Abstract
The Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC – www.nitrc.org) suite of services include a resources registry, image repository and a cloud computational environment to meet the needs of the neuroimaging researcher. NITRC provides image-sharing functionality through both the NITRC Resource Registry (NITRC-R), where bulk data files can be released through the file release system (FRS), and the NITRC Image Repository (NITRC-IR), a XNAT-based image data management system. Currently hosting 14 projects, 6845 subjects, and 8285 MRI imaging sessions, NITRC-IR provides a large array of structural, diffusion and resting state MRI data. Designed to be flexible about management of data access policy, NITRC provides a simple, free, NIH-funded service to support resource sharing in general, and image sharing in particular.
Introduction
The Neuroimaging Informatics Tools and Resources Clearinghouse (NITRC www.nitrc.org) suite of services include a resources registry, image repository and a cloud computational environment to meet the needs of the neuroimaging researcher. NITRC has been funded by contract with NIH Blueprint for Neuroscience Research as well as four NIH institutes1,2 since 2006.
Rationale
NITRC’s mission is to facilitate sharing, through the finding and comparing all ‘resources’ for neuroimaging research. Over the years the scope of these resources has expanded to support scientific domains from MR to PET, SPECT, CT, MEG/EEG, optical imaging, genetic imaging, clinical neuroinformatics and computational neuroscience. Resources, in this case, are broadly defined to include software, hardware, data, websites, community organizations, etc. By enhancing discoverability, NITRC strives to promote reuse, reduce duplication, and enhance quality in neuroscience research.
Overview
NITRC provides a triad of core services to the community. The key building block of NITRC is the Resources Registry (NITRC-R). Each NITRC project has a homepage that describes the resource, provides a standard set of resource characteristics (i.e. keywords, license, dependencies, etc.), and provides a standard set of links for the resource (i.e. download, documentation, support, etc.). Each resource page is maintained by the resource administrator, who is responsible for keeping it up to date. All resource content is fully customizable. While NITRC provides wikis, source code management, file hosting (via a File Release System (FRS)), trackers and forum support, etc. for every project, resource administrators are also free to enable/disable any functionality and redirect any content to other sources, as pertinent to the resource developers needs. Visitors to the NITRC site can search for resources based on keywords, free text and specific capabilities in order to find relevant resources
The NITRC Image Repository (NITRC-IR) provides an image database sharing infrastructure for images and related data that can be closely integrated with the NITRC-R resources in order to better, support, promote, and manage data sharing functions for NITRC-hosted projects. In order to support a database-centric mode of data sharing, we introduced the NITRC Image Repository (NITRC-IR) 3.
Completing the NITRC triad of services is the NITRC Computational Environment (NITRC-CE). NITRC-CE preinstalls a number of popular NITRC-listed neuroimaging tools (such as FreeSurfer (Dale, Fischl, and Sereno 1999), FSL (Smith et al. 2004), AFNI (Cox 1996), etc; see4 for a complete listing) into a standardized computational environment. This environment can be deployed in the cloud (using Amazon Web Services Elastic Compute Cloud (EC2) or the Microsoft Azure Cloud Computing Platform), or as a virtual machine for local use. The NITRC-CE lowers the barriers to cloud computing and provides a turn-key solution to scalability and replication of standardized and documented execution platforms.
