR4A family, NR4A1 (NUR77), NR4A2 (NURR1) and NR4A3 (NOR1) have been shown to play a redundant role in T-cell homeostasis and definite phenotypes are observed only when all three members of the NR4A family are deleted specifically in mouse T cells [51]. The triple knockout (TKO) of the complete NR4A family in mice shows that these receptors are strong suppressors of Th1- and Th2-cell differentiation and promoters of Foxp3 expression and Treg-cell differentiation [51]. Consequently, TKO-NR4A mice have no Treg cells and suffer from overt autoimmune disease similar to that observed in Foxp3−/− animals [51]. Given that NR4A receptors are widely expressed in tissues, specific studies are required to define the cell-specific functions of these receptors in each immune cell lineage. For example, aside from having a role in T-cell biology, NR4A1 has also been shown to be essential for the differentiation of mouse Ly6C− monocytes from bone marrow precursors [52]. Ly6C− monocytes circulate in the blood and spleen and patrol the blood vessels, scanning the endothelium. In the presence of TLR7 “danger signals” Ly6C− monocytes facilitate the neutrophil-driven necrosis of endothelial cells [53]. Another orphan NHR with cell lineage-restricted function is NR0B2 (SHP), which is expressed in macrophages, and which has been shown to suppress TLR signaling and protect mice from septic shock [54].
RORγt, a lymphoid tissue-specific isoform of RORγ (NR1F3) with an identical LBD to RORγ, is a master regulator for several lymphoid lineages. RORγt has been shown to be required for the differentiation of a specific innate lymphoid cell type 3 lineage (ILC3) – the lymphoid tissue inducer (Lti) cells – and thus regulates the development of secondary lymphoid organs such as lymph nodes [55, 56]. This role is represented here by the development of lymphoid aggregates induced by Lti-like cells in the adult mouse (Fig. 2A, bottom).
Figure 2.
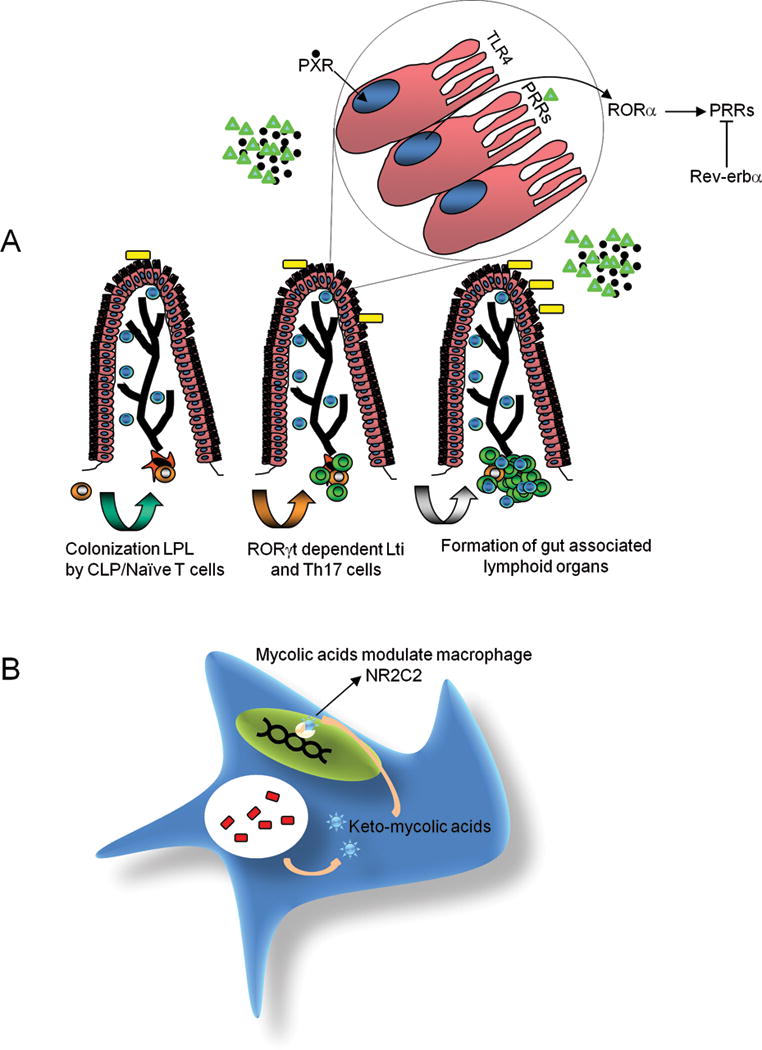
Examples of the role of non-classical NHRs in immunity. (A). Control of immune homeostasis in the mouse gut by NHRs. In epithelium of the gut (cells with villi) the expression of PRRs (pattern recognition receptors) is regulated by non-classic NHRs (Top). The microbiota (yellow squares) produce indole metabolites (black dots) and pathogen associated molecular patterns (PAMPs) (green triangles). The binding of indole metabolites to nuclear PXR results in the regulation of TLR4 expression (top). Furthermore, the expression of many PRRs like TLRs1-5, TLR9 and NOD2 is regulated by the 12 hour cycle of the circadian clock that contains two non-classic NHRs, RORα, which promotes transcription of PRRs and REV-ERBα which represses transcription of PRRs. RORγt regulates the differentiation of Th17 cells and Lti-like cells (bottom) in the intestinal lamina propria. In the intestinal lamina propria Lti-like cells most likely develop from common lymphoid progenitors (CLP) (brown cells) that interact with the stroma (orange fibroblasts) and induce them to differentiate into Lti-like cells (green cells), which then expand forming cryptopatches and recruiting other lymphoid cells like T cells (Blue cells). (B). Hijacking of the immune system by microbes using NHRs. Mycolic acid derivatives (blue stars) produced by Mycobacterium tuberculosis (red bars) in the phagosome of macrophages (blue cell) can act as NR2C2 (beige semicircle) ligands, promoting the formation of granulomas that protect the bacteria from immune clearance.
