Abstract
Genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in six genes that are associated with childhood acute lymphoblastic leukemia (ALL). A lead SNP was found to occur on chromosome 9p21.3, a region that is deleted in 30% of childhood ALLs, suggesting the presence of causal polymorphisms linked to ALL risk. We used SNP genotyping and imputation-based fine-mapping of a multiethnic ALL case-control population (Ncases=1464, Ncontrols=3279) to identify variants of large effect within 9p21.3. We identified a CDKN2A missense variant (rs3731249) with 2% allele frequency in controls that confers three-fold increased risk of ALL in children of European-ancestry (OR=2.99; P=1.51×10−9) and Hispanic children (OR=2.77; P=3.78×10−4). Moreover, of 17 patients whose tumors displayed allelic imbalance at CDKN2A, 14 preferentially retained the risk allele and lost the protective allele (PBinomial=0.006), suggesting that the risk allele provides a selective advantage during tumor growth. Notably, the CDKN2A variant was not significantly associated with melanoma, glioblastoma, or pancreatic cancer risk, implying that this polymorphism specifically confers ALL risk but not general cancer risk. Taken together, our findings demonstrate that coding polymorphisms of large effect can underlie GWAS “hits” and that inherited polymorphisms may undergo directional selection during clonal expansion of tumors.
INTRODUCTION
The etiology of childhood acute lymphoblastic leukemia (ALL) is multifactorial, influenced by environmental stimuli, immune development, and genetic factors (1). Recent genome-wide association studies (GWAS) have identified single nucleotide polymorphisms (SNPs) in six genes that modify ALL risk, including ARID5B, IKZF1, CEBPE, CDKN2A, PIP4K2A and GATA3 (2, 3). With the exception of rs3824662 in GATA3, which confers 3-fold increased risk of Philadelphia chromosome-like ALL (4), GWAS of ALL have not identified variants of large effect sizes (odds ratio range: 1.30-1.86)(2).
The lead SNP on 9p21.3 from previous ALL GWAS, rs3731217, is located in intron 1 of the tumor suppressor gene CDKN2A within a linkage block containing another tumor suppressor gene (CDKN2B) and a functional non-coding RNA (CDKN2B-AS1)(5). Inherited SNPs in this linkage block are associated with several additional cancers, including: melanoma, glioblastoma, nasopharyngeal carcinoma, and squamous cell lung cancer (6-9). Somatic alteration of 9p21.3 via focal and whole-arm deletions, as well as copy-neutral loss of heterozygosity (LOH), occurs in many cancers and in ~30% of childhood ALL cases (10). The co-occurrence of inherited (i.e. germline) and acquired (i.e. somatic) cancer-associated variants on 9p21.3 suggests that heritable risk alleles in this region may undergo selection during tumor evolution, a phenomenon known as preferential allelic imbalance (PAI).
We sought to identify causal polymorphisms underlying the childhood ALL association peak near CDKN2A by performing SNP genotyping and imputation-based fine-mapping of the region in a multi-ethnic case-control population. The discovery sample for our fine-mapping analysis included 321 Hispanic children with ALL and 454 Hispanic control children from the California Childhood Leukemia Study (CCLS). Validation fine-mapping was performed in an independent set of 980 European-ancestry children with ALL and 2624 control children from the Children's Oncology Group (COG) and the Wellcome Trust Case-Control Consortium (WTCCC). Following association testing and bioinformatics analyses of SNP function, the top SNP underwent validation genotyping in a third independent ALL case-control set. Using a novel application of droplet-digital PCR (ddPCR), we further assessed whether the newly identified risk allele underwent PAI in ALL tumor samples.
MATERIALS AND METHODS
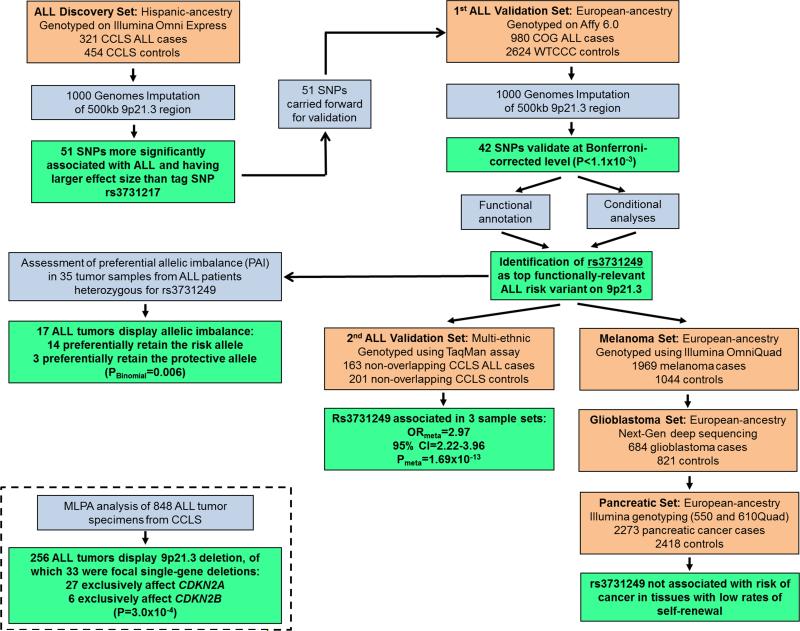
Figure 1 and Supplementary Table 1 summarize the study design and study populations, respectively. This study was approved and reviewed by all collaborating institutions, including the institutional review committees at the California Department of Public Health (CDPH) and the University of California, Berkeley.
Figure 1. Summary of study design and analyses.
The flowchart details progression of subjects and analyses through various stages of the study.
Subjects
CCLS Hispanic ALL discovery set
The discovery sample includes 321 Hispanic participants with ALL and 454 Hispanic controls from CCLS, whose recruitment has been described in detail previously (11). This population-based case-control study includes subjects from 35 California counties recruited from 1995-2008. Birth certificate information obtained from the CDPH was used to select 1-2 controls for each case, matching on date of birth, sex, Hispanic ethnicity, and maternal race.
COG/WTCCC ALL validation set
The ALL fine-mapping validation sample includes 980 European-ancestry children from COG protocols P9904 and P9905. Data were obtained from dbGaP study accession phs000638.v1.p1 (Genome-Wide Association Study of Relapse of Childhood Acute Lymphoblastic Leukemia), and have been described in detail (12, 13). Case-control comparisons were made with 2624 European-ancestry individuals accessed from the WTCCC (14). Demographic characteristics of COG cases and WTCCC controls appear in Supplementary Table 1.
CCLS multi-ethnic ALL validation set
An additional 111 African-American cases, 52 Hispanic cases, 154 African-American controls, and 47 Hispanic controls from CCLS were genotyped at rs3731249 using a TaqMan assay. These CCLS study participants do not overlap with individuals in the discovery sample or the COG/WTCCC validation set.
