Abstract
SchizConnect (www.schizconnect.org) is built to address the issues of multiple data repositories in schizophrenia neuroimaging studies. It includes a level of mediation—translating across data sources—so that the user can place one query, e.g. for diffusion images from male individuals with schizophrenia, and find out from across participating data sources how many datasets there are, as well as downloading the imaging and related data. The current version handles the Data Usage Agreements across different studies, as well as interpreting database-specific terminologies into a common framework. New data repositories can also be mediated to bring immediate access to existing datasets. Compared with centralized, upload data sharing models, SchizConnect is a unique, virtual database with a focus on schizophrenia and related disorders that can mediate live data as information are being updated at each data source. It is our hope that SchizConnect can facilitate testing new hypotheses through aggregated datasets, promoting discovery related to the mechanisms underlying schizophrenic dysfunction.
Keywords: Data mediation and integration, neuroinformatics, mega analysis, schizophrenia databases
1. Introduction
Schizophrenia is a complex psychiatric disease with heterogeneous clinical, behavioral, cognitive and genetic manifestations, and the literature, especially on neuroimaging studies, has yet to achieve a state of consistency and reproducibility. As a result, multi-site consortia have been created to coordinate the collection of large datasets in order to address these issues (Turner, 2014). Consortia such as the Functional Biomedical Informatics Research Network (FBIRN) (Friedman et al., 2008; Helmer et al., 2011a; Keator et al., 2008), the Mind Clinical Imaging Consortium (MCIC) (Gollub et al., 2013), North American Prodrome Longitudinal Study Consortium (NAPLS) (Addington et al., 2012) and Bipolar-Schizophrenia Network for Intermediate Phenotypes (B-SNIP) (Keshavan et al., 2011; Thaker, 2008) have led to the improvement of our understanding of brain circuitry, brain function and genetic variability in schizophrenia (Allen et al., 2011; Arnold et al., 2015; Cannon et al., 2015; Castro et al., 2014; Chen et al., 2012; Chen et al., 2013; Ehrlich et al., 2014; Hass et al., 2014; Hass et al., 2015; Kim et al., 2009; Kim et al., 2010; Mathew et al., 2014; Potkin et al., 2009). These efforts were successful in part because they were created with data sharing in mind, and they anticipated many of the difficulties in combining data from multiple sites in their design and schema. Yet, in building data repositories, many decisions were made that were specific to that particular repository or study, about what they would call different datatypes. In one study’s data a structural MRI scan may be listed in-formatively as “T1-weighted scan”, or something as complex as “5MPRAGE-AVG” or just “scan1”. Combining data from datasets that do not share the same protocols and data structures is challenging and it remains a barrier to aggregating mega-datasets. When combining data across different sources, individual investigators face the difficult and costly process of understanding the different imaging, subject, assessment, meta-data content (definitions, formats, organizations) and converting the data into standardized terms. A critical and unmet need exists to automate and virtualize this process so that appropriate data can be retrieved and combined from different databases regardless of differences in the their structure and terminology.
In this paper, we describe the initial deployment of “SchizConnect” (publicly available at http://www.schizconnect.org/), a new resource aimed at establishing mega-datasets (Wang et al., 2014). Building on the success of previous consortia efforts, SchizConnect creates a resource that supports structured querying and retrieval of neuroimaging and related data across these consortia repositories.
2. Methods
2.1. SchizConnect Architecture
SchizConnect follows the classical virtual data integration framework (Florescu et al., 1998; Halevy, 2001; Lenzerini, 2002; Ullman, 1997; Wiederhold, 1992), which saw its initial theoretical application on schizophrenia neuroimaging data within the FBIRN consortium (Ashish et al., 2010). A distinct feature of this framework is that the data can reside at and be maintained at the original data sources in their original formats and schemas (i.e., their own ways to define the structure, content, and semantics of data). SchizConnect consists the following three fundamental components (Figure 1): 1) the data sources provide the data, structured according to native schemas; 2) the mediation software (i.e., mediator) reconciles the semantic differences in the data across the different sources through domain modeling and inter-schema mapping; and 3) the web portal provides a user-friendly interface for querying and downloading data.
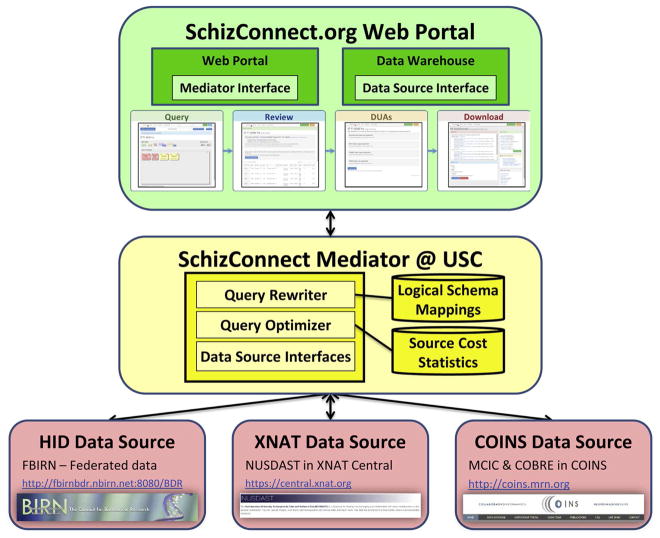
Figure 1. SchizConnect Architecture & Data Flow.
The SchizConnect architecture has the following 3 components: the federated data sources, the SchizConnect Mediator, and the SchizConnect.org web portal. After the user builds the query at the SchizConnect web portal, the query command is passed to the SchizConnect Mediator engine. The Mediator engine then relies on the SchizConnect domain model to translate the incoming query into source-specific schemas, then queries each data source directly and in parallel, using source-specific query mechanisms, languages, and variables. The returns from the sources are handled by the Mediator engine, which provides the SchizConnect web portal with a unified results table. The SchizConnect web portal then interacts with the user for further processing, including signing of data use agreements and data download.
The typical data flow begins with a user query constructed by dragging and dropping terms in a graphical user interface (GUI) at the SchizConnect web portal. The graphical query is translated to SQL and passed to the SchizConnect mediator. The mediator then relies on the SchizConnect domain model and inter-schema mappings to translate the user query into source-specific terms, and then queries each data source directly and concurrently, using the available source-specific query mechanisms, languages and variables. The results from the sources are collated and joined together by the mediator and provided to the SchizConnect web portal as a unified results table containing the mediated (harmonized) variables. The SchizConnect web portal then interacts with the user for further processing, including signing of data use agreements and data download. Below we describe each of the components in detail.
2.2. The data sources
SchizConnect accesses the data available at each of the sources in real time. It maintains a library of connectors (wrappers) to common source types, including relational databases, XML (Extensible Markup Language) or JSON (JavaScript Object Notation) databases, SOAP (Simple Object Access Protocol) or REST (Representation State Transfer) web services, or flat files. For previously unseen source types, the architecture is extensible to program new connectors. Each source runs their own server platforms with their own database schemas, formats and application programming interfaces (APIs). SchizConnect imposes no requirements on data sources with regard to databasing or access methods, but is flexible to special needs (see results below for specials case example).
2.3. SchizConnect Mediator
The SchizConnect mediator is built on the BIRN mediator (Ashish et al., 2010; Helmer et al., 2011b). The core idea is to define a common domain model/schema and inter-schema mapping rules that map between the domain schema and the different source schemas. The domain model defines an integrated view of the shared data in the application domain (i.e., SchizConnect). This concept is closely related to that of an ontology, but it is more limited in scope, in the sense that it only models the part of the application domain that is relevant to integrating a set of sources (Ashish et al., 2010). It can be viewed as an incremental, data-driven, pragmatic approach to domain ontology development. Inter-schema mappings are logical formulas, or rules, that relate the data models at each of the sources with the domain model, reconciling their semantic differences in the description of the data. The SchizConnect domain model consists of terms that are pertinent to schizophrenia neuroimaging research (such as those related to imaging protocol, disease severity or cognitive functioning). Definitions of these terms are provided via downloadable documents available on the SchizConnect web portal. During development, we have aligned the SchizConnect domain model terms with the standards provided by the ontology community: XCEDE (XML-Based Clinical Experiment Data Exchange Schema, http://www.xcede.org/XCEDE.html) (Gadde et al., 2012), NeuroLEX (the Neuroscience Lexicon, http://neurolex.org/) (Larson and Martone, 2013), NIF (the Neuroscience Information Framework, http://neuinfo.org/) (Gardner et al., 2008) and INCF (the International Neuroinformatics Coordinating Facility, http://incf.org/) (Bjaalie and Grillner, 2007; De Schutter, 2009), wherever appropriate. The domain terms with links to existing ontology can be found on the SchizConnect website documentations page. For example, the Positive and Negative Symptom Scale (PANSS) has an entry on NeuroLex (birnlex_3032, http://neurolex.org/wiki/Category:Positive_and_Negative_Symptom_Scale). In cases where no accepted ontology has been defined, such as the UPSA (University of California Performance-based Skills Assessment), we work with the neuroimaging and neuroinformatics community to define them. Although such effort is ongoing, since there doesn’t yet exist a standard process for ontological term contribution, we will work with data contributors and the neuroinformatics community on a case-by-case basis.
