Abstract
Increasing evidence identifies dicarbonyl stress from reactive glucose metabolites, such as methylglyoxal (MG), as a major pathogenic link between hyperglycemia and complications of diabetes. MG covalently modifies arginine residues, yet the site specificity of this modification has not been thoroughly investigated. Sites of MG adduction in the plasma proteome were identified using LC-MS/MS analysis in vitro following incubation of plasma proteins with MG. Treatment of plasma proteins with MG yielded 14 putative MG hotspots from five plasma proteins (albumin [nine hotspots], serotransferrin, haptoglobin [2 hotspots], hemopexin, and Ig lambda-2 chain C regions). The search results revealed two versions of MG-arginine modification, dihydroxyimidazolidine (R+72) and hydroimidazolone (R+54) adducts. One of the sites identified was R257 in human serum albumin, which is a critical residue located in drug binding site I. This site was validated as a target for MG modification by a fluorescent probe displacement assay, which revealed significant drug dissociation at 300 μM MG from a prodan-HSA complex (75 μM). Moreover, twelve human plasma samples (six male, six female, with two type 2 diabetic subjects from both genders) were analyzed using multiple reaction monitoring (MRM) tandem mass spectrometry and revealed the presence of the MG-modified albumin R257 peptide. These data provide insights into the nature of the site-specificity of MG modification of arginine, which may be useful for therapeutic treatments that aim to prevent MG-mediated adverse responses in patients.
Keywords: Adducts, glyco-oxidation, methylglyoxal, proteomics, type 2 diabetes mellitus
Introduction
Methylglyoxal (MG) is a reactive glucose metabolite formed by the spontaneous degradation of triosephosphates, oxidative metabolism of ketone bodies, and catabolism of threonine (Ahmed et al., 2005b). Accumulation of MG by increased formation and/or decreased metabolism creates a state of carbonyl stress (Xue et al., 2012). Excess MG is linked to the pathophysiology of many chronic diseases, including diabetes and complications associated with diabetes (Brownlee, 2001; Beisswenger et al., 2005). One of the critical pathogenic consequences of hyperglycemia in diabetes is a deficit in the removal of reactive dicarbonyls, such as MG (Sheetz and King, 2002). Many factors can trigger the accumulation of MG in vivo, including aging, hyperglycemia, inflammation, oxidative stress, and uremia (Ahmed and Thornalley, 2007).
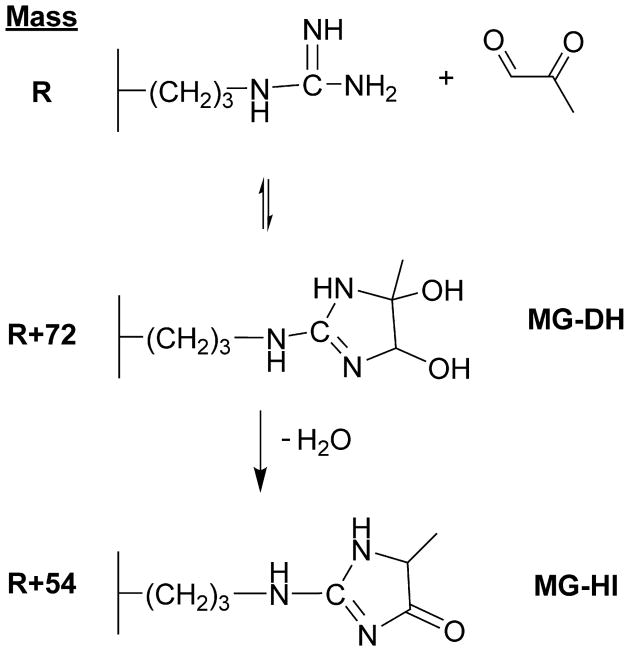
MG is a reactive dicarbonyl electrophile that forms adducts with guanidine and amino groups of proteins, guanosyl bases of nucleic acids, and amino groups of basic phospholipids. MG reacts up to 20 X 103 times faster with proteins than glucose (Ahmed and Thornalley, 2007). One of the most quantitatively and functionally important MG adducts is the methylglyoxal-derived hydroimidazolone (MG-HI) adduct on arginine, producing a loss of positive charge via formation of a neutral hydroimidazolone (Scheme 1). The pKa of MG-HI is 4.58, a drastic change from the arginine side chain pKa of 12.48 (Wang et al., 2012). MG will also adduct to the side chains on lysine and cysteine residues, but adducts formed on these residues are transient and exhibit faster off-rate kinetics (Lo et al., 1994). Depending on the protein, approximately 0.1% of arginines in plasma proteins bear the MG-HI modification, as determined by exhaustive digestion and quantitation using MS/MS (Duran-Jimenez et al., 2009). Moreover, the relative abundance of MG adducts is estimated to be as high as one MG-derived adduct on 3–13% of all proteins, assuming the average protein contains 26 arginines (Ahmed et al., 2005b). There remains debate over the exact physiological concentration range of MG, and since MG is a reactive electrophile, with >99% bound either reversibly or irreversibly with protein (Dhar et al., 2009). The rate of MG formation is estimated to be 120 μM/day under normoglycemic conditions (Thornalley, 1993). Different analytical platforms to measure the MG-derivatives yield widely different plasma concentrations, ranging from 40 nM to 4.5 μM (Khuhawar et al., 2008; Dhar et al., 2009; Rabbani and Thornalley, 2011; Rabbani and Thornalley, 2014; Thornalley and Rabbani, 2014). These reports also indicate that diabetic patients exhibit higher concentrations of MG relative to normoglycemic patients. Prolonged exposure to low concentrations of endogenous MG throughout the half-life of plasma proteins may have a profound adverse effect in situations of poor glycemic control and increased dicarbonyl stress.
Scheme 1. Chemistry of MG-Arginine Adduction.
MG primarily reacts with arginine residues to form neutral ring structures- MG-DH (dihydroxyimidazolidine, R+72) and MG-HI (hydroimidazolone, R+54).
