Abstract
The majority of normal animal cells are 10–20 µm in diameter. Many signaling mechanisms, notably PI3K/Akt/mTOR, Myc, and Hippo pathways, tightly control and coordinate cell growth, cell size, cell division, and cell number during homeostasis. These regulatory mechanisms are frequently deregulated during tumorigenesis resulting in wide variations in cell sizes and increased proliferation in cancer cells. Here, we first review the evidence that primitive stem cells in adult tissues are quiescent and generally smaller than their differentiated progeny, suggesting a correlation between small cell sizes with the stemness. Conversely, increased cell size positively correlates with differentiation phenotypes. We then discuss cancer stem cells (CSCs) and present some evidence that correlates cell sizes with CSC activity. Overall, the causal link between CSCs and cell size has been under-studied and remains to be rigorously assessed. In the future, optimizing methods for isolating cells based on size should help elucidate the connection between cancer cell size and CSC characteristics.
Keywords: stem cells, cancer, cancer stem cells, cell size
Stem cells and cell size
In multicellular organisms, homeostatic control mechanisms are regulated so that internal conditions ensuring cell number and size remain stable and relatively constant (reviewed in [1]). These control mechanisms are an integration of extracellular nutritional environments and multiple cell-specific growth, mitogenic, and survival signals that coalesce to create a balanced homeostatic state in terms of rates of synthesis and degradation of macromolecules, and thus cell size. The majority of animal cells are 10–20 µm in diameter and rarely vary more than 2-fold outside of this range suggesting that the mechanism for cell size regulation is highly conserved [2]. Nonetheless, the mechanisms that control cell size and the relationship between cell growth (cell mass increase over time), cell division, and cell lifespan remain poorly understood.
A correlation between size and lifespan was first observed in yeast [3] and the same observations have also been made in mammalian cells. For example, as yeast cells approach quiescence, proliferation slows but cell growth continues, and thus cells increase in size with age. Mammalian cells in vivo also steadily increase in size with age. In a recent study, a genetic link between cell size, growth rate and lifespan has been reported in yeast cells [4]. Authors show that mutations that increase cell size concomitantly increase growth rate and decrease life span. Thus, small cell mutants age slowly and are long-lived while large cells grow, divide and age dramatically faster in comparison. Specifically, intracellular RNA and protein contents increase with age, even though the synthesis of macromolecules decreases, and these elevations contribute to the increase in the cell size, numbers of inclusion bodies, and other cellular components [5].
Cell growth and proliferation are distinct processes that both require extensive instructive signals. It is unclear what types of mechanisms coordinate cellular growth and the cell cycle in metazoan cells. It has been suggested that commitment to proliferation is dependent upon the attainment of a minimum “critical cell size” [6, 7]. In support, large cells tend to divide faster than small cells [8]. One group found that both the expression and the activity of G1-phase cyclins are modulated by growth rate and cell size in yeast, suggesting that the proliferative capacity correlates with cell size and cell growth rates, such that the largest cells begin to proliferate five times faster than the smallest cells [9]. Other experiments in mammalian cells support the conclusion that cell size correlates closely with the proliferative potential of cells [10–12]. Another group examined cell size distributions in lymphoblasts and showed that growth rate is size-dependent throughout the cell cycle. Alternatively, authors concluded that cell division probability varies independently with cell size and age, indicating that mammalian cells have an intrinsic mechanism for cell size maintenance [13].
Mammalian adult stem cells are rare, long-living cells with the inherent traits of both indefinite self-renewal and multilineage differentiation capabilities [14]. Thus, stem cells normally divide asymmetrically into a new stem cell and a committed progenitor, the latter of which has limited self-renewing ability and can give rise to progeny that are more restricted in their differentiating potential and finally to functionally mature cells. Between the two, primitive stem cells are generally smaller than differentiated cells. Stem cells are also generally detected in a predominantly quiescent state, a reversible arrest in proliferation as determined by an integration of diverse antimitogenic signals. The proliferative and quiescent states have vastly different metabolic needs, the former requiring tremendous metabolic energy in order to synthesize DNA, protein, and lipids. Indeed, quiescent cells are widely reported to exhibit reduced nucleotide synthesis, as well as reduced metabolic activity and cell size. As expected, most studies that look at cell size have described stem cells to be much smaller in size than the more committed and highly proliferative cells. For example, in murine bone marrow (BM), hematopoietic stem cells (HSCs) are small measuring between 4 and 5 µm [15, 16]. Another group described human BM HSCs (Lin−CD34+c-Kit+) to be small at ~6 µm [17]. Neuroblasts (neural stem cells) and myoblasts are notably smaller than their differentiated daughter cells, i.e., neurons and skeletal muscle cells, respectively. The presence of heterogeneous CD34+CD45− nonhematopoietic tissue-committed, putative stem cells that measure 5–7 µm has also been described [18]. Finally, small pluripotent epiblastic-like cells of 8–10 µm from the rat skeletal muscle [19], very small embryonic-like (VSEL) stem cells of 3 µm from the bone marrow [20], very small stem cell-like cells (2–4 µm) that express embryonic markers such as SSEA-4, Oct-4, Nanog, Sox-2, and c-kit in the human ovarian surface epithelium [21] have been reported. Whether these latter, small-sized, putative stem cells truly possess stem cell properties (i.e., self-renewal and multi-lineage differentiation) has not been rigorously examined and remains somewhat controversial.
Identification of different sized subpopulations has been largely based on regular light or electron microscopy [15–18], fluorescence-activated cell sorting (FACS) [22], size-sieving methods [23], centrifugal elutriation, and long-term culture under specific conditions [24] (Figure 1). These methods have been instrumental in producing evidence that cell size is related to cell cycle [25], cell proliferation [26–28], and differentiation [29, 30]. For example, the differentiation marker involucrin has been reported to correlate with increasing cell size and terminal differentiation in human epidermal cultures [31]. In studies utilizing human epidermal keratinocytes, the smallest cells sorted by centrifugal elutriation expressed the highest levels of basal cell markers (p63 and basonuclin) and possessed the greatest clonogenicity in culture [26, 28, 30]. Vice versa, the proliferative potential of human fibroblasts and keratinocytes was shown to be inversely dependent on cell size [26, 27].