GENEVA melanoma case-control set
Melanoma case-control data were obtained from dbGaP Study Accession phs000187.v1.p1 (High Density SNP Association Analysis of Melanoma: Case-Control and Outcomes Investigation). Demographic characteristics of these 1969 adult melanoma cases and 1044 controls appear in Supplementary Table 1. These samples were genotyped as part of the Gene Environment Association Studies initiative (GENEVA) (15).
UCSF/Mayo glioblastoma case-control set
Glioblastoma data were generated via pooled deep-sequencing of 684 adults with glioblastoma and 821 controls from the UCSF Adult Glioma Study (AGS) and The Mayo Clinic. AGS cases were adults with incident histologically-confirmed glioblastoma. AGS controls were matched on age, sex and ethnicity (16). Mayo Clinic cases with incident glioblastoma were recruited from 2005-2012. Mayo controls were consented individuals who had a general medical exam at the Mayo Clinic, matched on sex, date of birth (within 2.5 years), self-identified race and residence (17). Demographic characteristics of glioblastoma cases and controls appear in Supplementary Table 1.
PanScan pancreatic cancer case-control set
Pancreatic cancer case-control data were obtained from dbGaP Study Accession phs000206.v4.p3 [Whole-Genome Scan for Pancreatic Cancer Risk in the Pancreatic Cancer Cohort Consortium (PanScan)]. Demographic characteristics of these 2273 pancreatic cancer cases and 2418 controls appear in Supplementary Table 1. These samples were genotyped as part of the Cancer Genetic Markers of Susceptibility (CGEMS) Project (18, 19).
Sample preparation and genotyping
Prediagnositic constitutive DNA for CCLS samples was extracted from neonatal bloodcards and genotyped using the Illumina Human OmniExpressV1 platform. DNA extraction was performed using the QIAamp DNA Mini Kit (QIAGEN, USA, Valencia, CA). Genotype reproducibility was verified using ten duplicate samples with average concordance >99.99%. Samples with genotyping call rates <98% were excluded. Samples were screened for cryptic relatedness using 10,000 unlinked SNPs and excluded if identity-by-descent was >0.15. Samples with discordant sex information (reported vs. genotyped sex) were excluded. SNPs with genotyping call rates <98% were excluded. Any SNP with a Hardy-Weinberg Equilibrium (HWE) P-value <1×10−5 in controls was excluded. DNA from an additional 163 CCLS cases and 201 CCLS controls was genotyped for rs3731249 using a TaqMan assay (Applied Biosystems: C__25611114_10). These samples were randomized on 96-well plates, containing HapMap trios and 5 duplicate samples per plate. All trio genotypes displayed Mendelian consistency and duplicates showed genotype concordance. Cluster plots were visually inspected.
Constitutive DNA from COG samples was extracted from remission blood, as previously detailed (12). DNA samples were genotyped on the Affymetrix 6.0 array and genotype data were downloaded from dbGaP accession phs000638.v1.p1. Control genotype data for European-ancestry control samples genotyped on the Affymetrix 6.0 array were downloaded from the Wellcome Trust Case-Control Consortium (14). Genotyping quality-control procedures were conducted independently in cases and in controls. Samples with genotyping call rates <98% in cases or controls were excluded. SNPs with genotyping call rates <98% were excluded. We excluded subjects showing evidence of non-European ancestry, samples with mismatched reported versus genotypes sex, and related subjects (IBD>0.15). SNP data were used to ensure no overlap between ALL cases included in validation analyses and those included in discovery analyses. COG and WTCCC genotype data were merged to create a final set of 980 cases and 2624 European-ancestry controls.
DNA for GENEVA melanoma case-control samples were genotyped using the Illumina HumanOmni1-Quad_v1-0_B array. Genotype data were downloaded from dbGaP accession phs000187.v1.p1. Samples were filtered based on a pre-computed sample filter provided by dbGaP. Subsequently, subjects with genotyping call-rate <95%, discordant genotyped versus reported sex, non-European ancestry, or cryptic relatedness were removed from analyses. SNPs with genotyping call rates <98% or HWE P-value <1×10−5 (among controls) were excluded. The final European-ancestry dataset contained 1969 cases and 1044 controls.
The glioma GWAS association peak on 9p21.3, between 21.930–22.135 Mb, underwent targeted deep-sequencing using pooled constitutive DNA from UCSF and Mayo samples. Four pools of DNA were prepared - two from glioblastoma cases (NUCSF=380; NMayo=304) and two from controls (NUCSF=547; NMayo=274). These DNA pools were subjected to long-range PCR covering the association peak, followed by next-generation sequencing to a target depth of 2,000X. NGS was performed by deCODE Genetics and subjected to quality-control measures as previously described (17). Variants were removed that had fewer than 1000 reads in a sequencing pool. Variants whose allele frequency estimates differed more than 10% between the two case pools or between the two control pools were removed.
DNA for PanScan pancreatic cancer case-control samples was genotyped using either the Illumina Human550v3 array or the 610QuadV1B array. Genotype data were downloaded from dbGaP accession phs000206.v4.p3. Quality-control was conducted separately by array. Subjects with genotyping call-rate <98%, discordant genotyped versus reported sex, non-European ancestry, or cryptic relatedness were removed. SNPs with genotyping call rates <98% or HWE P-value <1×10−5 (among controls) were excluded. The final set contained 2273 European-ancestry cases and 2418 controls.
Statistical analyses
Using Illumina OmniExpress array data (for CCLS discovery samples) or Affymetrix 6.0 data (COG validation samples), targeted SNP imputation was performed for a 500kb region on chromosome nine from 21.735Mb to 22.235Mb (GRCh37/hg19). The region is approximately centered on the original ALL GWAS hit in the region (rs3731217), first published by Sherborne et al (5). Imputation was performed using the IMPUTE v2.3.1 software and its standard Markov chain Monte Carlo algorithm and default settings for targeted imputation (20). All 1,000 Genomes Phase I integrated haplotypes were provided as the imputation reference panel (21). SNPs with imputation quality (info) scores <0.70 or posterior probabilities <0.90 were excluded. Association statistics for imputed and directly genotyped SNPs were calculated using logistic regression in SNPTESTv2, using an allelic additive model (22) and a missing-data likelihood score-test to account for additional uncertainty inherent in analysis of imputed genotypes. The first five ancestry-informative principal components from Eigenstrat were used as covariates in logistic regression analyses of both CCLS discovery and COG validation sets (23). Of note, the rs3731249 missense variant that is the focus of this manuscript was directly genotyped on-array and was not imputed in the CCLS discovery set.
Case-control associations for CCLS samples undergoing TaqMan genotyping were evaluated using logistic regression, assuming an allelic additive model, adjusting for self-reported ethnicity (African-American or Hispanic). We have previously demonstrated the validity of self-reported ethnicity as a measure of genetic ancestry in CCLS (24).
Associations from the CCLS Hispanic discovery set, the COG/WTCCC validation set, and the CCLS ALL validation set were combined using fixed effects meta-analysis in the META software package to generate a summary odds ratio and P-value for the combined ALL case-control comparisons (25).
Genome-wide SNP data for melanoma cases and controls were analyzed using logistic regression, adjusted for the first five ancestry-informative principal components, using Plink and Eigenstrat (23, 26). As in the CCLS Hispanic discovery set, rs3731249 was directly genotyped on-array in the melanoma dataset.
