Abstract
The nuclear receptor LRH-1 (Liver Receptor Homolog-1, NR5A2) is a transcription factor that regulates gene expression programs critical for many aspects of metabolism and reproduction. Although LRH-1 is able to bind phospholipids, it is still considered an orphan nuclear receptor (NR) with an unknown regulatory hormone. Our prior cellular and structural studies demonstrated that the signaling phosphatidylinositols PI(4,5)P2 (PIP2) and PI(3,4,5)P3 (PIP3) bind and regulate SF-1 (Steroidogenic Factor-1, NR5A1), a close homolog of LRH-1. Here, we describe the crystal structure of human LRH-1 ligand binding domain (LBD) bound by PIP3 - the first phospholipid with a head group endogenous to mammals. We show that the phospholipid hormone binds LRH-1 with high affinity, stabilizing the receptor LBD. While the hydrophobic PIP3 tails (C16/C16) are buried inside the LRH-1 ligand binding pocket, the negatively charged PIP3 head group is presented on the receptor surface, similar to the phosphatidylinositol binding mode observed in the PIP3-SF-1 structure. Thus, data presented in this work reinforce our earlier findings demonstrating that signaling phosphatidylinositols regulate the NR5A receptors LRH-1 and SF-1.
Keywords: Nuclear receptor, Liver Receptor Homolog-1, LRH-1, NR5A2, Crystal structure, PIP3, Ligand
1. Introduction
Members of the nuclear receptor (NR) superfamily of ligand-activated transcription factors bind regulatory hydrophobic ligands, which modulate NR transcriptional activity (Lin et al., 2009). Liver Receptor Homolog-1 (LRH-1, NR5A2) and Steroidogenic Factor-1 (SF-1, NR5A1) are highly homologous NRs that activate similar downstream target genes (Ruggiero et al., 2014), but exhibit non-overlapping expression patterns and thus, regulate distinct transcriptional programs in mammals (Nishimura et al., 2004). Previous studies by our group and others showed that LRH-1 and SF-1 bind phospholipids in their hydrophobic ligand-binding pockets (Krylova et al., 2005; Li et al., 2005b; Wang et al., 2005). Our work also suggested that the signaling phosphoinositides PI(4,5)P2 (PIP2) and PI(3,4,5)P3 (PIP3) might function as physiological NR5A ligands (Blind et al., 2012; Krylova et al., 2005; Mullaney et al., 2010; Sablin et al., 2009). Recently, we solved the crystal structures of SF-1 bound to PIP2 and PIP3 (Blind et al., 2014), and demonstrated that PIP3 activates SF-1 by creating a novel interaction site on the receptor surface that facilitates the recruitment of co-regulatory factors (Blind et al., 2014).
Several lines of evidence suggest that both LRH-1 and SF-1 are regulated by PIP3. First, these receptors bind to phosphoinositides with higher affinity than any other phospholipids tested in vitro (Blind et al., 2012; Blind et al., 2014; Krylova et al., 2005). Second, when purified from mammalian cells, SF-1 is found associated with the phosphoinositide PIP2 (Blind et al., 2012). Third, knockdown of the PIP3-kinase Inositol Polyphosphate Multikinase (IPMK) in cells decreases SF-1 transcriptional activity (Blind et al., 2012). Fourth, overexpressing the PIP3 phosphatase PTEN decreases SF-1 activity (Blind et al., 2012). Finally, activating global cellular PIP3 production via G protein-coupled estrogen receptor (GPER or GPR30) signaling correlates with increased SF-1 transcriptional activity (Lin et al., 2009). Taken together with the high homology between SF-1 and LRH-1 LBDs, these data suggest that the NR5A nuclear receptors are able to decode and interpret nuclear phosphoinositide signaling, altering their target gene expression output.
To date, only one phospholipid thought to be an exogenous activating receptor ligand - the short-chain phosphatidylcholine dilauryl phosphatidylcholine (DLPC) (Lee et al., 2011; Ingraham, 2011) has been co-crystalized and imaged bound to LRH-1 (Musille et al., 2012). However, no structure is reported for LRH-1 bound by a mammalian phospholipid that could function as the receptor’s native hormone. Here, we present the crystal structure of LRH-1 bound by the signaling phospholipid PIP3, which has a head group endogenous to mammals and functions as a potent ligand of NR5As (Blind et al., 2014; Krylova et al., 2005).