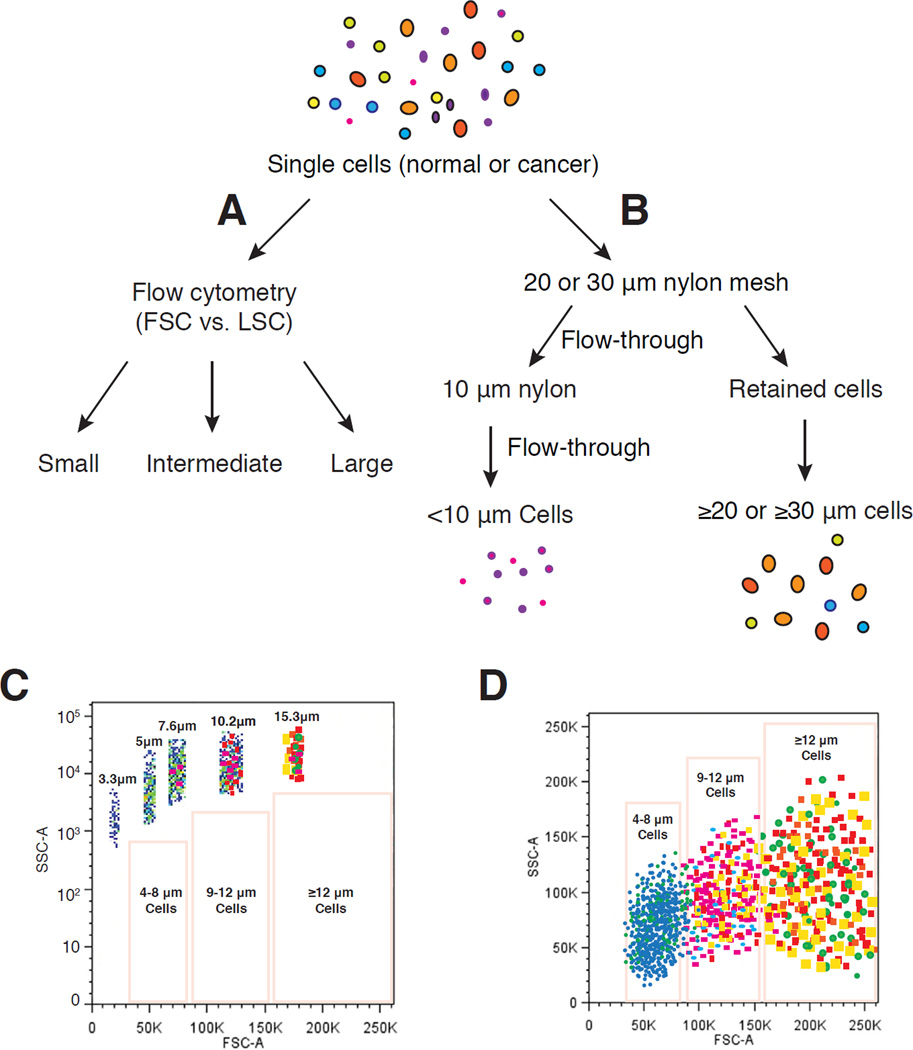
Figure 1.
Schematic illustration of strategies to enrich cell populations with varying sizes. (A) Single cells from normal or cancerous cultures or tissues can be separated into (relatively) small, intermediate and large cell populations based on the forward scatter (FSC) intensity value in flow cytometry (i.e., FSC-based FACS). (B) Size-sieving approach using nylon mesh filtration. Single cells are separated into small cells (<10 µm) and large cells (≥ 20 or 30 µm) using 20 or 30 µm nylon mesh. (C–D) Cell populations with different sizes can be more precisely purified via a beads-sizing method by FACS (i.e., beads sizing-based FACS) (modified from Figure 1-A of ref. 32).
When utilizing FACS, cells of different sizes can be separated based on forward scatter (FSC), which is an indirect measurement of size, versus light scatter (LSC), which is a measurement of the cell’s granularity (Figure 1A). Alternatively, a mixture of synthetic beads of predefined sizes can be employed in FACS to fractionate subpopulations of cells of varying sizes (Figure 1C–D). In one study, human corneal epithelial cells were sorted by FACS based on FSC [32]. Four fractions (A, B, C, and D) of cells ranging in size from 10 to 16, 17 to 23, 24 to 30, and >31 µm in diameter, respectively, were isolated. Cell size was shown to positively correlate with the expression of the differentiation markers keratin (K) 3, K12, and involucrin and inversely with the levels of stem cell–associated markers ΔNp63 and ABCG2 and with colonyforming efficiency and growth capacity. Cells with the smallest size contained the greatest number of BrdU label-retaining slow-cycling cells, displayed the highest percentage of cells immune-positive to p63 and ABCG2 and negative to K3 and involucrin, expressed the highest levels of ΔNp63 and ABCG2 mRNA and the lowest levels of K3, K12, and involucrin, and possessed the highest colony-forming efficiency and growth capacity [32].
As discussed above, when keratinocytes undergo terminal differentiation, both in vivo and in culture, they increase progressively in cellular size [31, 33]. In studying the p53/MDM2 regulatory loop in human epidermal differentiation, one group found that induction of MDM2 and downregulation of p53 characterized the transition from proliferation to differentiation in primary human keratinocytes. These changes correlated with an increase in cell size and an irreversible commitment to terminal differentiation [29]. The same group also found that upon differentiation, keratinocytes continued DNA replication after cell division was suppressed [34]. As a consequence of this phenomenon, referred to as endoreplication, cell growth results not in proliferation, but rather in an increased cellular size and polyploidy.
Size has also been interrogated in mesenchymal cells. In order to obtain homogeneous subpopulations of stem cells from human umbilical cord matrix, one group applied the counterflow centrifugal elutriation to separate cells with distinct characteristics with respect to size, morphology and proliferative activity [35]. In another study, both FACS and the elutriation method were used to identify a phenotypically distinct population of mesenchymal stem/progenitor cells (MSPCs) within human BM [36]. The MSPC activity resided within a population of rare, small CD45−CD73+CD90+CD105+ cells that lacked CD44. These rare MSPCs, which were between 5 and 12 µm in diameter, expanded rapidly in culture and demonstrated tri-lineage mesenchymal differentiation potential into osteoblasts, chondrocytes, and adipocytes [36]. Other studies utilizing normal human peripheral blood cells and hematopoietic progenitor cells have also uncovered a correlation between cell volume with stem cell marker expression, allowing for the identification of small stem cells [37].
Rapidly self-renewing mesenchymal stem cells (MSCs) of ~7 µm [22,39] or even smaller VSEL stem cells [20,38] have been described in the BM. Size-sieving based approach using smaller pores, for example, 3 µm, has also been employed to isolate very small BM MSCs with proliferation and self-renewal capacities that lack markers of osteoblastic differentiation and can serve as progenitors for all mesenchymal cell lineages, including osseous, adipose, and cartilaginous tissues [23]. In another study, equine umbilical cords were processed and cells separated into larger and smaller sieved populations using multi-dishes with 8-µm pore transwell inserts [40]. Cells from both populations (i.e., >8-µm and <8-µm) expressed MSC and pluripotency markers and were able to differentiate into mesodermic and ectodermic lineages. After sieving, both large intervascular and small perivascular cells were rapidly replicating cells. However, sieved cells (i.e., <8-µm) had more proliferative potential than un-sieved cells [40]. Interestingly, Katsube et al measured the proliferation and cellular thickness of human MSCs by atomic force microscopy and found that the MSCs with high proliferative activity were small and those with low proliferative activity were flat and large [41]. The MSCs with medium proliferative activity were of intermediate size.
The VESL stem cells described above [20,38] have the phenotype of CXCR4+Sca-1+CD45−Lin− in murine BM and are highly enriched at the mRNA and protein levels for markers in embryonic pluripotent stem cells. A transmission electron microscopy study showed that these cells were extremely small (2–4 µm) and had the ability to differentiate into cells from all three germ cell layers in vitro [42]. The same group later isolated a similar population of CXCR4+AC133+CD34+Lin−CD45− mononuclear cells from human cord blood (CB), which were very small (3–5 µm) and expressed embryonic transcription factors Oct-4 and Nanog [43]. The authors showed that murine VSELs could differentiate into the hematopoietic lineage after coculture over OP9 stromal cells [44]. Nevertheless, these observations on VSELs have recently become controversial because studies from an independent group found no evidence for VSELs in murine BM, and no molecular signatures associated with pluripotency in any mouse BM cells smaller than 7 µm across [45]. In addition, the small cells did not form spheres in vitro and differentiate into blood cells [45].
In contrast to the above studies that correlate small cell size with adult stem cell properties, a recent study reported an opposite correlation in mouse mammary stem cells (MaSCs) [46]. In addition to the CD24+CD49fhiCD29hiSca1− marker profile, adult MaSCs can be defined by the property of size. Based on FACS FSC, cells with a low FSC (approximately <10 µM) lacked outgrowth potential and failed to reconstitute the mammary gland when transplanted into the cleared fat pads of syngeneic mice. In contrast, cells >10 µM in size had increased outgrowth potential as compared with Lin− control cells. Limiting dilution transplantation assays indicated that the repopulating ability of Lin−CD24−CD29hi cells that were >10 µM in size was significantly increased as compared with cells marked by CD24 and CD29 alone [46].