The mediator performs two core functions. First, it uses the inter-schema mappings to rewrite the user query, e.g., “T1 images from individuals with schizophrenia older than 30 years old,” from the domain schema to the formats and languages specific to each source. It currently uses GAV (global-as-view) mappings (Ashish et al., 2010; Florescu et al., 1998; Halevy, 2001; Lenzerini, 2002; Ullman, 1997; Wiederhold, 1992), where each domain predicate is constructed as a query over the source predicates. Second, the mediator constructs, optimizes, and executes a distributed query evaluation plan, based on the rewritten source-level query, which provides the answers to the user query. The source queries are executed directly over the data sources in parallel through each source connector (which wraps the source APIs). Results from source queries are collated and joined together in the mediator, and served to the SchizConnect web portal as a unified table organized using the domain schema terms.
The query execution engine used by the mediator for source-level query evaluation is based on the Open Grid Services Architecture Data Access and Integration (OGSA-DAI) and Distributed Query Processing (OGSA-DQP) services (http://www.ogsadai.org.uk/) (Antonioletti et al., 2005; Grant et al., 2008; Lynden et al., 2009; Lynden et al., 2008). OGSA-DAI is a streaming workflow engine in which data resources and web services are wrapped through a library of connectors. OGSA-DQP is a query engine that optimizes and implements the query evaluation plan as an OGSA-DAI workflow, which accesses the multiple data sources.
The SchizConnect domain query language is SQL, expressed on the domain model terms. Each data source query is wrapped as an OGSA-DAI resource through its library of connectors to common data sources such as PostgreSQL, MySQL, or XML. For additional details on the mediator architecture, see our previous work on the BIRN mediator (Ashish et al., 2010).
2.4. SchizConnect Web Portal
SchizConnect has been conceptualized as a one-stop resource for querying and retrieving neuroimaging and related data from distributed, heterogeneous repositories. SchizConnect is therefore set up to facilitate user registration and compliance with data use agreement (DUA) with each data source. To maximize the usage and efficiency, while complying with NIH policies and best practices, the SchizConnect web portal is structured to have two query modes. The first is a preliminary search, for example, for the number of male subjects between the ages of 20 and 60 with diffusion MRI scans. This mode is completely open to the public, allowing for anyone to browse and immediately obtain useful, current information on a current summary count of available data without having the need to register. However, no subject-level data at this point are downloadable. In the second mode, the user registers and signs in, and with the same query is able to view subject-level data, sign DUAs and download the data. Only data for which DUA has been signed are downloadable. Signed DUAs are forwarded to each respective data source administrator for reviewing and filing. The administrators at SchizConnect as well as each data source will work with each user’s institution for data use and institutional review board (IRB) terms upon request. Regarding to participant privacy, we require that it is the responsibility of the data source to ensure that data are shared in accordance to rules and regulations set forth by their IRB. The responsibility of SchizConnect is to confirm with each data source that they agree to this responsibility.
3. Results
The initial deployment of SchizConnect makes available data on 1,129 subjects from its contributing sources. Sample demographics can be found in Table 1 as well as on the SchizConnect web portal (http://schizconnect.org/#subject-stats). Of these subjects, 1,029 have scan data, which include structural MRI (sMRI), resting-state functional MRI (fMRI), task-paradigm fMRI, and diffusion MRI (dMRI) scans, totaling 21,309 volumes (Table 2 and Table 3). Demographic, neuropsychological measures, and clinical assessments are either available or in the process of becoming available. As the data sources take in new subjects/scans, they can automatically become available for querying through SchizConnect, depending on the source data repository’s policies. At the writing of this manuscript, SchizConnect has 49 users and 37 downloads, and that statistic is steadily growing (http://schizconnect.org/#user-stats).
Table 1.
Sample Characteristics on Modeled Terms.
| Dx | No Known Disorder | Schizophre nia Broad | Schizophre nia Strict | Schizoaffective | Bipolar Disorder | Sibling of Schizophrenia Strict | Sibling of No Known Disorder |
|---|---|---|---|---|---|---|---|
| Number of Current Subjects* | 481 | 117 | 333 | 38 | 10 | 44 | 66 |
| Gender (m/f) | 295/186 | 87/30 | 242/91 | 22/16 | 5/5 | 21/23 | 16/50 |
| Age (years) (mean±s.d.) | 34.2±12.9 | 34.4±11.0 | 35.4±12.9 | 39.5±10.0 | 46.6±14.4 | 21.6±3.7 | 20.4±3.5 |
| Age range (years) min – max | 13 – 67 | 18 – 61 | 17 – 66 | 19 – 59 | 21 – 64 | 14 – 28 | 14 – 28 |
40 subjects from the FBIRN database contains no diagnosis information and are not listed here.
Table 2.
SchizConnect Data Sources, including Current and Planned Sample Sizes.
| Data Source | FBIRN | NU | MRN |
|---|---|---|---|
| Project Name | FBIRN Phase II | NUSDAST | MCIC & COBRE |
| Database (Language) | HID (PostgreSQL) | XNAT (XML) | COINS Data Exchange (*) |
| URL | http://fbirnbdr.nbirn.net:8080/BDR/ | https://central.xnat.org/REST/projects/NUDataSharing | http://coins.mrn.org/ |
| Number of Current Subjects (with Imaging Data) | 255 (251) | 451 (368) | 423 (410) |
| Image Type | Structural MRI Resting-state fMRI Task fMRI dMRI | Structural MRI | Structural MRI Resting-state fMRI Task fMRI dMRI |
| Number of Current Subjects with Multimodal Imaging$ | 208 | - | 404 |
| Number of New Subjects (Timeline) | 400 – fBIRN Phase 3 (2015) | 130 (2015) | 200 (2016) |
The native COINS architecture involves dynamic data packaging following the query, which does not allow for data to be immediately returned to the query engine. With permission from the COINS executive committee, an API was built to enable us to extract domain-model defined variables from the COINS databases and to duplicate them at the Mediator site. The database underlying the duplicated MCICShare/COBRE data is MySQL using the relational data model.
Multimodal imaging is defined as having any of the combination of the following: T1, diffusion MRI, task-paradigm fMRI, and resting-state fMRI.
Table 3.
Example Queries and Returns at SchizConnect.org.
| Query Terms | Number of Images & Distinct Subjects Returned
|
||||
|---|---|---|---|---|---|
| Total | fBIRN @ HID | COBRE @ COINS | MCICShare @ COINS | NUSDAST @ XNAT | |
| T1 SCZ+CON | 3,178 | 1,048 | 484 | 264 | 1,382 |
| 737 | 185 | 173 | 95 | 284 | |
|
| |||||
| 3T T1 20–60 Yr | 1,348 | 750 | 492 | 106 | - |
| 345 | 112 | 180 | 53 | ||
|
| |||||
| T1 Healthy Controls | 1,846 | 655 | 243 | 264 | 684 |
| 439 | 113 | 94 | 95 | 137 | |
|
| |||||
| Sternberg SCZ 3T<=30 Yr | 210 | 210 | - | - | - |
| 18 | 18 | ||||
|
| |||||
| Resting BP+CON | 103 | - | 103 | - | - |
| 101 | 101 | ||||
|
| |||||
| Sensory Motor + T1,SCZ+CON, 30–50 Yr (multimodal) | 1,310 | 1,140 | - | 170 | - |
| 119 | 83 | 36 | |||
|
| |||||
| All MRI | 21,309 | 13,552 | 1,596 | 4,347 | 1,814 |
| 1,029 | 251 | 198 | 212 | 368 | |
3.1. SchizConnect Data Sources
Current data sources include the following schizophrenia-related datasets, which are all publicly available themselves and have been extensively curated, documented, and subjected to quality assurance. Detailed explanations of data can be found in the accompanying special papers describing the data source repositories, as well as the following papers: (Cetin et al., 2014; Glover et al., 2012; Gollub et al., 2013; Wang et al., 2013).
FBIRN Phase II dataset @ UCI (http://fbirnbdr.nbirn.net:8080/BDR/)1 (Glover et al., 2012; Potkin and Ford, 2009), containing cross-sectional multisite data from 251 subjects, each with two visits. (See paper on FBIRN data in this special issue.) Data include sMRI and fMRI scans collected on a variety of 1.5T and 3T scanners, including Sternberg Item Recognition Paradigm (SIRP) and Auditory Oddball paradigms, breath-hold and sensorimotor tasks. The underlying database is HID (Keator et al., 2009; Ozyurt et al., 2010) in PostgreSQL using the relational data model. Access is provided via Java database connectivity technology (JDBC), a database-access API for the Java programming language. Results from the query are returned to the Mediator as an SQL ResultSet.