Technical Implementation
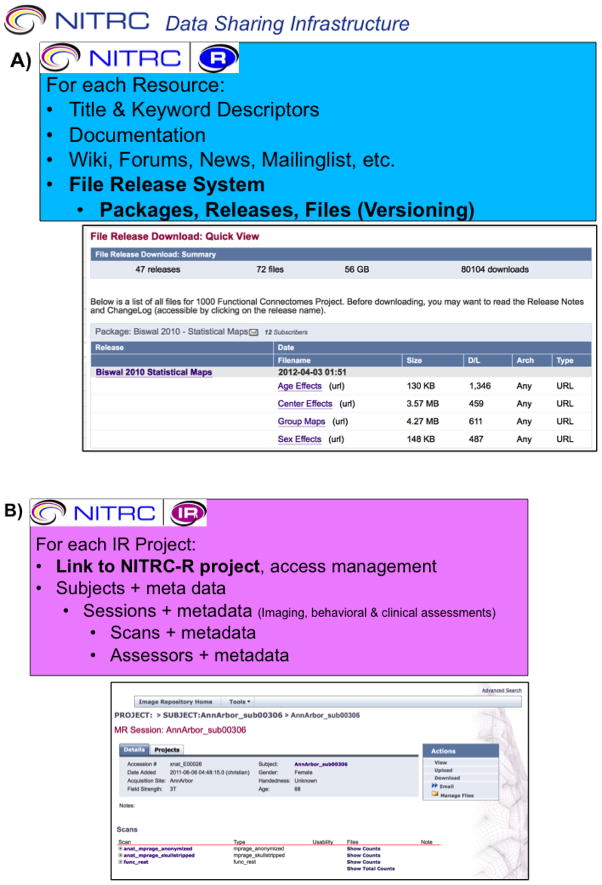
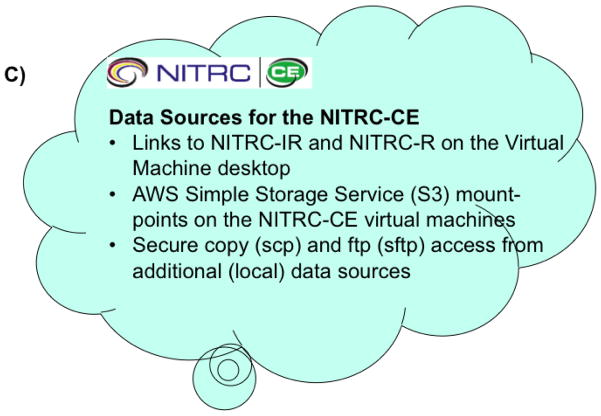
Throughout its development, NITRC has striven to reuse, as opposed to reinvent, core operational technologies. NITRC-R was designed to support a broad and integrated set of functions for neuroimaging resource projects through customization of a GForge5 base infrastructure. The NITRC-IR is built on XNAT (Marcus et al. 2007) and provides sharing infrastructure for images and related data that can be closely integrated with the NITRC-R resources in order to better, support, promote, and manage data sharing functions for NITRC-hosted projects. NITRC-R projects can be associated with NITRC-IR (XNAT) ‘projects’ and these can be interlinked. The NITRC-CE is built upon the NeuroDebian operating system framework (Halchenko and Hanke 2012; Hanke and Halchenko 2011). Figure 1 shows the general data-sharing framework for the overall NITRC implementation.
Figure 1. NITRC Data Sharing Infrastructure.
Data sharing infrastructure within the NITRC triad of services. A) Data sharing within the NITRC Resources system is principally supported by the file release system (FRS) which provides support for release of packages, releases and files. B) Data sharing within the NITRC Image Repository is build around projects, subjects, sessions and assessors. C) Data access for the NITRC Computational Environment is facilitated through access to NITRC-IR and NITRC-R.
Design Objectives for Data Sharing
NITRC provides two distinct mechanisms for data sharing. First, each NITRC-R project provides a very low-barrier to provider entry-point for data sharing within the community. Using a project’s file release system (FRS) file hosting capability, data files (either individual images or complete archived datasets) can be made available. Resource administrators can control access to data, by requiring (or not) project registration, and providing click-through data usage agreement forms. In addition, project administrators can optionally require that downloaders provide contact information in order to facilitate critical communication with the resource users, if needed. File downloads are counted in order to provide utilization information to the resource administrators. The FRS mode of data sharing through NITRC-R is relatively easy for the data provider, it places more of the data decoding burden the end user, particularly when the data for a project is released in multiple archive files each containing many subjects. In such situations it may be necessary to download numerous data files in order to access a specific desired subset of data.
