Abstract
DNA lesions arise from many endogenous and environmental agents, and they promote deleterious events leading to genomic instability and cell death. Base excision repair (BER) is the main DNA repair pathway responsible for repairing single strand breaks, base lesions and abasic sites in mammalian cells. During BER, DNA substrates and repair intermediates are channeled from one step to the next in a sequential fashion so that release of toxic repair intermediates is minimized. This includes handoff of the product of gap-filling DNA synthesis to the DNA ligation step. The conformational differences in DNA polymerase β (pol β) associated with incorrect or oxidized nucleotide (8-oxodGMP) insertion could impact channeling of the repair intermediate to the final step of BER, i.e., DNA ligation by DNA ligase I or the DNA Ligase III/XRCC1 complex. Thus, modified DNA ligase substrates produced by faulty pol β gap-filling could impair coordination between pol β and DNA ligase. Ligation failure is associated with 5′-AMP addition to the repair intermediate and accumulation of strand breaks that could be more toxic than the initial DNA lesions. Here, we provide an overview of the consequences of ligation failure in the last step of BER. We also discuss DNA-end processing mechanisms that could play roles in reversal of impaired BER.
Keywords: Base excision repair (BER); DNA polymerase β (pol β); DNA ligase; 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG); ligation failure; abortive ligation products
1. Repair of oxidant and environmental toxicant-induced DNA lesions by base excision repair
Environmental and endogenous stressors damage genomic DNA [1]. These stressors include radiation, base loss through spontaneous hydrolysis of the glycosidic bond and attack by reactive agents such as reactive oxygen and nitrogen species and alkylating agents [2]. One of the most abundant lesions in DNA is the abasic or apurinic/apyrimidinic (AP) site [3]. This lesion is mutagenic and can block DNA replication and transcription leading to cell death [4]. DNA bases also can become oxidized and one of the prominent oxidized bases is 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxoG) in DNA. Furthemore, the oxidized guanine base can be formed in the dNTP pool (8-oxodGTP), and the nucleotide pool can contain enough 8-oxodGTP to promote mutagenesis [5–8].
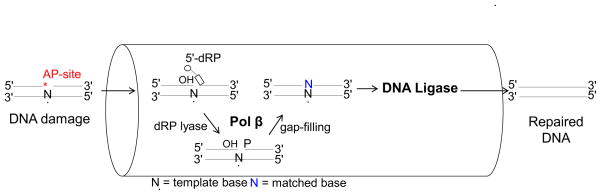
The primary defense mechanism against oxidative DNA damage and the AP-site lesion in cells is the DNA repair pathway known as base excision repair (BER) [9–11]. The overall BER process in mammalian cells consists of two sub-pathways: single-nucleotide (SN) or short patch BER, and long patch (LP) BER [12]. In SN-BER of the AP-site, the AP-site is recognized by apurinic/apyrimidinic endonuclease 1 (APE1), which cleaves the phosphodiester backbone leaving 3′-hydroxyl (3′-OH) and 5′-deoxyribose phosphate (5′-dRP) groups at the termini in a single-nucleotide gap (Fig. 1; *AP-site). DNA polymerase β (pol β), a bifunctional enzyme, removes the 5′-dRP group via its lyase activity, and then catalyzes single-nucleotide gap-filling DNA synthesis through its polymerase activity [13–15]. This generates a substrate for the final BER step accomplished by DNA ligase I or the XRCC1-DNA ligase III complex [16]. If the dRP group is modified so that it cannot be removed by the pol β lyase, BER switches to the alternate LP-BER sub-pathway. This involves damaged strand excision by flap endonuclease 1 (FEN1) and DNA polymerase replacement of several nucleotides ahead of the site of base damage [17].
Fig. 1.
Substrate channeling between pol β and DNA ligase in the BER pathway
BER repair of 8-oxoG is initiated by its removal by 8-oxoguanine DNA glycosylase (OGG1). Since the lyase activity of OGG1 is only weak, the resulting AP-site after base removal is processed by APE1 as described above [18, 19]. The negative cellular impact of 8-oxoG in DNA is mediated, in part, by replicative DNA polymerases [20–22]. These enzymes either fail to bypass the lesion when it persists in the template DNA or perform mutagenic repair by inserting a wrong, pro-mutagenic, nucleotide opposite the lesion [23–25]. In addition, during periods of oxidative stress, pol β can perform mutation-prone repair by inserting the oxidized dNTP pool nucleotide 8-oxodGTP (syn) opposite to template adenine base [26].