Allele frequencies from the deCODE NGS glioblastoma case-control data were calculated based on read counts; separately for reads generated from each case and control pool. Based on the derived allele frequencies, the number of chromosomes in the original pools carrying various alleles was estimated. Association tests were conducted using Fisher's exact test for the estimated number of chromosomes carrying the minor alleles in the corresponding case and control pools, with Mayo and UCSF data combined as previously described (17).
Genome-wide SNP data for pancreatic cancer cases and controls were stratified by array (550 versus 610Q). Data from each array underwent imputation using the IMPUTE v2.3.1 software (20) and all 1,000 Genomes Phase I integrated haplotypes (21). Allelic additive SNP association statistics were calculated using logistic regression in SNPTESTv2, stratified by array (22). Five ancestry-informative principal components from Eigenstrat were used as covariates in logistic regression analyses (23). Association statistics from the two arrays were combined using fixed effects meta-analysis in the META software package (25).
Calculation of ancestral components in CCLS Hispanics
Ancestral components were calculated in Hispanic study subjects using Human Genome Diversity Project samples as the reference founder populations (27). A total of 63,303 autosomal SNPs, common to both datasets, was used to evaluate ancestry using the program Structure (28), as previously described (29). Ancestral proportions were compared between individuals that carried the rs3731249 risk allele and those that did not carry the risk allele using a t-test.
MLPA
Multiplex ligation-dependent probe amplification (MLPA) was performed, as previously described, for 848 ALL patients with sufficient bone marrow DNA available(29). MLPA was carried out using the SALSA MLPA probemix P335-B1 ALL-IKZF1 (MRC Holland), which includes 2 probes within CDKN2A and 1 probe within CDKN2B. Single-gene deletions were identified when either the CDKN2A probes or the CDKN2B probe exclusively showed evidence of copy-number loss, with the other probe(s) remaining copy-neutral. Data analysis was carried out using “Coffalyser.Net” fragment analysis software (MRC Holland). In brief, peak height ratios were determined by intra-sample normalization using data from 13 reference probes in genomic regions known not to be somatically altered in childhood ALL, and by inter-sample normalization using data from constitutive DNA reference samples.
Assessment of rs3731249 allelic imbalance and copy-number using SMART-ddPCR
We assayed all ALL patients that were heterozygous for rs3731249 and had diagnostic bone marrow DNA available (N=37) using a novel application of Droplet DigitalTM PCR (ddPCR) (Bio-Rad), termed Somatic Mutation Allelic Ratio Test (SMART-ddPCR). SMART-ddPCR was used to assess preferential allelic imbalance (PAI) of the CDKN2A missense SNP rs3731249 in tumor DNA. This allowed assessment of the hypothesis that the protective allele of rs3731249 will be preferentially lost in tumors and the risk allele preferentially retained.
ddPCR was carried out as previously described (30). The Taqman SNP Genotyping Assay for rs3731249 (ABI: C__25611114_10) was used in ddPCR reactions, with FAM- and VIC-labeled probes for detection of the risk (T) and protective (C) alleles, respectively. For each of the samples, ddPCR was carried out in duplicate and data were analyzed using QuantaSoftTM Software (Bio-Rad). To determine presence of allelic imbalance, for each subject we calculated the proportion of risk allele compared to protective allele as follows:
This results in a proportion between 0 and 1, with an expected proportion of 0.50 for samples with no allelic imbalance. Two of the 37 samples were excluded due to low concentrations for both the risk and protective alleles in the ddPCR reactions.
To determine thresholds for calling the presence of allelic imbalance in a subject, we analyzed constitutive DNA from rs3731249 heterozygotes using ddPCR. We used 3 standard deviations above and below the mean risk allele proportion in these samples (0.491) to define the upper (0.524) and lower (0.458) thresholds, respectively. Tumor samples with allelic ratios falling outside this range were considered to demonstrate allelic imbalance. Among samples showing allelic imbalance (N=17), the number of tumor samples with PAI favoring the risk allele was compared to the number favoring the protective allele using a binomial test and assuming, under the null hypothesis, that a sample was equally likely to lose one allele as the other (i.e. p=q=0.50).
To assess copy number at the rs3731249 locus, a second ddPCR reaction was carried out using a Taqman assay targeting the SLC24A3 gene within a region not known to be copy number variable in ALL. Concentration of total CDKN2A relative to concentration of the genomic control was calculated as follows:
Tumor samples showing no evidence of copy-number loss but showing allelic imbalance were likely to have acquired copy-neutral LOH. Conversely, samples that presented with copy number loss but without allelic imbalance were presumed to have subclonal homozygous deletions.
RESULTS
Fine-mapping the 9p21.3 association with childhood ALL
Genotype data were generated for 321 Hispanic children with ALL and 454 Hispanic control children from CCLS using dried neonatal bloodspot DNA and the Illumina Omni-Express genotyping array (31). Imputation to 1000 Genomes was performed across a 500kb region on chromosome nine from 21.735Mb to 22.235 Mb (GRCh37/hg19), encompassing the MTAP, CDKN2A, CDKN2B and CDKN2B-AS1 genes and promoters. One-hundred thirty-seven SNPs in the region were directly genotyped on-array and an additional 1909 were successfully imputed. The region is approximately centered on the original ALL GWAS lead SNP (rs3731217), first published by Sherborne et al (5).
The tag SNP from the previous GWAS, rs3731217, was associated with ALL risk in the CCLS Hispanic case-control data (OR=0.65; P=0.020). Capitalizing on the reduced linkage disequilibrium (LD) in the genetically admixed Hispanic discovery set (Figure 2), we identified 51 SNPs with p-values <0.020 and effect sizes larger than that observed for rs3731217. These SNPs were more strongly associated with ALL than rs3731217, both in terms of statistical significance and magnitude of effect, and were carried forward for validation in a second, independent case-control set.
Figure 2. Ethnic-specific patterns of linkage disequilibrium in the 9p21.3 region.
(A) Haplotype structure in the California Childhood Leukemia Study Hispanic discovery set (Ncases=321, Ncontrols=454). (B) Haplotype structure in the Children's Oncology Group and Wellcome Trust Consortium validation set (Ncases=980, Ncontrols=2624). Darker shading indicates higher R2 values and greater correlation between SNPs.
Genotype data were generated for 980 European-ancestry children with ALL using remission blood samples and the Affymetrix 6.0 array by COG, as previously described (12). These 980 ALL cases were combined with 2624 European-ancestry control children from the WTCCC to form the validation case-control set (14). Imputation to 1000 Genomes was performed across the same 500kb region on chromosome nine. 127 SNPs in the region were directly genotyped on-array and an additional 1002 were successfully imputed. Of the 51 SNPs more strongly associated with ALL risk in the Hispanic discovery data than the rs3731217 tag SNP, 46 were successfully genotyped or imputed in the COG/WTCCC validation set. Applying a strict Bonferroni correction for 46 tests of association, 42 of these SNPs were associated at P<1.1×10−3 in the validation set.