In addition to directly regulating specific cellular lineages of the immune system, many non-classic NHRs also have indirect functions in immunity, for example, by inducing non-immune tissues to produce signaling molecules that affect immune cells, or by regulating cellular “niches” for the development of lymphoid organs. Specifically, NR3B3 (ESRRG), NR1I2 (PXR), NR1D1 (REV-ERBα) and NR1F1 (RORα) are examples of non-classic NHRs expressed in non-immune tissues which then drive immune phenotypes in those tissues [57–59]. The increased expression of the orphan NHR NR3B3 in the liver results in increased production of hepcidin by liver cells [57]; liver hepcidin is subsequently released in the blood where it circulates and binds to its receptor in macrophages and induces these cells to reduce their iron export [60]. This increases the iron load in macrophages, which can promote the growth of some intracellular microbes [60], suggesting that hepcidin induction by NR3B3 could act as a bacterial evasion mechanism for Salmonella thyphimurium infection in mice [57], the extension of these findings to humans could turn this NHR into a therapeutic target [57]. Two non-classic NHRs that are in the process of deorphanization, ROR(α,β,γ) and REV-ERB(α,β) (Table 1) are also involved in the regulation of the circadian clock, which contains a regulatory loop that consists on the BMAL1:CLOCK heterodimer driving the expression of the repressors period (PER1-3) and cryptochrome (CRY1-2) reviewed in [61]. RORs and REV-ERBs act by binding to the same ROR-response elements (RORE) in many genes including BMAL1 [61]. Binding of RORs promotes transcription and binding of REV-ERBs represses transcription of target containing RORE [61]. The expression of TLRs in mouse intestinal epithelial cells (IECs) is regulated by the circadian clock and the NHRs RORα and REV-ERBα are directly involved in the promotion and repression of expression of TLRs1-5, TLR9 and NOD2 during the day [58] (Fig. 2A, Top). Furthermore, IECs from PXR−/− mice have increased expression of TLRs which results in greater permeability of the intestinal epithelium and increased inflammation [59]. In contrast the TLR levels in PXR+/+ mice are lower and permeability of the intestinal barrier is lower [59]. This process seems to be regulated by binding of indole metabolites from bacterial symbionts to PXR [59] (Fig. 2A, Top). Whether PXR directly regulates the expression of TLR4 is still open to debate.
Several NHRs have been implicated in the regulation of the immune microenvironment in the form of stromal and vascular components, which are required for the maturation of different lymphoid organs. For example, the non-classic orphan NHR NR2F2 (COUP-TFII) has been shown to be essential for the development of the lymphatic endothelium in the fetus, which serves as a “niche” for the formation of primary and secondary lymphoid tissues [62]. Another NHR involved in immune “niche” formation is NR5A1 which may be a receptor for phosphatidyl-inositols [63] (Table 1), which regulates the development of the tubular framework required for the formation of the structured vasculature in the mouse spleen [64]. This function appears to be evolutionarily conserved, as in humans NR5A1 mutations have been associated with asplenia [65], although it is not yet confirmed whether the mechanism that leads to loss of spleen development in humans is associated with the defect in tubular framework formation observed in Nr5a1−/− animals.
Finally, it is important to point out recent findings suggesting that NHRs could be hijacked by pathogens as a mechanism to evade the immune response. For example, the non-classic orphan receptor NR2C2 (TR4) is expressed in macrophages and can bind oxygenated derivatives of mycolic acid produced by phagocytosed Mycobacterium tuberculosis [66] (Fig.2B). The engagement of NR2C2 with these oxygenated bacterial products results in the differentiation of the infected macrophages into foam cells and promotes the formation of granulomas, which protect the bacteria from the immune system [66].
These examples are just the “tip of the iceberg” of the involvement of NHRs in the immune system, since many NHRs, orphans both in ligand and function, are expressed in different lineages of human and mouse immune cells (see Table 1 for detail).
Deorphanization: Connecting NHRs to general cell metabolism
The identification of the physiological ligands for NHRs is crucial for understanding their function. Recent evidence that cholesterol biosynthetic intermediates (CBIs) serve as natural FXR, and Liver X Receptor (LXR) and RORγ/γt ligands (Table 1 and Fig.1) provides a framework with which to interrogate other NHRs [42, 67–70]. These pioneering studies suggest that CBIs have functions beyond their role as either cholesterol intermediates or regulators of cholesterol biosynthesis, a view that is now supported by a considerable body of work that will be discussed in detail below.
Specific CBIs have been assigned as NHR modulators. For example, desmosterol, which is the last intermediate before cholesterol in the Bloch pathway (Fig. 1), has been shown to be a physiological LXRα/β ligand in foam macrophages [68]. The binding of desmosterol to LXRα/β in mouse peritoneal foam macrophages “in vitro” results in the suppression of expression of pro-inflammatory genes induced by TLR4 [68]. Another example are C4-methylated CBIs, which are those comprehending the intermediates from lanosterol to 4α-methyl-zymosterol (Fig. 1), and that have been identified as physiological ligands for RORγ/γt [42]. Importantly, genetic deletion of enzymes involved in the synthesis of C4-methylated CBIs like Cyp51 and Sc4mol (Fig. 1), recapitulate the immune phenotype observed in RORγ/γt-deficient mice, including defects in the development of lymph nodes and Th17 cells, while sparing other RORγt-independent T-cell and hematopoietic lineages [42]. It therefore follows that in human patients with mutations in cholesterol biosynthetic enzyme SC4MOL [71], in which CBIs of the C4-methylsterol group accumulate, suffer from autoimmune psoriasis that may be associated with the ability of the accumulating C4-methylsterols to act as RORγ/γt ligands. Nonetheless, the identification of universal endogenous mammalian metabolites such as CBIs as ligands for NHRs, suggests that CBIs and other endogenous metabolites could be ligands for other orphan NHRs.
Challenges in the deorphanization of NHRs that bind endogenous metabolites
Since CBIs are endogenous ligands produced by all mammalian cells, NHRs that bind CBIs will be in a “constitutively active” state. This contrasts with the behavior of “classic” NHRs. The standard model for NHR-promoted transcription is based on hormone and vitamin receptors expressed in cells which do not produce the ligand themselves, and therefore are present as Apo (empty) receptors. The addition of exogenous ligand generates the Holo (ligand bound) receptor that is transcriptionally active. Thus, luciferase-based reporters can be used to screen for exogenous ligands since the background noise caused by NHRs present in cells in Apo configuration is low. Under these conditions, the addition of high affinity ligand produces a sharp increase in specific reporter transcription. While some deorphanized NHRs, such as DAF-12, which binds dafachronic acids in nematodes [72] fit into this model, ligands to other NHRs may not be identified based solely on luciferase assays or receptor binding and may require to link the identified ligand with the biological activity of the receptor. A very stringent criteria that still needs to be applied to many non-classic NHRs that have putative ligands (Table 1).