Adduction of proteins by MG is likely to be of physiological importance because of the high levels of MG residues in cellular and extracellular proteins relative to other advanced glycation endproducts (AGEs) (Ahmed et al., 2005a). Such adducts could be functionally important, especially since arginine residues occur at high frequency in ligand and substrate recognition sites of transport proteins, receptors and enzyme active sites (Thornalley et al., 2003; Kimzey et al., 2011; Bose et al., 2013; Morgan et al., 2013). Of all 20 amino acids, arginine has the highest probability of being located in these active sites, with approximately 20% of active sites containing at least one arginine (Gallet et al., 2000). Receptor binding domain analysis of 80,000 protein sequences estimate that this frequency is a 3.8-fold greater than a completely random distribution would predict (Gallet et al., 2000).
The current study represents a comprehensive proteomics analysis designed to identify potential MG modification sites in human plasma. The strategy permits the sensitive and high-throughput identification of arginine sites in the plasma of human patients. Knowledge of sites of MG modification in the plasma proteome is a prerequisite for determining possible biological and toxicological sequelae of such modifications.
Materials and Methods
Materials
HPLC grade solvents were purchased from Sigma-Aldrich unless otherwise noted. Sequencing grade trypsin was purchased from Promega (Fitchburg, WI). Prodan (6-Propionyl-2 – dimethylaminonaphthalene) was a product of Anaspec Inc (Fremont, CA, catalog #88212, lot #64774). Fatty acid-free human serum albumin (catalog # A3782) and 40% methylglyoxal solution were obtained from Sigma-Aldrich. Lipidex-1000 was acquired from PerkinElmer (Waltham, MA).
Subject selection
Study design and subject recruitment was approved by the University of Arizona Institutional Review Board through the Human Subjects Protection Program (project number 07-0812-01). All subjects provided informed consent. Subjects were recruited from the University Medical Center, University Physicians Healthcare-Kino, Southern Arizona VA Health Care System, and El Rio diabetes and primary care clinics. From this subject pool, a cross-section of 12 subjects were selected for this preliminary study. Six male and six female, with two type 2 diabetes subjects each, were selected. Subjects were not age-matched.
Sample handling and storage
Blood was collected into heparin coated vacutainer tubes and immediately placed on ice. Blood samples were centrifuged at 4 °C and plasma was aspirated and stored at −80 °C in 200 μL aliquots. The total time between blood collection and sample storage was less than one hour.
Plasma protein fractionation and modification
Plasma (50 μl) from two healthy subjects were diluted to 600 μl with TBS (Tris-buffered saline) pH 7.4 and centrifuged through a 0.2 μm pore size spin filter to remove particulates. The samples were incubated with a concentration of 500 μM MG at 37 °C for 24 hours in order to best detect MG-adducted peptides via LC/MS-MS methods. Unmodified plasma from the same patients were used as a baseline control. The samples were buffered exchanged into 100 mM ammonium bicarbonate pH 7.4 using Vivaspin centrifuge concentrators (MWCO 3K).
Tryptic digestion
Modified plasma was reduced with DTT (20 mM in 100 mM ammonium bicarbonate pH 7.4) for 30 minutes at 55°C and alkylated with iodoacetamide (55 mM in 100 mM ammonium bicarbonate pH 7.4) for 30 minutes at room temperature in the dark. Protein was then digested with trypsin (protein to trypsin at 50:1 w/w ratio) overnight at 37°C. Peptides were desalted using Hypersep C18 columns (Thermo Scientific), lyophilized, and re-suspended in 10 μL of 1% TFA immediately prior to LC-MS/MS.
LC-MS/MS for plasma proteins
LC-MS/MS analysis of in-solution trypsin digested-proteins (Shevchenko et al., 1996) was carried out using a LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an Advion nanomate ESI source (Advion, Ithaca, NY), following ZipTip (Millipore, Billerica, MA) C18 sample clean-up according to the manufacturer’s instructions. Peptides were eluted from a C18 precolumn (100-μm id × 2 cm, Thermo Fisher Scientific) onto an analytical column (75-μm ID × 10 cm, C18, Thermo Fisher Scientific) using a using a 5% hold of solvent B (acetonitrile, 0.1% formic acid) for 5 min, followed by a 5–7% gradient of solvent B over 5 min, 7–15% gradient of solvent B over 45 min, 15–35% gradient of solvent B over 60 min, 35–40% gradient of solvent B over 28 min, 40–85% gradient of solvent B over 5 min, 85% hold of solvent B for 10 min and finally a return to 5% in 1 minute and another 10 minute hold of 5% solvent B. All flow rates were 400 nl/min. Solvent A consisted of water and 0.1% formic acid. Data dependent scanning was performed by the Xcalibur v 2.1.0 software (Andon et al., 2002) using a survey mass scan at 60,000 resolution in the Orbitrap analyzer scanning m/z 350–1600, followed by collision-induced dissociation (CID) tandem mass spectrometry (MS/MS) of the fourteen most intense ions in the linear ion trap analyzer. Sequest was set up to search human proteins downloaded from UniProtKB on 08/06/2013. Variable modifications considered during the search included methionine oxidation (15.995 Da), cysteine carbamidomethylation (57.021 Da), as well as two products of the adduction of arginine residues by MG, MG-derived hydroimidazolone (MG-H1; 54.011 Da) and MG-derived dihydroxyimidazoldine (MG-DH; 72.021 Da). At the time of the search, the Human UniProt database contained 88,323 entries. Proteins were identified at 95% confidence with XCorr scores (Qian et al., 2005) as determined by a reversed database search using the Percolator algorithm (http://per-colator.com) (Spivak et al., 2009). Identified modified peptides were considered with a q-value < 0.01 (Kall et al., 2008). In order to manually validate sites for adduction, we employed the following criteria: B and Y ion series must provide coverage at the site of modification; tryptic peptides identified with a C-terminal MG modification were not included (false positives); and unmodified peptides from the protein must also be identified.
