Abstract
Cerebral cavernous malformations (CCM) are vascular dysplasias that usually occur in the brain and are associated with mutations in the KRIT1/CCM1, CCM2/MGC4607/OSM/Malcavernin, and PDCD10/CCM3/TFAR15 genes. Here we report the 2.9 Å crystal structure of the ankyrin repeat domain (ARD) and FERM domain of the protein product of KRIT1 (KRIT1; Krev interaction trapped 1). The crystal structure reveals that the KRIT1 ARD contains 4 ankyrin repeats. There is an unusual conformation in the ANK4 repeat that is stabilized by Trp-404, and the structure reveals a solvent exposed ankyrin groove. Domain orientations of the three copies within the asymmetric unit suggest a stable interaction between KRIT1 ARD and FERM domains, indicating a globular ARD-FERM module. This resembles the additional F0 domain found N-terminal to the FERM domain of talin. Structural analysis of KRIT1 ARD-FERM highlights surface regions of high evolutionary conservation, and suggests potential sites that could mediate interaction with binding partners. The structure therefore provides a better understanding of KRIT1 at the molecular level.
Keywords: crystal structure, X-ray crystallography, ankyrin repeat, protein complex, Cerebral Cavernous Malformations
1. Introduction
Cerebral cavernous malformations (CCM) are vascular dysplasias that usually occur in the brain and have been found in up to 0.5 % of the human population (Otten et al., 1989). Sporadic occurrence of the disease is mostly associated with a single lesion (Fischer et al., 2013), and can lead to hemorrhagic stroke, focal neurological defects and sometimes death. Familial occurrence of the disease is more serious than the sporadic form. Familial cases are associated with multiple lesions throughout life, and can have devastating effects (Cavalcanti et al., 2012). The biological basis for this disease is still not definitively defined, although in recent years there have been significant advances in understanding the basic signaling pathways and mechanisms that are disrupted in the disease (Draheim et al., 2014; Draheim et al., 2015; Faurobert et al., 2013; Fischer et al., 2013; Fisher and Boggon, 2014; Fisher et al., 2015b; Maddaluno et al., 2013; Zhou et al., 2015). Paramount to these studies were the discoveries that almost all familial CCM patients were heterozygous for mutations in one of three genes, termed KRIT1/CCM1 (Laberge-le Couteulx et al., 1999; Sahoo et al., 1999), CCM2/MGC4607/OSM/Malcavernin (Liquori et al., 2003), and PDCD10/CCM3/TFAR15 (Bergametti et al., 2005; Guclu et al., 2005). In these patients, lesions are associated with a ‘second hit’ mutation that usually results in ablated expression of the resulting protein (Akers et al., 2009; Gault et al., 2005; Pagenstecher et al., 2009). It is therefore clinically and biologically important to understand the functional roles of the protein products of these three genes, and why disruption of their expression results in destabilized neurovasculature.
The three proteins expressed by KRIT1, CCM2, and PDCD10 genes are termed KRIT1 (Krev interaction trapped 1; cerebral cavernous malformations 1, CCM1), CCM2 (cerebral cavernous malformations 2; osmosensing scaffold for MEKK3, OSM) and CCM3 (cerebral cavernous malformations 3; programmed cell death 10, PDCD10). These protein products are generally considered scaffolding proteins and can directly interact with one another (Draheim et al., 2015; Fisher and Boggon, 2014; Fisher et al., 2015a). Structural studies have been conducted for CCM2 and CCM3 that discovered novel domains in both proteins (Fisher et al., 2013; Li et al., 2010), and the structural basis for CCM2 and CCM3 interactions with binding partners (Draheim et al., 2015; Fisher et al., 2015b; Li et al., 2011; Xu et al., 2013; Zhang et al., 2013). Similarly, structural studies have been conducted for KRIT1, and have facilitated the discovery of a Nudix fold domain at the KRIT1 N-terminus (Liu et al., 2013), a novel PTB-binding site which interacts with the protein ICAP1 (integrin cytoplasmic associated protein 1) (Liu and Boggon, 2013; Liu et al., 2013), the mode of interactions with CCM2 (Fisher et al., 2015a) and (SNX17) sorting nexin 17 (Stiegler et al., 2014), and the distinct and unusual manner by which the C-terminal FERM (band 4-point-1, ezrin, radixin, moesin) domain binds to the small GTPase Rap1 (Ras related protein Rap1) and the cell adhesion molecule HEG1 (Heart of Glass 1) (Gingras et al., 2012; Gingras et al., 2013; Li et al., 2012). These structural studies have therefore provided insights into the molecular basis for the CCM complex proteins and their interactions (Fisher and Boggon, 2014), however gaps still remain in our structural understanding of KRIT1. The domain assignments of this protein include the crystallographically defined N-terminal Nudix domain, three NPxY/F motifs, a predicted ankyrin repeat domain (ARD) and a C-terminal FERM domain (Fig. 1A). The ARD and FERM domain folds are well described as mediators of protein-protein interactions (Frame et al., 2010; Sedgwick and Smerdon, 1999), however, until now, no structure contains the KRIT1 ARD. We therefore considered whether crystallographic studies could reveal the structure of the KRIT1 ARD, help us better understand how KRIT1 folds, and improve our understanding of KRIT1’s biological function.