2. Materials and Methods
2.1. Protein preparation and ligand exchange
Recombinant ligand binding domain (LBD) of LRH-1 was prepared as described previously (Krylova et al., 2005); this procedure generates LRH-1 LBD bound to phospholipids that are derived from the bacteria (bPL) (Krylova et al., 2005). PIP3-bound LRH-1 species were prepared using a protocol similar to that described previously for SF-1 (Blind et al., 2014; Sablin et al., 2009). Briefly, PI(3,4,5)P3 (C16:0/C16:0, Cayman Chemicals) was re-suspended in water, sonicated in a Branson Bioruptor™ sonicator, and added at concentration of 75 μM to the bPL-bound LRH-1 LBD to generate PIP3-bound receptor; PIP3-LRH-1 complex was then purified from free PIP3 and non-exchanged LRH-1 using a Mono Q column (GE Biosciences) (Blind et al., 2014; Sablin et al., 2009).
2.2. Direct binding assay
The apparent dissociation constant for PIP3-LRH-1 complex was determined by an electrophoretic mobility shift assay using apo-hLRH-1 LBD. The apo-hLRH-1 protein was generated using a dilution-washout procedure developed previously for SF-1 (Blind et al., 2014, see Supplementary data for details). PIP3 (at concentrations 5 nM–1000 nM) was added to binding reactions containing 1 μM of apo-hLRH-1 in 20 mM HEPES pH 8.0, 1 mM EDTA and 10 mM ammonium acetate. Binding reactions were incubated at 37 °C for 1 h, mixed with loading buffer (40% glycerol, 0.005% Ponceau S) and run on a 4–16% gradient polyacrylamide Bis-Tris NativePage™ gel (Invitrogen). After fixation and silver staining (BioRad), gels were scanned, the bands corresponding to PIP3-LRH-1 quantitated, and the apparent KD determined by non-linear curve fit to a single-site binding model in GraphPad Prism. All data were collected and processed in replicates of three, using four (n=4) independent measurements.
2.3. Differential Scanning Fluorimetry (DSF) assay
Thermal stability of hLRH-1 LBD in different ligand bound states (Apo-, bPL- and PIP3-bound as well as PIP3-LRH-1 bound by 15 mer DAX-1 peptide) was assessed by the DSF method, using MxPro3005P qRT-PCR Detection System (Stratagene), with FAM and ROX filters for fluorescence excitation (492 nm) and emission (610 nm). The DSF spectra for all samples were recorded in buffer containing 20 mM HEPES pH 8.0, 100 mM Ammonium Acetate and 1 mM EDTA, with Sypro-Orange dye added at 1/2000 dilution; DAX-1 peptide was added at 1:3 protein:peptide (5 μM : 15 μM) ratio. All samples were heated from 25 °C to 96 °C, and the corresponding fluorescence recorded following every 1 °C increase. The melting temperature for each sample was deduced by KaleidaGraph program (Synergy) from the first derivative of the corresponding denaturation curve generated by the MxPro QPCR software (Stratagene). All data were collected in replicates of six, using three (n=3) independent measurements.