SNP associations from the discovery and validation sets were combined using meta-analysis to generate a volcano plot of the joint associations, with effect size plotted on the x-axis and statistical significance plotted on the y-axis (Figure 3). The volcano plot highlights the 42 SNPs identified in discovery analyses and validated in COG data, and reveals three clear outliers in tight LD (R2>0.99): rs113650570, rs36228834 and rs3731249.
Figure 3. Volcano plot of SNP associations from case-control analyses of California Childhood Leukemia Study (CCLS) participants and Children's Oncology Group (COG) participants.
SNP associations from CCLS Hispanics and COG European-ancestry subjects were combined via meta-analysis then plotted with -log10(P-values) on the Y-axis and ln(odds ratio) on the X-axis. The open black circle denotes rs3731217, the ALL tag-SNP identified in previous GWAS. 42 SNPs having smaller p-values and larger effect sizes than GWAS tag SNP rs3731217 in CCLS discovery analyses, and also associated at P<1.1×10−3 in COG validation analyses, appear as black diamonds. Three outlier associations are labeled with their rsID.
Bioinformatic, in-silico functional genomic and conditional SNP analyses
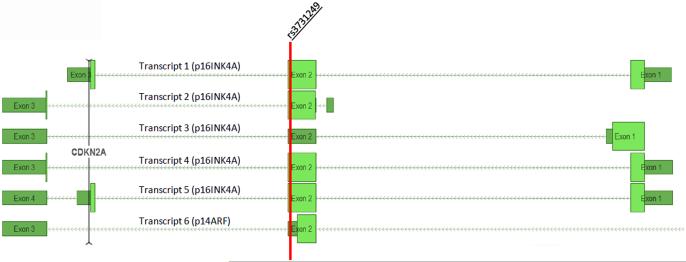
The 42 SNPs identified in discovery analyses which were associated at P<1.1×10−3 in the validation set appear in Supplementary Table 2. These SNPs were functionally annotated using ENCODE2 data, implemented in HaploRegV3 and RegulomeDB (32, 33), and were also investigated using data from the Genotype-Tissue Expression (GTEx) project to determine if they were cis eQTLs for MTAP, CDKN2A or CDKN2B (34). The three outliers identified by the volcano plot (rs113650570, rs36228834 and rs3731249) are more than three orders of magnitude more statistically significantly associated with ALL risk than the fourth-ranked variant, rs56018935 (Supplementary Table 2). While rs113650570 and rs36228834 are intronic SNPs with little direct evidence suggesting a regulatory function (Supplementary Table 2), rs3731249 is a p16 missense variant (A148T) in exon 2 of CDKN2A (Figure 4).
Figure 4. Representation of the coding sequences of the CDKN2A gene.
Transcripts 1-5 are p16 splice variants and transcript 6 is the p14ARF (Exon 1 not pictured). Light green blocks depict coding exons. Dark green blocks are untranslated. The red vertical line indicates the position of the rs3731249 missense variant. This polymorphism alters amino acid coding (Alanine → Threonine) in p16 splice variants 1, 2, 4, and 5, but not in splice variant 3 or in p14ARF. Gene transcript data were extracted from the NCBI Homo sapiens Annotation Release 105.
Numerous SNPs in the region have putative functional relevance based on their location within promoter histone marks, enhancer histone marks, DNase hypersensitivity sites, and canonical transcription factor binding motifs. Additionally, a number of linked SNPs are significant CDKN2B eQTLs (Supplementary Table 2). Thus, it is possible that a number of ALL-associated variants in the region are causally related to leukemogenesis.
Conditional analyses were performed in the discovery and validation datasets to determine whether rs3731249 could explain the majority of the ALL association signal in the 9p21.3 region. Because the three lead SNPs (rs113650570, rs36228834 and rs3731249) are in complete LD, conditional analyses are unable to assess their effects independent of each other. Adjusting for subject genotype at rs3731249 greatly attenuated SNP associations in the region in logistic regression models (Figure 5). After adjusting for rs3731249, the most significantly associated SNP in meta-analysis of the two datasets was rs2188127, an intergenic variant between MTAP and CDKN2A (P=1.1×10−4, OR=0.73, 95% CI=0.62-0.86). Prior to adjustments for rs3731249, 54 SNPs had smaller p-values in the meta-analysis. These conditional analyses indicate that rs3731249, or a variant in tight LD, underlies the CDKN2A association signal in the region.
Figure 5. SNP association plots for ALL risk at 9p21.3 in meta-analysis of CCLS Hispanic discovery set and the COG/WTCCC validation set, both with and without adjustment for missense variant rs3731249.
The strength of linkage disequilibrium between each SNP and missense variant rs3731249 (purple circle) is indicated by color. Recombination rates, plotted in light blue, are based on 1000 Genomes CEU samples. (A) Association plot for ALL risk in the combined CCLS Hispanic discovery set and COG validation set. (B) Association plot for ALL risk in the combined CCLS Hispanic discovery set and COG validation set, adjusted for rs3731249 genotype.
Association of rs3731249 with childhood ALL
In the CCLS Hispanic discovery set, the minor (T) allele of the missense SNP rs3731249 was associated with a nearly 3-fold increased risk of ALL (OR=2.77; 95% CI=1.58-4.85; P=3.78×10−4). Hispanic subjects carrying the rs3731249 risk allele had genomes that were more European than individuals not carrying the risk allele, among both cases and controls (63.1% European versus 56.2% European; P=5.6×10−3). This 7% increase in European genomic ancestry also corresponded to a 7% decrease in Native American genomic ancestry. The association between the rs3731249 risk allele and increased European ancestry suggests that this allele originated on a European haplotype. The higher risk allele frequency in European-ancestry genomes also suggests that rs3731249 may be an ALL risk factor in additional populations harboring European ancestry, including non-Hispanic whites and African-Americans.
In the European-ancestry COG validation set, the minor allele of missense SNP rs3731249 was also associated with a 3-fold increased risk of ALL (OR=2.99; 95% CI=2.10-4.26; P=1.51×10−9). Along with the two intronic SNPs rs113650570 and rs36228834, this was the most statistically significant association in the validation set.
TaqMan genotyping of a third ALL case-control set of Hispanic and African-American children (163 cases, 201 controls) again confirmed the 3-fold increased risk associated with rs3731249 (OR=3.59; 95% CI=1.22-10.59; P=8.8×10−3). Meta-analysis of the association between rs3731249 and ALL risk, across the three ALL case-control sets, reached genome-wide statistical significance (Pmeta=1.69×10−13; ORmeta=2.97; 95% CI=2.22-3.96).
There was no heterogeneity of effect between B-cell and T-cell ALL in CCLS subjects, but rs3731249 did have a larger effect size within certain molecularly-defined ALL subgroups (29). Rs3731249 was associated with a 4.5-fold increased risk of ALL harboring a somatic CDKN2A deletion (OR=4.5; P=6.4×10−3), a 6-fold increased risk of IKZF1-deleted ALL (OR=6.3; P=1.9×10−3) and a 6-fold increased risk of KRAS- or NRAS-mutated ALL (OR=6.4; P=6.1×10−4). Effect sizes were similar in high-hyperdiploid ALL as in the ETV6-RUNX1 fusion ALL subgroup (OR=2.7 and 2.0, respectively).