NUSDAST @ XNAT Central (https://central.xnat.org/REST/projects/NUDataSharing) (Wang et al., 2013), containing data from 368 subjects, the majority with longitudinal data (~2 years apart). (See paper on NU data in this special issue.) Data include sMRI scans collected on a single Siemens 1.5T Vision scanner. The underlying database is XNAT (Marcus et al., 2007), using the XML data model. Access is provided via XNAT REST API. The mediator constructs a query XML document conforming to the XNAT search web service specification. Results from the query are returned to the Mediator as an XML document.
COBRE & MCICShare @ COINS Data Exchange (http://coins.mrn.org/) (Bockholt et al., 2010; Scott et al., 2011; Wood et al., 2014), containing data from 198 and 212 subjects from COBRE (Cetin et al., 2014) and MCICShare (Gollub et al., 2013) projects, respectively. (See paper on COINS data in this special issue.) Data for COBRE include sMRI and rest-state fMRI scans collected on a single 3T scanner. Data for the multisite MCICShare include sMRI, rest-state fMRI and dMRI scans, collected on 1.5T and 3T scanners. COINS data required special handling in order to satisfy SchizConnect’s needs because the native COINS architecture involves dynamic data packaging following the query, which does not allow for data to be immediately returned to the query engine2. With permission from the COINS executive committee, an API was built to enable SchizConnect to extract domain-model defined variables (see below) from the COINS databases and to duplicate them at the Mediator site. The database underlying the duplicated MCICShare/COBRE data is MySQL using the relational data model. Access is provided via JDBC to the MySQL server. Results from the query are returned to the Mediator as an SQL ResultSet.
3.2. SchizConnect Mediator
The SchizConnect Mediator domain schema defines the following concepts: project, subject, imaging, cognitive, and clinical, described in detail below.
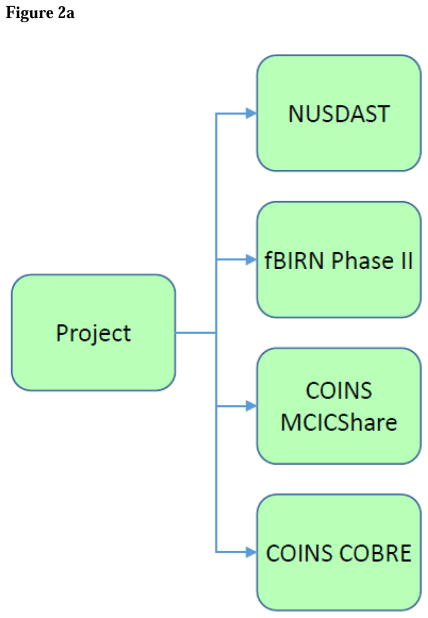
Project contains the name and description of the 4 projects from 3 data sources for which data are collected (Figure 2A).
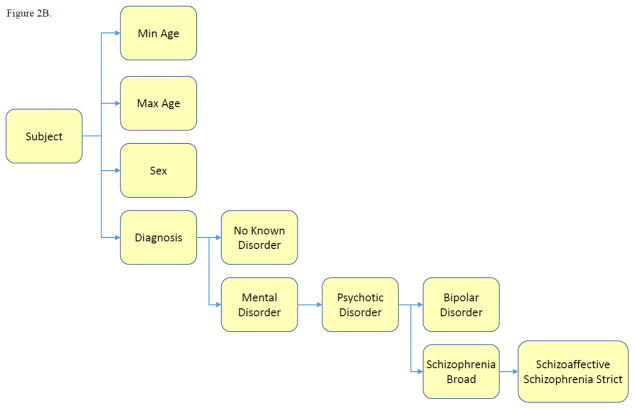
Subject contains demographic and diagnostic information for individual participants, including “subject id”, “age”, “sex” and “diagnosis” (Figure 2B). Current diagnosis categories are: “no known disorder” (for healthy controls), “bipolar disorder,” “schizophrenia broad,” which includes “schizophrenia strict” and “schizoaffective.” We note that the precise definition of “control” may depend on specific studies and study populations, we did not use the term “control” to label the healthy comparison subjects provided by each data source. Instead, the term “no known disorders” is used.
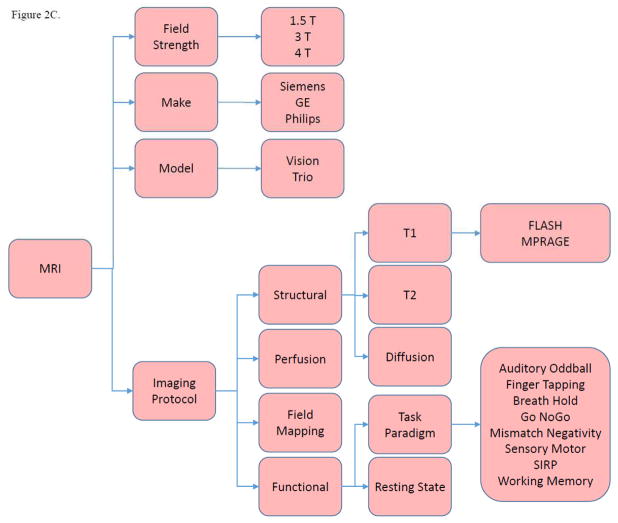
Imaging_Protocol (MRI) contains information on MRI scanner platforms and imaging protocols. “Imaging protocol” consists of “structural,” “functional,” “perfusion,” and “field mapping” categories. Both “structural” and “functional” protocols are subdivided for further distinctions, e.g., “T1,” “T2,” “resting state,” and “task paradigm” (Figure 2C).
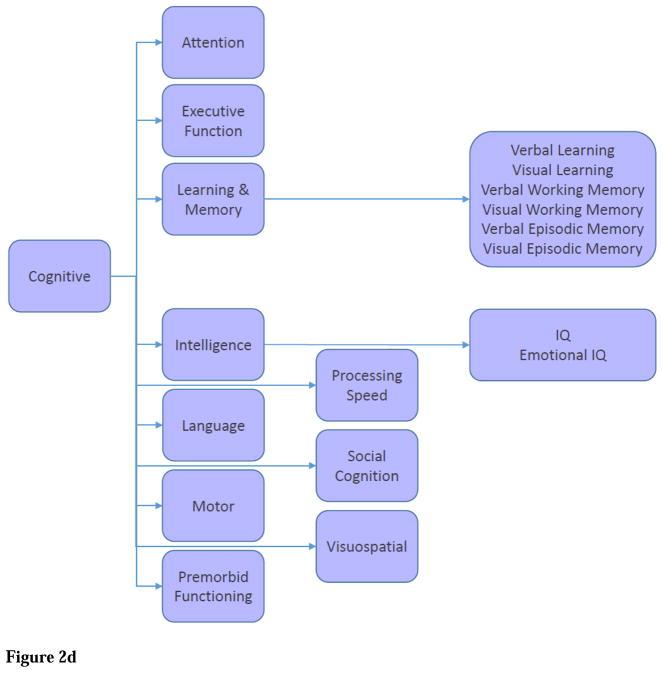
Cognitive domain contains neuropsychological assessments. The domain model uses the MATRICS Consensus Cognitive Battery (Nuechterlein et al., 2008) terms whenever appropriate, which are “attention,” “executive function,” “learning,” “working memory,” “episodic memory,” “intelligence,” “language,” “motor,” “premorbid functioning,” “processing speed,” “social cognition,” and “visuospatial” (Figure 2D).
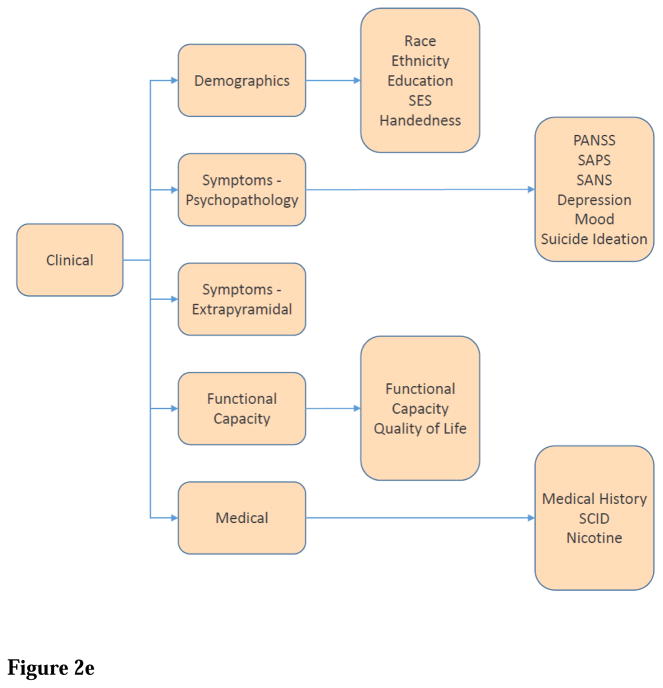
Clinical domain contains full demographics, assessments of symptoms and functional capacity, and medical information. Demographics information includes “Race,” “Ethnicity,” “Education,” “SES (socioeconomic status),” and “Handedness.” Symptom measures include “PANSS (Positive And Negative Symptoms Scales),” “SAPS (Scale for the Assessment of Positive Symptoms),” “SANS (Scale for the Assessment of Negative Symptoms),” using the Unified Medical Language System (UMLS) terms whenever appropriate. Symptom measures also include “Depression,” “Mood,” “Suicide Ideation,”, and “Extrapyramidal Symptoms.” Medical information includes “Medical History/Medication,” “SCID (Structured Clinical Interview for DSM Disorders),” and “Nicotine Addiction/Dependence” (Figure 2E).