2. DNA ligation coupled to pol β gap-filling DNA synthesis in BER
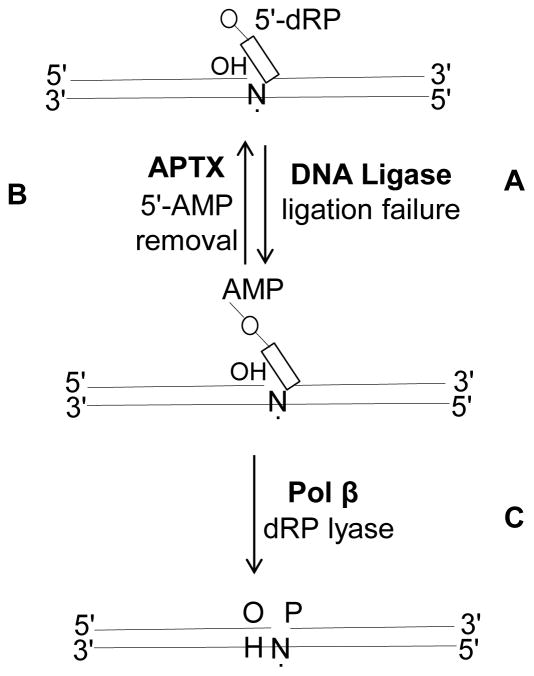
BER is a sequential multistep process that is coordinated by protein-protein and enzyme-DNA interactions. BER appears to involve channeling of DNA intermediates through the repair pathway [27–30]; this may prevent accumulation of toxic repair intermediates once repair has been initiated. DNA intermediates were channeled or handed off from one step to the next in vitro [31]. In more recent biochemical studies with purified human BER enzymes, substrate channeling between pol β and DNA ligase was revealed (Fig. 1). After pol β dRP removal and gap-filling steps, the nicked DNA product was channeled to the ligation step where DNA ligase catalyzes phosphodiester bond formation between the 3′-OH and 5′-phosphate (5′-P) groups of the nick. On the other hand, environmental and metabolic sources of DNA damage can result in failed BER when the ligation step is not successful [32]. This involves ligase termination, premature ligation, and formation of the abortive ligation product with the 5′-adenylate (5′-AMP) group at the nick [33, 34] (Fig. 2). Namely, DNA ligases fail when they engage damaged DNA structures including direct oxidative single-strand breaks, DNA nicks with 3′-AP-sites, and RNA-DNA junctions arising during ribonucleotide excision repair [35]. Moreover, during repair of AP-sites when the 5′-dRP group is not removed by pol β lyase prior to the ligation step, DNA ligases (i.e., DNA ligase I or DNA ligase III/XRCC1 complex) can fail and the abortive ligation product with the 5′-adenylated-dRP-containing BER intermediate can be formed [36, 37] (Fig. 2A).
Fig. 2.
Ligation failure on the 5′-dRP-containing BER intermediate and repair of abortive ligation product with the 5′-adenylated-dRP by APTX and pol β
3. Impact of pol β structural conformations on channeling DNA intermediates to ligation step in BER
DNA polymerases select the proper nucleoside triphosphate from a pool of similar molecules to preserve the integrity of the genome during DNA synthesis [38]. Structural and biochemical data support the hypothesis that some DNA polymerases discriminate between alternate dNTP substrates through an “induced fit” mechanism where binding of the correct nucleotide leads to substrate/protein conformational adjustments that align catalytic groups to optimize chemistry [39–43]. Recently, time-lapse X-ray crystallography studies using natural substrates revealed high-resolution structures of novel catalytic intermediates within the pol β active site [44–46]. These intermediates provided structural insight into roles of active site conformational changes for phosphodiester bond formation and subsequent product release events that accelerate or hinder nucleotide insertion. From these molecular snapshots of pol β inserting an incoming correct nucleotide, the pol β active site undergoes molecular adjustments that optimize correct nucleotide insertion. On the other hand, the structure of ternary mismatch complexes showed important structural differences compared to correct nucleotide insertion. The key differences involved a lack of the structural changes that pol β normally undergoes in response to the incoming correct nucleotide. In addition, pol β kinetic data and ternary complex crystal structures with gapped DNA indicated that pol β can insert 8-oxodGMP opposite both adenine and cytosine bases in the template position [22, 24, 47, 48]. Time-lapse crystallography snapshots of 8-oxodGTP insertion opposite cytosine revealed surprising structural features [49, 50]. For example, the inserted 8-oxodGMP modulates the pol β active site, such that the conformation of the active site opens after the insertion event and the Watson-Crick base pair observed prior to insertion is lost. This is in contrast to the picture after insertion of the normal guanine nucleotide opposite template cytosine, where the active site remains closed and the base pair is maintained after insertion.