Second, the NITRC-IR was designed as a fully curated federated XNAT neuroimaging database that could host data for NITRC projects in a searchable framework. We customized an XNAT instance to seamlessly interoperate with the NITRC-R environment. This included establishing a common user-base mapping between the NITRC and XNAT applications as we desired a ‘single sign-on’ and needed to manage project-level permissions via the NITRC-R users and projects. Each NITRC-R project can have an associated NITRC-IR (XNAT) project. The permissions for access to the XNAT project can be set to mirror the accessibility options of the NITRC-R project. Some NITRC projects are provided completely in the open, without login or registration. Other projects requite a user to be registered with the specific project in order to gain access to the data. The criteria for joining a project is determined at the discretion of the specific project administrators. NITRC-IR uses a standard XNAT data model for subjects and sessions. Assessors have been added on a per-project basis to accommodate additional information that the project is releasing.
It is important to note that the NITRC-IR is curated by NITRC staff in conjunction with the project administrators. This curation level permits homogenization of data elements across all NITRC-IR-hosted projects. Thus scan parameters, units, demographics, etc. can be guaranteed to be encoded in an identical fashion across projects, which is necessary in order to facilitate cross-project searching. The curation also permits available disk space to be managed.
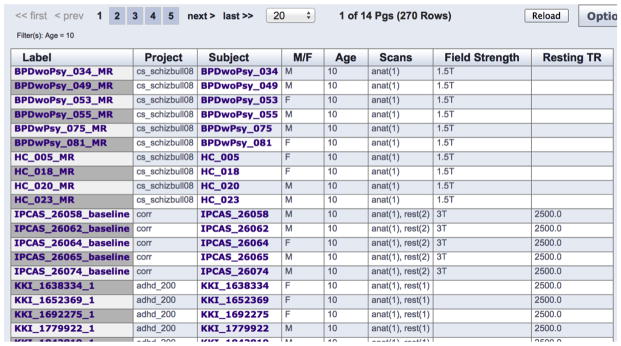
In addition to within-project viewing and searching functions, users can also interact with the entire NITRC-IR portfolio of data (that they have permission to access) at a level ‘above’ the individual projects. This greatly facilitates searches across the projects and permits ease of identification and aggregation of comparable data within multiple projects. Figure 2 shows an example of the search capability and results.
Figure 2. NITRC-IR Search.
Results of a search across all of the NITRC-IR projects for all subjects who are 10 years old. The result indicated 270 cases that match this query, and these subjects come from numerous different projects. Variables such as data acquisition, field strength, resting state TR, etc. are also available to further refine the search.
The NITRC site for sharing avoids issues with local institutional firewalls, and has a very high uptime, including a fail-over back up that can be invoked should the need arise. NITRC overall, and the NITRC-IR in particular, is continually being developed and improved; at least four major releases are scheduled each year. All releases are vetted through a test and stage facility, before moving into production. All features of each release are tested and documented. NITRC as a system is an open-source project and is released at http://www.nitrc.org/projects/nitrcext.
Both NITRC-R and –IR are dynamic, expanding data resources. Both support the archival and distribution of datasets for researchers who have on-going or completed studies. Data available through NITRC-R is completely at the design and timing of the project administrator, whereas data to be released in NITRC-IR needs to be scheduled for curation and release with the NITRC development team. NITRC is available as a resource to meet the data sharing mandate for many NIH grantees.
Content
As of April 2015, NITRC-R hosts 755 publically accessible projects and 12,575 registered users. Within the NITRC-IR there are 14 projects, 6845 subjects, and 8285 MRI imaging sessions available. Five of these datasets are available without restriction, whereas the remaining 9 datasets require some sort of project registration. Table 1 provides a summary of these data. While XNAT can support imaging domains in addition to MRI, NITRC-IR does not currently host imaging data from other domains. Each NITRC-IR project, as a collection, has a unique URI. Each project provides extended demographic information at the project website. The CANDIShare project6 provides manually segmented structural results available as well.
Table 1.