Figure 2. SchizConnect Mediator (Virtual) Domain Schema and Inter-schema Mappings.
(A) Project, containing description of the research project for which data are collected. Currently 4 projects from 3 data sources are included in the domain model. (B) Subject, containing demographic and diagnostic information for individual participants of the project. “Subject” is subsequently defined by “minimum age,” “maximum age,” “sex,” and “diagnosis.” Current diagnosis categories are: “no known disorder” (for healthy controls), “bipolar disorder,” “schizophrenia broad,” which includes “schizophrenia strict” and “schizoaffective.” We note that the precise definition of “control” may depend on specific studies and study populations, we did not use the term “control” to label the healthy comparison subjects provided by each data source. Instead, the term “no known disorders” is used. (C) Imaging_Protocol (MRI), containing information on MRI scanner platforms and imaging protocols under which subject imaging data were collected. “Imaging protocol” is subsequently defined by “structural,” “functional,” “perfusion,” and “field mapping.” Both “structural” and “functional” protocols are subdivided for further distinctions including “T1,” “T2,” “resting state,” and “task paradigm.” (D) Cognitive domain contains neuropsychological assessments. The domain model uses the MATRICS Consensus Cognitive Battery (Nuechterlein et al., 2008) terms whenever appropriate, which are “attention,” “executive function,” “learning,” “working memory,” “episodic memory,” “intelligence,” “language,” “motor,” “premorbid functioning,” “processing speed,” “social cognition,” and “visuospatial.” (E) Clinical domain contains full demographics, assessments of symptoms and functional capacity, and medical information. Demographics information includes “Race,” “Ethnicity,” “Education,” “SES (socioeconomic status),” and “Handedness.” Symptom measures include “PANSS (Positive And Negative Symptoms Scales),” “SAPS (Scale for the Assessment of Positive Symptoms),” “SANS (Scale for the Assessment of Negative Symptoms),” using the Unified Medical Language System (UMLS) terms whenever appropriate. Symptom measures also include “Depression,” “Mood,” “Suicide Ideation,”, and “Extrapyramidal Symptoms.” Medical information includes “Medical History/Medication,” “SCID (Structured Clinical Interview for DSM Disorders),” and “Nicotine Addiction/Dependence.”
(G) Examples of inter-schema mappings. For example, subject data for the NUSDAST source is obtained by calling the XNAT search web service and joining with a diagnostic code mapping table to harmonize the diagnoses; normalized diagnosis of Strict Schizophrenia for the subjects in the HID source is computed on the fly, by first joining three HID tables with subject and assessment data, then selecting the subjects according to a predefined algorithm on assessment values.
The SchizConnect Mediator relates the above domain model concepts to the terms in the source schemas using inter-schema mappings (see Figure 2F for sample mappings). For example, subject data for the NUSDAST source is obtained by calling the XNAT search web service and joining with a diagnostic code mapping table to harmonize the diagnoses; normalized diagnosis of Strict Schizophrenia for the subjects in the HID source is computed on the fly, by first joining three HID tables with subject and assessment data, then selecting the subjects according to a predefined algorithm on assessment values.
Currently, the SchizConnect model comprises 7 domain predicates, 17 source predicates (from the 4 sources HID, NUSDAST, COBRE and MCICShare, a local database that contains value mapping information, and utility functional sources), and 18 inter-schema mappings (10 for HID, 4 for NUSDAST, and 4 for COBRE/MCICShare). It is worth noting that since the inter-schema mappings are specified in a declarative rule language, the mediator is easily extensible to incorporate additional sources by writing the appropriate inter-schema mappings.
3.3. SchizConnect.org Web Portal and Example Queries
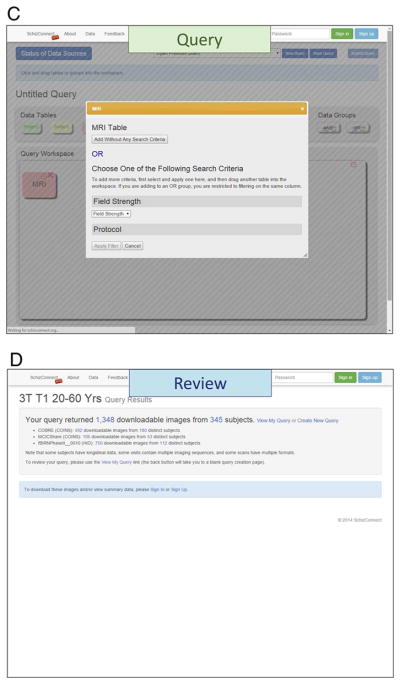
At the http://www.schizconnect.org/ web portal, the operating statuses of the data sources are displayed (Figure 3A). Queries are performed only on data sources that are operational. Data queries are constructed using a drag-and-drop GUI, in which a series of logical “AND” and “OR” operators can be concatenated filtering on domain model terms.
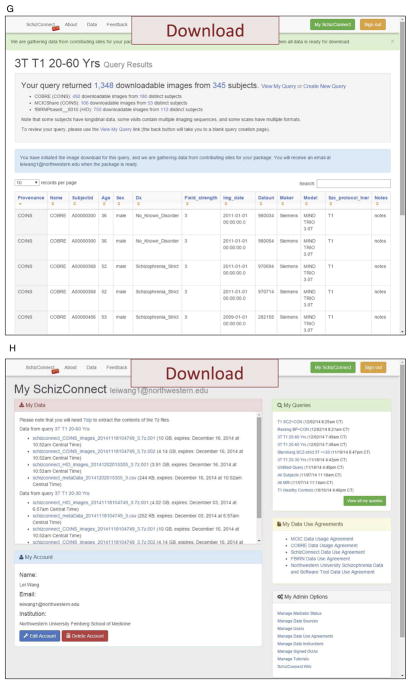
Figure 3. SchizConnect Web Portal, Query, and Return.
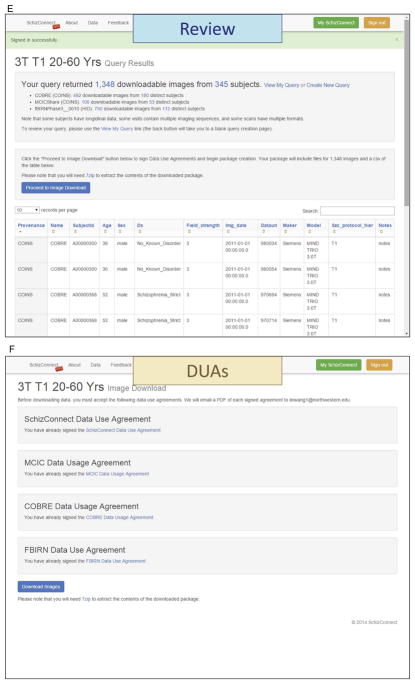
(A) SchizConnet.org At the web portal, the status of the data sources are shown. If one or more of the data sources are unavailable due to source site maintenance or whatever reason, since each data source is queried independent of the others, data sources that are operational can still be queried. The end user is warned of the down sites and therefore their lack of return. (B, C, D) Query and review without signing up. Even without signing up, any end user has the ability to query SchizConnect. Here an example is shown for a query of “T1-weighted 3T scans of subjects between the ages 20 and 60.” The query is built from a drag-and-drop user-friendly graphical user interface. This particular query would return 1,348 downloadable images from 345 distinct subjects from COBRE, MCICShare and fBIRNPhaseII datasets. (E) Query and review after signing up. After signing in, detailed subject-level information returned from the query are now provided as an on-screen table. This table lists sortable variables including provenance, age, sex, and scan parameters. The end user can review and use this information to decide whether to further refine the query. The user has the ability to modify, save and retrieve queries. A data download button is also active at this point, which will lead to the data use agreement page and then data downloading. In the case where multiple images are returned for the same subject, for example as the result of multiple acquisitions within the same MRI session, these images will be indicated with unique “Datauri”s (such as the first 4 entries seen here) fields. (F, G, H) DUA and data download. The user has the option of choosing which DUAs to agree with, and receive data only from data sources with which the DUAs have been agreed. The imaging data resulting from the queries are first transferred out of the data sources and staged at SchizConnect.org host together with the mediated meta-data table for a specified limited time period for downloading. Imaging data files are compressed and packaged into 10 GB individual segments, easily reconstructable with the 7-zip utility (http://www.7-zip.org/). They contain the original and/or preprocessed imaging data shared by each source, in DICOM, Analyze, or NIFTI formats. Links to these files along with unpacking instructions are sent to the user via email and are available on the “MySchizConnect” page of the website.