2.4. Crystallization and crystallographic analyses
The purified PIP3-LRH-1 complex was concentrated to 20 mg/ml and used fresh for crystallization. A 3:1 molar excess of a 15-mer peptide corresponding to the human transcriptional co-regulator DAX-1 (residues 140–154, NH2-PRQGSILYSLLTSSK-COOH) was added to hLRH-1-PIP3 in the presence of 10 μM excess of PIP3. hLRH-1-PIP3-DAX-1 complex was crystallized by the vapor diffusion method, using sitting drops and reservoir solution containing 20% PEG 4000, 0.2 M NaOAc and 0.1 M Tris pH 8.5. Crystals were cryo-preserved by adding 20% ethylene glycol to the drops, and flash-frozen in liquid nitrogen. Native data were collected at the Stanford Synchrotron Radiation Lightsource (SSRL) beamline BL11-1, at 100 K, at a wavelength of 1.0 Å, using the BLU-ICE data collection environment (McPhillips et al., 2002). Data were integrated and scaled to 1.86 Å resolution using programs XDS and XSCALE (Leslie and Powell, 2007), and structure factor amplitudes calculated using TRUNCATE (French, 1978). Crystals were of the space group C121 with cell dimensions of a=71.6 Å, b=50.2 Å, c=93.6 Å and α =900°, β =109.20°, γ =900°. Crystals contained one LRH-1-ligand complex in the asymmetric unit. The structure was determined by the molecular replacement method using PHASER implemented in PHENIX (Adams et al., 2010), with a search model derived from the atomic coordinates for hLRH-1 LBD (PDB ID 1YUC) (Ortlund et al., 2005). Initial electron-density maps were calculated from the phases of the search model. Subsequent rounds of model building and refinement were performed using PHENIX (Adams et al., 2010) and COOT (Emsley et al., 2010), respectively. Figures illustrating structural data were generated using the PyMOL molecular graphics system (Schrödinger).
2.5. Structure validation and deposition
The quality of the crystal structure was analyzed using the Joint Center for Structural Genomics (JCSG) Quality Control server (http://smb.slac.stanford.edu/jcsg/QC), which verifies the stereochemical quality of the model using AutoDepInputTool (Diederichs and Karplus, 1997), MolProbity (Chen et al., 2010) and PHENIX (Adams et al., 2010), the agreement between the atomic model and the data using RESOLVE (Terwilliger, 2000), the protein sequence using CLUSTALW (Larkin et al., 2007), and the ADP distribution using PHENIX (Adams et al., 2010). Protein quaternary structure analysis was performed using the PISA server (Krissinel and Henrick, 2007). The structure of PIP3-LRH-1-DAX-1 complex is deposited to the RCSB Protein Data Bank with the PDB accession number 4RWV.
3. Results and discussion
3.1. PIP3 binds to LRH-1 with high affinity and stabilizes the receptor LBD
Our previous study demonstrated high affinity binding interactions between human SF-1 receptor and the signaling phospholipid PI(3,4,5)P3 (estimated KD ~ 80 nM) (Blind et al., 2014). Binding of PIP3 to SF-1 was shown to stabilize the receptor LBD, as judged by its increased melting temperature in differential scanning fluorimetry (DSF) assay.
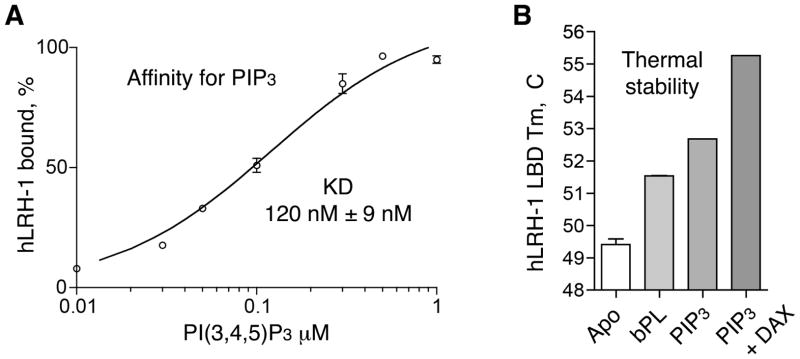
Here, using an electrophoretic mobility shift assay (EMSA), we show that PIP3 binds with high affinity to human LRH-1 receptor (apparent KD = 120 ± 9 nM, Fig. 1A; we acknowledge the limitations of this assay and the fact that a simple, single-site binding model oversimplifies the association of hydrophobic PIP3 ligand to the receptor in aqueous solutions). Binding of PIP3 to LRH-1 stabilizes the receptor LBD; the stabilization effect is evidenced by the upward shift in the melting temperature (Tm) of PIP3-LRH-1 complex in the DSF assay (Fig. 1B), compared to the Tm values observed for the Apo (ligand-free) receptor or the LBD bound by non-specific bacterial phospholipids (bPL, a mixture of phosphatidylglycerol (PG), phosphatidylcholines (PC) and phosphatidylethanolamines (PE) that co-purify with LRH-1 from the bacterial expression system; purified bPL-LRH-1 species might also include a small fraction of apo-LRH-1). We note that binding of the DAX-1 15 mer co-regulator peptide (residues 140–154, NR Box 3 motif) results in additional stabilization of the PIP3-LRH-1 complex, as judged by the upward shift (+2.5 °C, Fig. 1B) of its transition temperature. High affinity interactions (estimated KD = 90 ± 1 nM) between hLRH-1 and the very same DAX-1 peptide were analyzed by the Surface Plasmon Resonance (SPR) method in our previously published study (Benod et al., 2013). The stabilized trimeric PIP3-LRH-1-DAX-1 complex was crystallized and used for analyses by X-ray crystallography.