Prodan displacement assay
In order to verify MG modification of R257, an assay targeting functionality of albumin drug binding site I where R257 resides was utilized. A solution of human serum albumin (HSA) (20 ml at 10 mg/ml) in 1X PBS pH 7.4 was prepared. A slurry (10 ml) of Lipidex-1000 in methanol was buffer exchanged into 1X PBS and the buffer decanted from the aqueous slurry. The HSA solution was added to the slurry and the mixture was rotated at room temperature for 30 min. The Lipidex slurry was completely removed from HSA by plunging through a 0.22 μm filter using a 60 ml syringe. Prodan (25 mg) was dissolved in of 40% ethanol, 60% methanol (5 ml at a final concentration is 5 mg/ml). A portion (10 ml) of delipidated HSA (10 mg/ml) was incubated with 666.7 μl of the prodan solution (1:10 molar ratio HSA:prodan), and this mixture was rotated at room temperature for 30 minutes. The HSA-prodan mixture was placed on dry ice and acetone precipitated by adding 40 ml of acetone at −20°C, which was mixed and centrifuged at 3000xg for 10 min to pellet the precipitated HSA-prodan. Two additional washes of cold acetone were used to remove any free prodan. The HSA pellet was brought to 10 ml with PBS and mixed and resuspended.
A 2-fold serial dilution of MG in 1X PBS pH 7.4 was prepared starting from a stock solution of 10 mM. An equal volume of HSA-prodan and MG solution (200 μl each) was mixed in triplicate reactions for each MG concentration and incubated at 37°C for 30 min. An aliquot (300 μl) of each reaction was read using a Gemini XPS Fluorescence Microplate Reader (Molecular Devices) in endpoint mode with 360 nm excitation, 420 nm filter cut-off, and 465 nm emission.
Modification of peptides with MG and transition optimization of synthetic peptides
Tryptic peptide R257 was non-quantitatively synthesized, HPLC purified, lyophilized, and resuspended in 18Ω Milli-Q grade water to 5 mg/ml, with 150 μl aliquots used for each reaction. Iodoacetamide (200 mM; 20 μl) in 20 mM ammonium bicarbonate pH 7.4 was added, and the solution incubated in the dark at room temperature for 1 hr. MG (150 μl of 10 mM) in 2X PBS pH 7.4 was added to R257 peptide solution, and the reaction was incubated at 37°C for 2 hrs. The reaction was terminated by the addition of formic acid (10 μl), and the modified peptide was desalted with 50 mg Hypersep C18 RP cartridges (Thermo Scientific) and eluted with 80% acetonitrile containing 0.1% TFA. The peptides were diluted four-fold with a solution of 0.5% formic acid in water prior to infusion.
Peptide solution was infused at 3 μl/min into a 4000 QTRAP (Applied Biosystems/ MDS Sciex) equipped with a Turbo Spray ion source and the manual transition optimization was performed. The source temperature was set at 200°C, source voltage was 5000 volts, GS1 and GS2 were set to 0 and 25 psi, respectively, and the declustering potential was set to 70 volts for all peptide parent ions.
Theoretical peptide fragment ion masses were obtained using the MS-Product function in ProteinProspector (Clauser et al., 1999). While the individual peptide solutions were infused into the QTRAP, mass spectra of parent ions for each charge state (+2, +3, +4) were analyzed using enhanced MS (EMS) mode in Analyst v. 1.4 (AB Sciex). Parent ions from each charge state (noted above) were fragmented in enhanced product ion (EPI) mode (MS/MS) and a list of potential MRM transitions was generated. From all of the possible intense ions generated from the MS/MS spectra, only b or y ions were selected for MRM optimization that were in agreement with the site of modification. In MRM mode, transitions were monitored as the collision energy was ramped from 5 to 100 eV and 1–5 candidate transitions were chosen per peptide.
Nine transitions were optimized for the albumin peptide R257. Ultimately the four highest transitions were utilized in human sample analysis. The optimized transitions are shown in Table 1.
Table 1.
R257 peptide MRM transitions
| R257 Peptide Sequence | Parent | Daughter | CE | Charge | Ion |
|---|---|---|---|---|---|
| VHTEC(57)C(57)HGDLLEC(57)ADDR(54)ADLAK | 660.6 | 801.5 | 31 | 4 | y20+3 |
| 660.6 | 635.6 | 32 | 4 | y21+4 | |
| 660.6 | 237.1 | 35 | 4 | b2 | |
| 660.6 | 109.6 | 90 | 4 | y2+2 | |
| 660.6 | 421.5 | 33 | 4 | y7+2 | |
| 528.7 | 605.2 | 19 | 5 | b10+2 | |
| 528.7 | 594.7 | 19 | 5 | y10+2 | |
| 528.7 | 659.3 | 19 | 5 | y11+2 | |
| 528.7 | 661.7 | 19 | 5 | b11+2 |
Nine transitions were generated for peptide R257 by modifying a pure peptide with MG. The transitions in bold were the transitions selected for further use in analysis, as they had the best linear response to concentration changes in vitro by MG. CE: collision energy.
Tryptic digestion and LC-MRM of human samples
Twelve human plasma samples (200 μl aliquots) were snap thawed by immediately placing the frozen tubes into a 37°C water bath for 10 min. Plasma samples were then centrifuged at 14,000xg for 5 min to pellet precipitate. Cleared plasma (5 μl) was added to 100 μl of Lipidex-1000 slurry and buffer exchanged into an equal volume of 100 mM ammonium bicarbonate (Ambic) pH 7.4. Samples were rotated at room temperature for 30 min to allow for slurry mixing and delipidation of plasma. Plasma proteins were separated from slurry by centrifuging through 0.22 μm centrifugal filter units (Cat# UFC30GVNB, Millipore) for 5 min at 12,000xg. An aliquot (100 μl) of 20 mM tris(2-carboxyethyl)phosphine (TCEP) pH 7.4 was added to the filtered protein samples, and these solutions were incubated at 55°C for 30 min. To further denature the protein and cool the samples, they were subsequently sonicated at room temperature for 10 min. Aliquots (100 μl) of 25 mM iodoacetamide in 100 mM Ambic pH 7.4 were added to the cooled solutions and the samples incubated in the dark at room temperature for 30 min. Sequencing-grade trypsin (50 μl at 0.1 μg/μl in 100 mM Ambic pH 7.4) was added to each sample (5 μg of total trypsin) and the samples incubated at 37°C for 16 hrs. Formic acid (5 μl) was added to stop the reactions, and the peptide samples were C18 purified using 50 mg Hypersep C18 RP cartridges (Thermo Scientific) as follows. The cartridges were washed with 5 ml acetonitrile and then equilibrated with 3 X 5 ml of 1% formic acid in water. The acidified peptide solutions were slowly passed through the C18 resin at a rate of about 1 drop per second. The cartridges were washed 2 X 5 ml of 1% formic acid and eluted with 1 ml of 80% acetonitrile containing 1% formic acid. The peptide solutions were frozen at −80°C, lyophilized to dryness, and stored at −80°C until analysis by LC-MRM.