An example query is shown in Figure 3B, C, with summary return information presented in Figure 3D. Additional sample queries are listed in Table 3. Upon signing in, detailed subject-level information returned from the query is presented to the user as an on-screen table (Figure 3E). This table lists sortable domain model variables including provenance, age, sex, and scan parameters for each subject with downloadable images. The user can review and use this information to decide whether to further refine the query. The user also has the ability to modify, save and retrieve queries. A data download button is available that leads to the DUA page (Figure 3F). The user must sign the SchizConnect DUA, and has the option of choosing which participating source DUAs to agree with, and receives data only from data sources with which the DUAs have been agreed.
For downloading, the imaging data resulting from the queries are transferred out of the data sources and staged at the SchizConnect.org host, together with the returned meta-data table of the mediated variables. Transfer is performed via gridFTP, REST API, HTTP, or others, depending on the specific data transfer protocol at each source. Imaging data files are compressed and packaged into 10-GB easily reconstructable, individual 7-zip (http://www.7-zip.org/) segments, available for the user to download within a specified limited time period (currently 2 weeks). Depending on the size of the requested imaging data, it can take up to a few days for all data to be staged for download. These data files contain the original and/or preprocessed imaging data shared by each source, in DICOM, Analyze, or NIFTI formats. Links to these files along with unpacking instructions are sent to the user via email. The links are also available on the “My SchizConnect” page of the web portal (Figure 3G, H). Documentation for descriptions of image formats and directory structures can be found on the SchizConnect website (http://www.schizconnect.org/documentation). All cognitive and clinical data (see model description above) can be included in the download package along with the download imaging data. On the documentation page, the user can find useful information including: Tutorials on how to use the website, DUAs of all data sources, data descriptions, data dictionaries, and peer-reviewed journal papers related to the data. Since not all neuroimaging data (such as scan sequence parameters) have been modeled into the SchizConnect domain hierarchy, it is important to provide technical information on our website to facilitate research. In the Data Description section, the NUSDAST description lists structural parameters (e.g., 3D MPRAGE: TR=9.7 ms, TE=4 ms, flip=10°, ACQ=1, 256×256 matrix, 1xl mm in-plane resolution, 128 slices, slice thickness=1.25 mm, 5:36 min scan time each) and the COBRE description lists resting-state parameters (resting state scans consisting of 149 volumes of T2*-weighted functional images, acquired using a gradient-echo EPI sequence: TR=2 s, TE=29 ms, flip=75°, slice thickness=3.5 mm, slice gap=1.05 mm, field of view=240 mm, matrix size=64 × 64, voxel size=3.75 mm × 3.75 mm × 4.55 mm). In the Publications section, we have provided a link to a peer-reviewed journal publication that provide detailed technical descriptions on each data source. For example, Gollub et al (2013) describes “the MCIC collection: a shared repository of multi-modal, multi-site brain image data from a clinical investigation of schizophrenia.” Similar papers describing all the other data sources currently participating in SchizConnect can be found here.
4. Discussions and Future Plans
These initial results demonstrate that SchizConnect allows combining of neuroimaging data from different databases via mediation to form compatible mega-datasets with accuracy and fidelity. In SchizConnect, data remains with the source rather than warehoused in a central repository. Providers maintain control of their data and do not need to modify them for sharing. The user query is done over a single, consistent, well-defined model that is translated to the schemas of the sources. This approach unburdens the user and each source from having to make contact and helping to interpret the data each time, especially when new data are being continuously added to the data source repository. The web portal is user-friendly and intuitive, appearing to the user as a single, virtual database with real-time query and download performance. As an on-going project, we are continuing to define additional domain model terms for neuropsychological and psychopathological variables, make available additional imaging modalities and subjects, and identify and evaluate potential new data sources. Work is also under way integrating with clinical research databases such as Research Electronic Data Capture (REDCap) (Harris et al., 2009) where clinical and cognitive assessment data are stored, further enhancing the functionality of SchizConnect by searching across imaging and non-imaging databases for the same subject.
SchizConnect shows considerable potential for overcoming current barriers for creating large-scale datasets to increase statistical power, accelerating the testing of new hypotheses and methods, and creating a resource for developing advanced techniques to better integrate disparate data at low cost. As a proof-of-concept for combining datasets, we compared SAPS/SANS and neurocognition across the current data sources. For SAPS/SANS, we found that while fewer schizophrenia subjects in the NU data scored 2 or higher, the distributions were not statistically different between sites. For neurocognitive measures, we computed z-scores on episodic memory, working memory, attention and executive function domains using each site’s own controls as reference sets. No statistical difference between sites was observed (NU range: −1.23 to −0.71, FBIRN: −1.28 to −0.65), indicating that schizophrenia subjects across both consortia exhibit similar degrees of psychopathology and cognitive impairment. Nonetheless, subtle differences in cohort make-up may still exist upon closer inspection, but users will have the opportunity to explore the sources of potential heterogeneity and use such differences to benefit in developing new hypotheses.
SchizConnect was aimed at combining across different types of data repositories, and its current data sources are examples of classic types of data repository: The HID that houses the FBIRN data is a multisite study repository with prospectively collected data using predetermined protocols; The COINS Data Exchange that houses the MCICShare and COBRE data, and the XNAT Central that houses the NUSDAST data, are centralized data repositories where data from unrelated studies are uploaded and stored, and the database platforms provide the user with queryable data structures and interfaces. When owners of these datasets are willing to share subject-level data (i.e., scans, assessments), SchizConnect is designed to interact with such data repositories for mediation. When investigators are only willing to share measures, meta-analysis efforts such as the ENIGMA (Gupta et al., 2014; Thompson et al., 2014; Turner et al., 2015 (accepted); Wright et al., 2015 (accepted)) will be an excellent alternative for data sharing.
During the initial implementation of SchizConnect using the current data sources as test beds, we have established a series of domain models and their translations to the initial data source models. The SchizConnect mediation approach simply translates user queries into the query language of each data source (i.e., equivalent to the user performing the same query at the individual data source’s own web portal). The SchizConnect data models can be extended easily to accommodate new data structures of a new data source while placing no restrictions on what the data source wants to share. Although the process of domain understanding and data modeling can be labor intensive, we have laid the foundation for extending SchizConnect to other schizophrenia neuroimaging data sources, which will build on the existing schemas. A limitation of the current domain model work is that the understanding and modeling of source data remains a manual step. In SchizConnect, this data mediation process starts with domain understanding by experts on the specific data that is being integrated so that we have a clear knowledge on the data terms and their definition. The data mediation experts can then integrate this new knowledge into existing SchizConnect models to enable the data to be accessed without requiring it to be modified. We have provided a questionnaire on the schizconnect.org web portal that can initiate the process, http://schizconnect.org/questionnaires/1/responses/new. It is important as a next step for us to develop approaches that learn from existing data to facilitate more automated procedures for inclusion of new data sources (e.g., following our previous work (Knoblock et al., 2012)). Another limitation is query evaluation in the virtual integration approach, is generally slower that query evaluation on a warehouse-model database.
Finally, SchizConnect is the only data finder (data broker) of its kind for researchers interested in combining neuroimaging data on schizophrenia and related disorders from disparate sources. Existing data portals such as the COINS, FBIRN, and XNAT Central, data sources used in this project, contain schizophrenia data but serve as repositories of these types of data. When users intent to combine data across these sources, harmonization of data terms and mediation of these terms remain a critical challenge and barrier. SchizConnect is fulfilling this critical need, and the data mediation framework that SchizConnect created can be readily extended to other clinical domains including Bipolar Disorder. Recent developments on data harmonization have led to the creation of the Research Domain Criteria Database (RDoCdb) at the National Institute of Mental Health (NIMH) and its associated data repository, the National Database for Autism Research (NDAR) (Hall et al., 2012). It should be noted that SchizConnect is a virtual database that can mediate data sources that make ongoing updates to their repositories, and SchizConnect is focused on schizophrenia and related disorders. Administratively, the SchizConnect web portal is hosted at Northwestern University, the SchizConnect mediator is hosted at the Information Science Institute of the University of Southern California. For inclusion into SchizConnect, new data sources will be evaluated on completeness, quality, and enthusiasm to collaborate. The focus of the initial expansion will be the quickest return for the research community, so that larger, richer datasets that have been carefully collected and checked for completeness and quality (e.g., already analyzed and published) would be of the highest priority. Data repositories based on XNAT, or those are already in COINS can be more readily integrated, up to any new data models and new querying news. For valuable datasets without a database platform, we can help develop an XNAT, HID, or COINS.