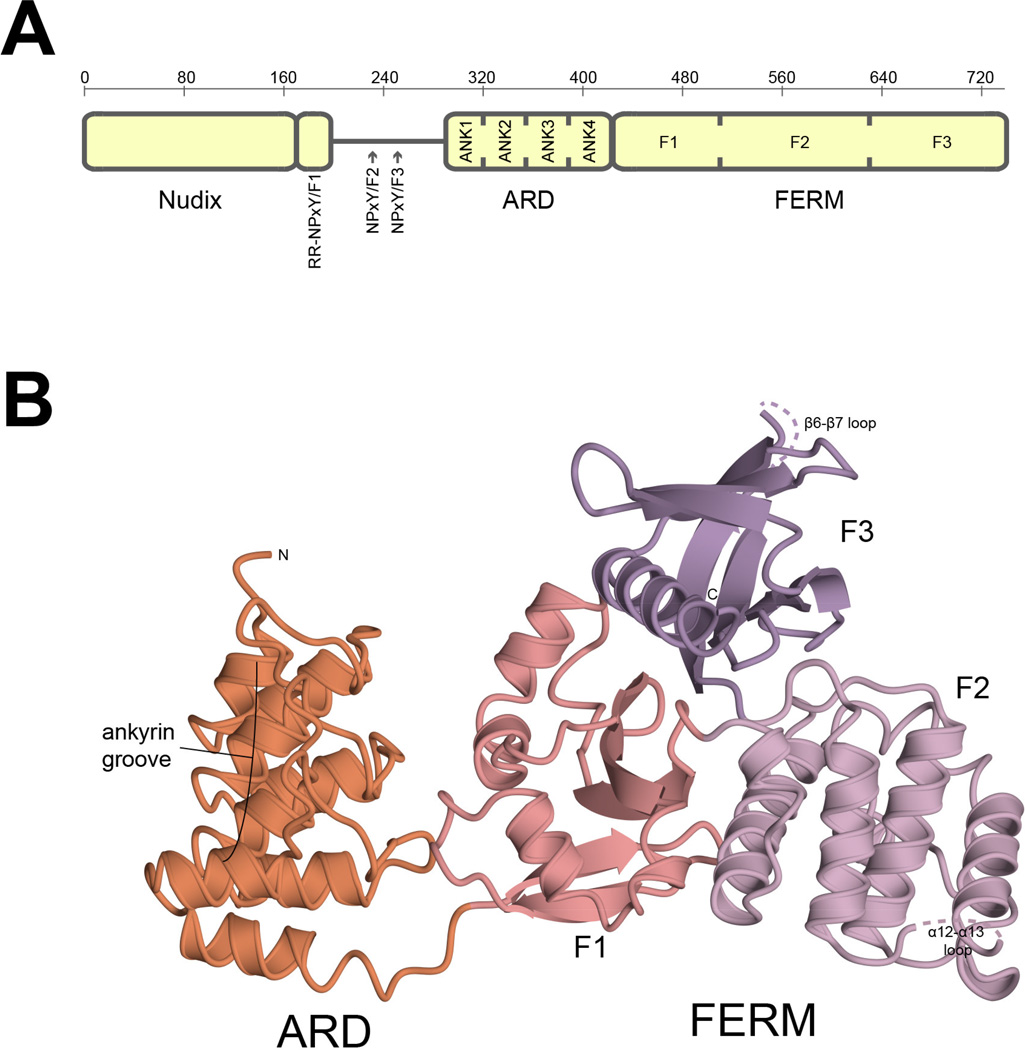
Figure 1. Schematic showing domain assignments for KRIT1 and cartoon diagram showing the structure of KRIT1ARD-FERM.
A) Schematic showing domain assignments for KRIT1. Amino acid numbering is indicated above. NPxY motifs are indicated by RR-NPxY/F1, NPxY/F2 and NPxY/F3. ARD indicates ankyrin repeat domain. ANK1-ANK4 indicate ankyrin repeats 1 through 4. FERM indicates band four-point-one, ezrin, radixin, moesin domain. F1, F2 and F3 indicate FERM subdomains. B) Overall structure of KRIT1ARD-FERM shown in cartoon format. The ankyrin repeat domain is colored orange and is indicated ARD. The band four-point-one, ezrin, radixin domain is indicated FERM and colored by sub-domain, F1 is salmon, F2 is lilac and F3 is lavender. The location of the ankyrin groove is indicated.
We report the crystal structure of human KRIT1 ankyrin repeat and FERM domains to 2.9 Å. This is the first crystal structure containing KRIT1 ARD. We find that KRIT1 contains four ankyrin repeats and that the ARD packs against KRIT1 FERM domain. This conformation is maintained over the three copies in the asymmetric unit, and because of its relationship to the FERM domain is reminiscent of the F0 domain found in talin. We therefore propose that the KRIT1 ARD is an F0-like domain. We also find that the ankyrin groove of the KRIT1 ARD is exposed and oriented away from the FERM domain, with a hydrophobic cleft and a conserved tryptophan residue (Trp-330) marking the center of the grove. We propose that the KRIT1 ARD is poised for protein-protein interactions at this site.
2. Materials and Methods
2.1. Sub-cloning, expression and purification of the ankyrin repeat and FERM domains of human KRIT1
Human KRIT1 (amino acids 259–736)(termed KRIT1ARD-FERM) (Uniprot ID: O00522) was subcloned into a modified pET32 vector with an N-terminal hexahistidine-tag. Recombinant KRIT1 was expressed in Escherichia coli Rosetta (DE3) cells (Novagen) and induced with 0.6 mM IPTG (OD600 0.6) overnight at 18 °C. Cells were harvested and resuspended in lysis buffer (20 mM Tris pH 8.0, 500 mM NaCl, 20 mM imidazole 10% glycerol and 1% triton X100) with protease inhibitors, lysozyme (100 µg/ml) and DNase I (10 µg/ml). Three cycles of freeze/thaw were conducted using ethanol/dry ice bath and cells were sonicated in 50 ml falcon tube for 2 minutes total. The samples were clarified by centrifugation at 20,000 rpm for 1 hour at 4 °C. Supernatant was applied to a 1 ml HisTrap (GE) nickel affinity column, and protein eluted with 5 ml lysis buffer supplemented with 400 mM imidazole. To cleave the affinity tag 300 µl of TEV protease (~1 mg/ml stock) was added to the protein samples, and samples were transferred to dialysis tubing and dialyzed overnight against 1 L lysis buffer at 4 °C. After the overnight dialysis, the sample was re-applied to a HisTrap column and the flow-through collected. Following filtration through a 0.2 µm filter, the protein sample was applied to a Superdex 200 (GE) preparative gel filtration column. Fractions were collected and analyzed by SDS-PAGE.