Cancer stem cells
Cancer is characterized by the excessive and uncontrolled expansion of abnormal, malignant cells that display morphological, proliferative, and functional heterogeneity. Morphological heterogeneity is further manifested in tumor cells of variegating size, shape, thickness, nucleus/cytoplasm ratio, etc. In order to explain this tumor cell heterogeneity, two models have been proposed, one being the cancer stem cell (CSC) concept [47,48]. This model postulates that, akin to growth of normal proliferative tissues, growth of tumors or expansion of a tumor clone is driven by a population of cells endowed with both self-renewal and differentiation capabilities [48]. CSCs, as with normal stem cells, are long-lasting and have self-renewal capabilities. Both human cancers (or tumor clones) and regenerating normal tissues are organized in a hierarchical manner according to stages of differentiation and proliferative potential with stem cells as the common denominator. However, this does not necessarily imply that CSCs are always derived from normal stem cells. Stem cells are often the target of genetic events that are necessary or sufficient for malignant transformation; however, restricted progenitors, due to their cycling feature, oftentimes represent the preferred transformation targets [47]. Even differentiated cells can undergo oncogenic reprogramming and dedifferentiation and be transformed [47]. Both normal stem cells and CSCs share the ability to self renew and produce differentiated progeny, and thus parallels can be found between signaling pathways that regulate these attributes. A CSC is set apart from a normal stem cell in that it has acquired the capacity for indefinite proliferation through accumulated genetic mutations and epigenetic alterations. In this case, when signaling pathways that regulate normal stem cell self-renewal are dysregulated, tumorigenesis occurs. Multiple approaches have been employed to identify, enrich, purify, and characterize CSCs in different tumor systems [47].
CSCs have now been reported in most human tumors. Multiple approaches have been employed to identify, enrich, purify, and characterize CSCs [47]. Cell surface biomarkers can be exploited to purify and analyze CSC populations by FACS or magnetic-activated cell sorting (MACS). Isolation of side population (SP) cells, defined by Hoechst dye exclusion in FACS, can also facilitate enrichment of CSCs. The SP cells are identified according to their ability to efflux the Hoechst dye at a higher pace than the remaining cells. The degree of efflux also correlates with maturation state, as cells with the highest efflux activity are less differentiated. Because the SP phenotype is mainly mediated by ABCG2, an ATP-binding cassette half-transporter associated with multidrug resistance, ABCG2 can also be used to enrich putative CSCs. High expression of aldehyde dehydrogenase (ALDH) is another marker for CSCs. Clonogenicity and sphere formation assays assess, to a certain degree, self-renewal properties in isolated cell populations. The former determines if one seeded cell has the ability to proliferative extensively, evidenced by the formation of large colonies. Additional serial seeding can further strengthen the clone’s self-renewal capacity. The latter employs low adherence plates and serum-free medium that specifically enrich for tumorspheres displaying CSC phenotypes. The application of genetic marking can be used to isolate CSCs. An example of this is use of a lentiviral reporter system that contains a specific promoter of interest specific to phenotypic characteristics of either stem cells or differentiated cells that drives the expression of a fluorescent tag. The same principle applies to lineage tracing whereby a single cell is marked, via a promoter of interest, and the label is transmitted to the cell’s progeny resulting in a set of labeled clones, thus providing information about the founder cell and its location. As mentioned previously, a common feature associated with stem cells is quiescence. Incorporation of DNA analogues (BrdU, EdU) during the S-phase is a type of label retention method for studying cell-cycle kinetics and thus enables detection of the slow-cycling, quiescent stem cells and CSCs.
Cancer stem cells and cell size
Tumors generally contain multiple clones, in which differentially sized tumor cells can be easily observed. It seems reasonable to speculate, a priori, that a certain population of cells in tumors with certain sizes might be endowed with particular characteristics to promote survival and longevity. In other words, can cell size be used as a determinant of CSCs vs. non-CSCs? Very few studies by far have been conducted to prospectively address this interesting question. A group recently generated a liver-derived progenitor cell (LDPC) line, RA1, by overexpressing the simian virus 40 (SV40) large T antigen (TAg) in primary LDPCs [49]. Interestingly, following transformation, LDPCs decreased in size significantly and the propagating cells measured 1 µm in diameter compared with the 10 µm size of the parental LDPCs. These small cells multiplied continuously and, after passage 36, they started to increase in size and reached a maximum size of 10–12 µm by passage 42. The authors speculated [49] that forced cell cycle entry by TAg might have been the trigger for the “reprogramming” of cells causing a change in their cell size, possibly via the process of ‘de-differentiation’, a feature observed in other stem cells and CSCs. To date, RA1 cells are the smallest mammalian cells to be reported in the literature.
Bortolomai et al investigated cancer stem/tumor initiating cells characteristics in the human epidermoid carcinoma cell line, A431, via growth as non-adherent spheres in specific media and ALDH enzymatic activity [50]. Spheres manifested increased stem-cell like properties including holoclone formation, high ALDH activity (the ALDH-positive fraction increased from 46% in adherent cultures to 65% in spheres), and a transient induction of stem cell markers such as Nanog, Nestin and Oct4. When compared to parental cells, spheres were greatly enriched in a podoplanin-positive subpopulation characterized by small cell sizes and the ability to propagate tumors in nude mice at a lower cell dose [50].
In contrast, Srivastava et al interrogated the DAOY medulloblastoma cell line with respect to the relationship between cell size and stem-like potential and observed opposing results [51]. They purified SP/non-SP DAOY cells, which were also sorted separately for viability, cell size, cell cycle status, and proliferative capacity evaluation. The SP, non-SP, CD133+, and CD133− fractions were all capable of reconstituting the original parental DAOY population. However, SP cells, which have been shown to enrich for CSCs in many tumor systems [47], differed from the non-SP cells in that they actually showed increased cell size, decreased S-phase, and slightly decreased proliferative capacity. Another example of stem-like cancer cells with increased cell size is polyploid giant cancer cells (PGCCs) that are frequently found in human solid tumors. These cells are large atypical cancer cells with multiple copies of DNAs and have been recently been studied in human ovarian cancer cell lines and primary ovarian cancer [52]. Of interest, these PGCCs are highly resistant to oxygen deprivation, express normal and CSC markers, divide asymmetrically and cycle slowly, and, surprisingly, can differentiate into adipose, cartilage and bone cells. A single PGCC can form cancer cell spheroids in vitro and generate tumors in immunodeficient mice, which manifest a mesenchymal phenotype with increased expression of CSC markers CD44 and CD133 and become more resistant to treatment with cisplatin [52].
Our laboratory, in the past 10 years, has been meticulously dissecting prostate cancer cell heterogeneity. Using cell surface markers, SP, holoclone and sphere formation, as well as tumor transplantation and serial transplantation assays, we have provided strong evidence for the presence of CSCs in long-term cultured prostate cancer cell lines and xenografts as well as in primary patient tumors [53–61]. We have recently made attempts to determine a correlation between CSCs and cell size in the most aggressive, fully undifferentiated prostate cancer cells PC3. PC3 cells completely lack differentiation markers such as androgen receptor (AR) and prostate-specific antigen (PSA). Virtually 100% of PC3 cells express commonly used CSC surface markers such as CD44, integrin α2β1; consequently, these markers would not differentiate between tumorigenic CSCs vs. non-CSCs. We have shown that PC3 holoclones harbor long-term self-renewing tumor-propagating cells [57].