The study of NHRs with endogenous ligands, such as RORγ/γt, will require the next generation of reporter systems, which are based on ligand-auxotroph cell lines and maintained in ligand-free synthetic media [42]. In the case of RORγ/γt, this was achieved by growing and maintaining insect cells, which are natural sterol auxotrophs, in sterol-free synthetic media [42, 73]. Insect cells are incapable of producing any type of sterol lipid, which has to be provided in the media. In such ligand-free systems the constitutive activity of RORγ/γt which is normally observed in mammalian cells is completely abolished, and the agonist activity of specific ligands for RORγ/γt can be directly assessed with luciferase reporters [42]. However, the findings from such ligand-free reporter systems need to be confirmed by genetic depletion or manipulation of the enzymes in the ligand biosynthetic pathway in order to map the exact origin of the endogenous ligand in mammalian cells [42]. One drawback of genetic studies is that they are less useful for NHRs with broad specificity for a whole class of ligands like RORγ. This is because genetic manipulation of an enzyme in the cholesterol biosynthetic pathway results in the depletion of CBIs downstream of the targeted enzyme, while it promotes accumulation and increases in the intracellular concentration of the CBI produced upstream the deleted enzyme. This is seen in Cyp51−/− fibroblasts, where precursor CBI (Fig. 1, Lanosterol) can therefore accumulate to reach concentrations hundreds of times above the physiological concentration found in non-manipulated cells [74]. When this approach is used to study NHRs with broad ligand specificity such as RORγ/γt, this accumulation of precursor CBI could result in the partial replacement of the physiological ligand by the accumulated cholesterol biosynthetic precursor [42].
Specificity of orphan NHRs for ligands
Classic hormone and vitamin receptors are exquisitely specific for their ligands but the example of RORγ/γt indicates that this is not necessarily the case for all NHRs. Experimental evidence suggests that NHR specificity correlates with ligand binding pocket (LBP) size [75]. Some orphan NHRs, such as NR4A2, have LBPs that are so small and filled up with aromatic amino acid side-chains that they are thought to be ligand-independent [76]. However, the finding that the small molecule 1,1-bis(3-indolyl)-1- (p-chlorophenyl)methane (DIM-C-pPhCl) is a NR4A2 agonist [77] suggests that NR4A receptors may be modulated by endogenous ligands, even though the binding site for these compounds within the NR4A2 LBD has not been yet identified. Generally, NHRs that bind hormones or vitamins have small LBPs with volumes <300 Å3, which can only accommodate a small number of potential molecules [75]. At the other extreme are receptors such as peroxisome proliferator-activated receptors (PPARs), liver receptor homolog-1 (LRH-1), PXR and constitutive androstane receptor (CAR), specialized in the catabolism of xenobiotics or ligands which are large fatty acids and phospholipids (Table 1). The LBPs of these receptors have volumes of >900 Å3 [75]. The LBPs of RORs and LXRs fall in the intermediate volume range between >300 and <900 Å3 [75]. Thus, one would expect RORs to be more specific than PPARs and less specific than vitamin or hormone receptors. As discussed above, experiments in ligand-free reporter systems show that RORγ/γt-dependent transcription can be induced by a broad range of sterol lipids, like CBIs, oxysterols, sterol acids and secosterols [42]. This explains why several groups have described different ligands for RORγ/γt, including CBIs [42, 67], oxysterols [78, 79] and vitamin D derivatives [80]. The broad recognition of sterol lipids by RORγ/γt may indicate that there is no highly specific ligand and that RORγ/γt, PPARs and other NHRs with medium to large LBPs may be similar in ligand binding to that of xeno/endobiotic receptors such as CAR and PXR, which can be modulated by a large number of structurally unrelated ligands [81–84]. The receptors with broad specificity are shown in Table 1. Conversely, the specificity of these receptors may be defined not by the receptor but by the abundance of a specific ligand present in a particular cell. In most mammalian cells RORγ/γt is semi-saturated by an abundant endogenous ligand like CBIs [42, 67]. It follows that it may not matter that RORγ/γt is promiscuous to sterol lipids, since the presence of an abundant local CBI will result in the occupancy of most intracellular RORγ/γt by the CBI, which outcompetes other exogenous sterols. In this case removing the endogenous ligand may result in an “apparent” broad specificity that does not exist under physiological conditions. Because of this effect, when an orphan NHR shows promiscuous binding to structurally related ligands in ligand-free, cell-based reporters or as recombinant protein, these studies need to be supported by genetic studies in which the ligand metabolic pathway is manipulated. Furthermore, future studies will require that NHR target gene expression is directly accessed by Chip-IPs to test whether the effect of a given metabolite on a cell is directly linked to its binding to a given NHR.
The specificity of an NHR could also be modulated by accessory proteins such as intracellular lipid binding proteins (iLBPs) that shuttle lipids from the cytoplasm to the nucleus in cells and in many cases transfer them to NHR [85–88]. The cellular retinoid acid binding protein II (CRABP-II), a iLBP, binds retinoic acid in the cytoplasm, which results in increased nuclear transport of retinoic acid-loaded CRABP-II and direct interaction with the retinoic acid receptor (RAR), a step that promotes the transfer of retinoic acid from CRABP-II to RAR [85, 86]. Similarly, fatty acid binding proteins (FABPs), another family of iLBPs, bind long-chain fatty acids (LCFA) and other PPAR ligands, which are then transported to the nucleus where they interact with PPARs and transfer the bound lipid to the NHR [87, 88]. For example, when FABP5 is loaded with retinoic acid, the retinoic acid becomes a PPARβ/δ ligand [89], suggesting that in these conditions FABP5 directly defines the ligand for PPARβ/δ. Furthermore, the competition of lipid binding proteins such as FABP5 and CRABP-II for retinoic acid defines whether retinoic acid will act as a RAR ligand or a PPAR ligand [89]. This has physiological consequences since activation of PPARβ/δ by retinoic acid has been shown to inhibit TNF-α-induced apoptosis in tumor and keratinocyte derived cell lines, while activation of RAR by retinoic acid rather enhances TNF-α-induced apoptosis in these cells [89]. Thus, changing the FABP5/CRABP-II ratio in cells can shift retinoic acid binding from one receptor to another and convert a pro-apoptotic signal mediated by RAR into an anti-apoptotic signal mediated by PPAR [89].
It is tempting to speculate that similarly to RARs and PPARs, RORs may also utilize iLBPs for ligand transport/loading. The need for a dedicated iLBP for particular biological outcomes in response to stimulation with broad activity NHR ligands would explain why CBIs provided in media can only partially reconstitute RORγ/γt reporter activity in mammalian cells with genetic deficiencies in sterol biosynthesis such as Fdft1−/− cells [42]. If this is the case, it is tempting to speculate that the specificity of RORγ/γt would depend on the interacting iLBP and depending on cell type and context, RORγ/γt could bind different sterols.
RORs: Metabolic regulators of cholesterol biosynthesis?