NITRC-IR Projects
| Project | N Subjects | N Sites | Data Types | Format |
|---|---|---|---|---|
| 1000 Functional Connectomes (“Classic”)10 | 1288, age 18–80 years | 24 | Str/RS | nii |
| ABIDE11 | 1112, age 6–64 years | 17 | Str/RS | nii |
| ADHD-20012 | 973, age 7–26 years | 8 | Str/RS | nii |
| Beijing Enhanced13 | 180, age 17–28 years | 1 | Str/RS/Diff | nii |
| Beijing Eyes Open Eyes Closed14 | 48, age 18–30 years | 1 | Str/RS/Diff | nii |
| Beijing Short TR15 | 28, age 18–46 years | 1 | Str/RS/Diff | nii |
| CANDIShare16 | 103, age 4–16 years | 1 | Str | nii |
| CoRR17 | 1286, age 6–88 years | 18 | Str/RS/Diff | nii |
| IXI18 | 584, age 19–86 years | 3 | Str/Diff/MRA | nii |
| KIN19 | 5 | |||
| NKI Rockland20 | 207, age 4–85 years | 1 | Str/RS/Diff | DCM, nii |
| Parkinson’s DTI21 | 53, age 47–81 years | 1 | Diff | nii |
| PING22 | 782, age 3–21 years | 9 | Str/Diff/RS | DCM |
| Study Forrest23 | 20, age 21–38 years | 1 | Str/Diff/fMRI | nii, HDF5, mat |
Abbreviations: Str – structural MRI; RS – resting state fMRI; Diff – diffusion MRI; MRA – Magnetic Resonance Angiography; nii – NifTI data format; DCM – DICOM data format; HDF5 – HDF5 data format; mat – MATLAB data format
The data collection holdings of NITRC-R are somewhat harder to characterize succinctly. A search for NITRC resources using the ‘data’ keyword generates 44 projects. 18 of these provide atlas data, while others host specific testing datasets. Investigators looking for data would have to refine their specific search criteria in order to identify NITRC projects that might contain relevant data for them. Because of the NITRC-IR project association with a NITRC Project, additional metadata is available per project and image. Table 2 depicts some of the metadata available for the NITRC datasets.
Table 2.
NITRC Metadata
| NITRC Metadata Availability | ||
|---|---|---|
| Dataset | Image file | |
| Title | ✓ | ✓ |
| Version | ✓ | ✓ |
| License | ✓ | |
| Description | ✓ | ✓ |
| Author Name | ✓ | ✓ |
| Author Affiliation | ✓ | ✓ |
| Keywords (MeSH) | ✓ | |
| Dependencies | ✓ | |
| Grant/Support Source | ✓ | |
| Acknowledgements | ✓ | |
| Publications | ✓ | |
Data from NITRC is being used within the community. Cited in Google Scholar over 1,900 times, NITRC has supported 1.3 million data downloads since its inception, with over 620,000 from the NITRC-IR. While the number of subjects currently held within the NITRC data archives may not be large (compared to the total number of NIH-funded MRI scan acquisitions, for example), these data have proven to be very useful to the community. NITRC currently has storage space available, and continuously seeks additional data providers as part of its outreach mission.
Quality Control
NITRC provides no specific quantitative quality control processing for the hosted data. The quality of the data provided in NITRC-R projects is completely at the discretion of the project administrators. Ultimately, data quality can be judged by data reuse. Data added to the NITRC-IR, while not undergoing a specific quantitative quality assurance, are added to the curation queue based upon the known quality and expected impact of the data for the field.
Access
Each NITRC project manages its own access policy. Data, in either NITRC-R or NITRC-IR can be completely open, not requiring any authentication; open to NITRC users; or open only to NITRC users who are registered with a specific project. Access policy is configurable to meet the needs of the investigator and their Institute Review Board’s requirements. While the XNAT system supports inclusion of imaging and phenotypic data, the majority of NITRC-IR data sets have their comprehensive phenotypic results available from the project websites.
NITRC offers data distributors the ability to distribute data freely, or craft their own Data Usage Agreements before allowing access to their data. For projects requiring accepted Data Use Agreements, project administrators can require users to register for membership in the project in order to access the data. The project administrators can require any level of authorization (from simple provision of email address, to provision of signed agreement) prior to granting project access. NITRC provides the capability to require a click-through acceptance the usage agreement prior to download of the data.
The NITRC team intentionally designed a broad framework to lower the barrier of data sharing by providing a range of workflows definable by the data resource administrator. While at the same time, NITRC provides a comprehensive set of potential access policy options that provides coverage for most simple or sensitive sharing needs.