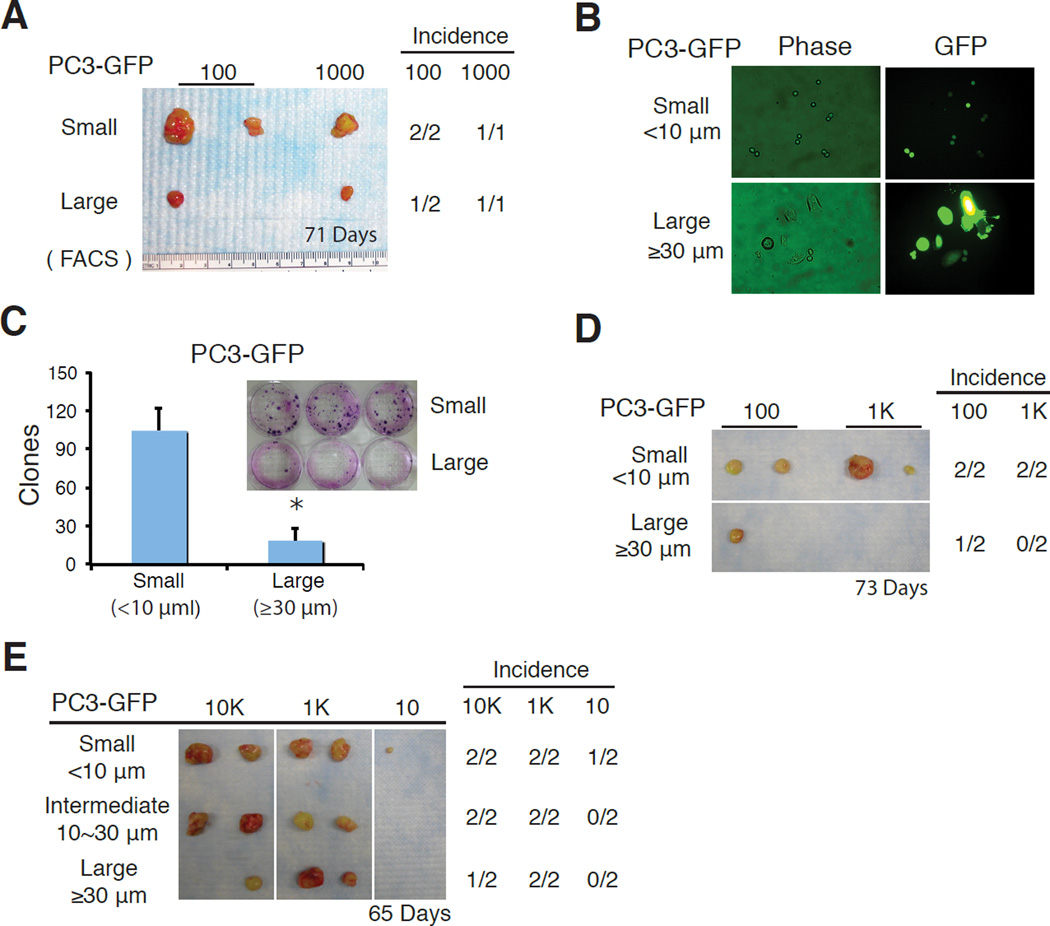
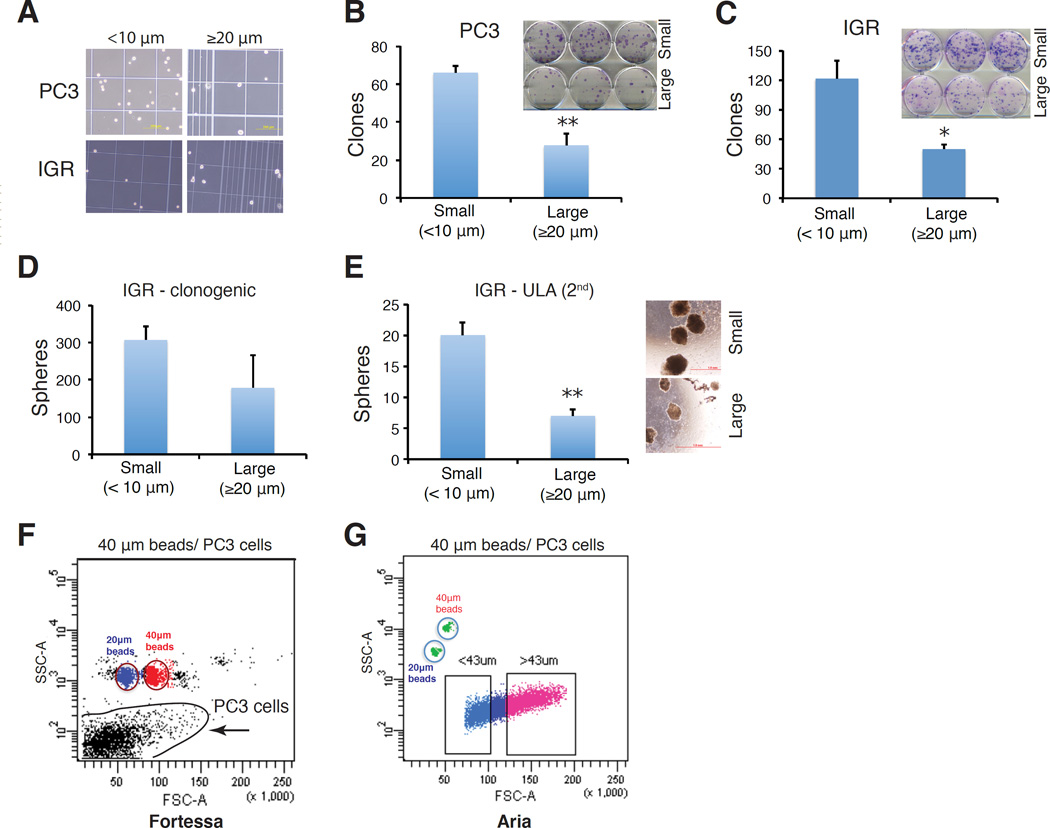
To address whether cell size is able to provide tumorigenic stratification in PC3 cells, we first utilized FSC-based FACS sorting (Figure 1A) to fractionate PC3 into, relatively, large and small sized populations and then implanted 100 and 1,000 cells, respectively, subcutaneously, in NOD/SCID mice. This experiment revealed a tendency of small cells being more tumorigenic manifested by more and larger tumors regenerated (Figure 2A). We then employed size-sieving approach by using nylon mesh of different pore sizes (Figure 1B) to separate PC3 cells into two cell populations varying in the cell sizes, i.e., small (< 10 µm) and large (≧ 20 or 30 µm) (Figure 2B–E; Figure 3A–B) followed by clonal (i.e., 2D) and clonogenic (i.e., 3D) assays as well as in vivo tumor regeneration. In two independent experiments, small PC3 cells demonstrated higher clonal capacity than large PC3 cells (Figure 2C; Figure 3B). Importantly, two separate tumor experiments again revealed the trend of small PC3 cells being more tumorigenic (Figure 2D–E). Similar studies in another AR−/PSA− prostate cancer cell line IGR also revealed that small IGR cells displayed higher clonal (Figure 3C) and clonogenic (Figure 3D–E) capacities than corresponding large cells.
Figure 2.
Small PC3 cells tend to be more clonal and tumorigenic than the isogenic large cells. (A) FSC-based FACS-purified small PC3-GFP cells exhibited a trend of increasing tumorigenicity than isogenic large cells. (B–E) PC3 cells were separated by size-sieving using nylon mesh into three (small, intermediate, and large) sizes and then used in various in vitro and in vivo assays. (B) Morphologic validation and GFP checking of small (<10 µm) and corresponding large (≥30 µm) cells. (C) Clonal assays. PC3-GFP cells of two different sizes were plated in 6-well plates (300 cells per well). Clones were counted 14 days after plating. Presented are the mean ± SD from triplicate wells (*P<0.05). (D–E) Two independent tumor experiments. PC3-GFP cells were separated by nylon mesh into the 3 sizes indicated and subcutaneously injected into NOD/SCID female mice at different cell doses. Presented are tumor images, tumor incidence, and the time (days; bottom) when tumors were harvested.