MLPA assessment of 9p21.3 focal deletions
We assessed frequency of somatic 9p21.3 deletions in 848 ALL tumor specimens from the CCLS using MLPA, and identified 256 with copy-number loss at 9p21.3 (30.2%). 33 of these samples had focal single-gene deletions, of which 27 exclusively affected CDKN2A and 6 exclusively affected CDKN2B, suggesting that CDKN2A is the principal target of 9p21.3 deletions in ALL (P=3.0×10−4).
Preferential allelic imbalance of the rs3731249 risk allele in ALL tumors
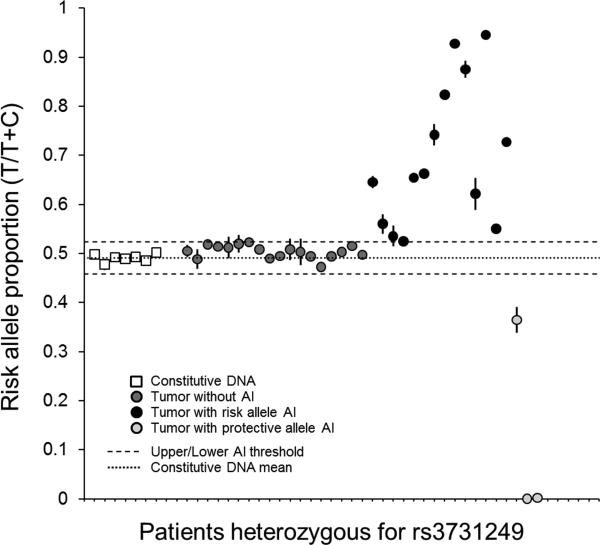
Because a heritable CDKN2A missense variant was strongly and robustly associated with increased ALL risk in three case-control sets, and ALL tumors frequently harbor somatic alterations of CDKN2A, we sought to determine if the risk allele is preferentially retained (and the protective allele preferentially lost) in ALL tumors. To investigate this interface of heritable and acquired CDKN2A variation in leukemogenesis, we assessed preferential allelic imbalance of rs3731249 using droplet digital PCR. This assay, termed “SMART-ddPCR” (see Methods), was used to measure the relative number of copies of the T and C alleles in constitutive DNA and in leukemia diagnostic bone marrow samples (i.e. tumor DNA) in a subset of CCLS cases. Copies of the rs3731249 risk (T) and protective (C) allele were approximately equal in diploid constitutive DNA from heterozygous patients, as anticipated, with a mean risk allele proportion of 0.491 and upper and lower thresholds of 0.524 and 0.458 respectively (Figure 6).
Figure 6. Risk allele proportions in constitutive and tumor DNA from ALL patients heterozygous for the rs3731249 missense variant.
Risk allele proportions are displayed as a fraction of total allelic copy number measured using ddPCR. Subjects were assayed in duplicate, and error bars represent standard error of the mean. Upper/lower thresholds of allelic imbalance (AI) were determined from repeat measurements of constitutive DNA samples (white squares). In tumor DNA, 14 patients showed AI favoring the rs3731249 risk allele versus 3 patients with AI favoring the protective allele (PBinomial=0.006).
A total of 57 ALL patients from CCLS were heterozygous for the rs3731249 risk allele in Illumina GWAS genotyping or targeted TaqMan genotyping. Sufficient tumor tissue was available for 35 of these patients, of which 17 showed evidence of allelic imbalance due to hemizygous deletion (N=11) or copy-neutral LOH (N=6). Of these 17 tumors with allelic imbalance, 14 preferentially retained the risk allele and 3 preferentially retained the protective allele (PBinomial=0.006) (Figure 6). This indicates that the rs3731249 missense SNP is subject to directional selection, wherein the ALL risk allele is favored during clonal evolution of the tumor. The 18 tumor samples that did not display allelic imbalance either had no detectable 9p21.3 lesions (N=13) or had subclonal homozygous deletion (N=5), suggesting a complex assortment of somatic CDKN2A alterations in leukemogenesis.
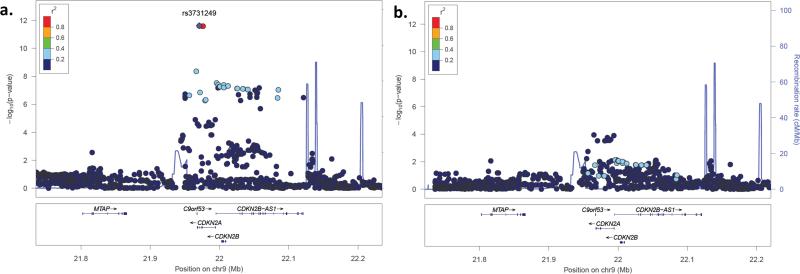
9p21.3 association mapping in additional cancers
Germline mutations in CDKN2A are associated with familial melanoma-astrocytoma syndrome (35) and familial melanoma-pancreatic cancer syndrome (36). Additionally, the cancer-associated GWAS hits on 9p21.3 span 320kb from a melanoma-associated variant at rs7023329 near MTAP(6) to a glioma-associated variant at rs4977756 in CDKN2B-AS1(7). The ALL association peak in CDKN2A partially overlaps the melanoma and glioma peaks, suggesting that pan-cancer risk alleles may reside in the region (Figure 7). To determine if the putatively hypomorphic rs3731249 variant is associated with adult melanoma, glioblastoma, or pancreatic cancer risk, we used data from three large case-control studies of European-ancestry populations. Among 1969 adults with melanoma and 1044 controls genotyped on-array, strong association was detected in MTAP (Pmin=5.5×10−7. However, no significant associations were detected in CDKN2A, including at missense variant rs3731249 (P=0.52) (Figure 7a). Pooled deep-sequencing of the 9p21.3 region in 684 adults with glioblastoma and 821 controls identified strong association in CDKN2B-AS1 (Pmin=8.2×10−8. However, rs3731249 was not significantly associated with glioblastoma risk (P=0.52) (Figure 7c). Among 2273 adults with pancreatic cancer and 2418 controls genotyped on-array, no 9p21.3 SNPs had P-values <0.05, including rs3731249 (P=0.15) (data not shown in figure).