2.2. Crystallization of KRIT1ARD-FERM
For crystallization, KRIT1 was concentrated to 2.4 mg/ml in 20 mM Tris pH 8.0, 250 mM NaCl with 2 mM TCEP. Sparse matrix and grid screens were conducted, and an initial hit was obtained from JCSG screen (Qiagen) against precipitant conditions 0.1 M HEPES pH 7.5, 10% PEG 8000, 8% Ethylene glycol. Crystallization was optimized by grid screening and the best crystals were obtained overnight at room temperature using hanging drop vapor diffusion methodology. 2 µl drops containing 1 µl protein and 1 µl precipitant were used to obtain optimal crystals against precipitant conditions of 0.05 M HEPES pH 7.3, 8% ethylene glycol, 8% PEG 8000. Crystals were cryoprotected by supplementing the precipitant with 10% glycerol and flash freezing in liquid nitrogen.
2.3. Data collection and structure determination
180° of data were collected from a single crystal at beamline 24-ID-E (NECAT) at the Advanced Photon Source. Data were integrated and scaled using the HKL package (Otwinowski and Minor, 1997). The structure was determined by molecular replacement using Phaser (McCoy et al., 2007). Matthews coefficient analysis suggested three copies per asymmetric unit and a solvent content of 61%. Two domains were searched for, the FERM domain of KRIT1 (PDB ID 4DXA)(Li et al., 2012) and residues 39 to 158 of an ankyrin repeat domain with high sequence identity (40% over 123 residues) to the predicted KRIT1 ARD region (4HB5, unpublished). Three copies of the KRIT1 FERM domain were found with final TFZ scores of 15.1, 39.7, 36.4 and three copies of the ankyrin repeat domain were found with final TFZ scores of 23.8, 21.3 and 17.3. The initial R/Rfree following the first round of refinement in Refmac5 (Murshudov et al., 2011) was 43.0/42.3. Following sequence correction and standard rounds of model building and refinement using Refmac5 (Murshudov et al., 2011), Phenix using TLS and NCS (Adams et al., 2010) and Coot (Emsley et al., 2010) the final R/Rfree values for the model are 20.6/24.6. The structure is deposited in the PDB under accession code 5D68.
3. Results and Discussion
3.1. Expression and crystallization of KRITARD-FERM
We began by sampling the expression and purification of KRIT1 constructs to determine the structure of KRIT1 ARD. We found that we could express and purify a construct of KRIT1 that encompassed both the predicted ARD and the FERM domain (residues 259–736) (KRIT1ARD-FERM) and that this protein could be purified to crystallization quality and concentrated to approximately 2.4 mg/ml. On conducting extensive crystallization trials with KRIT1ARD-FERM we were able to obtain diffraction-quality crystals and collected X-ray diffraction data to 2.9 Å resolution. We determined the structure by molecular replacement using the KRIT1 FERM domain (Li et al., 2012) and an engineered ankyrin repeat protein with high sequence similarity (40% identity over 123 residues) to the KRIT1 predicted ARD (PDB ID: 4HB5). We deposited the final structure in the Protein Data Bank under accession code 5D68. Data collection and refinement statistics are shown in Table 1.
Table 1.
Data collection and refinement statistics for KRIT1 ARD-FERM.
| Data Collection | |
| Space Group | C2 |
| X-ray source | APS 24-ID-E |
| Cell dimensions, a, b, c (Å) | 125.7, 80.2, 213.1 |
| α, β, γ (°) | 90, 91.7, 90 |
| Wavelength (Å) | 0.97918 |
| Resolution range (Å)* | 50 – 2.9 (3.0 – 2.9) |
| No. unique reflections | 46892 (4654) |
| Degrees of data (°) | 180 |
| Completeness (%)* | 100 (100) |
| Rpim (%)*+ | 11.6 (53.8) |
| I/σ(I)* | 7.5 (1.7) |
| Redundancy* | 3.7 (3.8) |
| Wilson B-factor (Å2) | 42.4 |
| Refinement | |
| Resolution Range (Å)* | 49.37 – 2.91 (2.97 – 2.91) |
| Rfactor (%)* | 20.6 (29.9) |
| Free Rfactor (%)* | 24.6 (29.9) |
| No. Free R reflections* | 2376 (127) |
| Free R reflections (%)* | 5.1 (5.0) |
| Residue range built | A/ 288–610, 615–647, 654–730 B/ 288–610,615–646,655–736 C/ 288–610,615–646, 654–729 |
| No. non-hydrogen protein atoms | 10652 |
| No. water molecules | 66 |
| Model Quality | |
| RMSD bond lengths (Å) | 0.004 |
| RMSD bond angles (°) | 0.788 |
| Overall B (all atoms) | 46.0 |
| Ramachandran plot (%) Favored/Allowed/Outliers |
97 / 3 / 0 |
| MolProbity clashscore/Percentile | 2.83/100th percentile |
Parentheses indicate highest resolution shell
Rpim indicates ‘Precision-indicating merging R factor’, a measure of the precision of the averaged intensity measurements (Weiss, 2001), where Rpim = •hkl [1/(N − 1)]½ •j |Ihkl,j − 〈Ihkl〉| / •hkl •j Ihkl,j
3.2. Overall structure
The 2.9 Å structure of KRIT1ARD-FERM shows that KRIT1 contains four ankyrin repeats that fold into an ankyrin repeat domain (ARD) between residues 288 and 419. It also shows that the KRIT1 FERM domain maintains its trefoil composition of F1 (ubiquitin-like), F2 (acyl-CoA-binding protein) and F3 (phosphotyrosine binding/pleckstrin homology) fold subdomains (Li et al., 2012) between residues 420 and 736 in the context of a longer construct (Fig. 1B)(Table 1). The overall structure comprises 3 copies of KRIT1ARD-FERM in the asymmetric unit. Residues 288 to 729 are visible in the electron density for each of the 3 copies, with portions of the α12–α13 loop (residues 610 to 615) and the β6–β7 loop (residues 646–655) within FERM F2 and F3 lobes, respectively, not observed. Within the crystal lattice there is an interaction between the FERM F2 lobe (helix α12) and the concave surface of the ARD (repeats 1 and 2). This interaction appears to be crystallographic as we do not observe higher-order oligomers on purification of KRIT1ARD-FERM by size exclusion chromatography (not shown). A compact configuration of the KRIT1 FERM domain and ARD occurs in each copy whereby the ARD stacks against the F1-lobe of the FERM domain. The three copies of KRIT1ARD-FERM within the asymmetric unit are experimentally identical with root-mean-square deviations below 0.75 Å over 429 carbon alpha atoms for each of the chain pairings. The structure represents the first visualization of the KRIT1 ARD and how it interacts with the KRIT1 FERM domain.