Peptide solutions were loaded onto a ZORBAX 300SB-C18 capillary column (5 μm, 0.5 x 150 mm, Agilent) using a Famos autosampler (LC Packings) with a 10 μl injection loop. Peptides were loaded and eluted from the column using an LC Packings Ultimate II HPLC (Dionex) into a 4000 QTRAP at a flow rate of 40 μl/min of solvent A (0.01% TFA, 0.5% formic acid) with a 30 min linear gradient from 5% to 45% of solvent B (acetonitrile containing 0.01% TFA, 0.5% formic acid). The 4000 QTRAP was operated in MRM mode with the optimal R257 transitions. Dwell time was set to 40 ms, Q1 resolution was set to low, and Q3 resolution was set to unit. The source temperature was set at 200°C, source voltage was 5000 volts, GS1 and GS2 were set to 0 and 25 psi, respectively, and the declustering potential (DP) was set to 70 volts for all peptide parent ions. There were two technical replicates per sample.
Results
MG sites in the plasma proteome
MG primarily reacts with arginine residues to form relatively stable ring structures- dihydroxyimidazolidine (MG-DH; R+72) and hydroimidazolone (MG-HI; R+54). This adduction results in a net loss of positive charge from the arginine site, as arginine is positively charged under physiological conditions, whereas the ring adducts are uncharged. In order for MG adducts to be detected by MS/MS, they must not be labile during ion activation. Upon in vitro treatment of plasma protein with MG, several MG hotspots were identified, revealing candidate sites on HSA (including R257), serotransferrin, haptoglobin, hemopexin, and Ig lambda-2 chain C regions (Table 2). The relative modification to total parent protein was identified by spectral counting for HSA (5.9%), serotransferrin (1.7%), haptoglobin (5.7%), hemopexin (3.1%), and Ig lambda-2 chain C regions (8.9%) The sites identified for HSA are consistent with previous studies performed with pure HSA protein, though fewer total sites were recognized due to the complexity of the plasma sample (Kimzey et al., 2011). The identified proteins that harbor sites for MG modification are considered abundant within the plasma proteome (Kuzyk et al., 2009). Data-dependent MS/MS sequencing in typical shotgun proteomics workflows tends to preferentially identify highly abundant peptides relative to less abundant peptides. As such, the identification of modified abundant proteins and abundant peptides is expected. These peptides contained arginine (R) with atomic mass unit increases of R+54 and R+72 that fragment along the peptide backbone with minimal neutral loss of the modified arginine moiety. Our findings corroborate other studies demonstrating that low-energy collision induced activation is appropriate for detection of MG hotspots (Gao and Wang, 2006; Brock et al., 2007).
Table 2.
Site-specific MG modifications in vitro.
| Protein | Accession | Site | Peptide Sequence | MG-HI (R+54) | MG-DH (R+72) |
|---|---|---|---|---|---|
| Albumin | P02768 | R81 | LCTVATLRETYGEMADCCAK | X | X |
| R117 | LVRPEVDVMCTAFHDNEETFLK[K] | X | X | ||
| R186 | LDELRDEG[K]ASSAK | X | X | ||
| R218 | AWAVARLSQR | X | X | ||
| R257 | VHTECCHGDLLECADDRADLAK | X | X | ||
| R410 | FQNALLVRYTK | X | X | ||
| R428 | [K]VPQVSTPTLVEVSRNLGK | X | X | ||
| R472 | TPVSDRVTK | X | |||
| R445 | RMPCAEDYLSVVLNQLCVLHEK | X | |||
| Serotransferrin | P02787 | R124 | SCHTGLGRSAGWNIPIGLLYCDLPEPR | X | |
| Haptoglobin | P00738 | R41 | LRTEGDGVYTLNDK[K] | X | X |
| R243 | VSVNERVMPICLPS[K]DYAEVGR | X | |||
| Hemopexin | P02790 | R185 | YYCFQGNQFLRFDPVR | X | |
| Ig lambda-2 | P0CG05 | R83 | SHRSYSCQVTHEGSTVEK | X | |
| chain C regions |
Spectra were identified for the R+54 adduct using human plasma or pure protein incubated with 500 μM MG. Spectra containing the intermediate R+72 dihydroimidazolidine adduct are indicated. All cysteines are carbamidomethylated (C+57) prior to digestion. Brackets indicate alternate cleavage peptides detected.
MG-DH: methylglyoxal-derived dihydroxyimidazolidine
MG-HI: methylglyoxal-derived hydroimidazolone
When adding potential modifications to database searches, false positives can result and it is therefore necessary to manually validate the MG-modified spectra by using criteria specific for this modification. Working under the notion that trypsin would have inhibited cleavage at MG-modified arginine, any results that gave a sequence with a C-terminal modified arginine were treated as false positives. The sequences of peptides where MG modification was identified with the intrapeptide arginine (R) modification site are listed in Table 2. As expected, no MG-modified peptides were detected in the untreated plasma control samples by LC-MS/MS.