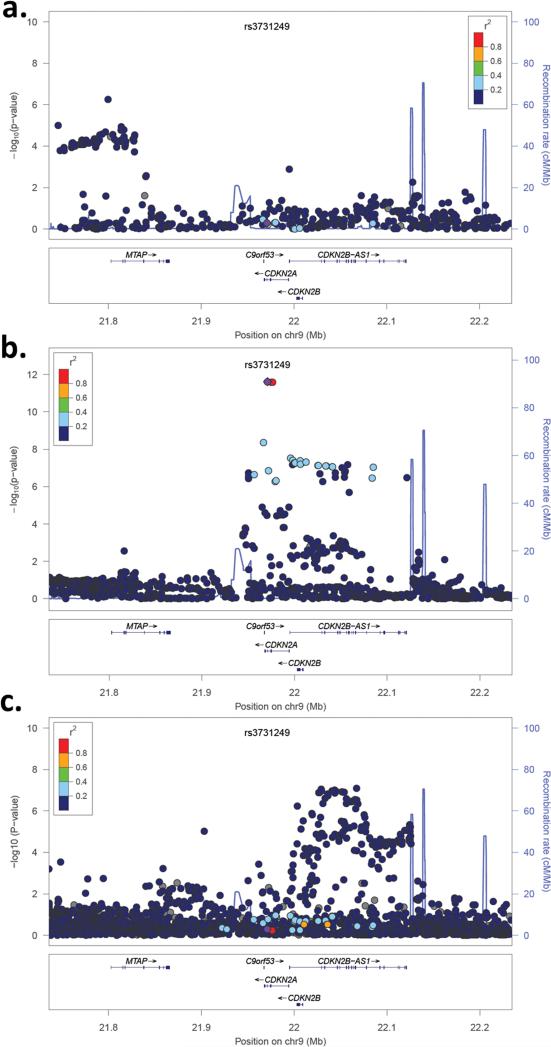
Figure 7. SNP association plots for risk of melanoma, childhood ALL, and glioblastoma at 9p21.3.
The strength of LD between each SNP and missense variant rs3731249 (purple circle) is indicated by color. Recombination rates, plotted in light blue, are based on 1000 Genomes samples. (A) SNP association plot for melanoma risk among 1969 cases and 1044 controls of European-ancestry. (B) SNP association plot for childhood ALL risk from a meta-analysis of 321 cases and 454 controls from the CCLS Hispanic GWAS and 980 cases and 2624 controls from COG/WTCCC. (C) SNP association plot for glioblastoma risk among 684 patients and 821 controls of European-ancestry from the UCSF Adult Glioma Study and The Mayo Clinic.
We also sought to determine if the lead SNPs from melanoma and glioblastoma case-control analyses are associated with ALL risk. The lead melanoma SNP from previous GWAS, rs7023329, was not associated with ALL risk in meta-analysis of the CCLS and COG datasets (OR=0.93; P=0.15). Similarly, the lead glioblastoma SNP from previous GWAS, rs4977756, was not associated with ALL risk in meta-analysis of the CCLS and COG datasets (OR=0.99; P=0.79).
DISCUSSION
We have refined the childhood ALL association peak at chromosome 9p21.3 via genotyping and fine-mapping, revealing a heritable p16 missense variant rs3731249 (A148T) that confers a 3-fold increased risk of childhood ALL, consistent across three ALL case-control sets with European ancestry or admixture. Adjusting for rs3731249 removed the majority of the association signal at chr9p21.3. Identifying a missense variant with such a large effect on ALL risk, and one that has not been previously reported, demonstrates the utility of fine-mapping known association loci through a combination of genotyping and imputation with whole-genome sequencing datasets, as well as the value of genetically admixed populations for identifying putatively causal variants.
Using several online tools, we assessed whether the amino acid change from alanine to threonine affects the protein function of p16, but found no evidence of any strong deleterious effects (37, 38). However, evidence from previous studies suggests that p16(A148T) may result in reduced efficiency of p16 as a cell cycle inhibitor. The variant protein has been shown to have diminished ability to compete with cyclin D for CDK4 binding (39) and has altered subcellular localization and expression relative to wild-type p16 (40). The 2% risk allele frequency observed in healthy controls suggests that the effect of A148T on p16 activity is likely subtle and may only become important in the context of a pre-cancerous lymphoblast. Furthermore, rs3731249 was not associated with risk of melanoma, glioblastoma, or pancreatic cancer. This suggests rs3731249 may be associated with risk of cancer only in tissues that constantly self-renew, such as the bone marrow, but not associated with risk of cancer in tissues with low rates of self-renewal, such as glia and melanocytes (41, 42).
SNPs on 9p21.3 have been associated with multiple cancers and as a variety of other medical conditions, including heart disease, stroke, diabetes, and glaucoma (14, 43-45). The widely assorted localization of these associations suggests multiple functional genetic polymorphisms exist in the region, and also highlights the core pleotropic functionality of genes on 9p21.3.
The second exon of CDKN2A encodes portions of both the p14 and p16 proteins. Although the rs3731249 polymorphism is within exon two, it is positioned outside of the p14 coding region and is therefore unlikely to affect function of that protein (Figure 4). This suggests that p16, and not p14 or p15, is the critical mediator of leukemogenesis in the 9p21.3 region. This is further supported by the MLPA data which demonstrated that CDKN2A, and not CDKN2B, was the target of focal single-gene deletions in the region.
Heritable genetic variants have recently been associated with specific subtypes of childhood ALL, with the GATA3 rs3824662 risk allele associated exclusively with genomic alterations that underlie Philadelphia chromosome-like ALL (4) and PIP4K2A SNPs associated exclusively with high-hyperdiploid ALL (46). However, the effects of these variants on subtype-specific somatic alterations have yet to be elucidated. In this study, we identify a CDKN2A missense variant that confers high risk of ALL and even higher risk of CDKN2A-deleted ALL, suggesting that the risk variant may initiate a molecular cascade enhanced by subsequent deletion. Furthermore, we demonstrate a direct interaction between heritable and somatic genetic variation in CDKN2A, made evident in the form of PAI.
We developed a novel methodology, SMART-ddPCR, to investigate PAI of cancer-associated heritable variants via absolute measurement of risk and protective allelic copy number. We demonstrate that in childhood ALL tumor samples, there is preferential retention of the risk allele and loss of the protective allele among rs3731249 heterozygotes with somatic CDKN2A alterations, suggesting an important role for this missense SNP in leukemia etiology and progression. Of twenty-one heterozygotes that did not display PAI for the rs3731249 risk allele, five were observed to have subclonal homozygous deletion of CDKN2A, thereby inactivating both alleles. Another three subjects had deletion of RB1, suggesting that subjects not displaying PAI may have CDKN2A point mutations or alterations to other genes involved in control of cell cycle.
The complexity and subclonality of somatic CDKN2A alterations as revealed by our SMART-ddPCR data support current evolutionary theories of cancer development. It has recently been shown that CDKN2A deletions can be caused by off-target RAG-mediated mutational processes (47), which could result in hemizygous or homozygous CDKN2A loss, or in copy-neutral LOH via attempted repair of hemizygous deletions. The initial event may be random (i.e. deletion of the rs3731249 risk allele is just as likely as the protective allele), but tumor cells retaining the risk allele have a competitive advantage during tumor evolution. Our results provide evidence that heritable genetic variation can act as an additional substrate for selection during clonal evolution of childhood ALL.