Another distinct feature of metabolic NHR-regulated transcription is the “feedback” mechanisms whereby the genetic program induced by NHRs directly modulates the ligand biosynthetic pathway. For example, in the liver, binding of oxysterols to LXRα promotes the conversion of oxysterols into bile acids [90], while binding of bile acids to FXR results in inhibition of bile acid biosynthesis in the liver and increased transport of bile acids from intestine to liver [91]. However, for other orphan NHRs the connection between NHR activation and ligand metabolism is not so obvious. CBIs are ligands for RORγ/γt but as yet, there is no known direct link between the genes and cellular processes regulated by RORγ/γt and cholesterol biosynthesis or catabolism. RORγt plays an important role in immunity, particularly in the development of lymph nodes [55, 56], gut-associated lymphoid tissues (GALT) [92] and Th17 cells [93]. The lymph node and GALT deficiencies in RORγ/γt-deficient animals are attributed to the absence of specific cell lineages, lymphoid tissue inducer cells (Lti) and Lti-like cells [55, 56, 92]. Lti cells are thought to be the terminal differentiation products of an Id2-dependent precursor derived from common lymphoid progenitors (CLPs) [94]. In the T-cell lineage, RORγt is important in the differentiation of Th17 cells but not for other T helper cell lineages [93]. Surprisingly for a transcription factor with such dramatic effects on Th17-cell differentiation, RORγt directly regulates the expression of only a small number of genes, including some interleukins and chemokines (Il17a, Il17f, Ccl20), interleukin receptors (Il23r, Il1r1, Ltb4r1) and proteases and membrane proteins (Furin, Fam124b, Tmem176a, Tmem176b)[95]. None of these RORγt targets have been shown to be directly related to the metabolism of sterol lipids. RORγt does play a role in Th17-cell differentiation as a positive and negative modulator of genetic programs initiated by other key transcription factors including IRF4, BATF and STAT3 [95] but a direct connection between cholesterol metabolism and the genetic programs initiated by these transcription factors has not been reported so far.
Conclusions
NHRs play an important role in the development and function of the immune system. The deorphanization of NHRs is not a trivial process. Some NHRs bind common endogenous metabolites present in all mammalian cells. For these NHRs it is necessary to develop new technologies such as ligand-free, cell-based reporter systems and genetic manipulation of metabolic pathways to define candidate ligands. If the NHR is highly specific for one metabolite, the technologies discussed in this Review maybe enough to identify a natural ligand. However, when receptors are of broad specificity, genetic manipulation of the ligand biosynthetic pathway may restrict the number of potential ligands. Further support for the ligand has to be obtained by identification of ligand carriers such as iLBPs that are natural protein partners of the NHR and assessment of effects on NHR target gene expression after manipulation of ligand concentration in cells. Almost half of NHRs have been shown to bind post-cholesterol derivatives of cholesterol [1]. In the last years, pre-cholesterol CBIs have been identified as ligands for at least two NHRs, LXR and RORγ/γt [42, 67, 68, 96]. As many canonical and non-canonical CBIs are being identified, an exciting possibility is that these CBIs could act like intracellular hormones, serving as messengers that drive the communication between different cellular compartments.
Acknowledgments
I thank Natalia B. Ivanova for critical reading of the manuscript. The work performed by FRS (Santori et al 2015) and cited in this review was supported by NIH grant R01-AI080885-01A1 to Dan R. Littman. FRS was also supported by NIH grant T32HL007151.
Footnotes
Conflict of interest:
The author declares no commercial or financial conflict of interest.
References
- 1.Germain P, Staels B, Dacquet C, Spedding M, Laudet V. Overview of nomenclature of nuclear receptors. Pharmacol Rev. 2006;58:685–704. doi: 10.1124/pr.58.4.2. [DOI] [PubMed] [Google Scholar]
- 2.Huang P, Chandra V, Rastinejad F. Structural overview of the nuclear receptor superfamily: insights into physiology and therapeutics. Annu Rev Physiol. 2010;72:247–272. doi: 10.1146/annurev-physiol-021909-135917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Starling EH. The Croonian Lectures. I. On the chemical correlation of the functions of the body. Lancet. 1905;166:339–341. [Google Scholar]
- 4.Funk C. The etiology of the deficiency diseases. Beri-beri, polyneuritis in birds, epidemic dropsy, scurvy, experimental scurvy in animals, infantile scurvy, ship beri-beri, pellagra. J State Med. 1912;20:341–368. [Google Scholar]
- 5.Kendall EC. The isolation in crystalline form of the compound containing iodin, which occurs in the thyroid. Its chemical nature and physiological activity. Trans Ass Am Physicians. 1915;30:420–449. [PubMed] [Google Scholar]
- 6.McCollum EV, Davis M. The nature of the dietary deficiencies of rice. J Biol Chem. 1915;23:181–230. [Google Scholar]
- 7.McCollum EV, Simmonds N, Becker JE, Shipley PG. Studies on experimental rickets: XXI. An experimental demonstration of the existence of a vitamin which promotes calcium deposition. 1922;53:293–312. 1922. [PubMed] [Google Scholar]
- 8.Harington CR, Barger G. Chemistry of Thyroxine: Constitution and Synthesis of Thyroxine. Biochem J. 1927;21:169–183. doi: 10.1042/bj0210169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, et al. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giguere V, Ong ES, Segui P, Evans RM. Identification of a receptor for the morphogen retinoic acid. Nature. 1987;330:624–629. doi: 10.1038/330624a0. [DOI] [PubMed] [Google Scholar]