Functional assay to confirm MG modification of R257 within HSA-drug binding site I
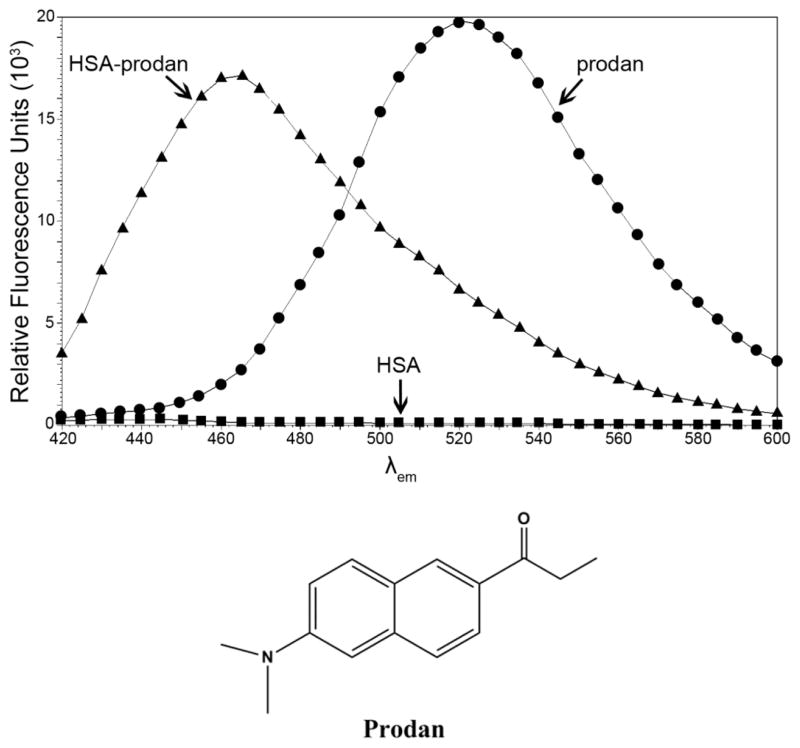
In order to determine the possible effects of R257 modification, which resides in drug site I of HSA (Petitpas et al., 2001), prodan was utilized. The unique spectral properties of prodan make it an effective tool for the study of drug site I (Figure 1). When prodan is free in solution it exhibits a fluorescent maximum at 520 nm when excited at 380 nm. However, when prodan is bound in drug site I of HSA, it undergoes a blue shift of 55 nm and this complex absorbs light at 380 nm yielding a fluorescent maximum at 465 nm (Figure 1). This phenomenon may be the result of radiation-less energy transfer between W214 and bound prodan (Moreno and Gonzalez-Jimenez, 1999). Additionally, prodan is non-nucleophilic and MG alone does not alter the spectral properties of prodan fluorescence. The prodan-HSA complex can be monitored spectrophometrically because HSA alone does not exhibit autofluorescence in this range.
Figure 1. Stoke’s shift of HSA-prodan complex yields fluorescent maxima at 465 nm.
HSA was incubated with a 10-fold excess of prodan and allowed to equilibrate for 30 minutes. Unbound prodan was removed by acetone precipitation and the emission spectra of free prodan, HSA-prodan and HSA were obtained. Excitation λ is 380 nm and upon binding HSA, prodan λmax blue-shifts from 520 nm to 465 nm. HSA without prodan (bottom spectra) shows negligible spectral emission. The endpoint values to monitor prodan displacement from the HSA-prodan complex are 380 nm excitation, 465 nm emission.
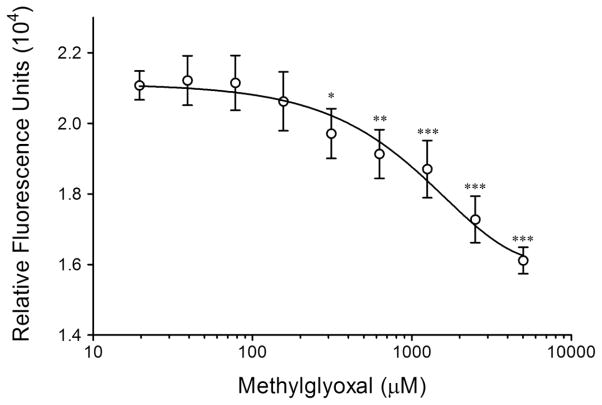
Using the fluorescent endpoint of 465 nm for the prodan-HSA complex permitted the study of the impact of MG adduction on drug site I. Any perturbations of this site can be indirectly measured by the displacement of prodan, which will ultimately decrease fluorescence at 465 nm. Therefore, maximum signal indicates unaffected prodan-HSA binding. Free prodan was purified from the HSA-prodan complex, and this complex was treated with a dilution of MG in triplicate reactions. Over time, prodan is naturally displaced from the HSA drug site I pocket, and the time point of 30 min was chosen because it permits sufficient time for MG to react with the protein complex. The “leakiness” of prodan from the complex does not permit analysis of sophisticated kinetic measurements, however the measure of the complex can be determined relative to MG treatment at any given time point.
The effect of MG on the prodan-HSA complex is illustrated in Figure 2. Relative to the unmodified control, where no MG was added to prodan-HSA (75 μM), MG treatment displaces prodan at concentrations above 300 μM in vitro. The MG concentration necessary to observe such an effect on prodan displacement from HSA conceivably is over estimated as MG is unstable and it is anticipated that a significant amount of MG would be degraded following the addition of MG to initiate the in vitro reaction. The data indicate that drug site I is targeted by MG in terms of binding to the prodan fluorescent probe. Currently, there is no available crystal structure of HSA bound to prodan, but from this data it seems that arginine side chains are not the only residues which influence the prodan interaction with drug site I. Thus, MG-modification of arginine reduces, but does not completely abolish, prodan binding in a dose dependent manner. Nevertheless, eventual displacement of prodan from this site at higher MG concentrations is further evidence that drug site I containing R257 is a target for MG adduction.
Figure 2. Prodan is displaced from drug site I by MG modification of HSA.