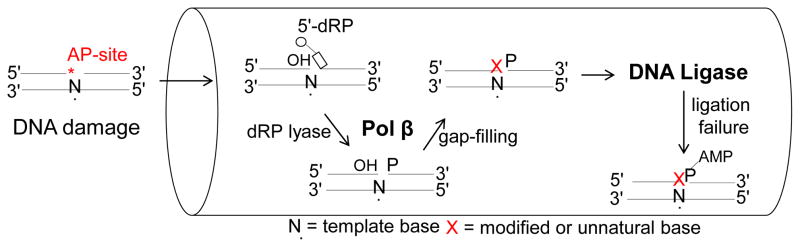
After an incorrect or oxidized (8-oxodGMP) nucleotide insertion into the single nucleotide gapped DNA intermediate by pol β, the resulting nicked product should be passed to the ligation step where DNA ligase would be responsible for nick sealing (Fig. 3). However, the presence of the modified or unnatural base pair at the 3′-margin of a nick could lead to ligation failure and formation of abortive ligation products with the 5′-AMP group at the resulting nicked DNA intermediate (Fig. 3). This would result in a lack of substrate channeling from the gap-filling DNA synthesis step to the ligation step in the BER pathway and subsequent impairment of normal coordination between pol β and DNA ligase. These 5′-adenylated BER intermediates with 3′-modified or unnatural bases could potentially become cytotoxic and lead to abnormal DNA replication and double-strand breaks. Therefore, repair of the 5′-adenylated BER intermediates by DNA-end processing enzymes is critical to cell viability and genomic stability [35, 51].
Fig. 3.
Impairment of substrate channeling from the gap-filling DNA synthesis to the ligation steps in the BER pathway
4. Reversal of impaired BER by DNA-end processing mechanisms
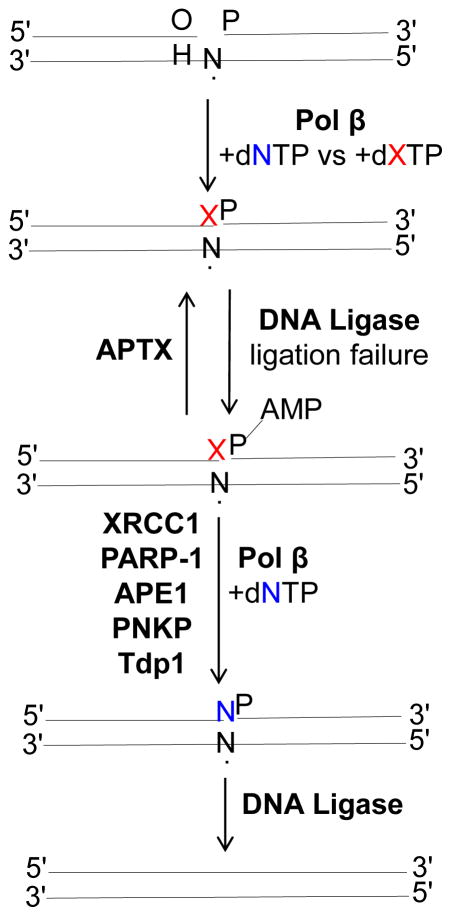
The presence of a modified or unnatural base pair at the 3′-margin of a nick after pol β gap-filling and then ligation failure involving addition of the adenylate group at the 5′-phosphate of the nicked substrate could result in BER pausing. This type of paused intermediate could serve as a signaling mechanism triggering action by DNA-end processing enzymes, like 5′-end processing enzymes for AMP removal (Figs. 2B and 4) or 3′-end trimming for removal of problematic 3′-ends (Fig. 4). Therefore, after trimming, the gap-filling step may be allowed to start over again so that DNA ligase would be able to the join 5′-P and 3′-OH groups.
Fig. 4.