3.3. The structure of the KRIT1 ankyrin repeat domain
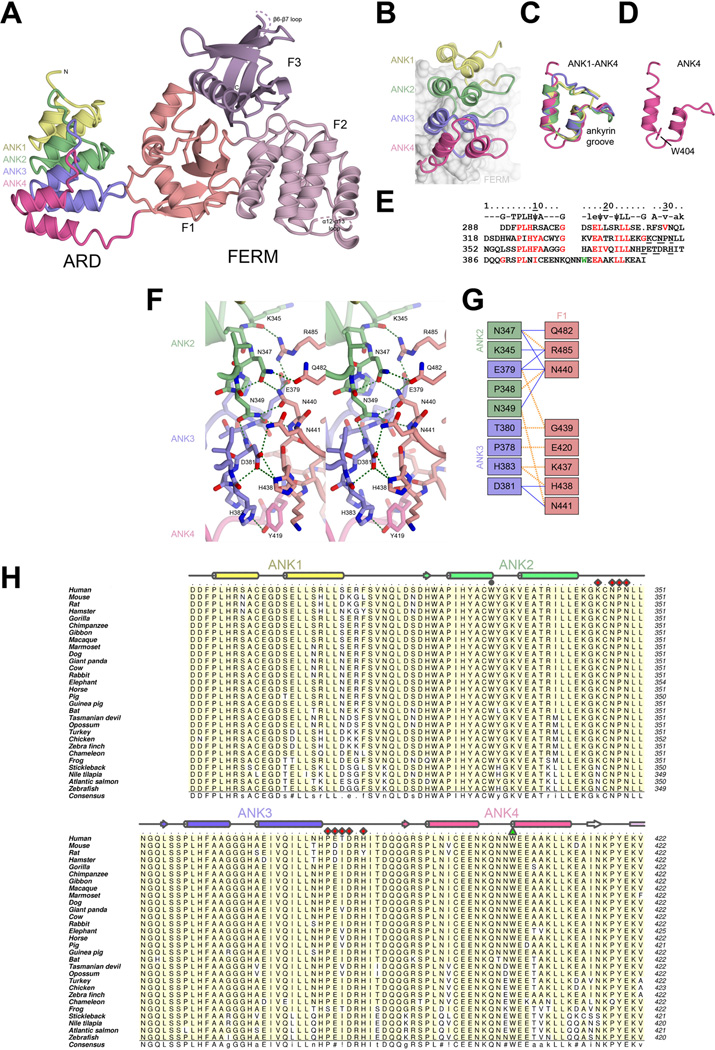
The ARD region of KRIT1 was predicted to contain an ankyrin repeat domain comprising four ankyrin repeats when the KRIT1 gene was identified (Serebriiskii et al., 1997), but until now this has not been confirmed at the structural level. We find that the KRIT1 ARD contains four ankyrin repeats (Fig. 2A). Ankyrin repeats are canonically 33 amino acids in length and comprise two α-helices and a β-hairpin that links to the next repeat. The repeats stack on top of one another in an ‘L’-like fold. Ankyrin repeat domains usually function in protein recognition by interacting with binding partners using the concave ankyrin groove between the ‘palm’ (formed by the α-helices) and the ‘fingers’ (formed by the β-hairpins)(Chiswell et al., 2008; Sedgwick and Smerdon, 1999). The KRIT1 ARD follows the canonical fold, with each of the repeats comprising the expected α-helices and β-hairpin-like secondary structure elements. The fourth ankyrin repeat, ANK4, however, contains an insert between its α-helices comprising three residues, Gln-401, Asn-402 and Asn-403. This insert seems to be stabilized by the highly conserved Trp-404, which interposes between the α-helices and may be required for this particular conformation (Fig. 2B–E).
Figure 2. Structure of KRIT1 ARD-FERM.
A) Overall structure of KRIT1 ARD-FERM shown in cartoon format. ARD is colored showing each ankyrin repeat (ANK1–ANK4). FERM domain colored as per Figure 1. B–E) Analysis of the KRIT1 ARD. B) Close-up of the ARD showing each ankyrin repeat. The FERM domain is shown as a surface in the background. C) Superposition of each of the ankyrin repeats. D) The fourth ankyrin repeat alone, showing residue W404 which stabilizes an insert (Q401, N402, N403). E) Alignment of KRIT1 ankyrin repeats. Top row shows consensus ankyrin repeat sequence with well conserved ankyrin repeat residues indicated by capitals, moderately conserved ankyrin repeat residues in lower case, and hydrophobic residues in the consensus indicated by ψ (Mosavi et al., 2004; Sedgwick and Smerdon, 1999). KRIT1 residues that conform to the consensus are colored red. W404 is colored green. Residues that interact with the FERM domain are underlined. F) Stereoview of the intramolecular interaction between KRIT1 ARD and FERM domains. ARD ankyrin repeats colored as in panel B, FERM F1 colored as in panel A. Hydrogen bonds are indicated in green. Interacting residues are shown in stick format with oxygen atoms colored red and nitrogen atoms colored blue. G) Schematic map of the ARD-FERM interface. Hydrophobic interactions are indicated by orange dashed lines and hydrogen bonds by solid blue lines. H) Sequence alignment of KRIT1 ARD. Secondary structure elements colored per ankyrin repeat with α-helices shown as cylinders and β-strands as arrows. ARD residues that interact with KRIT1 FERM domain are indicated with red diamonds, and W404 is shown with a green triangle. W330 is indicated with a grey circle. Uniprot or GenBank accession number are followed by the Latin name and label in parentheses: O00522 Homo sapiens (Human), Q6S5J6 Mus musculus (Mouse), B5DF47 Rattus norvegicus (Rat), XP_003496996 Cricetulus griseus (Hamster), G3QHP7 Gorilla gorilla (Gorilla), XP_001166592 Pan troglodytes (Vhimpanzee), G1RZ72 Nomascus leucogenys (Gibbon), F6TVZ5 Macaca mulatta (Macaque), F7I3T9 Callithrix jacchus (Marmoset), E2RAA5 Canis familiaris (Dog), XP_002925462 Ailuropoda melanoleuca (Giant panda), Q6TNJ1 Bos taurus (Cow), G1SM20 Oryctolagus cuniculus (Rabbit), G3TAH0 Loxodonta africana (Elephant), XP_001491579 Equus caballus (Horse), F1SFD0 Sus scrofa (Pig), XP_003475143 Cavia porcellus (Guinea pig), G1PWA1 Myotis lucifugus (Bat), G3X3N7 Sarcophilus harrisii (Tasmanian devil), F7CJF5 Monodelphis domestica (Opossum), G1N921 Meleagris gallopavo (Turkey), NP_001026144 Gallus gallus (Chicken), XP_002196126 Taeniopygia guttata (Zebra finch), G1KRC6 Anolis carolinensis (Chameleon), F7CJC5 Xenopus tropicalis (Frog), G3NJI7 Gasterosteus aculeatus (Stickleback), XP_003452511 Oreochromis niloticus (Nile tilapia), C0H9F0 Salmo salar (Atlantic salmon), B8JIZ5 Danio rerio (Zebrafish). Alignment was conducted using ClustalW (Larkin et al., 2007). Figure was generated using Aline (Bond and Schuttelkopf, 2009). Interacting residues were defined using the PISA server (Krissinel and Henrick, 2007).