- 11.Petkovich M, Brand NJ, Krust A, Chambon P. A human retinoic acid receptor which belongs to the family of nuclear receptors. Nature. 1987;330:444–450. doi: 10.1038/330444a0. [DOI] [PubMed] [Google Scholar]
- 12.Evans RM. The steroid and thyroid hormone receptor superfamily. Science. 1988;240:889–895. doi: 10.1126/science.3283939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Weinberger C, Thompson CC, Ong ES, Lebo R, Gruol DJ, Evans RM. The c-erb-A gene encodes a thyroid hormone receptor. Nature. 1986;324:641–646. doi: 10.1038/324641a0. [DOI] [PubMed] [Google Scholar]
- 14.Clarke ND, Berg JM. Zinc fingers in Caenorhabditis elegans: finding families and probing pathways. Science. 1998;282:2018–2022. doi: 10.1126/science.282.5396.2018. [DOI] [PubMed] [Google Scholar]
- 15.Goldstein JL, Brown MS. Regulation of the mevalonate pathway. Nature. 1990;343:425–430. doi: 10.1038/343425a0. [DOI] [PubMed] [Google Scholar]
- 16.Gober HJ, Kistowska M, Angman L, Jeno P, Mori L, De Libero G. Human T cell receptor gammadelta cells recognize endogenous mevalonate metabolites in tumor cells. J Exp Med. 2003;197:163–168. doi: 10.1084/jem.20021500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kistowska M, Rossy E, Sansano S, Gober HJ, Landmann R, Mori L, De Libero G. Dysregulation of the host mevalonate pathway during early bacterial infection activates human TCR gamma delta cells. Eur J Immunol. 2008;38:2200–2209. doi: 10.1002/eji.200838366. [DOI] [PubMed] [Google Scholar]
- 18.Dunn SE, Youssef S, Goldstein MJ, Prod’homme T, Weber MS, Zamvil SS, Steinman L. Isoprenoids determine Th1/Th2 fate in pathogenic T cells, providing a mechanism of modulation of autoimmunity by atorvastatin. J Exp Med. 2006;203:401–412. doi: 10.1084/jem.20051129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Houten SM, Kuis W, Duran M, de Koning TJ, van Royen-Kerkhof A, Romeijn GJ, Frenkel J, et al. Mutations in MVK, encoding mevalonate kinase, cause hyperimmunoglobulinaemia D and periodic fever syndrome. Nat Genet. 1999;22:175–177. doi: 10.1038/9691. [DOI] [PubMed] [Google Scholar]
- 20.Drenth JP, Cuisset L, Grateau G, Vasseur C, van de Velde-Visser SD, de Jong JG, Beckmann JS, et al. Mutations in the gene encoding mevalonate kinase cause hyper-IgD and periodic fever syndrome. International Hyper-IgD Study Group. Nat Genet. 1999;22:178–181. doi: 10.1038/9696. [DOI] [PubMed] [Google Scholar]
- 21.Bloch K. The biological synthesis of cholesterol. Science. 1965;150:19–28. doi: 10.1126/science.150.3692.19. [DOI] [PubMed] [Google Scholar]
- 22.Kandutsch AA, Russell AE. Preputial gland tumor sterols. 3. A metabolic pathway from lanosterol to cholesterol. J Biol Chem. 1960;235:2256–2261. [PubMed] [Google Scholar]
- 23.Nelson JA, Steckbeck SR, Spencer TA. Biosynthesis of 24,25-epoxycholesterol from squalene 2,3;22,23-dioxide. J Biol Chem. 1981;256:1067–1068. [PubMed] [Google Scholar]
- 24.Janowski BA, Grogan MJ, Jones SA, Wisely GB, Kliewer SA, Corey EJ, Mangelsdorf DJ. Structural requirements of ligands for the oxysterol liver X receptors LXRalpha and LXRbeta. Proc Natl Acad Sci U S A. 1999;96:266–271. doi: 10.1073/pnas.96.1.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lehmann JM, Kliewer SA, Moore LB, Smith-Oliver TA, Oliver BB, Su JL, Sundseth SS, et al. Activation of the nuclear receptor LXR by oxysterols defines a new hormone response pathway. J Biol Chem. 1997;272:3137–3140. doi: 10.1074/jbc.272.6.3137. [DOI] [PubMed] [Google Scholar]
- 26.Bae SH, Paik YK. Cholesterol biosynthesis from lanosterol: development of a novel assay method and characterization of rat liver microsomal lanosterol delta 24-reductase. Biochem J. 1997;326(Pt 2):609–616. doi: 10.1042/bj3260609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Alloza I, Otaegui D, de Lapuente AL, Antiguedad A, Varade J, Nunez C, Arroyo R, et al. ANKRD55 and DHCR7 are novel multiple sclerosis risk loci. Genes Immun. 2012;13:253–257. doi: 10.1038/gene.2011.81. [DOI] [PubMed] [Google Scholar]
- 28.Cooper JD, Smyth DJ, Walker NM, Stevens H, Burren OS, Wallace C, Greissl C, et al. Inherited variation in vitamin D genes is associated with predisposition to autoimmune disease type 1 diabetes. Diabetes. 2011;60:1624–1631. doi: 10.2337/db10-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holick SA, Lezin MS, Young D, Malaikal S, Holick MF. Isolation and identification of 24-dehydroprovitamin D3 and its photolysis to 24-dehydroprevitamin D3 in mammalian skin. J Biol Chem. 1985;260:12181–12184. [PubMed] [Google Scholar]
- 30.Kuehnle K, Crameri A, Kalin RE, Luciani P, Benvenuti S, Peri A, Ratti F, et al. Prosurvival effect of DHCR24/Seladin-1 in acute and chronic responses to oxidative stress. Mol Cell Biol. 2008;28:539–550. doi: 10.1128/MCB.00584-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Saucier SE, Kandutsch AA, Gayen AK, Nelson JA, Spencer TA. Oxygenation of desmosterol and cholesterol in cell cultures. J Lipid Res. 1990;31:2179–2185. [PubMed] [Google Scholar]
- 32.Spann NJ, Glass CK. Sterols and oxysterols in immune cell function. Nat Immunol. 2013;14:893–900. doi: 10.1038/ni.2681. [DOI] [PubMed] [Google Scholar]
- 33.Krisans SK. Cell compartmentalization of cholesterol biosynthesis. Ann N Y Acad Sci. 1996;804:142–164. doi: 10.1111/j.1749-6632.1996.tb18614.x. [DOI] [PubMed] [Google Scholar]
- 34.Acimovic J, Rozman D. Steroidal triterpenes of cholesterol synthesis. Molecules. 2013;18:4002–4017. doi: 10.3390/molecules18044002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Silve S, Dupuy PH, Ferrara P, Loison G. Human lamin B receptor exhibits sterol C14-reductase activity in Saccharomyces cerevisiae. Biochim Biophys Acta. 1998;1392:233–244. doi: 10.1016/s0005-2760(98)00041-1. [DOI] [PubMed] [Google Scholar]
- 36.Wassif CA, Brownson KE, Sterner AL, Forlino A, Zerfas PM, Wilson WK, Starost MF, et al. HEM dysplasia and ichthyosis are likely laminopathies and not due to 3beta-hydroxysterol Delta14-reductase deficiency. Hum Mol Genet. 2007;16:1176–1187. doi: 10.1093/hmg/ddm065. [DOI] [PubMed] [Google Scholar]