Figure 3.
Small prostate cancer cells (<10 µm) are more clonal and clonogenic than isogenic large cells (≥20 µm) in both PC3 (A–B) and IGR-CaP1 (IGR; C–E) cell lines. (A) Morphologic validation of different cell sizes separated by nylon mesh. (B) Clonal assay in PC3 cells. PC3 cells were sorted into small (<10 µm) and large (≥20 µm) sized populations by nylon mesh, and plated in 6-well plates (300 cells per well). Clones were counted 14 days after plating. Presented are the mean ± SD from triplicate wells (**P<0.01). Shown in the inset is the Giemsa-stained image. (C) Clonal assay in IGR. Sorted small and large IGR-CaP1 cells were plated in 6-well plates (200 cells per well) and clones were enumerated 14 days after plating. Presented are the mean ± SD from triplicate cells (*P<0.05). (D) Clonogenic assay in IGR-CaP1 cells. Small and large IGR cells were mixed with Matrigel and plated in 12-well plates (1,000 cells/well) and colonies were counted in 2 weeks. The large cells showed reduced colony-forming activity in comparison to small cells although this difference was not statistically significant. (E) Secondary (2°) sphere formation assays in IGR-CaP1 cells. Nylon mesh-separated cells were plated in 6-well ultra-low attachment (ULA) plates (1,000 cells per well), and cultured in serum-free medium for 2 weeks. 1° spheres were harvested, digested into single-cell suspension, re-sorted into small cell (<10 µm) and large cells (≥20 µm) by nylon mesh and plated into the ULA plates (1,000 cells per well) for 2° sphere formation assays. Spheres were counted after 2 weeks (*P<0.05). (F–G) Ongoing trials of beads sizing-based FACS protocol in PC3. Different FACS machines exhibited distinct sorting profiles.
The above studies suggest that in two undifferentiated prostate cancer cell models, small-sized cells possess higher CSC-associated properties (i.e., higher clonal, clonogenic, and tumorigenic capacities). Serial tumor transplantations are needed to verify the true CSC traits in small prostate cancer cells. Current experimental strategies in fractionating cancer cells into different sizes have obvious pros and cons. Nylon mesh-based size sieving represents a cheap and facile method that is gentle on cells leading to high viability; but purity is a concern and precise cell sizes cannot be determined. FACS produces populations with higher purity; however, the high speed at which cells are sorted leads to low cell viability. Also, the use of forward angle light scatter is not an accurate measure of cell size as light scatter is influenced by a number of factors. Finally, although FACS with beads sizing has been used to fractionate normal cells into cell populations of different sizes [32,46], our preliminary studies in PC3 cells, which vary widely in sizes in culture, demonstrate that this approach might not be readily applicable to cultured human cancer cells as two flow cytometers give completely different flow profiles (Figure 3F–G).
Perspectives
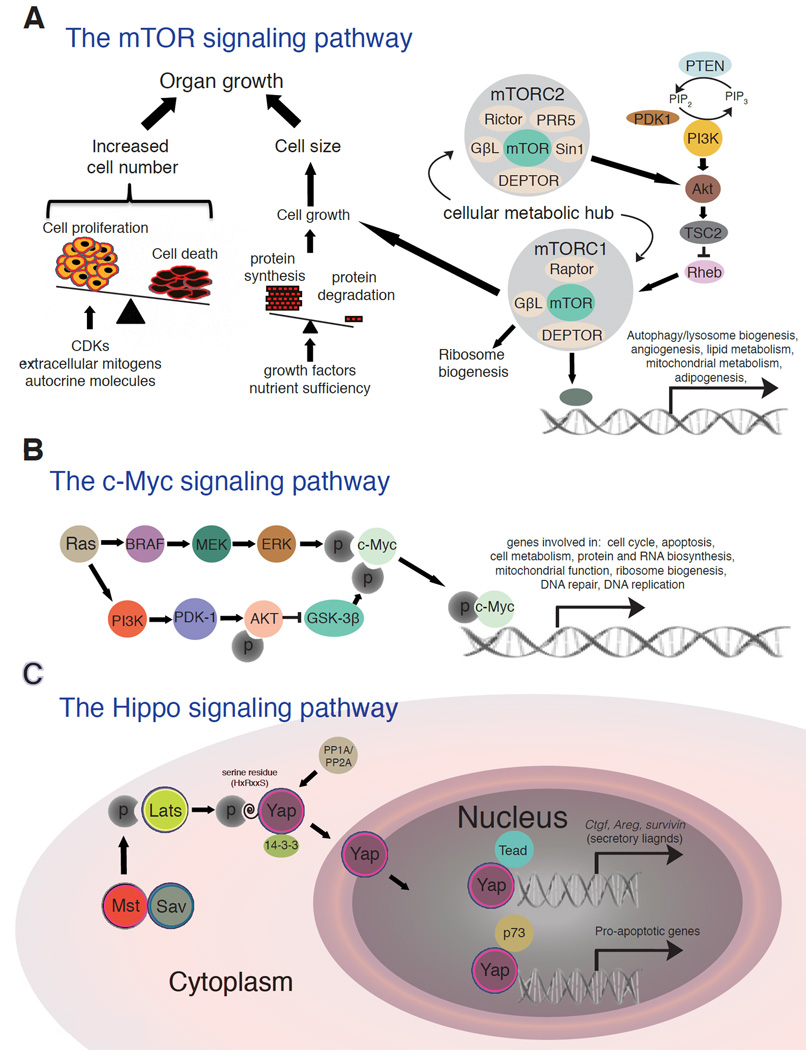
Significant progress has been made in the identification of three key and inter-connected regulatory pathways, i.e., mTOR, Myc, and Hippo, that control normal cell growth, and this has given us clues as to how cell size is controlled in homeostasis and how cancer cells might have abnormal cell size control mechanisms (Figure 4). The PI3K/AKT/mTOR signaling pathway is a major regulator of cell growth and thus a key determinant of cell size [62]. Critical inputs regulating this pathway include growth factors, amino acids, stress, energy status, and oxygen. The activated pathway promotes protein synthesis, lipogenesis, and energy metabolism, activities that directly relate to cell size (Figure 4A). Many of the components of the PI3K signaling pathway, which is upstream of both mTOR complexes, are mutated in human caners. Additionally, the loss of p53 promotes mTOR complex activation. Another major regulator is Myc, a transcription factor that increases cell growth and cell size in multiple tissues and organisms (Figure 4B). Myc is frequently overexpressed as a consequence of genomic amplification and heightened growth or mitogenic signaling from, e.g., Ras activation (Figure 4B). Endoreplication and cell enlargement is stimulated in keratinocytes by continuous activation of c-Myc [34]. Continuous activity of c-Myc also results in increased cellular size and loss of the cell cycle control in other cell types when mitosis is impaired [63–66]. When c-Myc is knocked out in the epidermis, there is a loss of the proliferative compartment and premature differentiation. The keratinocyte cells size, growth and endoreplication are all reduced and the stem cell amplification is compromised [66]. The Hippo pathway controls tissue/organ size via regulating cell number and cell size [67] (Figure 4C). YAP, the main downstream target of this pathway promotes organ growth via activation of mTOR. PTEN, an upstream regulator of mTOR, is a critical mediator of YAP regulation. Therefore, YAP is the functional link between the mTOR and HIPPO pathways that regulates cell size, tissue growth and hyperplasia [68]. Both Myc and YAP are frequently overexpressed in human cancers.