Depending on the project and its data access workflow, there are a variety of ways to update users if data is withdrawn, revised, or added to. For researchers that downloaded data without having to register on a particular project, the data administrators can post a news bulletin or forum update. The users would only receive such notifications if they had opted into monitoring either of these communication channels. For the researchers that downloaded data using the workflow of registering on the data distributor’s project, they are more likely to receive updates as they are known to the resource. Again, the resource would post a news bulletin or forum update, but has the additional capability to email all users who have accessed the data.
Access to shared imaging data (from NITRC and other sources) is also facilitated for use with the NITRC Computational Environment. While downloading data to ones own computer system is the more common current data usage scenario, as data volume gets larger, transmission bandwidth may become rate limiting. Thus, the NITRC-CE helps to promote the concept of computing ‘local’ to data hosting facility. As highlighted in Figure 1c, the NITRC-CE includes data access options for easy access to NITRC-IR and NITRC-R, secure file transfer from outside sources, and file system mounting of AWS S3 resources.
Service Design
Data downloads are conducted through HTTP/HTTPS (using range requests to support paused and interrupted connections) and FTP/SFTP with continuation support. In addition, for NITRC-IR-hosted studies, we provide subject search, data packaging, and compression through the open source XNAT image repository framework.
Service Implementation
NITRC-R and IR are housed at the UCSD Center for Research in Biological Systems (CRBS) San Diego Supercomputer Center (SDSC). The SDSC uses a VMWare VSphere Cloud virtual machine farm with a cloud data storage system with redundant copies and continuous error checking across the 100% disk-based storage system. Load-balancing and automated failover ensure continued access. 10Gb Ethernet switching provides sustained read rates of 8+ gigabytes (GB) per second.
Submission
Anyone with a valid email can register for NITRC, create a NITRC-R project and contribute new data to NITRC. The only requirements are that the IRB has approved their distributing the data on NITRC, and that the data has been de-identified. Projects with shared data can then request curation and inclusion in the NITRC-IR. NITRC-IR requests are prioritized relative to level of effort, available effort and anticipated community impact.
Sustainability Plan and Future Perspectives
NITRC has been supported by the NIH since 2006 through a series of contracts. NITRC continues to seek re-funding of the contracting agreements, as well as explore other funding opportunities. The NIH has not demonstrated a history of long-term support of extramural sharing infrastructure. New planning in the area of ‘big data’7, the Data Discovery Index8 and the Data Commons9 may present more commercial solutions to future NIH data sharing mandates and funding of required data sharing as part of the routine grant process (akin to the evolution of the PubMed Central publication sharing mandate). Clearly, the NIH, and other funding agencies, and publishers, who are beginning to require data availability as part of publication, will play a part in the evolution of the sustainability of all data sharing resources. As the needs for data hosting continue to grow, resources like NITRC will be critical to sustaining the data infrastructure of reproducible neuroimaging. NITRC continues to seek institutional funding and develop cost-sharing sustainability plans that may include sponsorship and fee-for-service (data hosting) models that would be commensurate with proper sustainable research funding support for the individual investigators and their research grants.
Conclusions
Since 2006, NITRC has been successful in creating a home for the neuroimaging community to share resources. From the sharing of software, data, and knowhow, to the generation of integrated computational environments that link software, data and execution, NITRC continues to provide a vital set of resources for the neuroimaging community.
Highlights.