Reversal of impaired BER by DNA-end processing enzymes
Aprataxin (APTX), a member of the histidine triad (HIT) superfamily, resolves the abortive DNA ligation products by 5′-AMP removal and thereby restores the 5′-P group at the 5′-terminus of the nicked DNA, and this will allow another attempt at ligation [52]. Another mechanism of removing 5′-end blocking lesions is the alternate BER sub-pathway, LP-BER. In this case, the role of FEN1 in processing of the 5′-adenylated BER intermediates via its flap excision activity is well known [36, 37]. Other blocked 5′-end reversal mechanisms, including polynucleotide kinase phosphatase (PNKP) or the Ku70/80 lyase activity, could play roles in 5′-DNA end-trimming as well [53, 54].
Regarding a blocked 3′-terminus, many repair mechanisms could serve to resolve a variety of problematic 3′-ends with modified or unnatural bases (Fig. 4). These enzymes include DNA glycosylases, APE1, APE2, and tyrosyl-DNA phosphodiesterase 1 (Tdp1), among others [55–57]. For example, OGG1 and nei endonuclease VIII-like 1 (NEIL1) can remove the 8-oxoG base lesion [58, 59] and APE1 can correct a 3′-mispaired nucleotide via its 3′-5′ exonuclease activity [60]. APE1 is known to interact with DNA ligase I and to stimulate its activity in BER [61]. In addition, a role of APE1 in the repair of DNA strand breaks with 3′-blocking damage has been shown in human cell extracts [56]. Therefore, APE1 appears to be actively involved in coordinating steps and proofreading errors during BER. Tdp1 is a general 3′-end-processing DNA repair enzyme that can function on mismatched 3′-end DNA [62].
Many examples of protein-protein interactions in the BER pathway have been reported, as discussed above. X-ray repair cross-complementing protein 1 (XRCC1) has been considered to be a scaffold protein facilitating multiprotein complex assemblies required in the BER pathway [63]. This role of XRCC1 involves its ability to form stable complexes with itself and other BER proteins, including DNA ligase III, PARP-1, PNKP and pol β [64–66]. Thus, XRCC1 could play a role in the recruitment of DNA-end processing proteins and factors involved in reversal of impaired BER due to lack of normal coordination between pol β and DNA ligase in the last step of the BER pathway. The coordination between BER proteins could also facilitate the removal of blocked DNA-ends after ligation failure (Fig. 4). For example, the BER enzymes pol β, APTX and FEN1 can coordinate in repairing blocked DNA intermediates [36, 37]. These include 5′-adenylated-dRP either through 5′-AMP removal by APTX, excision of the AMP blocked 5′-dRP group plus one-to-two nucleotides by FEN1, or removal of the 5′-AMP-dRP group by the pol β lyase activity (Fig. 2C). These roles involving removal of blocked DNA-ends were found to be especially critical in biochemical studies of APTX-deficient cells isolated from Ataxia with oculomotor apraxia type 1 (AOA1) patients.
5. Concluding remarks and future directions
DNA ligases play important roles in maintaining genomic integrity by catalyzing the joining of breaks in the phosphodiester backbone of double-stranded DNA during repair, replication and recombination [67]. The final step in BER involves DNA strand sealing by DNA ligase, which indeed is a terminal or near-terminal step of almost all types of DNA repair pathways [68]. High-resolution crystallography has revealed that pol β shows different structural conformations upon correct versus incorrect or oxidized (8-oxodGMP) nucleotide insertion [50]. The question of how these structural adjustments could affect the pol β and DNA ligase interaction and the efficiency of BER is still unclear. Ligation failure in the last step of BER could be an important source of genomic instability and cytotoxicity in mammalian cells. The biochemical and cytotoxic effects of premature ligation during BER after pol β-dependent insertion of incorrect or modified nucleotides could mediate mutagenesis, influence cancer therapeutics, and impact bacterial antibiotic development [21]. Moreover, the cytotoxic nicked BER intermediate generated following ligation failure could increase the probability for apoptotic cell signaling [69, 70].
Finally, we highlight the well-known concept that DNA repair defects have been linked to many types of cancer, and inhibition of repair enzymes in tumors with DNA repair defects is of great interest [71]. Therefore, development of targeted DNA repair inhibitors is a therapeutic strategy toward selectively killing cancer cells. Because of the involvement of DNA ligases in replication and repair, inhibitors for DNA ligases have potential as cancer therapeutic agents [72, 73]. The development of DNA ligase inhibitors could provide for cancer specificity because of the high level of intrinsic oxidative stress in cancer cells and the attendant BER. In addition, such inhibitors will be useful in understanding the biological implications of DNA ligation failure in BER compromised by environmental toxicant-induced effects. Moreover, this could serve a strategy for understanding neurological disorders caused by deficiency in enzymes that play roles in repairing blocked DNA-ends after ligation failure in the BER pathway.