Figure 4.
Three major signaling pathways that regulate cell size in normal cells.
A. The PI3K/AKT/mTOR pathway. An increase in cell number and cell size both can lead to organ growth. Cell number is dependent on the intricate balance between cell proliferation, which is controlled by extracellular mitogens and inhibitory molecules, and cell death, which is initiated in response to developmental cues or lack of survival factors. Cell size is dependent on cell growth, which is controlled by a balance between protein synthesis and protein degradation based on extracellular growth factors and nutrient sufficiency. Central to the regulation of cell growth, is the mTOR pathway. The core components of the mTOR pathway are shown.
B. The c-Myc pathway. The Ras/PI3K/ERK pathway induces c-Myc. Myc mainly functions as a transcription factor to regulate target genes including those involved in cell cycle, apoptosis, cell metabolism and protein and RNA biosynthesis. Myc is also frequently upregulated in cancer cells due to genomic amplification (not shown).
C. The Hippo pathway. The pathway is thought to sense cell density and to regulate gene expression for control of organ size. The key downstream effector of the mammalian Hippo pathway is the Yes-associated protein (YAP), which function as a transcription co-activator. Mst is a mammalian homolog of Drosophila Hippo and encodes a kinase that phosphorylates Lats with cooperation from Sav. Phosphorylated Lats then exerts kinase activity on YAP. Phosphorylated YAP is then trapped in the cytoplasm by a 14-3-3 protein, until de-phosphorylation by a phosphatase such as PP1A allows translocation to the nucleus, thereby facilitating transcriptional regulation.
Other factors have also been found that regulate cell size. One example is the transcription factor myostatin, a negative regulator of skeletal muscle size that inhibits muscle cell differentiation. Myostatin inhibits activation of the Akt/mTOR/p70S6 protein synthesis pathway, which mediates both differentiation in myoblasts and hypertrophy in myotubes [69]. Ion channel activity has been shown to simultaneously affect cell cycle and cell volume in the S phase of the cell cycle in embryonic stem cells [70]. Also, the Erg channel is critical in controlling cell volume during cell cycle in embryonic stem cells. In support of this, cell death following Erg inhibition is a consequence of the inability to regulate cell volume [71]. The Notch pathway has been studied in relationship to cell size. Recently, type II neural stem cells in Drosophila are used for studying CSC-initiated tumorigenesis [72]. These cells, marked by a transcriptional target of Notch involved in their self-renewal and the absence of a differentiation-promoting transcription factor, give rise to immature progenitors that are small in size. Notch signaling is required for the maintenance of these normal stem cells although the specific mechanisms are unclear. When Notch signaling is inhibited, these neural stem cells exhibit reduced cell growth and cell size [72]. Finally, ectopic expression of the p21 cyclin-dependent kinase inhibitor has been shown to induce hypertrophy, increase cell size and reduced the replicative lifespan of cells [73].
Cell size is a reflection of the balance between anabolic and catabolic processes that are initiated by various signaling pathways (Figure 4). Cell size thus can dynamically change based on the total net input of these signals. Clear correlations between cell size and particular phenotypes such as stem cell features have been discovered in normal cell lineages. Limited reports and our preliminary studies also suggest that small cancer cells appear to be more tumorigenic and possess more CSC properties. However, whether a relationship truly exists between cell sizes and all CSCs is less clear, may likely be tumor cell type-dependent, and requires more thorough investigations. This is due in part to the various cell fractionation methods that may favor certain cell sizes over others or that are simply not technically adept. Recent advances in computer science, microfabrication, and microfluidic devices have spurred the rapid development of precision mass-quantifying approaches, allowing more precise quantification in cell size. Future studies should aim to further relate cell size (a phenotype) to functional properties such as stemness based on a combination of next-generation techniques.
Acknowledgements
Work in the authors’ lab was supported, in part, by grants from NIH (NCI R01-CA155693), DOD (W81XWH-13-1-0352 and W81XWH-14-1-0575), CPRIT (RP120380), and MDACC Center for Cancer Epigenetics (all to D.G.T). X. Chen was supported, in part, by a DOD postdoc fellowship PC141581. We apologize to the colleagues whose work was not cited due to space constraint.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
None.
References
- 1.Lloyd AC. The regulation of cell size. Cell. 2013;154:1194–1205. doi: 10.1016/j.cell.2013.08.053. [DOI] [PubMed] [Google Scholar]
- 2.Conlon I, Raff M. Size control in animal development. Cell. 1999;96:235–244. doi: 10.1016/s0092-8674(00)80563-2. [DOI] [PubMed] [Google Scholar]
- 3.Mortimer RK, Johnston JR. Life span of individual yeast cells. Nature. 1959;183:1751–1752. doi: 10.1038/1831751a0. [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Dungrawala H, Hua H, Manukyan A, Abraham L, Lane W, et al. Cell size and growth rate are major determinants of replicative lifespan. Cell Cycle. 2011;10:144–155. doi: 10.4161/cc.10.1.14455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Phipps SM, Berletch JB, Andrews LG, Tollefsbol TO. Aging cell culture: methods and observations. Methods Mol Biol. 2007;371:9–19. doi: 10.1007/978-1-59745-361-5_2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Coelho CM, Leevers SJ. Do growth and cell division rates determine cell size in multicellular organisms? J Cell Sci. 2000;113(Pt 17):2927–2934. doi: 10.1242/jcs.113.17.2927. [DOI] [PubMed] [Google Scholar]
- 7.Stocker H, Hafen E. Genetic control of cell size. Curr Opinion Genet Dev. 2000;10:529–535. doi: 10.1016/s0959-437x(00)00123-4. [DOI] [PubMed] [Google Scholar]
- 8.A M. Hunt: The Cell Cycle. Oxford: University Press; 1993. [Google Scholar]
- 9.Zhang J, Del Aguila R, Schneider C, Schneider BL. The importance of being big. J Invest Dermatol. 2005;10:131–141. doi: 10.1111/j.1087-0024.2005.200414.x. [DOI] [PubMed] [Google Scholar]
- 10.Son S, Tzur A, Weng Y, Jorgensen P, Kim J, Kirschner MW, et al. Direct observation of mammalian cell growth and size regulation. Nat Methods. 2012;9:910–912. doi: 10.1038/nmeth.2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rouzaire-Dubois B, Malo M, Milandri JB, Dubois JM. Cell size-proliferation relationship in rat glioma cells. Glia. 2004;45:249–257. doi: 10.1002/glia.10320. [DOI] [PubMed] [Google Scholar]
- 12.Dolznig H, Grebien F, Sauer T, Beug H, Mullner EW. Evidence for a size-sensing mechanism in animal cells. Nat Cell Biol. 2004;6:899–905. doi: 10.1038/ncb1166. [DOI] [PubMed] [Google Scholar]
- 13.Tzur A, Kafri R, LeBleu VS, Lahav G, Kirschner MW. Cell growth and size homeostasis in proliferating animal cells. Science. 2009;325:167–171. doi: 10.1126/science.1174294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weissman IL. Stem cells: units of development, units of regeneration, and units in evolution. Cell. 2000;100:157–168. doi: 10.1016/s0092-8674(00)81692-x. [DOI] [PubMed] [Google Scholar]