3.4. Intramolecular interaction between KRIT1 ARD and KRIT1 FERM domains
In the structure an unexpected interface is observed between KRIT1 ARD and KRIT1 FERM domain. This interaction is mediated by the convex surface of ankyrin repeats 2 and 3 and predominantly the region between strand β2 and helix α2 of the F1 lobe of the FERM domain. The interaction buries on average 993 Å2 (503 Å2 from the ARD and 490 Å2 from the FERM domain) (calculated using PISA server (Krissinel and Henrick, 2007)). It is a mostly polar interaction, comprising 10–12 hydrogen bonds (Figure 2F,G) and is well conserved over evolution (Figure 2H and (Li et al., 2012)). The compactness of the ARD-FERM interface and the consistence of this arrangement over the three copies found in the asymmetric unit suggest that the ARD could be considered a ‘F0’ lobe of the KRIT1 FERM domain, similar to F0 domains observed in talin and kindlin (Goult et al., 2009; Goult et al., 2010).
3.5. Comparison with other KRIT1 structures
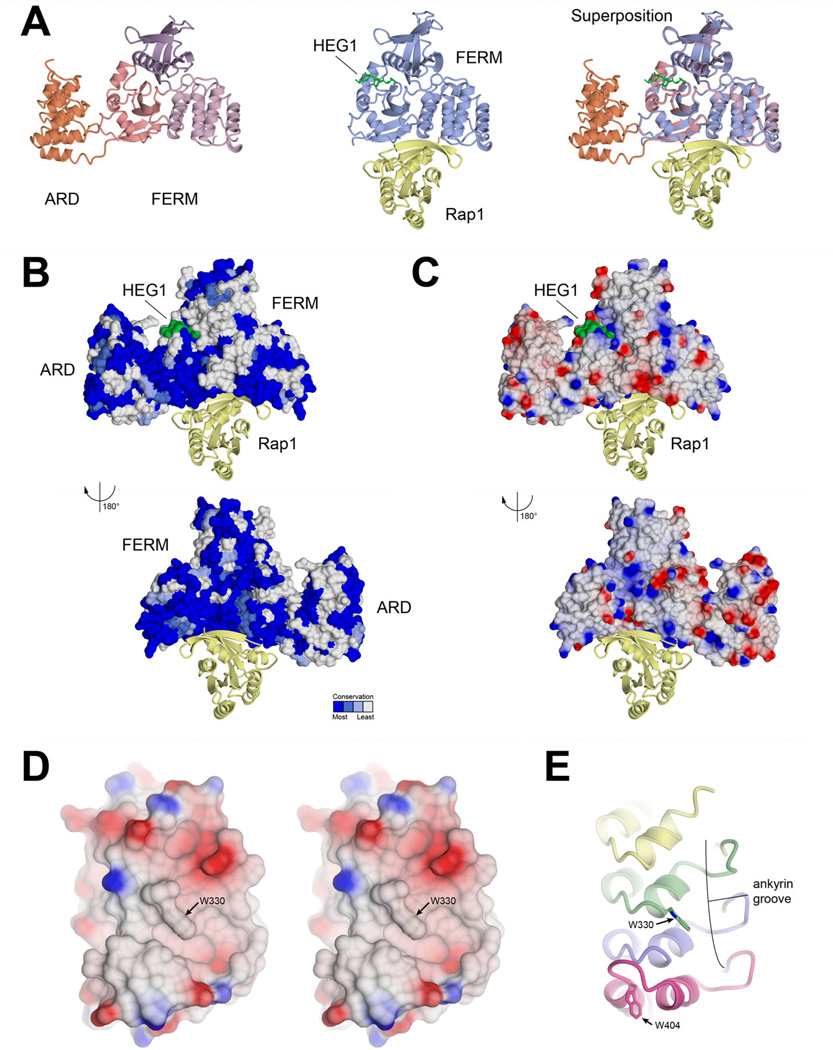
The evolutionary conservation of KRIT1 has the potential to indicate regions of the protein important for intermolecular interactions. Previously, we demonstrated that the interaction between KRIT1 and Rap1 small GTPase takes place using a highly conserved surface (Li et al., 2012). We therefore superposed the current structure with the previously determined structures of KRIT1 in complex with its binding partners Rap1 and HEG1 to investigate potential binding sites for other partners (Gingras et al., 2012; Gingras et al., 2013; Li et al., 2012). We find that, based on the superpositions, no conformational change is required in KRIT1 ARD to accommodate binding to Rap1 (Fig 3A). We also find that structural comparison of each of the FERM domains from the different complexes yields maximal root-mean-squared deviations of 0.72 Å over 297 carbon alpha residues, indicating very little conformational change on either inclusion of the ARD or binding of Rap1 or HEG1.
Figure 3. Structural insight into KRIT1 interactions with binding partners.