Highlights.
BER is the DNA repair pathway responsible for repairing single strand breaks, base lesions and abasic sites in mammalian cells.
BER intermediates are channeled during the pathway so that release of toxic repair intermediates is minimized.
Handoff of repair intermediates from the pol β gap-filling to DNA ligation steps during BER pathway is important for genome stability.
Structural differences that pol β shows after incorrect or oxidized nucleotide insertion could affect accuracy of BER.
Acknowledgments
This research was supported by grants Z01-ES050158 and Z01-ES050159 from the Intramural Research Program of the NIH, National Institute of Environmental Health Sciences.
Footnotes
Conflict of interest statement
None
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362(6422):709–15. doi: 10.1038/362709a0. [DOI] [PubMed] [Google Scholar]
- 2.Lutz WK. Endogenous genotoxic agents and processes as a basis of spontaneous carcinogenesis. Mutat Res. 1990;238(3):287–95. doi: 10.1016/0165-1110(90)90020-c. [DOI] [PubMed] [Google Scholar]
- 3.Lindahl T, Ljungquist S. Apurinic and apyrimidinic sites in DNA. Basic Life Sci. 1975;5a:31–8. doi: 10.1007/978-1-4684-2895-7_5. [DOI] [PubMed] [Google Scholar]
- 4.Dianov GL, et al. Repair of abasic sites in DNA. Mutat Res. 2003;531(1–2):157–63. doi: 10.1016/j.mrfmmm.2003.09.003. [DOI] [PubMed] [Google Scholar]
- 5.Ames BN, Gold LS. Endogenous mutagens and the causes of aging and cancer. Mutat Res. 1991;250(1–2):3–16. doi: 10.1016/0027-5107(91)90157-j. [DOI] [PubMed] [Google Scholar]
- 6.Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc Natl Acad Sci U S A. 1993;90(17):7915–22. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Topal MD, Baker MS. DNA precursor pool: a significant target for N-methyl-N-nitrosourea in C3H/10T1/2 clone 8 cells. Proc Natl Acad Sci U S A. 1982;79(7):2211–5. doi: 10.1073/pnas.79.7.2211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fraga CG, et al. Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci U S A. 1990;87(12):4533–7. doi: 10.1073/pnas.87.12.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krokan HE, et al. Base excision repair of DNA in mammalian cells. FEBS Lett. 2000;476(1–2):73–7. doi: 10.1016/s0014-5793(00)01674-4. [DOI] [PubMed] [Google Scholar]
- 10.Lindahl T. Keynote: past, present, and future aspects of base excision repair. Prog Nucleic Acid Res Mol Biol. 2001;68:xvii–xxx. doi: 10.1016/s0079-6603(01)68084-x. [DOI] [PubMed] [Google Scholar]
- 11.Parikh SS, Mol CD, Tainer JA. Base excision repair enzyme family portrait: integrating the structure and chemistry of an entire DNA repair pathway. Structure. 1997;5(12):1543–50. doi: 10.1016/s0969-2126(97)00303-1. [DOI] [PubMed] [Google Scholar]
- 12.Srivastava DK, et al. Mammalian abasic site base excision repair. Identification of the reaction sequence and rate-determining steps. J Biol Chem. 1998;273(33):21203–9. doi: 10.1074/jbc.273.33.21203. [DOI] [PubMed] [Google Scholar]
- 13.Mol CD, Hosfield DJ, Tainer JA. Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat Res. 2000;460(3–4):211–29. doi: 10.1016/s0921-8777(00)00028-8. [DOI] [PubMed] [Google Scholar]
- 14.Prasad R, et al. Human DNA polymerase beta deoxyribose phosphate lyase. Substrate specificity and catalytic mechanism. J Biol Chem. 1998;273(24):15263–70. doi: 10.1074/jbc.273.24.15263. [DOI] [PubMed] [Google Scholar]
- 15.Beard WA, Prasad R, Wilson SH. Activities and mechanism of DNA polymerase beta. Methods Enzymol. 2006;408:91–107. doi: 10.1016/S0076-6879(06)08007-4. [DOI] [PubMed] [Google Scholar]
- 16.Lindahl T, Barnes DE. Mammalian DNA ligases. Annu Rev Biochem. 1992;61:251–81. doi: 10.1146/annurev.bi.61.070192.001343. [DOI] [PubMed] [Google Scholar]
- 17.Klungland A, Lindahl T. Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1) Embo j. 1997;16(11):3341–8. doi: 10.1093/emboj/16.11.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fortini P, et al. 8-Oxoguanine DNA damage: at the crossroad of alternative repair pathways. Mutat Res. 2003;531(1–2):127–39. doi: 10.1016/j.mrfmmm.2003.07.004. [DOI] [PubMed] [Google Scholar]