Figure 1. Analyses of PIP3 binding to LRH-1 LBD.
(A) Determination of binding affinity of PIP3 for LRH-1 by EMSA; data are shown as average of four (n=4) independent measurements, with experimental errors indicated. (B) DSF assay shows stabilization of LRH-1 upon PIP3 and DAX-1 peptide binding. Melting temperatures (Tm) of the Apo-, bPL- and PIP3-bound LRH-1 are shown as average of three (n=3) independent measurements.
3.2. Overview of the PIP3-LRH-1 complex
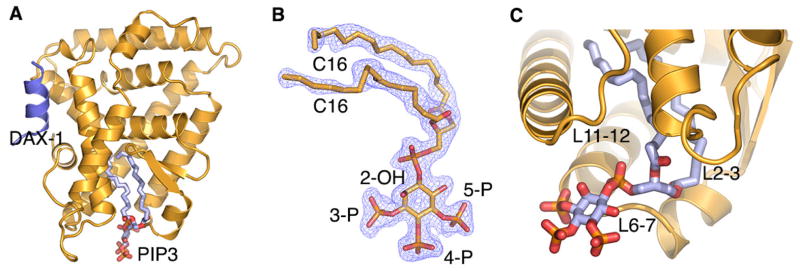
The PIP3-LRH-1-DAX-1 structure was determined using the molecular replacement method. The current model includes LRH-1 residues 297–539 and DAX-1 amino acids 140–154 and is refined to 1.86 Å with Rfree/Rwork values of 0.21/0.17 (Fig. 2, Table S1). The structure of PIP3-bound LRH-1 (yellow ribbon model in Fig. 2A) adopts the general NR LBD fold consisting of 12 α-helices, which are distributed in the three core layers, and two short antiparallel β-strands that form a hairpin loop near the receptor ligand-binding pocket; the extended helix H2 forms an additional, fourth layer on the surface of the LBD (Krylova et al., 2005). The co-regulator DAX-1 peptide (dark blue in Fig. 2A) is bound in the Activation Function (AF) 2 site of LRH-1.
Figure 2. Structure of PIP3 bound LRH-1.
(A) The polypeptide chain of LRH-1 is shown as yellow cartoon model. PIP3 (in light blue) is bound in the receptor’s hormone pocket; co-regulator DAX-1 peptide (dark blue) is bound in the receptor AF-2 cleft. (B) Electron density map (2Fo−Fc, blue mesh, contoured at 1.5 σ) corresponding to the bound PIP3. (C) A magnified view of the PIP3 ligand. Receptor surface loops surrounding the exposed ligand head group are indicated.
The difference Fourier (Fo − Fc) map for the LRH-1 protein model revealed a continuous electron density corresponding to the bound PIP3 ligand. The density allowed for unambiguous modeling of the two acyl chains (C16/C16 tails), the inositol phosphate (IP) head, and the bridging phosphate group at the tail-to-head junction of the ligand (Fig. 2B). The hydrophobic tails of the bound phospholipid reside inside the receptor hormone pocket, while the polar, negatively charged head group of PIP3 is fully exposed to the solvent (Fig. 2C). The bridging phosphate group of PIP3 is coordinated at the opening of the ligand-binding pocket, making contacts with conserved LRH-1 residues G421 from helix H7 and Y516 and K520 from helix H11. High resolution electron density corresponding to the ligand head is consistent with the inositol ring of PIP3 adopting a preferred, biologically relevant D-myo configuration, with the three phosphate groups (3-P, 4-P and 5-P) in the equatorial positions, and the 2-hydroxyl group (2-OH) assuming an axial position relative to the ring; this configuration of the head group is analogous to that previously described for PIP3 bound to SF-1 (Blind et al., 2014).