A) Superposition on r.h.s. of KRIT1ARD-FERM, shown on l.h.s., with KRIT1 FERM domain in complex with Rap1 small GTPase and Heart of Glass receptor tail (HEG1)(PDB ID: 4HDQ)(Gingras et al., 2013) shown in middle. B) Conservation and (C) surface electrostatics of KRIT1ARD-FERM. The superposed Rap1 and HEG1 are shown as per panel A. Dark blue indicates complete conservation and white indicates no conservation in panel B, as defined by the consurf server (Landau et al., 2005). Red indicates negative potential and blue positive potential in panel C. D) Stereoview showing electrostatic potential of KRIT1 ARD ankyrin groove. E) Ribbon diagram in same orientation as D. The location of tryptophans 330 and 404 are indicated.
Conservation analysis of KRIT1ARD-FERM shows that the most extensive completely conserved surface is the Rap1 binding site on FERM domain F1 and F2 lobes (Li et al., 2012) (Fig 3B). There are also extensive completely conserved regions on the ARD, and these fall in both the ankyrin groove and the back side of the ARD (Fig 3B). The conserved regions of the ARD are largely hydrophobic, with some regions of negative electrostatic potential (Fig 3C). On analysis of the KRIT1ARD we noticed that a well-conserved tryptophan residue (Trp-330; indicated with a grey circle in Figure 2H). This tryptophan sits in a hydrophobic patch of the ankyrin groove, and in concert with the ANK4 insert stabilized by Trp-404 seems to define a structural feature with potential to mediate protein-protein interactions (Figure 3D,E). It will be interesting to discover in future studies whether the ankyrin groove of KRIT1 mediates protein-protein interactions.
3.6. Comparison of KRIT1 ARD with other ARD structures
The KRIT1 ARD contains 4 ankyrin repeats. On submission of the structure to the Dali server (Holm and Rosenstrom, 2010) high similarity is observed with designed ankyrin repeat proteins, termed DARPins. For example, a Dali Z-score of 20.5, root-mean-squared deviation of 1.3 Å over 121 amino acids and sequence identity of 31% over 155 amino acids is observed for a DARPin designed to interact with β-tubulin (PDB ID: 4DRX) (Pecqueur et al., 2012). Although KRIT1 has a known function as a tubulin binding protein (Beraud-Dufour et al., 2007; Gunel et al., 2002), structural superposition suggested that the interaction would generate a clash, and microtubule binding assays confirmed that the ARD does not bind microtubules (not shown).
3.7. Discussion
Mutations in the KRIT1 gene are strongly associated with the cerebral cavernous malformations disease. We set out to better understand the structure and function of KRIT1 by determination of the crystal structure for KRIT1ARD-FERM. Our analysis of the 2.9 Å structure shows that KRIT1 contains an ankyrin repeat domain of 4 ankyrin repeats which is held in a stable conformation adjacent to the KRIT1 FERM domain.
The structure of KRIT1ARD-FERM suggests surface regions that are completely conserved over evolution (Fig 3B), and of these the ankyrin groove is hydrophobic (Fig 3D). The arrangement of completely conserved Trp-330 along with surrounding residues, particularly the unusual loop in ANK4 stabilized by Trp-404, seems to define a potential interaction surface, although the binding partner is not yet known. Our extensive studies to probe for the ability of binding partners to interact at this surface have, thus far, been inconclusive and will be followed up in a future study. We have also been hampered by our finding that KRIT1FERM strongly binds by pull-down to a wide range of proteins (not shown); we would therefore caution against the use of the KRIT1 FERM domain alone for protein interaction studies. Nonetheless, the ankyrin groove of KRIT1 ARD seems tailor-made as a macromolecular interaction site.
The presence of additional domains adjacent to FERM domains has been well documented for the integrin-binding proteins, talin and kindlin (Goult et al., 2009; Goult et al., 2010) where “F0” precedes the F1, F2 and F3 lobes of their FERM domains. For KRIT1, the ARD seems to fulfill a similar structural role. It is conformationally in a very similar arrangement in each of the three copies of the asymmetric unit of our crystal structure. Therefore, instead of considering the ARD and FERM domains as ‘beads-on-a-string’, we propose that the ARD-FERM region should be considered a globular module. In addition, the orientation of the ARD with respect to the FERM domain generates a conserved V-shaped wedge, and it will also be interesting to see if this mediates a site of inter-molecular interaction.
Overall, the determination of the KRITARD-FERM structure provides a clear definition of the KRIT1 ARD, which contains 4 ankyrin repeats. The structure suggests a stable orientation for the ARD and FERM domains, and it also implicates potential sites for binding partner interactions.
Acknowledgements
We thank the staff at NE-CAT at the APS. We thank Stephanie Hamill, Amy Stiegler and Byung Hak Ha for helpful discussions. This work is funded by National Institutes of Health grants P41 GM103403 (NE-CAT) and R01NS085078 (T.J.B.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author contributions
RZ and TJB conceived of the study and wrote the paper. RZ: sub-cloned, expressed, purified, crystallized and refined the structure. XL: data reduction, structure solution and refinement. TJB: data collection, structure refinement, structure analysis.