- 19.Sung JS, Demple B. Roles of base excision repair subpathways in correcting oxidized abasic sites in DNA. Febs j. 2006;273(8):1620–9. doi: 10.1111/j.1742-4658.2006.05192.x. [DOI] [PubMed] [Google Scholar]
- 20.Brown JA, et al. Single-turnover kinetic analysis of the mutagenic potential of 8-oxo-7,8-dihydro-2′-deoxyguanosine during gap-filling synthesis catalyzed by human DNA polymerases lambda and beta. J Mol Biol. 2007;367(5):1258–69. doi: 10.1016/j.jmb.2007.01.069. [DOI] [PubMed] [Google Scholar]
- 21.Cabelof DC, et al. Induction of DNA polymerase beta-dependent base excision repair in response to oxidative stress in vivo. Carcinogenesis. 2002;23(9):1419–25. doi: 10.1093/carcin/23.9.1419. [DOI] [PubMed] [Google Scholar]
- 22.Shibutani S, Takeshita M, Grollman AP. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature. 1991;349(6308):431–4. doi: 10.1038/349431a0. [DOI] [PubMed] [Google Scholar]
- 23.Batra VK, et al. Mutagenic conformation of 8-oxo-7,8-dihydro-2′-dGTP in the confines of a DNA polymerase active site. Nat Struct Mol Biol. 2010;17(7):889–90. doi: 10.1038/nsmb.1852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Batra VK, et al. Binary complex crystal structure of DNA polymerase beta reveals multiple conformations of the templating 8-oxoguanine lesion. Proc Natl Acad Sci U S A. 2012;109(1):113–8. doi: 10.1073/pnas.1112235108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beard WA, V, Batra K, Wilson SH. DNA polymerase structure-based insight on the mutagenic properties of 8-oxoguanine. Mutat Res. 2010;703(1):18–23. doi: 10.1016/j.mrgentox.2010.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Differing conformational pathways before and after chemistry for insertion of dATP versus dCTP opposite 8-oxoG in DNA polymerase beta. Biophys J. 2007;92(9):3063–70. doi: 10.1529/biophysj.106.092106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prasad R, et al. Substrate channeling in mammalian base excision repair pathways: passing the baton. J Biol Chem. 2010;285(52):40479–88. doi: 10.1074/jbc.M110.155267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Prasad R, et al. Pol beta associated complex and base excision repair factors in mouse fibroblasts. Nucleic Acids Res. 2012;40(22):11571–82. doi: 10.1093/nar/gks898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilson SH, Kunkel TA. Passing the baton in base excision repair. Nat Struct Biol. 2000;7(3):176–8. doi: 10.1038/73260. [DOI] [PubMed] [Google Scholar]
- 30.Liu Y, et al. Coordination of steps in single-nucleotide base excision repair mediated by apurinic/apyrimidinic endonuclease 1 and DNA polymerase beta. J Biol Chem. 2007;282(18):13532–41. doi: 10.1074/jbc.M611295200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prasad R, et al. A review of recent experiments on step-to-step “hand-off” of the DNA intermediates in mammalian base excision repair pathways. Mol Biol (Mosk) 2011;45(4):586–600. [PMC free article] [PubMed] [Google Scholar]
- 32.Tomkinson AE, et al. DNA ligases: structure, reaction mechanism, and function. Chem Rev. 2006;106(2):687–99. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 33.Rass U, Ahel I, West SC. Defective DNA repair and neurodegenerative disease. Cell. 2007;130(6):991–1004. doi: 10.1016/j.cell.2007.08.043. [DOI] [PubMed] [Google Scholar]
- 34.Reynolds JJ, et al. Defective DNA ligation during short-patch single-strand break repair in ataxia oculomotor apraxia 1. Mol Cell Biol. 2009;29(5):1354–62. doi: 10.1128/MCB.01471-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andres SN, et al. Recognition and repair of chemically heterogeneous structures at DNA ends. Environ Mol Mutagen. 2015;56(1):1–21. doi: 10.1002/em.21892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Caglayan M, et al. Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair. Nat Struct Mol Biol. 2014;21(5):497–9. doi: 10.1038/nsmb.2818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Caglayan M, et al. Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Res. 2015;43(4):2271–81. doi: 10.1093/nar/gkv079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase beta. Mutat Res. 2000;460(3–4):231–44. doi: 10.1016/s0921-8777(00)00029-x. [DOI] [PubMed] [Google Scholar]