Provides an overview of the NITRC services, in general
Provides details on NITRC data-sharing services (NITRC-R & NITRC-IR)
Reviews rationale, technical design, implementation and content of NITRC
Comments on sustainability of NITRC as a data sharing resource
Footnotes
National Institute of Biomedical Imaging and Bioengineering, National Institute of Mental Health, National Institute of Neurological Disorders and Stroke, and the National Institute on Drug Abuse
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Cox RW. AFNI: Software for Analysis and Visualization of Functional Magnetic Resonance Neuroimages. Computers and Biomedical Research. 1996;29:162–73. doi: 10.1006/cbmr.1996.0014. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis. I. Segmentation and Surface Reconstruction. NeuroImage. 1999;9(2):179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Halchenko Yaroslav O, Michael Hanke. Open Is Not Enough. Let’s Take the Next Step: An Integrated, Community-Driven Computing Platform for Neuroscience. Frontiers in Neuroinformatics. 2012;6(January):22. doi: 10.3389/fninf.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke Michael, Halchenko Yaroslav O. Neuroscience Runs on GNU/Linux. Frontiers in Neuroinformatics. 2011;5(January):8. doi: 10.3389/fninf.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Daniel S, Olsen Timothy R, Ramaratnam Mohana, Buckner Randy L. The Extensible Neuroimaging Archive Toolkit: An Informatics Platform for Managing, Exploring, and Sharing Neuroimaging Data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Smith Stephen M, Jenkinson Mark, Woolrich Mark W, Beckmann Christian F, Behrens Timothy EJ, Johansen-Berg Heidi, Bannister Peter R, et al. Advances in Functional and Structural MR Image Analysis and Implementation as FSL. NeuroImage. 2004;23(Suppl 1)(January):S208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis. I. Segmentation and Surface Reconstruction. NeuroImage. 1999;9(2):179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Halchenko Yaroslav O, Hanke Michael. Open Is Not Enough. Let’s Take the Next Step: An Integrated, Community-Driven Computing Platform for Neuroscience. Frontiers in Neuroinformatics. 2012;6(January):22. doi: 10.3389/fninf.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke Michael, Halchenko Yaroslav O. Neuroscience Runs on GNU/Linux. Frontiers in Neuroinformatics. 2011;5(January):8. doi: 10.3389/fninf.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Daniel S, Olsen Timothy R, Ramaratnam Mohana, Buckner Randy L. The Extensible Neuroimaging Archive Toolkit: An Informatics Platform for Managing, Exploring, and Sharing Neuroimaging Data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Smith Stephen M, Jenkinson Mark, Woolrich Mark W, Beckmann Christian F, Behrens Timothy EJ, Johansen-Berg Heidi, Bannister Peter R, et al. Advances in Functional and Structural MR Image Analysis and Implementation as FSL. NeuroImage. 2004;23(Suppl 1)(January):S208–19. doi: 10.1016/j.neuroimage.2004.07.051. [DOI] [PubMed] [Google Scholar]
- Dale AM, Fischl B, Sereno MI. Cortical Surface-Based Analysis. I. Segmentation and Surface Reconstruction. NeuroImage. 1999;9(2):179–94. doi: 10.1006/nimg.1998.0395. [DOI] [PubMed] [Google Scholar]
- Halchenko Yaroslav O, Hanke Michael. Open Is Not Enough. Let’s Take the Next Step: An Integrated, Community-Driven Computing Platform for Neuroscience. Frontiers in Neuroinformatics. 2012;6(January):22. doi: 10.3389/fninf.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke Michael, Halchenko Yaroslav O. Neuroscience Runs on GNU/Linux. Frontiers in Neuroinformatics. 2011;5(January):8. doi: 10.3389/fninf.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Daniel S, Olsen Timothy R, Ramaratnam Mohana, Buckner Randy L. The Extensible Neuroimaging Archive Toolkit: An Informatics Platform for Managing, Exploring, and Sharing Neuroimaging Data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Halchenko Yaroslav O, Hanke Michael. Open Is Not Enough. Let’s Take the Next Step: An Integrated, Community-Driven Computing Platform for Neuroscience. Frontiers in Neuroinformatics. 2012;6(January):22. doi: 10.3389/fninf.2012.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke Michael, Halchenko Yaroslav O. Neuroscience Runs on GNU/Linux. Frontiers in Neuroinformatics. 2011;5(January):8. doi: 10.3389/fninf.2011.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus Daniel S, Olsen Timothy R, Ramaratnam Mohana, Buckner Randy L. The Extensible Neuroimaging Archive Toolkit: An Informatics Platform for Managing, Exploring, and Sharing Neuroimaging Data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Marcus Daniel S, Olsen Timothy R, Ramaratnam Mohana, Buckner Randy L. The Extensible Neuroimaging Archive Toolkit: An Informatics Platform for Managing, Exploring, and Sharing Neuroimaging Data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5(1):11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]