Prodan bound to HSA (prodan-HSA complex, 75 μM) was treated with a dilution of MG and the fluorescence (excitation 380 nm, emission 465 nm, filter cut-off 420 nm) was measured after 30 minutes. Means ± SD for three separate experiments are given. Significant (*p<0.05; ** p<0.01; *** p<0.001) as compared to control (HSA without MG treatment)
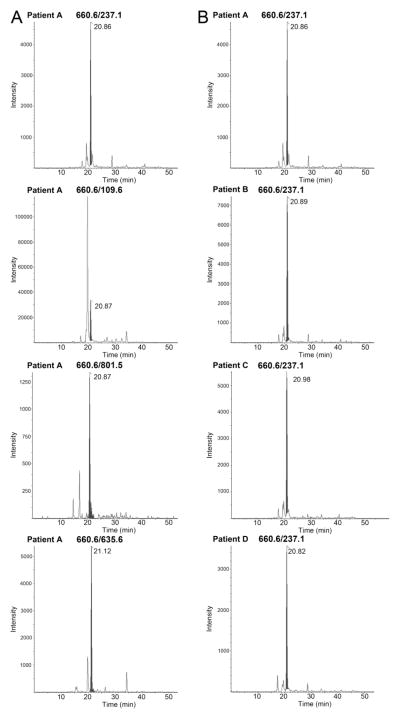
Identification of MG-modified R257 in human plasma
All 12 human plasma samples analyzed, from both diabetic and non-diabetic patients, showed the presence of R257 transitions. Four of the representative human samples, and the presence of transition 660.6/237.1, are shown in Figure 3. All samples exhibited peaks for R257 transitions, although differences in area under the curve varied between samples. The specificity of four distinct R257 transitions were found to be a reliable indicator of peptide presence (Figure 3A), and the retention time of the R257 transitions did not vary drastically within each batch of samples analyzed (Figure 3B). Further quantitative analyses of these transitions to establish potential differences in R257 transition levels between diabetic and non-diabetic individuals are ongoing.
Figure 3. Human plasma samples indicate the presence of modified R257.
(A) Multiple-reaction monitoring transitions (660.6/237.1; 660.6/109.6; 660.6/801.5; 660.6/635.6) indicate the presence of a modified R257-containing peptide of albumin in the plasma of human patients, shown in a representative diabetic patient sample. (B) Four separate patient plasma samples, representative of all 12 patients analyzed, showed the presence of 660.6/237.1 transition.
Discussion
Several arginine residues were identified in abundant plasma proteins that are susceptible to MG modification. Most of these arginine residues contained both the +54 and +72 modifications. The dihydroxyimidazolidine and open ring intermediates are isobaric, and indistinguishable by mass spectrometry. However, the presence of the +72 adduct further validates these sites as bona fide targets for MG, because arginine must form these structures prior to the subsequent loss of water to generate the +54 adduct. While 500 μM MG appears to exceed the concentration of MG observed in human plasma, this concentration was utilized to ensure maximal detection of all sites within the proteome susceptible to any amount of MG, anticipating the chemical instability of MG in solution. Identification of multiple MG-modified peptides by untargeted LC-MS/MS methods at these concentrations permits, the subsequent application of more selective methods, such as MRM, to identify sensitive sites in vivo. As only select sites were identified, it appears that 500 μM MG did not overwhelm and occupy every arginine site, but rather modified only arginine sites with particular sensitivity. In particular, HSA possesses 27 arginine residues, but only nine were found to be modified by MG. The chemical basis for the selectivity of these nine sites for MG modification is currently under investigation.
Our work has identified numerous preferential sites of MG adduction (Table 2) beyond those previously reported (Baraka-Vidot et al., 2012; Schmidt et al., 2015), and provide a basis to further determine the role such modification play in altered protein function. Thus, it is possible that some of the sites identified within the plasma proteome are critical in terms of function for these particular proteins. Previous work has established that modification of R410 within drug binding site II in HSA results in inhibited ketoprofen binding and esterase activity (Ahmed et al., 2005b). Another important MG-modified arginine identified in HSA was R257, which resides in drug site I. This site was not identified by Ahmed et al. (2005b) possibly due to interferences from multiple overlapping peaks with their LC-MS only approach. Tandem MS/MS is not affected by such interferences, because co-eluting peptides are separated and individually fragmented. Using this approach the R485 MG site in HSA was another site that was uniquely identified. The proximity effect of charged residues in promoting glycation has been proposed (Venkatraman et al., 2001; Rabbani and Thornalley, 2012) and the results herein support and expand upon these findings. Thus, we identify several MG modified targets in the plasma proteome in efforts to develop a structure-activity model for MG adduction.
Following identification of sites of modification we validated that drug site I in HSA (R218 and R257) is a target for MG modification using the prodan binding assay. A benefit of using prodan to study drug site I is that, unlike warfarin, the emission peak of the bound prodan is distinct from the emission peak of free prodan, a useful property for detecting small changes in free versus bound conformations. In addition to warfarin and prodan, additional probes used to target drug site I include 5-dimethylaminonaphtalene-1-sulfonamide (DNSA), dansylamide, dansyl-L-glutamine, dansyl-L-asparagine, dansyl-L-lysine, n-butyl p-aminobenzoate, and phenol red (Kragh-Hansen et al., 2002). In a displacement assay to study MG, any one of these probes would likely exhibit similar behavior to prodan. Our results confirmed that arginine residues in drug site I are targets for MG adduction. However, while R218 and R257 reside within the drug site I, modification of the protein likely occurs at all nine sites identified, and therefore changes in tertiary structure affecting protein function due to arginine adduction distal from drug site I cannot be ruled out.
The importance of demonstrating potential functional changes in HSA due to MG adduction is emphasized by our ability to identify a MRM transition specific to the R257 peptide in each of the 12 human samples analyzed (Figure 3). This clearly establishes that sufficient MG is present in circulating blood to adduct HSA. Indeed, MG formation is estimated to be 120 μM/day under normoglycemic conditions (Thornalley, 1993) and given a half-life of ~20 days this would equate exposure to 2.4 mM MG; even higher amounts of MG would be anticipated in diabetic patients. However, because this initial human study was not designed to establish absolute levels of MG-modified R257 peptide, but rather its presence (or otherwise) in human plasma, differences between diabetic and non-diabetic subjects were not assessed; though subjects from both were utilized to confirm the presence of modified R-257 in each cohort. Work is ongoing to determine quantitative differences between R257 (and other modified arginine peptides) MRM transition levels between cohorts.
It is necessary to elucidate the site-specificity of MG adduction, as critical residues involved in protein function could be susceptible to modification. Moreover, particularly reactive MG-binding sites could serve as biomarkers for MG-mediated dysregulation, similar to the use of glycated hemoglobin (HbA1C) as a marker for glucose exposure. Such high-affinity sites of MG adduction may offer a better measure of the in vivo exposure to reactive dicarbonyls. It is equally important to characterize the sites of modification so that specific antibodies can be designed for future high throughput analysis.