- 15.Matsuoka T, Tavassoli M. Electron microscopic identification of hemopoietic progenitor cells by exploiting their sugar-recognizing receptors using a newly developed minibead technique. Exp Hematol. 1989;17:326–329. [PubMed] [Google Scholar]
- 16.Radley JM, Ellis S, Palatsides M, Williams B, Bertoncello I. Ultrastructure of primitive hematopoietic stem cells isolated using probes of functional status. Exp Hematol. 1999;27:365–369. doi: 10.1016/s0301-472x(98)00017-4. [DOI] [PubMed] [Google Scholar]
- 17.Berardi AC, Wang A, Levine JD, Lopez P, Scadden DT. Functional isolation and characterization of human hematopoietic stem cells. Science. 1995;267:104–108. doi: 10.1126/science.7528940. [DOI] [PubMed] [Google Scholar]
- 18.Vacanti MP, Roy A, Cortiella J, Bonassar L, Vacanti CA. Identification and initial characterization of spore-like cells in adult mammals. J Cell Biochem. 2001;80:455–460. [PubMed] [Google Scholar]
- 19.Young HE, Duplaa C, Yost MJ, Henson NL, Floyd JA, Detmer K, et al. Clonogenic analysis reveals reserve stem cells in postnatal mammals. II. Pluripotent epiblastic-like stem cells. Anat Records Part A. 2004;277:178–203. doi: 10.1002/ar.a.20000. [DOI] [PubMed] [Google Scholar]
- 20.Zuba-Surma EK, Kucia M, Abdel-Latif A, Dawn B, Hall B, Singh R, et al. Morphological characterization of very small embryonic-like stem cells (VSELs) by ImageStream system analysis. J Cell Mol Med. 2008;12:292–303. doi: 10.1111/j.1582-4934.2007.00154.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Virant-Klun I, Zech N, Rozman P, Vogler A, Cvjeticanin B, Klemenc P, et al. Putative stem cells with an embryonic character isolated from the ovarian surface epithelium of women with no naturally present follicles and oocytes. Differentiation. 2008;76:843–856. doi: 10.1111/j.1432-0436.2008.00268.x. [DOI] [PubMed] [Google Scholar]
- 22.Smith JR, Pochampally R, Perry A, Hsu SC, Prockop DJ. Isolation of a highly clonogenic and multipotential subfraction of adult stem cells from bone marrow stroma. Stem Cells. 2004;22:823–831. doi: 10.1634/stemcells.22-5-823. [DOI] [PubMed] [Google Scholar]
- 23.Hung SC, Chen NJ, Hsieh SL, Li H, Ma HL, Lo WH. Isolation and characterization of size-sieved stem cells from human bone marrow. Stem Cells. 2002;20:249–258. doi: 10.1634/stemcells.20-3-249. [DOI] [PubMed] [Google Scholar]
- 24.D'Ippolito G, Howard GA, Roos BA, Schiller PC. Sustained stromal stem cell self-renewal and osteoblastic differentiation during aging. Rejuvenation Res. 2006;9:10–19. doi: 10.1089/rej.2006.9.10. [DOI] [PubMed] [Google Scholar]
- 25.Gao FB, Raff M. Cell size control and a cell-intrinsic maturation program in proliferating oligodendrocyte precursor cells. J Cell Biol. 1997;138:1367–1377. doi: 10.1083/jcb.138.6.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrandon Y, Green H. Cell size as a determinant of the clone-forming ability of human keratinocytes. Proc Natl Acad Sci USA. 1985;82:5390–5394. doi: 10.1073/pnas.82.16.5390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Angello JC, Pendergrass WR, Norwood TH, Prothero J. Proliferative potential of human fibroblasts: an inverse dependence on cell size. J Cell Physiol. 1987;132:125–130. doi: 10.1002/jcp.1041320117. [DOI] [PubMed] [Google Scholar]
- 28.Parsa R, Yang A, McKeon F, Green H. Association of p63 with proliferative potential in normal and neoplastic human keratinocytes. J Invest Dermatol. 1999;113:1099–1105. doi: 10.1046/j.1523-1747.1999.00780.x. [DOI] [PubMed] [Google Scholar]
- 29.Dazard JE, Piette J, Basset-Seguin N, Blanchard JM, Gandarillas A. Switch from p53 to MDM2 as differentiating human keratinocytes lose their proliferative potential and increase in cellular size. Oncogene. 2000;19:3693–3705. doi: 10.1038/sj.onc.1203695. [DOI] [PubMed] [Google Scholar]
- 30.Tseng H, Green H. Association of basonuclin with ability of keratinocytes to multiply and with absence of terminal differentiation. J Cell Biol. 1994;126:495–506. doi: 10.1083/jcb.126.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Watt FM, Green H. Involucrin synthesis is correlated with cell size in human epidermal cultures. J Cell Biol. 1981;90:738–742. doi: 10.1083/jcb.90.3.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.De Paiva CS, Pflugfelder SC, Li DQ. Cell size correlates with phenotype and proliferative capacity in human corneal epithelial cells. Stem Cells. 2006;24:368–375. doi: 10.1634/stemcells.2005-0148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Banks-Schlegel S, Green H. Involucrin synthesis and tissue assembly by keratinocytes in natural and cultured human epithelia. J Cell Biol. 1981;90:732–737. doi: 10.1083/jcb.90.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gandarillas A, Davies D, Blanchard JM. Normal and c-Myc-promoted human keratinocyte differentiation both occur via a novel cell cycle involving cellular growth and endoreplication. Oncogene. 2000;19:3278–3289. doi: 10.1038/sj.onc.1203630. [DOI] [PubMed] [Google Scholar]
- 35.Majore I, Moretti P, Hass R, Kasper C. Identification of subpopulations in mesenchymal stem cell-like cultures from human umbilical cord. Cell Commun Signal. 2009;7:6. doi: 10.1186/1478-811X-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hall SR, Jiang Y, Leary E, Yavanian G, Eminli S, O'Neill DW, et al. Identification and isolation of small CD44-negative mesenchymal stem/progenitor cells from human bone marrow using elutriation and polychromatic flow cytometry. Stem Cells Tranl Med. 2013;2:567–578. doi: 10.5966/sctm.2012-0155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sharma S, Cabana R, Shariatmadar S, Krishan A. Cellular volume and marker expression in human peripheral blood apheresis stem cells. Cytometry Part A. 2008;73:160–167. doi: 10.1002/cyto.a.20524. [DOI] [PubMed] [Google Scholar]
- 38.Ratajczak MZ, Zuba-Surma EK, Wysoczynski M, Wan W, Ratajczak J, Wojakowski W, et al. Hunt for pluripotent stem cell -- regenerative medicine search for almighty cell. J. Autoimmun. 2008;30:151–162. doi: 10.1016/j.jaut.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Colter DC, Class R, DiGirolamo CM, Prockop DJ. Rapid expansion of recycling stem cells in cultures of plastic-adherent cells from human bone marrow. Proc Natl Acad Sci USA. 2000;97:3213–3218. doi: 10.1073/pnas.070034097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Corradetti B, Lange-Consiglio A, Barucca M, Cremonesi F, Bizzaro D. Size-sieved subpopulations of mesenchymal stem cells from intervascular and perivascular equine umbilical cord matrix. Cell Prolif. 2011;44:330–342. doi: 10.1111/j.1365-2184.2011.00759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katsube Y, Hirose M, Nakamura C, Ohgushi H. Correlation between proliferative activity and cellular thickness of human mesenchymal stem cells. Biochem Biophys Res Commun. 2008;368:256–260. doi: 10.1016/j.bbrc.2008.01.051. [DOI] [PubMed] [Google Scholar]
- 42.Kucia M, Reca R, Campbell FR, Zuba-Surma E, Majka M, Ratajczak J, et al. A population of very small embryonic-like (VSEL) CXCR4(+)SSEA-1(+)Oct-4+ stem cells identified in adult bone marrow. Leukemia. 2006;20:857–869. doi: 10.1038/sj.leu.2404171. [DOI] [PubMed] [Google Scholar]