The exposed ligand head group makes extensive, direct and water mediated interactions with the receptor LBD in the vicinity of the opening to the ligand pocket. In particular, residues from the N-terminal part of helix H7 (T423, L424) and the preceding loop L6-7 (Q419, A420, G421, A422) contact the solvent exposed part of the bound ligand. Direct interactions between bound PIP3 and hLRH-1 loop L6-7 are notable, as amino acid sequence variations in L6-7 of human, mouse and Drosophila receptor orthologs (hLRH-1, mLRH-1 and dmFTZ-f1) were shown to differentially tune the sensitivity of NR5A receptors to phospholipid ligand binding (Krylova et al., 2005; Musille et al., 2013; Sablin et al., 2003). In total, 10 hydrogen bonds, excluding those mediated by water molecules, coordinate and organize the ligand-protein interface outside the receptor hormone pocket (see Table S2 for details); these electrostatic contacts are formed in addition to the extensive van der Waals interactions that dominate ligand-receptor association inside the hydrophobic pocket. The combined ligand-protein interactions inside and outside the pocket result in an estimated binding free energy of ΔG = −16.5 kcal/mol (all calculations characterizing the receptor-ligand and receptor-peptide interfaces were performed using PISA server; http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver). The calculated estimate of free energy of this association is consistent with high affinity interactions observed between PIP3 and hLRH-1 LBD (apparent KD = 120 ± 9 nM, Fig. 1A). The extensive ligand-receptor interactions also explain the increased thermal stability of PIP3-bound LRH-1 LBD (Tm = 52.7 ± 0.1 °C) compared to its Apo (ligand-free) state (Tm = 49.2 ± 0.1 °C) (Fig. 1B). For comparison, free energy of a bPL-hLRH-1 association (PDB ID 1YUC) (Ortlund et al., 2005) is calculated to be −6.5 kcal/mol; consistent with the lower binding energy, only three hydrogen bonds are formed between the receptor LBD and the bound bPL ligand. We attribute an additional gain in free binding energy in PIP3-LRH-1 complex to direct electrostatic interactions of the exposed inositol phosphate head group of the ligand with the receptor LBD. In contrast, association between bPL and human LRH-1 is driven mostly by the hydrophobic interactions of the ligand inside the receptor hormone pocket. As expected, an increased thermal stability is observed for the PIP3-LRH-1 complex (Tm = 52.7 ± 0.1 °C) compared to the bPL bound receptor (Tm = 51.5 ± 0.1 °C) (Fig. 1B).
3.3. Comparative analysis between PIP3-LRH-1 and PIP3-SF-1 structures
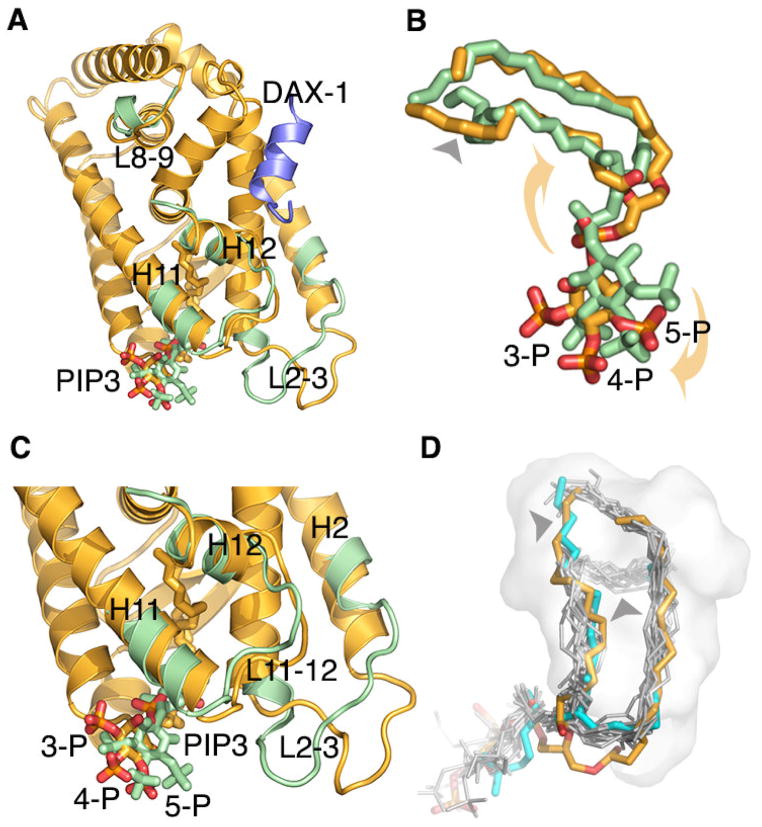
Whereas the overall configuration of the bound phospholipid in PIP3-LRH-1 complex is similar to that observed in PIP3-SF-1 structure (Blind et al., 2014), the exact positions of the ligand head groups differ between the two complexes. We note a rotational shift in the position of the head group of PIP3 bound to LRH-1 compared to that observed in PIP3-SF-1 structure (yellow and green stick models in Fig. 3A–C). While we attribute this shift to a steric interference imposed by the PIP3-LRH-1 crystal lattice, we note that other contributing factors, including possible allosteric effects of bound co-regulator peptides (PGC1α in PIP3-SF-1 and DAX-1 in PIP3-LRH-1) might influence the exact molecular architecture of PIP3-NR5A complexes.