- 39.Arndt JW, et al. Insight into the catalytic mechanism of DNA polymerase beta: structures of intermediate complexes. Biochemistry. 2001;40(18):5368–75. doi: 10.1021/bi002176j. [DOI] [PubMed] [Google Scholar]
- 40.Batra VK, et al. Magnesium-induced assembly of a complete DNA polymerase catalytic complex. Structure. 2006;14(4):757–66. doi: 10.1016/j.str.2006.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Beard WA, et al. Enzyme-DNA interactions required for efficient nucleotide incorporation and discrimination in human DNA polymerase beta. J Biol Chem. 1996;271(21):12141–4. doi: 10.1074/jbc.271.21.12141. [DOI] [PubMed] [Google Scholar]
- 42.Beard WA, et al. DNA polymerase beta substrate specificity: side chain modulation of the “A-rule”. J Biol Chem. 2009;284(46):31680–9. doi: 10.1074/jbc.M109.029843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Beard WA, et al. Substrate-induced DNA polymerase beta activation. J Biol Chem. 2014;289(45):31411–22. doi: 10.1074/jbc.M114.607432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Freudenthal BD, et al. Observing a DNA polymerase choose right from wrong. Cell. 2013;154(1):157–68. doi: 10.1016/j.cell.2013.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Freudenthal BD, Beard WA, Wilson SH. Structures of dNTP intermediate states during DNA polymerase active site assembly. Structure. 2012;20(11):1829–37. doi: 10.1016/j.str.2012.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freudenthal BD, Beard WA, Wilson SH. Watching a DNA polymerase in action. Cell Cycle. 2014;13(5):691–2. doi: 10.4161/cc.27789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sampoli Benitez B, et al. How DNA polymerase X preferentially accommodates incoming dATP opposite 8-oxoguanine on the template. Biophys J. 2013;105(11):2559–68. doi: 10.1016/j.bpj.2013.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Duarte V, Muller JG, Burrows CJ. Insertion of dGMP and dAMP during in vitro DNA synthesis opposite an oxidized form of 7,8-dihydro-8-oxoguanine. Nucleic Acids Res. 1999;27(2):496–502. doi: 10.1093/nar/27.2.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freudenthal BD, Beard WA, Wilson SH. DNA polymerase minor groove interactions modulate mutagenic bypass of a templating 8-oxoguanine lesion. Nucleic Acids Res. 2013;41(3):1848–58. doi: 10.1093/nar/gks1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Freudenthal BD, et al. Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide. Nature. 2015;517(7536):635–9. doi: 10.1038/nature13886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.El-Khamisy SF. To live or to die: a matter of processing damaged DNA termini in neurons. EMBO Mol Med. 2011;3(2):78–88. doi: 10.1002/emmm.201000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ahel I, et al. The neurodegenerative disease protein aprataxin resolves abortive DNA ligation intermediates. Nature. 2006;443(7112):713–6. doi: 10.1038/nature05164. [DOI] [PubMed] [Google Scholar]
- 53.Weinfeld M, et al. Tidying up loose ends: the role of polynucleotide kinase/phosphatase in DNA strand break repair. Trends Biochem Sci. 2011;36(5):262–71. doi: 10.1016/j.tibs.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doherty AJ, Jackson SP. DNA repair: how Ku makes ends meet. Curr Biol. 2001;11(22):R920–4. doi: 10.1016/s0960-9822(01)00555-3. [DOI] [PubMed] [Google Scholar]