- 37.Hoffmann K, Dreger CK, Olins AL, Olins DE, Shultz LD, Lucke B, Karl H, et al. Mutations in the gene encoding the lamin B receptor produce an altered nuclear morphology in granulocytes (Pelger-Huet anomaly) Nat Genet. 2002;31:410–414. doi: 10.1038/ng925. [DOI] [PubMed] [Google Scholar]
- 38.Shultz LD, Lyons BL, Burzenski LM, Gott B, Samuels R, Schweitzer PA, Dreger C, et al. Mutations at the mouse ichthyosis locus are within the lamin B receptor gene: a single gene model for human Pelger-Huet anomaly. Hum Mol Genet. 2003;12:61–69. doi: 10.1093/hmg/ddg003. [DOI] [PubMed] [Google Scholar]
- 39.Hoffmann K, Sperling K, Olins AL, Olins DE. The granulocyte nucleus and lamin B receptor: avoiding the ovoid. Chromosoma. 2007;116:227–235. doi: 10.1007/s00412-007-0094-8. [DOI] [PubMed] [Google Scholar]
- 40.Gaines P, Tien CW, Olins AL, Olins DE, Shultz LD, Carney L, Berliner N. Mouse neutrophils lacking lamin B-receptor expression exhibit aberrant development and lack critical functional responses. Exp Hematol. 2008;36:965–976. doi: 10.1016/j.exphem.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Subramanian G, Chaudhury P, Malu K, Fowler S, Manmode R, Gotur D, Zwerger M, et al. Lamin B receptor regulates the growth and maturation of myeloid progenitors via its sterol reductase domain: implications for cholesterol biosynthesis in regulating myelopoiesis. J Immunol. 2012;188:85–102. doi: 10.4049/jimmunol.1003804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Santori FR, Huang P, van de Pavert SA, Douglass EF, Jr, Leaver DJ, Haubrich BA, Keber R, et al. Identification of Natural RORgamma Ligands that Regulate the Development of Lymphoid Cells. Cell Metab. 2015;21:286–297. doi: 10.1016/j.cmet.2015.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Andreyev AY, Fahy E, Guan Z, Kelly S, Li X, McDonald JG, Milne S, et al. Subcellular organelle lipidomics in TLR-4-activated macrophages. J Lipid Res. 2010;51:2785–2797. doi: 10.1194/jlr.M008748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rahier A, Darnet S, Bouvier F, Camara B, Bard M. Molecular and enzymatic characterizations of novel bifunctional 3beta-hydroxysteroid dehydrogenases/C-4 decarboxylases from Arabidopsis thaliana. J Biol Chem. 2006;281:27264–27277. doi: 10.1074/jbc.M604431200. [DOI] [PubMed] [Google Scholar]
- 45.Gachotte D, Barbuch R, Gaylor J, Nickel E, Bard M. Characterization of the Saccharomyces cerevisiae ERG26 gene encoding the C-3 sterol dehydrogenase (C-4 decarboxylase) involved in sterol biosynthesis. Proc Natl Acad Sci U S A. 1998;95:13794–13799. doi: 10.1073/pnas.95.23.13794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Song BL, Javitt NB, DeBose-Boyd RA. Insig-mediated degradation of HMG CoA reductase stimulated by lanosterol, an intermediate in the synthesis of cholesterol. Cell Metab. 2005;1:179–189. doi: 10.1016/j.cmet.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 47.Ashwell JD, Lu FW, Vacchio MS. Glucocorticoids in T cell development and function*. Annu Rev Immunol. 2000;18:309–345. doi: 10.1146/annurev.immunol.18.1.309. [DOI] [PubMed] [Google Scholar]
- 48.Mora JR, Iwata M, von Andrian UH. Vitamin effects on the immune system: vitamins A and D take centre stage. Nat Rev Immunol. 2008;8:685–698. doi: 10.1038/nri2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hermann-Kleiter N, Gruber T, Lutz-Nicoladoni C, Thuille N, Fresser F, Labi V, Schiefermeier N, et al. The nuclear orphan receptor NR2F6 suppresses lymphocyte activation and T helper 17-dependent autoimmunity. Immunity. 2008;29:205–216. doi: 10.1016/j.immuni.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michalek RD, Gerriets VA, Nichols AG, Inoue M, Kazmin D, Chang CY, Dwyer MA, et al. Estrogen-related receptor-alpha is a metabolic regulator of effector T-cell activation and differentiation. Proc Natl Acad Sci U S A. 2011;108:18348–18353. doi: 10.1073/pnas.1108856108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekiya T, Kashiwagi I, Yoshida R, Fukaya T, Morita R, Kimura A, Ichinose H, et al. Nr4a receptors are essential for thymic regulatory T cell development and immune homeostasis. Nat Immunol. 2013;14:230–237. doi: 10.1038/ni.2520. [DOI] [PubMed] [Google Scholar]
- 52.Hanna RN, Carlin LM, Hubbeling HG, Nackiewicz D, Green AM, Punt JA, Geissmann F, et al. The transcription factor NR4A1 (Nur77) controls bone marrow differentiation and the survival of Ly6C- monocytes. Nat Immunol. 2011;12:778–785. doi: 10.1038/ni.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carlin LM, Stamatiades EG, Auffray C, Hanna RN, Glover L, Vizcay-Barrena G, Hedrick CC, et al. Nr4a1-dependent Ly6C(low) monocytes monitor endothelial cells and orchestrate their disposal. Cell. 2013;153:362–375. doi: 10.1016/j.cell.2013.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yuk JM, Shin DM, Lee HM, Kim JJ, Kim SW, Jin HS, Yang CS, et al. The orphan nuclear receptor SHP acts as a negative regulator in inflammatory signaling triggered by Toll-like receptors. Nat Immunol. 2011;12:742–751. doi: 10.1038/ni.2064. [DOI] [PubMed] [Google Scholar]
- 55.Kurebayashi S, Ueda E, Sakaue M, Patel DD, Medvedev A, Zhang F, Jetten AM. Retinoid-related orphan receptor gamma (RORgamma) is essential for lymphoid organogenesis and controls apoptosis during thymopoiesis. Proc Natl Acad Sci U S A. 2000;97:10132–10137. doi: 10.1073/pnas.97.18.10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sun Z, Unutmaz D, Zou YR, Sunshine MJ, Pierani A, Brenner-Morton S, Mebius RE, et al. Requirement for RORgamma in thymocyte survival and lymphoid organ development. Science. 2000;288:2369–2373. doi: 10.1126/science.288.5475.2369. [DOI] [PubMed] [Google Scholar]
- 57.Kim DK, Jeong JH, Lee JM, Kim KS, Park SH, Kim YD, Koh M, et al. Inverse agonist of estrogen-related receptor gamma controls Salmonella typhimurium infection by modulating host iron homeostasis. Nat Med. 2014;20:419–424. doi: 10.1038/nm.3483. [DOI] [PubMed] [Google Scholar]