- 43.Kucia M, Halasa M, Wysoczynski M, Baskiewicz-Masiuk M, Moldenhawer S, Zuba-Surma E, et al. Morphological and molecular characterization of novel population of CXCR4+ SSEA-4+ Oct-4+ very small embryonic-like cells purified from human cord blood: preliminary report. Leukemia. 2007;21:297–303. doi: 10.1038/sj.leu.2404470. [DOI] [PubMed] [Google Scholar]
- 44.Ratajczak J, Wysoczynski M, Zuba-Surma E, Wan W, Kucia M, Yoder MC, et al. Adult murine bone marrow-derived very small embryonic-like stem cells differentiate into the hematopoietic lineage after coculture over OP9 stromal cells. Exp Hematol. 2011;39:225–237. doi: 10.1016/j.exphem.2010.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Miyanishi M1, Mori Y, Seita J, Chen JY, Karten S, Chan CK, et al. Do pluripotent stem cells exist in adult mice as very small embryonic stem cells? Stem Cell Reports. 2013;1:198–208. doi: 10.1016/j.stemcr.2013.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Machado HL, Kittrell FS, Edwards D, White AN, Atkinson RL, Rosen JM, et al. Separation by cell size enriches for mammary stem cell repopulation activity. Stem Cell Transl Med. 2013;2:199–203. doi: 10.5966/sctm.2012-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tang DG. Understanding cancer stem cell heterogeneity and plasticity. Cell Res. 2012;22:457–472. doi: 10.1038/cr.2012.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 49.Aravalli RN, Behnan Sahin M, Cressman EN, Steer CJ. Establishment and characterization of a unique 1 microm diameter liver-derived progenitor cell line. Biochem Biophys Res Commun. 2010;391:56–62. doi: 10.1016/j.bbrc.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 50.Bortolomai I, Canevari S, Facetti I, De Cecco L, Castellano G, Zacchetti A, et al. Tumor initiating cells: development and critical characterization of a model derived from the A431 carcinoma cell line forming spheres in suspension. Cell Cycle. 2010;9:1194–1206. doi: 10.4161/cc.9.6.11108. [DOI] [PubMed] [Google Scholar]
- 51.Srivastava VK, Nalbantoglu J. Flow cytometric characterization of the DAOY medulloblastoma cell line for the cancer stem-like phenotype. Cytometry Part A. 2008;73:940–948. doi: 10.1002/cyto.a.20633. [DOI] [PubMed] [Google Scholar]
- 52.Zhang S, Mercado-Uribe I, Xing Z, Sun B, Kuang J, Liu J. Generation of cancer stem-like cells through the formation of polyploid giant cancer cells. Oncogene. 2014;33:116–128. doi: 10.1038/onc.2013.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Patrawala L, et al. Side population (SP) is enriched in tumorigenic, stem-like cancer cells whereas ABCG2+ and ABCG2− cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- 54.Patrawala L, et al. Highly purified CD44+ prostate cancer cells from xenograft human tumors are enriched in tumorigenic and metastatic progenitor cells. Oncogene. 2006;25:1696–1708. doi: 10.1038/sj.onc.1209327. [DOI] [PubMed] [Google Scholar]
- 55.Patrawala L, Calhoun-Davis T, Schneider-Broussard R, Tang DG. Hierarchical organization of prostate cancer cells in xenograft tumors: The CD44+α2β1+ cell population is enriched in tumor-initiating cells. Cancer Res. 2007;67:6796–6805. doi: 10.1158/0008-5472.CAN-07-0490. [DOI] [PubMed] [Google Scholar]
- 56.Tang DG, et al. Prostate cancer stem/progenitor cells: Identification, characterization, and implications. Mol. Carcinogenesis. 2007;46:1–14. doi: 10.1002/mc.20255. [DOI] [PubMed] [Google Scholar]
- 57.Li HW, Chen X, Calhoun-Davis T, Claypool K, Tang DG. PC3 Human prostate carcinoma cell holoclones contain self-renewing tumor-initiating cells. Cancer Res. 2008;68:1820–1825. doi: 10.1158/0008-5472.CAN-07-5878. [DOI] [PubMed] [Google Scholar]
- 58.Li HW, et al. Methodologies in assaying prostate cancer stem cells. Methods Mol Biol. 2009;569:85–138. doi: 10.1007/978-1-59745-280-9_7. [DOI] [PubMed] [Google Scholar]
- 59.Liu C, et al. The microRNA miR-34a inhibits prostate cancer stem cells and metastasis by directly repressing CD44. Nature Med. 2011;17:211–215. doi: 10.1038/nm.2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qin J, et al. The PSA−/lo prostate cancer cell population harbors self-renewing long-term tumor-propagating cells that resist castration. Cell Stem Cell. 2012;10:556–569. doi: 10.1016/j.stem.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jeter CR, Yang T, Wang J, Chao H, Tang DG. Nanog in cancer stem cells and tumor development: An update and outstanding questions. Stem Cells. 2015 doi: 10.1002/stem.2007. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149:274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li Q, Dang CV. c-Myc overexpression uncouples DNA replication from mitosis. Mol Cell Biol. 1999;19:5339–5351. doi: 10.1128/mcb.19.8.5339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Johnston LA, Prober DA, Edgar BA, Eisenman RN, Gallant P. Drosophila myc regulates cellular growth during development. Cell. 1999;98:779–790. doi: 10.1016/s0092-8674(00)81512-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Iritani BM, Eisenman RN. c-Myc enhances protein synthesis and cell size during B lymphocyte development. Proc Natl Acad Sci USA. 1999;96:13180–13185. doi: 10.1073/pnas.96.23.13180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zanet J, Pibre S, Jacquet C, Ramirez A, de Alboran IM, Gandarillas A. Endogenous Myc controls mammalian epidermal cell size, hyperproliferation, endoreplication and stem cell amplification. J Cell Sci. 2005;118:1693–1704. doi: 10.1242/jcs.02298. [DOI] [PubMed] [Google Scholar]
- 67.Tumaneng K, Russell RC, Guan KL. Organ size control by Hippo and TOR pathways. Curr Biol. 2012;22:R368–R379. doi: 10.1016/j.cub.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tumaneng K, Schlegelmilch K, Russell RC, Yimlamai D, Basnet H, Mahadevan N, et al. YAP mediates crosstalk between the Hippo and PI(3)K-TOR pathways by suppressing PTEN via miR-29. Nat Cell Biol. 2012;14:1322–1329. doi: 10.1038/ncb2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trendelenburg AU, Meyer A, Rohner D, Boyle J, Hatakeyama S, Glass DJ. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am J Physiol. 2009;296:C1258–C1270. doi: 10.1152/ajpcell.00105.2009. [DOI] [PubMed] [Google Scholar]
- 70.Andang M, Hjerling-Leffler J, Moliner A, Lundgren TK, Castelo-Branco G, Nanou E, et al. Histone H2AX-dependent GABA(A) receptor regulation of stem cell proliferation. Nature. 2008;451:460–464. doi: 10.1038/nature06488. [DOI] [PubMed] [Google Scholar]
- 71.Abdelhady S, Kitambi SS, Lundin V, Aufschnaiter R, Sekyrova P, Sinha I, et al. Erg channel is critical in controlling cell volume during cell cycle in embryonic stem cells. PloS one. 2013;8:e72409. doi: 10.1371/journal.pone.0072409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Song Y, Lu B. Regulation of cell growth by Notch signaling and its differential requirement in normal vs. tumor-forming stem cells in Drosophila. Genes & Dev. 2011;25:2644–2658. doi: 10.1101/gad.171959.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Demidenko ZN, Blagosklonny MV. Growth stimulation leads to cellular senescence when the cell cycle is blocked. Cell Cycle. 2008;7:3355–3361. doi: 10.4161/cc.7.21.6919. [DOI] [PubMed] [Google Scholar]