The rs3731249 risk allele is found at a frequency of 2% in healthy control children. Such common variants do not often have such large effect sizes. However, a SNP in the 3’-UTR of TP53 was recently shown to confer 3-fold increased risk of basal cell carcinoma, neuroblastoma, and glioma (48-50). Our data reveal that p16 missense SNP rs3731249 confers a 3-fold increased risk of ALL in three independent and ethnically diverse case-control sample sets. Furthermore, this variant is subject to directional selection during tumor evolution. These results demonstrate, for the first time, a direct interaction between heritable and somatic CDKN2A variation underlying leukemogenesis.
Supplementary Material
ACKNOWLEDGEMENTS
This work was financially supported by NCI R01CA155461 (JLW, XM), NCI R25CA112355 (KMW), NIEHS and EPA P01ES018172 (CM, JLW), NIEHS R01ES09137 (CM, JLW) and grants from the Swiss Cancer League and the SICPA Foundation (SG). Work at The University of California, San Francisco Department of Neurological Surgery was supported by the National Institutes of Health (grant numbers R01CA52689, P50CA097257), as well as the National Brain Tumor Foundation, the Stanley D. Lewis and Virginia S. Lewis Endowed Chair in Brain Tumor Research, the Robert Magnin Newman Endowed Chair in Neuro-oncology, and by donations from families and friends of John Berardi, Helen Glaser, Elvera Olsen, Raymond E. Cooper, and William Martinusen. Work at the Mayo Clinic was supported by the National Institutes of Health (grant numbers P50CA108961 and P30 CA15083), the National Institute of Neurological Disorders and Stroke (grant number RC1NS068222Z), the Bernie and Edith Waterman Foundation, and the Ting Tsung and Wei Fong Chao Family Foundation.
This study makes use of data generated by the Wellcome Trust Case-Control Consortium. A full list of the investigators who contributed to the generation of the data is available from www.wtccc.org.uk. Funding for the project was provided by the Wellcome Trust under award 076113 and 085475. Melanoma case-control data were obtained from dbGaP Study Accession phs000187.v1.p1 (High Density SNP Association Analysis of Melanoma: Case-Control and Outcomes Investigation). Research support to collect data and develop an application to support this project was provided by 3P50CA093459, 5P50CA097007, 5R01ES011740, and 5R01CA133996. Pancreatic cancer case-control data were obtained from dbGaP Study Accession phs000206.v4.p3 (CGEMS Pancreatic Cancer (PanScan)). The ALL Relapse GWAS dataset used for the analyses described in this manuscript were obtained from dbGaP study accession phs000638.v1.p1 (Genome-Wide Association Study of Relapse of Childhood Acute Lymphoblastic Leukemia). The ALL Relapse GWAS dataset was generated at St. Jude Children's Research Hospital and by the Children's Oncology Group, supported by NIH grants CA142665, CA21765, CA158568, CA156449, CA36401, CA98543, CA114766, CA140729, and U01GM92666, Jeffrey Pride Foundation, the National Childhood Cancer Foundation, and by ALSAC.
The authors gratefully acknowledge the clinical investigators at the following collaborating hospitals for help in recruiting patients: University of California Davis Medical Center (Dr. Jonathan Ducore), University of California San Francisco (Drs. Mignon Loh and Katherine Matthay), Children's Hospital of Central California (Dr. Vonda Crouse), Lucile Packard Children's Hospital (Dr. Gary Dahl), Children's Hospital Oakland (Dr. James Feusner), Kaiser Permanente Roseville (formerly Sacramento) (Drs. Kent Jolly and Vincent Kiley), Kaiser Permanente Santa Clara (Drs. Carolyn Russo, Alan Wong, and Denah Taggart), Kaiser Permanente San Francisco (Dr. Kenneth Leung), and Kaiser Permanente Oakland (Drs. Daniel Kronish and Stacy Month). Bioinformatic analysis of next-gen sequencing data from glioma cases and controls was performed with extensive support from Yuanyuan Xiao. Paige M. Bracci contributed helpful analytic advice for calculating agreement between MLPA and ddPCR assays via Bland-Altman plots.
The collection of cancer incidence data used in this study was supported by the California Department of Public Health as part of the statewide cancer reporting program mandated by California Health and Safety Code Section 103885; the National Cancer Institute's Surveillance, Epidemiology and End Results Program under contract HHSN261201000140C awarded to the Cancer Prevention Institute of California, contract HHSN261201000035C awarded to the University of Southern California, and contract HHSN261201000034C awarded to the Public Health Institute; and the Centers for Disease Control and Prevention's National Program of Cancer Registries, under agreement # U58DP003862-01 awarded to the California Department of Public Health. The ideas and opinions expressed herein are those of the author(s) and endorsement by the State of California Department of Public Health, the National Cancer Institute, and the Centers for Disease Control and Prevention or their Contractors and Subcontractors is not intended nor should be inferred.
Footnotes
CONFLICT OF INTEREST STATEMENT:
The authors declare no competing financial interests.
REFERENCES
- 1.Greaves M. Infection, immune responses and the aetiology of childhood leukaemia. Nat Rev Cancer. 2006;6(3):193–203. doi: 10.1038/nrc1816. [DOI] [PubMed] [Google Scholar]
- 2.Xu H, Yang W, Perez-Andreu V, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst. 2013;105(10):733–42. doi: 10.1093/jnci/djt042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Migliorini G, Fiege B, Hosking FJ, et al. Variation at 10p12.2 and 10p14 influences risk of childhood B-cell acute lymphoblastic leukemia and phenotype. Blood. 2013;122(19):3298–307. doi: 10.1182/blood-2013-03-491316. [DOI] [PubMed] [Google Scholar]
- 4.Perez-Andreu V, Roberts KG, Harvey RC, et al. Inherited GATA3 variants are associated with Ph-like childhood acute lymphoblastic leukemia and risk of relapse. Nat Genet. 2013;45(12):1494–8. doi: 10.1038/ng.2803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sherborne AL, Hosking FJ, Prasad RB, et al. Variation in CDKN2A at 9p21.3 influences childhood acute lymphoblastic leukemia risk. Nat Genet. 2010;42(6):492–4. doi: 10.1038/ng.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett JH, Iles MM, Harland M, et al. Genome-wide association study identifies three new melanoma susceptibility loci. Nat Genet. 2011;43(11):1108–13. doi: 10.1038/ng.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wrensch M, Jenkins RB, Chang JS, et al. Variants in the CDKN2B and RTEL1 regions are associated with high-grade glioma susceptibility. Nat Genet. 2009;41(8):905–8. doi: 10.1038/ng.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bei JX, Li Y, Jia WH, et al. A genome-wide association study of nasopharyngeal carcinoma identifies three new susceptibility loci. Nat Genet. 2010;42(7):599–603. doi: 10.1038/ng.601. [DOI] [PubMed] [Google Scholar]