- 58.Mukherji A, Kobiita A, Ye T, Chambon P. Homeostasis in intestinal epithelium is orchestrated by the circadian clock and microbiota cues transduced by TLRs. Cell. 2013;153:812–827. doi: 10.1016/j.cell.2013.04.020. [DOI] [PubMed] [Google Scholar]
- 59.Venkatesh M, Mukherjee S, Wang H, Li H, Sun K, Benechet AP, Qiu Z, et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity. 2014;41:296–310. doi: 10.1016/j.immuni.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ganz T. Systemic iron homeostasis. Physiol Rev. 2013;93:1721–1741. doi: 10.1152/physrev.00008.2013. [DOI] [PubMed] [Google Scholar]
- 61.Curtis AM, Bellet MM, Sassone-Corsi P, O’Neill LA. Circadian clock proteins and immunity. Immunity. 2014;40:178–186. doi: 10.1016/j.immuni.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 62.Lin FJ, Chen X, Qin J, Hong YK, Tsai MJ, Tsai SY. Direct transcriptional regulation of neuropilin-2 by COUP-TFII modulates multiple steps in murine lymphatic vessel development. J Clin Invest. 2010;120:1694–1707. doi: 10.1172/JCI40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, et al. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- 64.Morohashi K, Tsuboi-Asai H, Matsushita S, Suda M, Nakashima M, Sasano H, Hataba Y, et al. Structural and functional abnormalities in the spleen of an mFtz-F1 gene-disrupted mouse. Blood. 1999;93:1586–1594. [PubMed] [Google Scholar]
- 65.Zangen D, Kaufman Y, Banne E, Weinberg-Shukron A, Abulibdeh A, Garfinkel BP, Dweik D, et al. Testicular differentiation factor SF-1 is required for human spleen development. J Clin Invest. 2014;124:2071–2075. doi: 10.1172/JCI73186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dkhar HK, Nanduri R, Mahajan S, Dave S, Saini A, Somavarapu AK, Arora A, et al. Mycobacterium tuberculosis keto-mycolic acid and macrophage nuclear receptor TR4 modulate foamy biogenesis in granulomas: a case of a heterologous and noncanonical ligand-receptor pair. J Immunol. 2014;193:295–305. doi: 10.4049/jimmunol.1400092. [DOI] [PubMed] [Google Scholar]
- 67.Hu X, Wang Y, Hao LY, Liu X, Lesch CA, Sanchez BM, Wendling JM, et al. Sterol metabolism controls TH17 differentiation by generating endogenous RORgamma agonists. Nat Chem Biol. 2015;11:141–147. doi: 10.1038/nchembio.1714. [DOI] [PubMed] [Google Scholar]
- 68.Spann NJ, Garmire LX, McDonald JG, Myers DS, Milne SB, Shibata N, Reichart D, et al. Regulated accumulation of desmosterol integrates macrophage lipid metabolism and inflammatory responses. Cell. 2012;151:138–152. doi: 10.1016/j.cell.2012.06.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Otte K, Kranz H, Kober I, Thompson P, Hoefer M, Haubold B, Remmel B, et al. Identification of farnesoid X receptor beta as a novel mammalian nuclear receptor sensing lanosterol. Mol Cell Biol. 2003;23:864–872. doi: 10.1128/MCB.23.3.864-872.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yang C, McDonald JG, Patel A, Zhang Y, Umetani M, Xu F, Westover EJ, et al. Sterol intermediates from cholesterol biosynthetic pathway as liver X receptor ligands. J Biol Chem. 2006;281:27816–27826. doi: 10.1074/jbc.M603781200. [DOI] [PubMed] [Google Scholar]
- 71.He M, Kratz LE, Michel JJ, Vallejo AN, Ferris L, Kelley RI, Hoover JJ, et al. Mutations in the human SC4MOL gene encoding a methyl sterol oxidase cause psoriasiform dermatitis, microcephaly, and developmental delay. J Clin Invest. 2011;121:976–984. doi: 10.1172/JCI42650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Motola DL, Cummins CL, Rottiers V, Sharma KK, Li T, Li Y, Suino-Powell K, et al. Identification of ligands for DAF-12 that govern dauer formation and reproduction in C. elegans. Cell. 2006;124:1209–1223. doi: 10.1016/j.cell.2006.01.037. [DOI] [PubMed] [Google Scholar]
- 73.Huh JR, Leung MW, Huang P, Ryan DA, Krout MR, Malapaka RR, Chow J, et al. Digoxin and its derivatives suppress TH17 cell differentiation by antagonizing RORgammat activity. Nature. 2011;472:486–490. doi: 10.1038/nature09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Keber R, Motaln H, Wagner KD, Debeljak N, Rassoulzadegan M, Acimovic J, Rozman D, et al. Mouse knockout of the cholesterogenic cytochrome P450 lanosterol 14alpha-demethylase (Cyp51) resembles Antley-Bixler syndrome. J Biol Chem. 2011;286:29086–29097. doi: 10.1074/jbc.M111.253245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Benoit G, Malewicz M, Perlmann T. Digging deep into the pockets of orphan nuclear receptors: insights from structural studies. Trends Cell Biol. 2004;14:369–376. doi: 10.1016/j.tcb.2004.05.007. [DOI] [PubMed] [Google Scholar]
- 76.Wang Z, Benoit G, Liu J, Prasad S, Aarnisalo P, Liu X, Xu H, et al. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature. 2003;423:555–560. doi: 10.1038/nature01645. [DOI] [PubMed] [Google Scholar]
- 77.Inamoto T, Papineni S, Chintharlapalli S, Cho SD, Safe S, Kamat AM. 1,1-Bis(3′-indolyl)-1-(p-chlorophenyl)methane activates the orphan nuclear receptor Nurr1 and inhibits bladder cancer growth. Mol Cancer Ther. 2008;7:3825–3833. doi: 10.1158/1535-7163.MCT-08-0730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Soroosh P, Wu J, Xue X, Song J, Sutton SW, Sablad M, Yu J, et al. Oxysterols are agonist ligands of RORgammat and drive Th17 cell differentiation. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1322807111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jin L, Martynowski D, Zheng S, Wada T, Xie W, Li Y. Structural basis for hydroxycholesterols as natural ligands of orphan nuclear receptor RORgamma. Mol Endocrinol. 2010;24:923–929. doi: 10.1210/me.2009-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Slominski AT, Kim TK, Takeda Y, Janjetovic Z, Brozyna AA, Skobowiat C, Wang J, et al. RORalpha and ROR gamma are expressed in human skin and serve as receptors for endogenously produced noncalcemic 20-hydroxy- and 20,23-dihydroxyvitamin D. FASEB J. 2014 doi: 10.1096/fj.13-242040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Dussault I, Yoo HD, Lin M, Wang E, Fan M, Batta AK, Salen G, et al. Identification of an endogenous ligand that activates pregnane X receptor-mediated sterol clearance. Proc Natl Acad Sci U S A. 2003;100:833–838. doi: 10.1073/pnas.0336235100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xie W, Evans RM. Orphan nuclear receptors: the exotics of xenobiotics. J Biol Chem. 2001;276:37739–37742. doi: 10.1074/jbc.R100033200. [DOI] [PubMed] [Google Scholar]