Highlights.
Methylglyoxal (MG) selectively modifies arginine sites in human plasma proteome.
Dihydroxyimidazolidine and hydroimidazolone adducts on serum albumin identified.
MG modification on albumin R257 associated with loss of drug site I binding capacity.
MRM-tandem mass spectrometry enables sensitive detection of albumin MG-R257.
Site-specific MG modification may represent a useful monitor of effective therapy of T2D.
Acknowledgments
Mass spectrometric data were acquired by the Arizona Proteomics Consortium supported by P30ES06694, P30CA023074 and by the BIO5 Institute of the University of Arizona.
Funding Information
This work was supported by the National Institutes of Health [RO1GM070890 to S.S.L.; R24DK083948 to R.N. and S.S.L.; P30ES006694 Pilot Project to C.S and S.S.L.; T32ES016652 to M.J.K.; T32ES007091 to O.R.K., K23HL107389 to H.N.Y., 1S10RR028868-01 to G.T.]; TRIF-BIO5/Biodesign Collaborative Project to R.N. and S.S.L.; Arizona Biomedical Research Commission 10–100 to S.S.L.; and American Heart Association grant 12CRP11750017 to H.N.Y.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmed N, Babaei-Jadidi R, Howell SK, Beisswenger PJ, Thornalley PJ. Degradation products of proteins damaged by glycation, oxidation and nitration in clinical type 1 diabetes. Diabetologia. 2005a;48:1590–1603. doi: 10.1007/s00125-005-1810-7. [DOI] [PubMed] [Google Scholar]
- Ahmed N, Dobler D, Dean M, Thornalley PJ. Peptide mapping identifies hotspot site of modification in human serum albumin by methylglyoxal involved in ligand binding and esterase activity. J Biol Chem. 2005b;280:5724–5732. doi: 10.1074/jbc.M410973200. [DOI] [PubMed] [Google Scholar]
- Ahmed N, Thornalley PJ. Advanced glycation endproducts: what is their relevance to diabetic complications? Diabetes Obes Metab. 2007;9:233–245. doi: 10.1111/j.1463-1326.2006.00595.x. [DOI] [PubMed] [Google Scholar]
- Andon NL, Hollingworth S, Koller A, Greenland AJ, Yates JR, 3rd, Haynes PA. Proteomic characterization of wheat amyloplasts using identification of proteins by tandem mass spectrometry. Proteomics. 2002;2:1156–1168. doi: 10.1002/1615-9861(200209)2:9<1156::AID-PROT1156>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- Baraka-Vidot J, Guerin-Dubourg A, Bourdon E, Rondeau P. Impaired drug-binding capacities of in vitro and in vivo glycated albumin. Biochimie. 2012;94:1960–1967. doi: 10.1016/j.biochi.2012.05.017. [DOI] [PubMed] [Google Scholar]
- Beisswenger PJ, Drummond KS, Nelson RG, Howell SK, Szwergold BS, Mauer M. Susceptibility to diabetic nephropathy is related to dicarbonyl and oxidative stress. Diabetes. 2005;54:3274–3281. doi: 10.2337/diabetes.54.11.3274. [DOI] [PubMed] [Google Scholar]
- Bose T, Bhattacherjee A, Banerjee S, Chakraborti AS. Methylglyoxal-induced modifications of hemoglobin: structural and functional characteristics. Arch Biochem Biophys. 2013;529:99–104. doi: 10.1016/j.abb.2012.12.001. [DOI] [PubMed] [Google Scholar]
- Brock JW, Cotham WE, Thorpe SR, Baynes JW, Ames JM. Detection and identification of arginine modifications on methylglyoxal-modified ribonuclease by mass spectrometric analysis. J Mass Spectrom. 2007;42:89–100. doi: 10.1002/jms.1144. [DOI] [PubMed] [Google Scholar]
- Brownlee M. Biochemistry and molecular cell biology of diabetic complications. Nature. 2001;414:813–820. doi: 10.1038/414813a. [DOI] [PubMed] [Google Scholar]
- Clauser KR, Baker P, Burlingame AL. Role of accurate mass measurement (+/− 10 ppm) in protein identification strategies employing MS or MS/MS and database searching. Anal Chem. 1999;71:2871–2882. doi: 10.1021/ac9810516. [DOI] [PubMed] [Google Scholar]
- Dhar A, Desai K, Liu J, Wu L. Methylglyoxal, protein binding and biological samples: are we getting the true measure? J Chromatogr B Analyt Technol Biomed Life Sci. 2009;877:1093–1100. doi: 10.1016/j.jchromb.2009.02.055. [DOI] [PubMed] [Google Scholar]
- Duran-Jimenez B, Dobler D, Moffatt S, Rabbani N, Streuli CH, Thornalley PJ, Tomlinson DR, Gardiner NJ. Advanced glycation end products in extracellular matrix proteins contribute to the failure of sensory nerve regeneration in diabetes. Diabetes. 2009;58:2893–2903. doi: 10.2337/db09-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet X, Charloteaux B, Thomas A, Brasseur R. A fast method to predict protein interaction sites from sequences. J Mol Biol. 2000;302:917–926. doi: 10.1006/jmbi.2000.4092. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wang Y. Site-selective modifications of arginine residues in human hemoglobin induced by methylglyoxal. Biochemistry. 2006;45:15654–15660. doi: 10.1021/bi061410o. [DOI] [PubMed] [Google Scholar]
- Kall L, Storey JD, MacCoss MJ, Noble WS. Posterior error probabilities and false discovery rates: two sides of the same coin. J Proteome Res. 2008;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- Khuhawar MY, Zardari LA, Laghari AJ. Capillary gas chromatographic determination of methylglyoxal from serum of diabetic patients by precolumn derivatization with 1,2-diamonopropane. J Chromatogr B Analyt Technol Biomed Life Sci. 2008;873:15–19. doi: 10.1016/j.jchromb.2008.04.048. [DOI] [PubMed] [Google Scholar]