PDB accession code
The coordinates and structure factors of KRIT1ARD-FERM have been deposited in the protein data bank under accession code 5D68.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akers AL, Johnson E, Steinberg GK, Zabramski JM, Marchuk DA. Biallelic somatic and germline mutations in cerebral cavernous malformations (CCMs): evidence for a two-hit mechanism of CCM pathogenesis. Hum Mol Genet. 2009;18:919–930. doi: 10.1093/hmg/ddn430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beraud-Dufour S, Gautier R, Albiges-Rizo C, Chardin P, Faurobert E. Krit 1 interactions with microtubules and membranes are regulated by Rap1 and integrin cytoplasmic domain associated protein-1. Febs J. 2007;274:5518–5532. doi: 10.1111/j.1742-4658.2007.06068.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergametti F, Denier C, Labauge P, Arnoult M, Boetto S, Clanet M, Coubes P, Echenne B, Ibrahim R, Irthum B, Jacquet G, Lonjon M, Moreau JJ, Neau JP, Parker F, Tremoulet M, Tournier-Lasserve E. Mutations within the programmed cell death 10 gene cause cerebral cavernous malformations. Am J Hum Genet. 2005;76:42–51. doi: 10.1086/426952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bond CS, Schuttelkopf AW. ALINE: a WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr D Biol Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- Cavalcanti DD, Kalani MY, Martirosyan NL, Eales J, Spetzler RF, Preul MC. Cerebral cavernous malformations: from genes to proteins to disease. J Neurosurg. 2012;116:122–132. doi: 10.3171/2011.8.JNS101241. [DOI] [PubMed] [Google Scholar]
- Chiswell BP, Zhang R, Murphy JW, Boggon TJ, Calderwood DA. The structural basis of integrin-linked kinase-PINCH interactions. Proc Natl Acad Sci U S A. 2008;105:20677–20682. doi: 10.1073/pnas.0811415106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draheim KM, Fisher OS, Boggon TJ, Calderwood DA. Cerebral cavernous malformation proteins at a glance. J Cell Sci. 2014;127:701–707. doi: 10.1242/jcs.138388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draheim KM, Li X, Zhang R, Fisher OS, Villari G, Boggon TJ, Calderwood DA. CCM2–CCM3 interaction stabilizes their protein expression and permits endothelial network formation. J Cell Biol. 2015;208:987–1001. doi: 10.1083/jcb.201407129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faurobert E, Rome C, Lisowska J, Manet-Dupe S, Boulday G, Malbouyres M, Balland M, Bouin AP, Keramidas M, Bouvard D, Coll JL, Ruggiero F, Tournier-Lasserve E, Albiges-Rizo C. CCM1-ICAP-1 complex controls beta1 integrin-dependent endothelial contractility and fibronectin remodeling. J Cell Biol. 2013;202:545–561. doi: 10.1083/jcb.201303044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A, Zalvide J, Faurobert E, Albiges-Rizo C, Tournier-Lasserve E. Cerebral cavernous malformations: from CCM genes to endothelial cell homeostasis. Trends Mol Medicine. 2013;19:302–308. doi: 10.1016/j.molmed.2013.02.004. [DOI] [PubMed] [Google Scholar]
- Fisher OS, Boggon TJ. Signaling pathways and the cerebral cavernous malformations proteins: lessons from structural biology. Cell Mol Life Sci. 2014;71:1881–1892. doi: 10.1007/s00018-013-1532-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher OS, Zhang R, Li X, Murphy JW, Demeler B, Boggon TJ. Structural studies of cerebral cavernous malformations 2 (CCM2) reveal a folded helical domain at its C-terminus. FEBS Lett. 2013;587:272–277. doi: 10.1016/j.febslet.2012.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher OS, Liu W, Zhang R, Stiegler AL, Ghedia S, Weber JL, Boggon TJ. Structural basis for the disruption of the cerebral cavernous malformations 2 (CCM2) interaction with Krev interaction trapped 1 (KRIT1) by disease-associated mutations. J Biol Chem. 2015a;290:2842–2853. doi: 10.1074/jbc.M114.616433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher OS, Deng H, Liu D, Zhang Y, Wei R, Deng Y, Zhang F, Louvi A, Turk BE, Boggon TJ, Su B. Structure and vascular function of MEKK3-cerebral cavernous malformations 2 complex. Nature communications. 2015b;6:7937. doi: 10.1038/ncomms8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frame MC, Patel H, Serrels B, Lietha D, Eck MJ. The FERM domain: organizing the structure and function of FAK. Nat Rev Mol Cell Biol. 2010;11:802–814. doi: 10.1038/nrm2996. [DOI] [PubMed] [Google Scholar]
- Gault J, Shenkar R, Recksiek P, Awad IA. Biallelic somatic and germ line CCM1 truncating mutations in a cerebral cavernous malformation lesion. Stroke. 2005;36:872–874. doi: 10.1161/01.STR.0000157586.20479.fd. [DOI] [PubMed] [Google Scholar]
- Gingras AR, Liu JJ, Ginsberg MH. Structural basis of the junctional anchorage of the cerebral cavernous malformations complex. J Cell Biol. 2012;199:39–48. doi: 10.1083/jcb.201205109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gingras AR, Puzon-McLaughlin W, Ginsberg MH. The Structure of the Ternary Complex of Krev Interaction Trapped 1 (KRIT1) bound to both the Rap1 GTPase and the Heart of Glass (HEG1) cytoplasmic tail. J Biol Chem. 2013;288:23639–23649. doi: 10.1074/jbc.M113.462911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Bouaouina M, Harburger DS, Bate N, Patel B, Anthis NJ, Campbell ID, Calderwood DA, Barsukov IL, Roberts GC, Critchley DR. The structure of the N-terminus of kindlin-1: a domain important for alphaiibbeta3 integrin activation. J Mol Biol. 2009;394:944–956. doi: 10.1016/j.jmb.2009.09.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goult BT, Bouaouina M, Elliott PR, Bate N, Patel B, Gingras AR, Grossmann JG, Roberts GC, Calderwood DA, Critchley DR, Barsukov IL. Structure of a double ubiquitin-like domain in the talin head: a role in integrin activation. Embo J. 2010;29:1069–1080. doi: 10.1038/emboj.2010.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guclu B, Ozturk AK, Pricola KL, Bilguvar K, Shin D, O'Roak BJ, Gunel M. Mutations in apoptosis-related gene, PDCD10, cause cerebral cavernous malformation 3. Neurosurgery. 2005;57:1008–1013. doi: 10.1227/01.neu.0000180811.56157.e1. [DOI] [PubMed] [Google Scholar]