- 83.Wei P, Zhang J, Egan-Hafley M, Liang S, Moore DD. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nature. 2000;407:920–923. doi: 10.1038/35038112. [DOI] [PubMed] [Google Scholar]
- 84.Forman BM, Tzameli I, Choi HS, Chen J, Simha D, Seol W, Evans RM, et al. Androstane metabolites bind to and deactivate the nuclear receptor CAR-beta. Nature. 1998;395:612–615. doi: 10.1038/26996. [DOI] [PubMed] [Google Scholar]
- 85.Budhu AS, Noy N. Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol Cell Biol. 2002;22:2632–2641. doi: 10.1128/MCB.22.8.2632-2641.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Dong D, Ruuska SE, Levinthal DJ, Noy N. Distinct roles for cellular retinoic acid-binding proteins I and II in regulating signaling by retinoic acid. J Biol Chem. 1999;274:23695–23698. doi: 10.1074/jbc.274.34.23695. [DOI] [PubMed] [Google Scholar]
- 87.Hostetler HA, McIntosh AL, Atshaves BP, Storey SM, Payne HR, Kier AB, Schroeder F. L-FABP directly interacts with PPARalpha in cultured primary hepatocytes. J Lipid Res. 2009;50:1663–1675. doi: 10.1194/jlr.M900058-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan NS, Shaw NS, Vinckenbosch N, Liu P, Yasmin R, Desvergne B, Wahli W, et al. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol Cell Biol. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Schug TT, Berry DC, Shaw NS, Travis SN, Noy N. Opposing effects of retinoic acid on cell growth result from alternate activation of two different nuclear receptors. Cell. 2007;129:723–733. doi: 10.1016/j.cell.2007.02.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Peet DJ, Turley SD, Ma W, Janowski BA, Lobaccaro JM, Hammer RE, Mangelsdorf DJ. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 91.Makishima M, Okamoto AY, Repa JJ, Tu H, Learned RM, Luk A, Hull MV, et al. Identification of a nuclear receptor for bile acids. Science. 1999;284:1362–1365. doi: 10.1126/science.284.5418.1362. [DOI] [PubMed] [Google Scholar]
- 92.Eberl G, Littman DR. Thymic origin of intestinal alphabeta T cells revealed by fate mapping of RORgammat+ cells. Science. 2004;305:248–251. doi: 10.1126/science.1096472. [DOI] [PubMed] [Google Scholar]
- 93.Ivanov II, McKenzie BS, Zhou L, Tadokoro CE, Lepelley A, Lafaille JJ, Cua DJ, et al. The orphan nuclear receptor RORgammat directs the differentiation program of proinflammatory IL-17+ T helper cells. Cell. 2006;126:1121–1133. doi: 10.1016/j.cell.2006.07.035. [DOI] [PubMed] [Google Scholar]
- 94.Cherrier M, Sawa S, Eberl G. Notch, Id2, and RORgammat sequentially orchestrate the fetal development of lymphoid tissue inducer cells. J Exp Med. 2012;209:729–740. doi: 10.1084/jem.20111594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ciofani M, Madar A, Galan C, Sellars M, Mace K, Pauli F, Agarwal A, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Janowski BA, Willy PJ, Devi TR, Falck JR, Mangelsdorf DJ. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 1996;383:728–731. doi: 10.1038/383728a0. [DOI] [PubMed] [Google Scholar]
- 97.Arpin C, Pihlgren M, Fraichard A, Aubert D, Samarut J, Chassande O, Marvel J. Effects of T3R alpha 1 and T3R alpha 2 gene deletion on T and B lymphocyte development. J Immunol. 2000;164:152–160. doi: 10.4049/jimmunol.164.1.152. [DOI] [PubMed] [Google Scholar]
- 98.van de Pavert SA, Ferreira M, Domingues RG, Ribeiro H, Molenaar R, Moreira-Santos L, Almeida FF, et al. Maternal retinoids control type 3 innate lymphoid cells and set the offspring immunity. Nature. 2014;508:123–127. doi: 10.1038/nature13158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chakravarthy MV, Lodhi IJ, Yin L, Malapaka RR, Xu HE, Turk J, Semenkovich CF. Identification of a physiologically relevant endogenous ligand for PPARalpha in liver. Cell. 2009;138:476–488. doi: 10.1016/j.cell.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Nagy L, Szanto A, Szatmari I, Szeles L. Nuclear hormone receptors enable macrophages and dendritic cells to sense their lipid environment and shape their immune response. Physiol Rev. 2012;92:739–789. doi: 10.1152/physrev.00004.2011. [DOI] [PubMed] [Google Scholar]
- 101.Raghuram S, Stayrook KR, Huang P, Rogers PM, Nosie AK, McClure DB, Burris LL, et al. Identification of heme as the ligand for the orphan nuclear receptors REV-ERBalpha and REV-ERBbeta. Nat Struct Mol Biol. 2007;14:1207–1213. doi: 10.1038/nsmb1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Traversari C, Sozzani S, Steffensen KR, Russo V. LXR-dependent and -independent effects of oxysterols on immunity and tumor growth. Eur J Immunol. 2014;44:1896–1903. doi: 10.1002/eji.201344292. [DOI] [PubMed] [Google Scholar]
- 103.Yuan X, Ta TC, Lin M, Evans JR, Dong Y, Bolotin E, Sherman MA, et al. Identification of an endogenous ligand bound to a native orphan nuclear receptor. PLoS One. 2009;4:e5609. doi: 10.1371/journal.pone.0005609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Heyman RA, Mangelsdorf DJ, Dyck JA, Stein RB, Eichele G, Evans RM, Thaller C. 9-cis retinoic acid is a high affinity ligand for the retinoid X receptor. Cell. 1992;68:397–406. doi: 10.1016/0092-8674(92)90479-v. [DOI] [PubMed] [Google Scholar]
- 105.Levin AA, Sturzenbecker LJ, Kazmer S, Bosakowski T, Huselton C, Allenby G, Speck J, et al. 9-cis retinoic acid stereoisomer binds and activates the nuclear receptor RXR alpha. Nature. 1992;355:359–361. doi: 10.1038/355359a0. [DOI] [PubMed] [Google Scholar]