- Kimzey MJ, Yassine HN, Riepel BM, Tsaprailis G, Monks TJ, Lau SS. New site(s) of methylglyoxal-modified human serum albumin, identified by multiple reaction monitoring, alter warfarin binding and prostaglandin metabolism. Chem Biol Interact. 2011;192:122–128. doi: 10.1016/j.cbi.2010.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kragh-Hansen U, Chuang VT, Otagiri M. Practical aspects of the ligand-binding and enzymatic properties of human serum albumin. Biol Pharm Bull. 2002;25:695–704. doi: 10.1248/bpb.25.695. [DOI] [PubMed] [Google Scholar]
- Kuzyk MA, Smith D, Yang J, Cross TJ, Jackson AM, Hardie DB, Anderson NL, Borchers CH. Multiple reaction monitoring-based, multiplexed, absolute quantitation of 45 proteins in human plasma. Mol Cell Proteomics. 2009;8:1860–1877. doi: 10.1074/mcp.M800540-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo TW, Westwood ME, McLellan AC, Selwood T, Thornalley PJ. Binding and modification of proteins by methylglyoxal under physiological conditions. A kinetic and mechanistic study with N alpha-acetylarginine, N alpha-acetylcysteine, and N alpha-acetyllysine, and bovine serum albumin. J Biol Chem. 1994;269:32299–32305. [PubMed] [Google Scholar]
- Moreno F, Gonzalez-Jimenez J. Binding of the Promen fluorescent probe to human serum albumin: a fluorescence spectroscopic study. Chem Biol Interact. 1999;121:237–252. doi: 10.1016/s0009-2797(99)00111-8. [DOI] [PubMed] [Google Scholar]
- Morgan PE, Sheahan PJ, Pattison DI, Davies MJ. Methylglyoxal-induced modification of arginine residues decreases the activity of NADPH-generating enzymes. Free Radic Biol Med. 2013;61:229–242. doi: 10.1016/j.freeradbiomed.2013.03.025. [DOI] [PubMed] [Google Scholar]
- Petitpas I, Bhattacharya AA, Twine S, East M, Curry S. Crystal structure analysis of warfarin binding to human serum albumin: anatomy of drug site I. J Biol Chem. 2001;276:22804–22809. doi: 10.1074/jbc.M100575200. [DOI] [PubMed] [Google Scholar]
- Qian WJ, Liu T, Monroe ME, Strittmatter EF, Jacobs JM, Kangas LJ, Petritis K, Camp DG, 2nd, Smith RD. Probability-based evaluation of peptide and protein identifications from tandem mass spectrometry and SEQUEST analysis: the human proteome. J Proteome Res. 2005;4:53–62. doi: 10.1021/pr0498638. [DOI] [PubMed] [Google Scholar]
- Rabbani N, Thornalley PJ. Glyoxalase in diabetes, obesity and related disorders. Semin Cell Dev Biol. 2011;22:309–317. doi: 10.1016/j.semcdb.2011.02.015. [DOI] [PubMed] [Google Scholar]
- Rabbani N, Thornalley PJ. Methylglyoxal, glyoxalase 1 and the dicarbonyl proteome. Amino Acids. 2012;42:1133–1142. doi: 10.1007/s00726-010-0783-0. [DOI] [PubMed] [Google Scholar]
- Rabbani N, Thornalley PJ. Dicarbonyl proteome and genome damage in metabolic and vascular disease. Biochem Soc Trans. 2014;42:425–432. doi: 10.1042/BST20140018. [DOI] [PubMed] [Google Scholar]
- Schmidt R, Bohme D, Singer D, Frolov A. Specific tandem mass spectrometric detection of AGE-modified arginine residues in peptides. J Mass Spectrom. 2015;50:613–624. doi: 10.1002/jms.3569. [DOI] [PubMed] [Google Scholar]
- Sheetz MJ, King GL. Molecular understanding of hyperglycemia's adverse effects for diabetic complications. JAMA. 2002;288:2579–2588. doi: 10.1001/jama.288.20.2579. [DOI] [PubMed] [Google Scholar]
- Shevchenko A, Wilm M, Vorm O, Mann M. Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal Chem. 1996;68:850–858. doi: 10.1021/ac950914h. [DOI] [PubMed] [Google Scholar]
- Spivak M, Weston J, Bottou L, Kall L, Noble WS. Improvements to the percolator algorithm for Peptide identification from shotgun proteomics data sets. J Proteome Res. 2009;8:3737–3745. doi: 10.1021/pr801109k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ. The glyoxalase system in health and disease. Mol Aspects Med. 1993;14:287–371. doi: 10.1016/0098-2997(93)90002-u. [DOI] [PubMed] [Google Scholar]
- Thornalley PJ, Battah S, Ahmed N, Karachalias N, Agalou S, Babaei-Jadidi R, Dawnay A. Quantitative screening of advanced glycation endproducts in cellular and extracellular proteins by tandem mass spectrometry. Biochem J. 2003;375:581–592. doi: 10.1042/BJ20030763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornalley PJ, Rabbani N. Assay of methylglyoxal and glyoxal and control of peroxidase interference. Biochem Soc Trans. 2014;42:504–510. doi: 10.1042/BST20140009. [DOI] [PubMed] [Google Scholar]
- Venkatraman J, Aggarwal K, Balaram P. Helical peptide models for protein glycation: proximity effects in catalysis of the Amadori rearrangement. Chem Biol. 2001;8:611–625. doi: 10.1016/s1074-5521(01)00036-9. [DOI] [PubMed] [Google Scholar]
- Wang T, Kartika R, Spiegel DA. Exploring post-translational arginine modification using chemically synthesized methylglyoxal hydroimidazolones. J Am Chem Soc. 2012;134:8958–8967. doi: 10.1021/ja301994d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue M, Rabbani N, Momiji H, Imbasi P, Anwar MM, Kitteringham N, Park BK, Souma T, Moriguchi T, Yamamoto M, Thornalley PJ. Transcriptional control of glyoxalase 1 by Nrf2 provides a stress-responsive defence against dicarbonyl glycation. Biochem J. 2012;443:213–222. doi: 10.1042/BJ20111648. [DOI] [PubMed] [Google Scholar]