- Gunel M, Laurans MS, Shin D, DiLuna ML, Voorhees J, Choate K, Nelson-Williams C, Lifton RP. KRIT1, a gene mutated in cerebral cavernous malformation, encodes a microtubule-associated protein. Proc Natl Acad Sci U S A. 2002;99:10677–10682. doi: 10.1073/pnas.122354499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Laberge-le Couteulx S, Jung HH, Labauge P, Houtteville JP, Lescoat C, Cecillon M, Marechal E, Joutel A, Bach JF, Tournier-Lasserve E. Truncating mutations in CCM1, encoding KRIT1, cause hereditary cavernous angiomas. Nat Genet. 1999;23:189–193. doi: 10.1038/13815. [DOI] [PubMed] [Google Scholar]
- Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Li X, Ji W, Zhang R, Folta-Stogniew E, Min W, Boggon TJ. Molecular recognition of LD motifs by the FAT-homology domain of cerebral cavernous malformation 3 (CCM3) J Biol Chem. 2011;286:26138–26147. doi: 10.1074/jbc.M110.211250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Draheim KM, Liu W, Calderwood DA, Boggon TJ. Structural Basis for Small G Protein Effector Interaction of Ras-related Protein 1 (Rap1) and Adaptor Protein Krev Interaction Trapped 1 (KRIT1) J Biol Chem. 2012;287:22317–22327. doi: 10.1074/jbc.M112.361295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Zhang R, Zhang H, He Y, Ji W, Min W, Boggon TJ. Crystal structure of CCM3, a cerebral cavernous malformation protein critical for vascular integrity. J Biol Chem. 2010;285:24099–24107. doi: 10.1074/jbc.M110.128470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liquori CL, Berg MJ, Siegel AM, Huang E, Zawistowski JS, Stoffer T, Verlaan D, Balogun F, Hughes L, Leedom TP, Plummer NW, Cannella M, Maglione V, Squitieri F, Johnson EW, Rouleau GA, Ptacek L, Marchuk DA. Mutations in a gene encoding a novel protein containing a phosphotyrosine-binding domain cause type 2 cerebral cavernous malformations. Am J Hum Genet. 2003;73:1459–1464. doi: 10.1086/380314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Boggon TJ. Cocrystal structure of the ICAP1 PTB domain in complex with a KRIT1 peptide. Acta Crystallogr Sect F. 2013;69:494–498. doi: 10.1107/S1744309113010762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W, Draheim KM, Zhang R, Calderwood DA, Boggon TJ. Mechanism for KRIT1 Release of ICAP1-Mediated Suppression of Integrin Activation. Mol Cell. 2013;49:719–729. doi: 10.1016/j.molcel.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddaluno L, Rudini N, Cuttano R, Bravi L, Giampietro C, Corada M, Ferrarini L, Orsenigo F, Papa E, Boulday G, Tournier-Lasserve E, Chapon F, Richichi C, Retta SF, Lampugnani MG, Dejana E. EndMT contributes to the onset and progression of cerebral cavernous malformations. Nature. 2013;498:492–496. doi: 10.1038/nature12207. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. Journal of applied crystallography. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten P, Pizzolato GP, Rilliet B, Berney J. 131 cases of cavernous angioma (cavernomas) of the CNS, discovered by retrospective analysis of 24,535 autopsies. Neurochirurgie. 1989;35:82–83. 128–131. [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. In: Carter CW, Sweet RM, editors. Methods in Enzymology. San Diego: Academic Press (New York); 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- Pagenstecher A, Stahl S, Sure U, Felbor U. A two-hit mechanism causes cerebral cavernous malformations: complete inactivation of CCM1, CCM2 or CCM3 in affected endothelial cells. Hum Mol Genet. 2009;18:911–918. doi: 10.1093/hmg/ddn420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pecqueur L, Duellberg C, Dreier B, Jiang Q, Wang C, Pluckthun A, Surrey T, Gigant B, Knossow M. A designed ankyrin repeat protein selected to bind to tubulin caps the microtubule plus end. Proc Natl Acad Sci U S A. 2012;109:12011–12016. doi: 10.1073/pnas.1204129109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahoo T, Johnson EW, Thomas JW, Kuehl PM, Jones TL, Dokken CG, Touchman JW, Gallione CJ, Lee-Lin SQ, Kosofsky B, Kurth JH, Louis DN, Mettler G, Morrison L, Gil-Nagel A, Rich SS, Zabramski JM, Boguski MS, Green ED, Marchuk DA. Mutations in the gene encoding KRIT1, a Krev-1/rap1a binding protein, cause cerebral cavernous malformations (CCM1) Hum Mol Genet. 1999;8:2325–2333. doi: 10.1093/hmg/8.12.2325. [DOI] [PubMed] [Google Scholar]
- Sedgwick SG, Smerdon SJ. The ankyrin repeat: a diversity of interactions on a common structural framework. Trends Biochem Sci. 1999;24:311–316. doi: 10.1016/s0968-0004(99)01426-7. [DOI] [PubMed] [Google Scholar]
- Serebriiskii I, Estojak J, Sonoda G, Testa JR, Golemis EA. Association of Krev-1/rap1a with Krit1, a novel ankyrin repeat-containing protein encoded by a gene mapping to 7q21–22. Oncogene. 1997;15:1043–1049. doi: 10.1038/sj.onc.1201268. [DOI] [PubMed] [Google Scholar]
- Stiegler AL, Zhang R, Liu W, Boggon TJ. Structural Determinants for Binding of Sorting Nexin 17 (SNX17) to the Cytoplasmic Adaptor Protein Krev Interaction Trapped 1 (KRIT1) J Biol Chem. 2014;289:25362–25373. doi: 10.1074/jbc.M114.584011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss M. Global indicators of X-ray data quality. Journal of applied crystallography. 2001;34:130–135. [Google Scholar]
- Xu X, Wang X, Zhang Y, Wang DC, Ding J. Structural Basis for the Unique Heterodimeric Assembly between Cerebral Cavernous Malformation 3 and Germinal Center Kinase III. Structure. 2013;21:1059–1066. doi: 10.1016/j.str.2013.04.007. [DOI] [PubMed] [Google Scholar]
- Zhang M, Dong L, Shi Z, Jiao S, Zhang Z, Zhang W, Liu G, Chen C, Feng M, Hao Q, Wang W, Yin M, Zhao Y, Zhang L, Zhou Z. Structural mechanism of CCM3 heterodimerization with GCKIII kinases. Structure. 2013;21:680–688. doi: 10.1016/j.str.2013.02.015. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Rawnsley DR, Goddard LM, Pan W, Cao XJ, Jakus Z, Zheng H, Yang J, Arthur JS, Whitehead KJ, Li D, Zhou B, Garcia BA, Zheng X, Kahn ML. The cerebral cavernous malformation pathway controls cardiac development via regulation of endocardial MEKK3 signaling and KLF expression. Dev Cell. 2015;32:168–180. doi: 10.1016/j.devcel.2014.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]