It is our hope that SchizConnect can facilitate the testing of new hypotheses that are hitherto not possible, thus greatly promote discovery related to the mechanisms underlying schizophrenia. We aim to expand to repositories on psychosis and related disorders, thus facilitating large-scale dimensional research. We hope that SchizConnect will become the prototype for the study of psychiatric disorders, serving as a model for broader efforts for the integration and sharing of biomedical information across the greater scientific community.
Highlights.
Query and combine imaging data from different databases within a single interface.
A single, virtual database where live data remains at the sources.
Web portal is user-friendly and intuitive, performing real-time query and download.
It is the only data broker of its kind for schizophrenia and related disorders.
SchizConnect currently contains data from 3 sources, and is extending to others.
Acknowledgments
This work was supported in part by NIH grants 1U01 MH097435, 1R01 MH084803, P50 MH071616, R01 MH056584, U24 RR025736-01, U24-RR021992, U24GM10420, P20 GM103472.
Footnotes
This link will be replaced by the SchizConnect web portal in the near future.
On the COINS website, after user builds a query, data are packaged offline and a link is provided for downloading. Therefore the user never sees the actual data until the data are downloaded via the link.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Addington J, Cadenhead KS, Cornblatt BA, Mathalon DH, McGlashan TH, Perkins DO, Seidman LJ, Tsuang MT, Walker EF, Woods SW, Addington JA, Cannon TD. North American Prodrome Longitudinal Study (NAPLS 2): overview and recruitment. Schizophr Res. 2012;142:77–82. doi: 10.1016/j.schres.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen EA, Erhardt EB, Damaraju E, Gruner W, Segall JM, Silva RF, Havlicek M, Rachakonda S, Fries J, Kalyanam R, Michael AM, Caprihan A, Turner JA, Eichele T, Adelsheim S, Bryan AD, Bustillo J, Clark VP, Feldstein Ewing SW, Filbey F, Ford CC, Hutchison K, Jung RE, Kiehl KA, Kodituwakku P, Komesu YM, Mayer AR, Pearlson GD, Phillips JP, Sadek JR, Stevens M, Teuscher U, Thoma RJ, Calhoun VD. A baseline for the multivariate comparison of resting-state networks. Front Syst Neurosci. 2011;5:2. doi: 10.3389/fnsys.2011.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antonioletti M, Atkinson M, Baxter R, Borley A, Chue Hong NP, Collins B, Hardman N, Hume AC, Knox A, Jackson M, Krause A, Laws S, Magowan J, Paton NW, Pearson D, Sugden T, Watson P, Westhead M. The design and implementation of Grid database services in OGSA-DAI. Concurrency and Computation: Practice and Experience. 2005;17:357–376. [Google Scholar]
- Arnold SJ, Ivleva EI, Gopal TA, Reddy AP, Jeon-Slaughter H, Sacco CB, Francis AN, Tandon N, Bidesi AS, Witte B, Poudyal G, Pearlson GD, Sweeney JA, Clementz BA, Keshavan MS, Tamminga CA. Hippocampal Volume Is Reduced in Schizophrenia and Schizoaffective Disorder But Not in Psychotic Bipolar I Disorder Demonstrated by Both Manual Tracing and Automated Parcellation (FreeSurfer) Schizophr Bull. 2015;41:233–249. doi: 10.1093/schbul/sbu009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashish N, Ambite JL, Muslea M, Turner JA. Neuroscience Data Integration through Mediation: An (F)BIRN Case Study. Front Neuroinformatics. 2010;4:118. doi: 10.3389/fninf.2010.00118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjaalie JG, Grillner S. Global neuroinformatics: the International Neuroinformatics Coordinating Facility. J Neurosci. 2007;27:3613–3615. doi: 10.1523/JNEUROSCI.0558-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bockholt HJ, Scully M, Courtney W, Rachakonda S, Scott A, Caprihan A, Fries J, Kalyanam R, Segall JM, de la Garza R, Lane S, Calhoun VD. Mining the mind research network: a novel framework for exploring large scale, heterogeneous translational neuroscience research data sources. Front Neuroinformatics. 2010;3:36. doi: 10.3389/neuro.11.036.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon TD, Chung Y, He G, Sun D, Jacobson A, van Erp TG, McEwen S, Addington J, Bearden CE, Cadenhead K, Cornblatt B, Mathalon DH, McGlashan T, Perkins D, Jeffries C, Seidman LJ, Tsuang M, Walker E, Woods SW, Heinssen R North American Prodrome Longitudinal Study C. Progressive reduction in cortical thickness as psychosis develops: a multisite longitudinal neuroimaging study of youth at elevated clinical risk. Biol Psychiatry. 2015;77:147–157. doi: 10.1016/j.biopsych.2014.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro E, Gupta CN, Martinez-Ramon M, Calhoun VD, Arbabshirani MR, Turner J. Identification of patterns of gray matter abnormalities in schizophrenia using source-based morphometry and bagging. Conf Proc IEEE Eng Med Biol Soc. 2014;2014:1513–1516. doi: 10.1109/EMBC.2014.6943889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cetin MS, Christensen F, Abbott CC, Stephen JM, Mayer AR, Canive JM, Bustillo JR, Pearlson GD, Calhoun VD. Thalamus and posterior temporal lobe show greater inter-network connectivity at rest and across sensory paradigms in schizophrenia. Neuroimage. 2014;97:117–126. doi: 10.1016/j.neuroimage.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Calhoun VD, Pearlson GD, Ehrlich S, Turner JA, Ho BC, Wassink TH, Michael AM, Liu J. Multifaceted genomic risk for brain function in schizophrenia. Neuroimage. 2012;61:866–875. doi: 10.1016/j.neuroimage.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen JY, Calhoun VD, Pearlson GD, Perrone-Bizzozero N, Sui J, Turner JA, Bustillo JR, Ehrlich S, Sponheim SR, Canive JM, Ho BC, Liu JY. Guided exploration of genomic risk for gray matter abnormalities in schizophrenia using parallel independent component analysis with reference. Neuroimage. 2013;83:384–396. doi: 10.1016/j.neuroimage.2013.05.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Schutter E. The International Neuroinformatics Coordinating Facility: evaluating the first years. Neuroinformatics. 2009;7:161–163. doi: 10.1007/s12021-009-9054-1. [DOI] [PubMed] [Google Scholar]
- Ehrlich S, Geisler D, Yendiki A, Panneck P, Roessner V, Calhoun VD, Magnotta VA, Gollub RL, White T. Associations of White Matter Integrity and Cortical Thickness in Patients With Schizophrenia and Healthy Controls. Schizophr Bull. 2014;40:665–674. doi: 10.1093/schbul/sbt056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florescu D, Levy AY, Mendelzon A. Database techniques for the world-wide web: a survey. SIGMOD Record. 1998;27:59–74. [Google Scholar]