- 9.Timofeeva MN, Hung RJ, Rafnar T, et al. Influence of common genetic variation on lung cancer risk: meta-analysis of 14 900 cases and 29 485 controls. Hum Mol Genet. 2012;21(22):4980–95. doi: 10.1093/hmg/dds334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schwab CJ, Chilton L, Morrison H, et al. Genes commonly deleted in childhood B-cell precursor acute lymphoblastic leukemia: association with cytogenetics and clinical features. Haematologica. 2013;98(7):1081–8. doi: 10.3324/haematol.2013.085175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Walsh KM, Chokkalingam AP, Hsu LI, et al. Associations between genome-wide Native American ancestry, known risk alleles and B-cell ALL risk in Hispanic children. Leukemia. 2013;27(12):2416–9. doi: 10.1038/leu.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang JJ, Cheng C, Devidas M, et al. Genome-wide association study identifies germline polymorphisms associated with relapse of childhood acute lymphoblastic leukemia. Blood. 2012;120(20):4197–204. doi: 10.1182/blood-2012-07-440107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang JJ, Cheng C, Devidas M, et al. Ancestry and pharmacogenomics of relapse in acute lymphoblastic leukemia. Nat Genet. 2011;43(3):237–41. doi: 10.1038/ng.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Amos CI, Wang LE, Lee JE, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Hum Mol Genet. 2011;20(24):5012–23. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walsh KM, Codd V, Smirnov IV, et al. Variants near TERT and TERC influencing telomere length are associated with high-grade glioma risk. Nat Genet. 2014;46(7):731–5. doi: 10.1038/ng.3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jenkins RB, Xiao Y, Sicotte H, et al. A low-frequency variant at 8q24.21 is strongly associated with risk of oligodendroglial tumors and astrocytomas with IDH1 or IDH2 mutation. Nat Genet. 2012;44(10):1122–5. doi: 10.1038/ng.2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petersen GM, Amundadottir L, Fuchs CS, et al. A genome-wide association study identifies pancreatic cancer susceptibility loci on chromosomes 13q22.1, 1q32.1 and 5p15.33. Nat Genet. 2010;42(3):224–8. doi: 10.1038/ng.522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amundadottir L, Kraft P, Stolzenberg-Solomon RZ, et al. Genome-wide association study identifies variants in the ABO locus associated with susceptibility to pancreatic cancer. Nat Genet. 2009;41(9):986–90. doi: 10.1038/ng.429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5(6):e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Abecasis GR, Altshuler D, Auton A, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467(7319):1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010;11(7):499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 23.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 24.Chokkalingam AP, Aldrich MC, Bartley K, et al. Matching on Race and Ethnicity in Case-Control Studies as a Means of Control for Population Stratification. Epidemiology (Sunnyvale) 2011;1:101. doi: 10.4172/2161-1165.1000101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu JZ, Tozzi F, Waterworth DM, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nat Genet. 2010;42(5):436–40. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81(3):559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li JZ, Absher DM, Tang H, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 28.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164(4):1567–87. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh KM, de Smith AJ, Welch TC, et al. Genomic ancestry and somatic alterations correlate with age at diagnosis in Hispanic children with B-cell acute lymphoblastic leukemia. Am J Hematol. 2014;89(7):721–5. doi: 10.1002/ajh.23727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hindson BJ, Ness KD, Masquelier DA, et al. High-throughput droplet digital PCR system for absolute quantitation of DNA copy number. Anal Chem. 2011;83(22):8604–10. doi: 10.1021/ac202028g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walsh KM, de Smith AJ, Chokkalingam AP, et al. GATA3 risk alleles are associated with ancestral components in Hispanic children with ALL. Blood. 2013;122(19):3385–7. doi: 10.1182/blood-2013-08-524124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boyle AP, Hong EL, Hariharan M, et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012;22(9):1790–7. doi: 10.1101/gr.137323.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40(Database issue):D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.The Genotype-Tissue Expression (GTEx) project. Nat Genet. 2013;45(6):580–5. doi: 10.1038/ng.2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bahuau M, Vidaud D, Jenkins RB, et al. Germ-line deletion involving the INK4 locus in familial proneness to melanoma and nervous system tumors. Cancer Res. 1998;58(11):2298–303. [PubMed] [Google Scholar]
- 36.Goldstein AM, Fraser MC, Struewing JP, et al. Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333(15):970–4. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 37.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat Protoc. 2009;4(7):1073–81. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 38.Schwarz JM, Rodelsperger C, Schuelke M, Seelow D. MutationTaster evaluates disease-causing potential of sequence alterations. Nat Methods. 2010;7(8):575–6. doi: 10.1038/nmeth0810-575. [DOI] [PubMed] [Google Scholar]
- 39.Reymond A, Brent R. p16 proteins from melanoma-prone families are deficient in binding to Cdk4. Oncogene. 1995;11(6):1173–8. [PubMed] [Google Scholar]
- 40.Walker GJ, Gabrielli BG, Castellano M, Hayward NK. Functional reassessment of P16 variants using a transfection-based assay. Int J Cancer. 1999;82(2):305–12. doi: 10.1002/(sici)1097-0215(19990719)82:2<305::aid-ijc24>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 41.Killela PJ, Reitman ZJ, Jiao Y, et al. TERT promoter mutations occur frequently in gliomas and a subset of tumors derived from cells with low rates of self-renewal. Proc Natl Acad Sci U S A. 2013;110(15):6021–6. doi: 10.1073/pnas.1303607110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tomasetti C, Vogelstein B. Cancer etiology. Variation in cancer risk among tissues can be explained by the number of stem cell divisions. Science. 2015;347(6217):78–81. doi: 10.1126/science.1260825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Burdon KP, Macgregor S, Hewitt AW, et al. Genome-wide association study identifies susceptibility loci for open angle glaucoma at TMCO1 and CDKN2B-AS1. Nat Genet. 2011;43(6):574–8. doi: 10.1038/ng.824. [DOI] [PubMed] [Google Scholar]
- 44.Helgadottir A, Thorleifsson G, Manolescu A, et al. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007;316(5830):1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 45.Yasuno K, Bilguvar K, Bijlenga P, et al. Genome-wide association study of intracranial aneurysm identifies three new risk loci. Nat Genet. 2010;42(5):420–5. doi: 10.1038/ng.563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Walsh KM, de Smith AJ, Chokkalingam AP, et al. Novel childhood ALL susceptibility locus BMI1-PIP4K2A is specifically associated with the hyperdiploid subtype. Blood. 2013;121(23):4808–9. doi: 10.1182/blood-2013-04-495390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papaemmanuil E, Rapado I, Li Y, et al. RAG-mediated recombination is the predominant driver of oncogenic rearrangement in ETV6-RUNX1 acute lymphoblastic leukemia. Nat Genet. 2014;46(2):116–25. doi: 10.1038/ng.2874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walsh KM, Anderson E, Hansen HM, et al. Analysis of 60 reported glioma risk SNPs replicates published GWAS findings but fails to replicate associations from published candidate-gene studies. Genet Epidemiol. 2013;37(2):222–8. doi: 10.1002/gepi.21707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Diskin SJ, Capasso M, Diamond M, et al. Rare variants in TP53 and susceptibility to neuroblastoma. J Natl Cancer Inst. 2014;106(4):dju047. doi: 10.1093/jnci/dju047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stacey SN, Sulem P, Jonasdottir A, et al. A germline variant in the TP53 polyadenylation signal confers cancer susceptibility. Nat Genet. 2011;43(11):1098–103. doi: 10.1038/ng.926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.