- 55.Parsons JL, Dianova, Dianov GL. APE1-dependent repair of DNA single-strand breaks containing 3′-end 8-oxoguanine. Nucleic Acids Res. 2005;33(7):2204–9. doi: 10.1093/nar/gki518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Izumi T, et al. Requirement for human AP endonuclease 1 for repair of 3′-blocking damage at DNA single-strand breaks induced by reactive oxygen species. Carcinogenesis. 2000;21(7):1329–34. [PubMed] [Google Scholar]
- 57.Pommier Y, et al. Tyrosyl-DNA-phosphodiesterases (TDP1 and TDP2) DNA Repair (Amst) 2014;19:114–29. doi: 10.1016/j.dnarep.2014.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vik ES, et al. Biochemical mapping of human NEIL1 DNA glycosylase and AP lyase activities. DNA Repair (Amst) 2012;11(9):766–73. doi: 10.1016/j.dnarep.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 59.Bjoras M, et al. Opposite base-dependent reactions of a human base excision repair enzyme on DNA containing 7,8-dihydro-8-oxoguanine and abasic sites. Embo j. 1997;16(20):6314–22. doi: 10.1093/emboj/16.20.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dyrkheeva NS, et al. 3′-5′ exonuclease activity of human apurinic/apyrimidinic endonuclease 1 towards DNAs containing dNMP and their modified analogs at the 3 end of single strand DNA break. Biochemistry (Mosc) 2006;71(2):200–10. doi: 10.1134/s0006297906020131. [DOI] [PubMed] [Google Scholar]
- 61.Ranalli TA, Tom S, Bambara RA. AP endonuclease 1 coordinates flap endonuclease 1 and DNA ligase I activity in long patch base excision repair. J Biol Chem. 2002;277(44):41715–24. doi: 10.1074/jbc.M207207200. [DOI] [PubMed] [Google Scholar]
- 62.El-Khamisy SF, Caldecott KW. TDP1-dependent DNA single-strand break repair and neurodegeneration. Mutagenesis. 2006;21(4):219–24. doi: 10.1093/mutage/gel024. [DOI] [PubMed] [Google Scholar]
- 63.Vidal AE, et al. XRCC1 coordinates the initial and late stages of DNA abasic site repair through protein-protein interactions. Embo j. 2001;20(22):6530–9. doi: 10.1093/emboj/20.22.6530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Caldecott KW, et al. XRCC1 polypeptide interacts with DNA polymerase beta and possibly poly (ADP-ribose) polymerase, and DNA ligase III is a novel molecular ‘nick-sensor’ in vitro. Nucleic Acids Res. 1996;24(22):4387–94. doi: 10.1093/nar/24.22.4387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Caldecott KW, et al. An interaction between the mammalian DNA repair protein XRCC1 and DNA ligase III. Mol Cell Biol. 1994;14(1):68–76. doi: 10.1128/mcb.14.1.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marintchev A, et al. Domain specific interaction in the XRCC1-DNA polymerase beta complex. Nucleic Acids Res. 2000;28(10):2049–59. doi: 10.1093/nar/28.10.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bray CM, et al. DNA ligase--a means to an end joining. SEB Exp Biol Ser. 2008;59:203–17. [PubMed] [Google Scholar]
- 68.Doherty AJ, Suh SW. Structural and mechanistic conservation in DNA ligases. Nucleic Acids Res. 2000;28(21):4051–8. doi: 10.1093/nar/28.21.4051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reynolds JJ, El-Khamisy SF, Caldecott KW. Short-patch single-strand break repair in ataxia oculomotor apraxia-1. Biochem Soc Trans. 2009;37(Pt 3):577–81. doi: 10.1042/BST0370577. [DOI] [PubMed] [Google Scholar]
- 70.Harris JL, et al. Aprataxin, poly-ADP ribose polymerase 1 (PARP-1) and apurinic endonuclease 1 (APE1) function together to protect the genome against oxidative damage. Hum Mol Genet. 2009;18(21):4102–17. doi: 10.1093/hmg/ddp359. [DOI] [PubMed] [Google Scholar]
- 71.Kelley MR, Logsdon D, Fishel ML. Targeting DNA repair pathways for cancer treatment: what’s new? Future Oncol. 2014;10(7):1215–37. doi: 10.2217/fon.14.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tomkinson AE, Howes TR, Wiest NE. DNA ligases as therapeutic targets. Transl Cancer Res. 2013;2(3) [PMC free article] [PubMed] [Google Scholar]
- 73.Chen X, et al. Rational design of human DNA ligase inhibitors that target cellular DNA replication and repair. Cancer Res. 2008;68(9):3169–77. doi: 10.1158/0008-5472.CAN-07-6636. [DOI] [PMC free article] [PubMed] [Google Scholar]