- Friedman L, Stern H, Brown GG, Mathalon DH, Turner J, Glover GH, Gollub RL, Lauriello J, Lim KO, Cannon T, Greve DN, Bockholt HJ, Belger A, Mueller B, Doty MJ, He J, Wells W, Smyth P, Pieper S, Kim S, Kubicki M, Vangel M, Potkin SG. Test-retest and between-site reliability in a multicenter fMRI study. Hum Brain Mapp. 2008;29:958–972. doi: 10.1002/hbm.20440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gadde S, Aucoin N, Grethe JS, Keator DB, Marcus DS, Pieper S, Fbirn MBC. XCEDE: an extensible schema for biomedical data. Neuroinformatics. 2012;10:19–32. doi: 10.1007/s12021-011-9119-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner D, Akil H, Ascoli GA, Bowden DM, Bug W, Donohue DE, Goldberg DH, Grafstein B, Grethe JS, Gupta A, Halavi M, Kennedy DN, Marenco L, Martone ME, Miller PL, Muller HM, Robert A, Shepherd GM, Sternberg PW, Van Essen DC, Williams RW. The neuroscience information framework: a data and knowledge environment for neuroscience. Neuroinformatics. 2008;6:149–160. doi: 10.1007/s12021-008-9024-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover GH, Mueller BA, Turner JA, van Erp TG, Liu TT, Greve DN, Voyvodic JT, Rasmussen J, Brown GG, Keator DB, Calhoun VD, Lee HJ, Ford JM, Mathalon DH, Diaz M, O’Leary DS, Gadde S, Preda A, Lim KO, Wible CG, Stern HS, Belger A, McCarthy G, Ozyurt B, Potkin SG. Function biomedical informatics research network recommendations for prospective multicenter functional MRI studies. J Magn Reson Imaging. 2012;36:39–54. doi: 10.1002/jmri.23572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gollub RL, Shoemaker JM, King MD, White T, Ehrlich S, Sponheim SR, Clark VP, Turner JA, Mueller BA, Magnotta V, O’Leary D, Ho BC, Brauns S, Manoach DS, Seidman L, Bustillo JR, Lauriello J, Bockholt J, Lim KO, Rosen BR, Schulz SC, Calhoun VD, Andreasen NC. The MCIC collection: a shared repository of multimodal, multi-site brain image data from a clinical investigation of schizophrenia. Neuroinformatics. 2013;11:367–388. doi: 10.1007/s12021-013-9184-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant A, Antonioletti M, Hume AC, Krause A, Dobrzelecki B, Jackson MJ, Parsons M, Atkinson MP, Theocharopoulos E. OGSA-DAI: Middleware for Data Integration: Selected Applications. IEEE Fourth International Conference on eScience, 2008. eScience ’08; 2008. pp. 343–343. [Google Scholar]
- Gupta CN, Calhoun VD, Rachakonda S, Chen J, Patel V, Liu J, Segall J, Franke B, Zwiers MP, Arias-Vasquez A, Buitelaar J, Fisher SE, Fernandez G, van Erp TG, Potkin S, Ford J, Mathalon D, McEwen S, Lee HJ, Mueller BA, Greve DN, Andreassen O, Agartz I, Gollub RL, Sponheim SR, Ehrlich S, Wang L, Pearlson G, Glahn DC, Sprooten E, Mayer AR, Stephen J, Jung RE, Canive J, Bustillo J, Turner JA. Patterns of Gray Matter Abnormalities in Schizophrenia Based on an International Mega-analysis. Schizophr Bull. 2014 doi: 10.1093/schbul/sbu177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halevy AY. Answering queries using views: a survey. VLDB Journal. 2001;10:270–294. [Google Scholar]
- Hall D, Huerta MF, McAuliffe MJ, Farber GK. Sharing heterogeneous data: the national database for autism research. Neuroinformatics. 2012;10:331–339. doi: 10.1007/s12021-012-9151-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass J, Walton E, Kirsten H, Turner J, Wolthusen R, Roessner V, Sponheim SR, Holt D, Gollub R, Calhoun VD, Ehrlich S. Complexin2 modulates working memory-related neural activity in patients with schizophrenia. Eur Arch Psychiatry Clin Neurosci. 2014 doi: 10.1007/s00406-014-0550-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass J, Walton E, Wright C, Beyer A, Scholz M, Turner J, Liu J, Smolka MN, Roessner V, Sponheim SR, Gollub RL, Calhoun VD, Ehrlich S. Associations between DNA methylation and schizophrenia-related intermediate phenotypes - A gene set enrichment analysis. Prog Neuropsychopharmacol Biol Psychiatry. 2015 doi: 10.1016/j.pnpbp.2015.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmer KG, Ambite JL, Ames J, Ananthakrishnan R, Burns G, Chervenak AL, Foster I, Liming L, Keator D, Macciardi F, Madduri R, Navarro JP, Potkin S, Rosen B, Ruffins S, Schuler R, Turner JA, Toga A, Williams C, Kesselman C. Enabling collaborative research using the Biomedical Informatics Research Network (BIRN) J Am Med Inform Assoc. 2011a doi: 10.1136/amiajnl-2010-000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmer KG, Ambite JL, Ames J, Ananthakrishnan R, Burns G, Chervenak AL, Foster I, Liming L, Keator D, Macciardi F, Madduri R, Navarro JP, Potkin S, Rosen B, Ruffins S, Schuler R, Turner JA, Toga A, Williams C, Kesselman C Biomedical Informatics Research N. Enabling collaborative research using the Biomedical Informatics Research Network (BIRN) J Am Med Inform Assoc. 2011b;18:416–422. doi: 10.1136/amiajnl-2010-000032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keator DB, Grethe JS, Marcus D, Ozyurt B, Gadde S, Murphy S, Pieper S, Greve D, Notestine R, Bockholt HJ, Papadopoulos P. A national human neuroimaging collaboratory enabled by the Biomedical Informatics Research Network (BIRN) IEEE Trans Inf Technol Biomed. 2008;12:162–172. doi: 10.1109/TITB.2008.917893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keator DB, Wei D, Gadde S, Bockholt J, Grethe JS, Marcus D, Aucoin N, Ozyurt IB. Derived Data Storage and Exchange Workflow for Large-Scale Neuroimaging Analyses on the BIRN Grid. Front Neuroinform. 2009;3:30. doi: 10.3389/neuro.11.030.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keshavan MS, Morris DW, Sweeney JA, Pearlson G, Thaker G, Seidman LJ, Eack SM, Tamminga C. A dimensional approach to the psychosis spectrum between bipolar disorder and schizophrenia: the Schizo-Bipolar Scale. Schizophr Res. 2011;133:250–254. doi: 10.1016/j.schres.2011.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim DI, Manoach DS, Mathalon DH, Turner JA, Mannell M, Brown GG, Ford JM, Gollub RL, White T, Wible C, Belger A, Bockholt HJ, Clark VP, Lauriello J, O’Leary D, Mueller BA, Lim KO, Andreasen N, Potkin SG, Calhoun VD. Dysregulation of working memory and default-mode networks in schizophrenia using independent component analysis, an fBIRN and MCIC study. Hum Brain Mapp. 2009;30:3795–3811. doi: 10.1002/hbm.20807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MA, Tura E, Potkin SG, Fallon JH, Manoach DS, Calhoun VD, Turner JA. Working memory circuitry in schizophrenia shows widespread cortical inefficiency and compensation. Schizophr Res. 2010;117:42–51. doi: 10.1016/j.schres.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblock CA, Szekely P, Jos #233, Ambite L, Goel A, Gupta S, Lerman K, Muslea M, Taheriyan M, Mallick P. Semi-automatically mapping structured sources into the semantic web. Proceedings of the 9th international conference on The Semantic Web: research and applications; Heraklion, Crete, Greece: Springer-Verlag; 2012. pp. 375–390. [Google Scholar]
- Larson SD, Martone ME. NeuroLex.org: an online framework for neuroscience knowledge. Front Neuroinform. 2013;7:18. doi: 10.3389/fninf.2013.00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzerini M. Data integration: a theoretical perspective. Proceedings of ACM Symposium on Principles of Database Systems; Madison, Wisconsin. 2002. [Google Scholar]
- Lynden S, Mukherjee A, Hume AC, Fernandes AAA, Paton NW, Sakellariou R, Watson P. The design and implementation of OGSA-DQP: A service-based distributed query processor. Future Generation Computer Systems. 2009;25:224–236. [Google Scholar]
- Lynden S, Pahlevi SM, Kojima I. Service-based data integration using OGSA-DQP and OGSA-WebDB. 2008 9th IEEE/ACM International Conference on Grid Computing; 2008. pp. 160–167. [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Mathew I, Gardin TM, Tandon N, Eack S, Francis AN, Seidman LJ, Clementz B, Pearlson GD, Sweeney JA, Tamminga CA, Keshavan MS. Medial temporal lobe structures and hippocampal subfields in psychotic disorders: findings from the Bipolar-Schizophrenia Network on Intermediate Phenotypes (B-SNIP) study. JAMA Psychiatry. 2014;71:769–777. doi: 10.1001/jamapsychiatry.2014.453. [DOI] [PubMed] [Google Scholar]
- Nuechterlein KH, Green MF, Kern RS, Baade LE, Barch DM, Cohen JD, Essock S, Fenton WS, Frese FJ, 3rd, Gold JM, Goldberg T, Heaton RK, Keefe RS, Kraemer H, Mesholam-Gately R, Seidman LJ, Stover E, Weinberger DR, Young AS, Zalcman S, Marder SR. The MATRICS Consensus Cognitive Battery, part 1: test selection, reliability, and validity. Am J Psychiatry. 2008;165:203–213. doi: 10.1176/appi.ajp.2007.07010042. [DOI] [PubMed] [Google Scholar]
- Ozyurt IB, Keator DB, Wei D, Fennema-Notestine C, Pease KR, Bockholt J, Grethe JS. Federated web-accessible clinical data management within an extensible neuroimaging database. Neuroinformatics. 2010;8:231–249. doi: 10.1007/s12021-010-9078-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Ford JM. Widespread cortical dysfunction in schizophrenia: the FBIRN imaging consortium. Schizophr Bull. 2009;35:15–18. doi: 10.1093/schbul/sbn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potkin SG, Turner JA, Brown GG, McCarthy G, Greve DN, Glover GH, Manoach DS, Belger A, Diaz M, Wible CG, Ford JM, Mathalon DH, Gollub R, Lauriello J, O’Leary D, van Erp TG, Toga AW, Preda A, Lim KO, Fbirn Working memory and DLPFC inefficiency in schizophrenia: the FBIRN study. Schizophr Bull. 2009;35:19–31. doi: 10.1093/schbul/sbn162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott A, Courtney W, Wood D, de la Garza R, Lane S, King M, Wang R, Roberts J, Turner JA, Calhoun VD. COINS: An Innovative Informatics and Neuroimaging Tool Suite Built for Large Heterogeneous Datasets. Front Neuroinform. 2011;5:33. doi: 10.3389/fninf.2011.00033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thaker G. Psychosis endophenotypes in schizophrenia and bipolar disorder. Schizophr Bull. 2008;34:720–721. doi: 10.1093/schbul/sbn055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson PM, Stein JL, Medland SE, Hibar DP, Vasquez AA, Renteria ME, Toro R, Jahanshad N, Schumann G, Franke B, Wright MJ, Martin NG, Agartz I, Alda M, Alhusaini S, Almasy L, Almeida J, Alpert K, Andreasen NC, Andreassen OA, Apostolova LG, Appel K, Armstrong NJ, Aribisala B, Bastin ME, Bauer M, Bearden CE, Bergmann O, Binder EB, Blangero J, Bockholt HJ, Boen E, Bois C, Boomsma DI, Booth T, Bowman IJ, Bralten J, Brouwer RM, Brunner HG, Brohawn DG, Buckner RL, Buitelaar J, Bulayeva K, Bustillo JR, Calhoun VD, Cannon DM, Cantor RM, Carless MA, Caseras X, Cavalleri GL, Chakravarty MM, Chang KD, Ching CR, Christoforou A, Cichon S, Clark VP, Conrod P, Coppola G, Crespo-Facorro B, Curran JE, Czisch M, Deary IJ, de Geus EJ, den Braber A, Delvecchio G, Depondt C, de Haan L, de Zubicaray GI, Dima D, Dimitrova R, Djurovic S, Dong H, Donohoe G, Duggirala R, Dyer TD, Ehrlich S, Ekman CJ, Elvsashagen T, Emsell L, Erk S, Espeseth T, Fagerness J, Fears S, Fedko I, Fernandez G, Fisher SE, Foroud T, Fox PT, Francks C, Frangou S, Frey EM, Frodl T, Frouin V, Garavan H, Giddaluru S, Glahn DC, Godlewska B, Goldstein RZ, Gollub RL, Grabe HJ, Grimm O, Gruber O, Guadalupe T, Gur RE, Gur RC, Goring HH, Hagenaars S, Hajek T, Hall GB, Hall J, Hardy J, Hartman CA, Hass J, Hatton SN, Haukvik UK, Hegenscheid K, Heinz A, Hickie IB, Ho BC, Hoehn D, Hoekstra PJ, Hollinshead M, Holmes AJ, Homuth G, Hoogman M, Hong LE, Hosten N, Hottenga JJ, Hulshoff Pol HE, Hwang KS, Jack CR, Jr, Jenkinson M, Johnston C, Jonsson EG, Kahn RS, Kasperaviciute D, Kelly S, Kim S, Kochunov P, Koenders L, Kramer B, Kwok JB, Lagopoulos J, Laje G, Landen M, Landman BA, Lauriello J, Lawrie SM, Lee PH, Le Hellard S, Lemaitre H, Leonardo CD, Li CS, Liberg B, Liewald DC, Liu X, Lopez LM, Loth E, Lourdusamy A, Luciano M, Macciardi F, Machielsen MW, Macqueen GM, Malt UF, Mandl R, Manoach DS, Martinot JL, Matarin M, Mather KA, Mattheisen M, Mattingsdal M, Meyer-Lindenberg A, McDonald C, McIntosh AM, McMahon FJ, McMahon KL, Meisenzahl E, Melle I, Milaneschi Y, Mohnke S, Montgomery GW, Morris DW, Moses EK, Mueller BA, Munoz Maniega S, Muhleisen TW, Muller-Myhsok B, Mwangi B, Nauck M, Nho K, Nichols TE, Nilsson LG, Nugent AC, Nyberg L, Olvera RL, Oosterlaan J, Ophoff RA, Pandolfo M, Papalampropoulou-Tsiridou M, Papmeyer M, Paus T, Pausova Z, Pearlson GD, Penninx BW, Peterson CP, Pfennig A, Phillips M, Pike GB, Poline JB, Potkin SG, Putz B, Ramasamy A, Rasmussen J, Rietschel M, Rijpkema M, Risacher SL, Roffman JL, Roiz-Santianez R, Romanczuk-Seiferth N, Rose EJ, Royle NA, Rujescu D, Ryten M, Sachdev PS, Salami A, Satterthwaite TD, Savitz J, Saykin AJ, Scanlon C, Schmaal L, Schnack HG, Schork AJ, Schulz SC, Schur R, Seidman L, Shen L, Shoemaker JM, Simmons A, Sisodiya SM, Smith C, Smoller JW, Soares JC, Sponheim SR, Sprooten E, Starr JM, Steen VM, Strakowski S, Strike L, Sussmann J, Samann PG, Teumer A, Toga AW, Tordesillas-Gutierrez D, Trabzuni D, Trost S, Turner J, Van den Heuvel M, van der Wee NJ, van Eijk K, van Erp TG, van Haren NE, van’t Ent D, van Tol MJ, Valdes Hernandez MC, Veltman DJ, Versace A, Volzke H, Walker R, Walter H, Wang L, Wardlaw JM, Weale ME, Weiner MW, Wen W, Westlye LT, Whalley HC, Whelan CD, White T, Winkler AM, Wittfeld K, Woldehawariat G, Wolf C, Zilles D, Zwiers MP, Thalamuthu A, Schofield PR, Freimer NB, Lawrence NS, Drevets W the Alzheimer’s Disease Neuroimaging Initiative, EC.IC.SY.SG. The ENIGMA Consortium: large-scale collaborative analyses of neuroimaging and genetic data. Brain Imaging Behav. 2014 doi: 10.1007/s11682-013-9269-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner J, Theo van Erp DH, Rasmussen Jerod, Glahn David, Pearlson Godfrey, Andreassen Ole, Agartz Ingrid, Westlye Lars, Haukvik Unn Kristin, Dale Anders, Hartberg Cecilie, Gruber Oliver, Krämer Bernd, Zilles David, Donohoe Gary, Kelly Sinead, McDonald Colm, Morris Derek, Cannon Dara M, Corvin Aiden, Machielsen Marise, Koenders Laura, de Haan Lieuwe, Veltman Dick, Satterthwaite Theodore, Wolf Daniel, Gur Ruben, Gur Raquel, Potkin Steven, Mathalon Daniel, Mueller Bryon, Preda Adrian, Macciardi Fabio, Ehrlich Stefan, Walton Esther, Hass Johanna, Calhoun Vince, Bockholt Henry, Sponheim Scott, Shoemaker Jody, Haren Neeltje van, Pol Hilleke Hulshoff, Ophoff Roel, Kahn Réne S, Roiz-Santiañez Roberto, Crespo-Facorro Benedicto, Wang Lei, Alpert Kathryn, Jonsson Erik, Dimitrova Rali, Bois Cathrine, Whalley Heather, McIntosh Andrew, Lawrie Stephen, Hashimoto Ryota, Thompson Paul, Melle I. Subcortical Brain Volume Abnormalities in 2,028 Individuals with Schizophrenia and 2,540 Healthy Controls via the ENIGMA Consortium. Molecular Psychiatry. 2015 doi: 10.1038/mp.2015.63. (accepted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JA. The rise of large-scale imaging studies in psychiatry. GigaScience. 2014;3 doi: 10.1186/2047-217X-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullman JD. Information integration using logical views. Proceedings of the Sixth International Conference on Database Theory; Delphi, Greece. 1997. pp. 19–40. [Google Scholar]
- Wang L, Alpert KI, Calhoun V, Keator D, King M, Kogan A, Landis D, Tallis M, Potkin SG, Turner JA, Ambite JL. SchizConnect: Large-Scale Schizophrenia Neuroimaging Data Integration and Sharing. Annual Meeting of the American College of Neuropsychopharmacology (ACNP); Phoenix, Arizona. 2014. [Google Scholar]
- Wang L, Kogan A, Cobia D, Alpert K, Kolasny A, Miller MI, Marcus D. Northwestern University Schizophrenia Data and Software Tool (NUSDAST) Front Neuroinform. 2013;7:25. doi: 10.3389/fninf.2013.00025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederhold G. Mediators in the Architecture of Future Information Systems. IEEE Computer. 1992;25:38–49. [Google Scholar]
- Wood D, King M, Landis D, Courtney W, Wang R, Kelly R, Turner JA, Calhoun VD. Harnessing modern web application technology to create intuitive and efficient data visualization and sharing tools. Front Neuroinform. 2014;8:71. doi: 10.3389/fninf.2014.00071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright C, Calhoun VD, Ehrlich S, Wang L, Turner JA, Perrone-Bizzozero NI. Meta Gene Set Enrichment Analyses Link miR-137-regulated Pathways with Schizophrenia Risk. Frontiers in Genetics, section Behavioral and Psychiatric Genetics. 2015 doi: 10.3389/fgene.2015.00147. accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]