Figure 3. Structures of PIP3-LRH-1 and PIP3-SF-1 compared.
(A) Superposition of PIP3-bound LRH-1 (yellow) and SF-1 (green) LBDs; the polypeptide chains are drawn as ribbons, the corresponding phospholipids as stick models; only divergent regions of SF-1 are shown. (B) A magnified view of the superposed LRH-1 and SF-1 ligands. PIP3 head group bound to LRH-1 is rotated (indicated by yellow arrows) compared to that observed in SF-1; the conformations of the lipid tails also differ (indicated by black arrow) between PIP3 bound LRH-1 and SF-1 structures. (C) Different position of PIP3 in LRH-1 structure is associated with rearrangements in the receptor loops L2-3 and L11-12. (D) The configuration of PIP3 tails (orange) bound into LRH-1 pocket (off-white surface) resembles that described for LRH-1 bound phospholipid DLPC (cyan); this conformation differs from that observed in other structures of phospholipid-bound NR5As (gray); conformational differences are indicated by black arrows.
The rotational movement of the ligand head group in PIP3-LRH-1 structure is associated with a notable rearrangement of the lipid tails buried in the pocket (Fig. 3B). We note that the configuration of PIP3 tails (in orange, Fig. 3D) resembles that described previously for LRH-1 bound phospholipid DLPC (PDB ID 4DOS (Musille et al., 2012), cyan in Fig. 3D); the packing mode of the DLPC and PIP3 tails differs from the configuration of the ligand tails observed in other structures of phospholipid-bound NR5A receptors (gray, Fig. 3D). This comparative analysis demonstrates that flexible acyl chains of the bound phospholipids can assume different conformations in the hydrophobic core of the receptor LBD. The conformational variations noted for the phospholipid ligands bound to NR5As could be an important feature for design of therapeutic small molecules that target these receptors.
Although superficial in its nature, the rotational movement of the bound ligand in PIP3-LRH-1 structure causes a few notable conformational rearrangements in the molecular surface of the receptor. In the PIP3-SF-1 complex, the exposed ligand head group is poised for direct interactions with the SF-1 loops L2-3, L6-7 and L11-12 that surround the entrance to the receptor hormone pocket. Consequently, the SF-1 structure in the vicinity of the exposed ligand head is well ordered (Blind et al., 2014). In PIP3-LRH-1 structure, the ligand head group retains its direct contacts with L6-7, however, is positioned further away from L2-3 and L11-12 and disengaged from interactions with these structural elements. As a result, loops L2-3 and L11-12 in PIP3-LRH-1 structure are more dynamic, as judged by their higher B-factors (average B-factors for L2-3 and L11-12 are 72.8 Å2 and 51.1 Å2 compared to 37.6 Å2 for the entire LBD, Table S1). Loops L2-3 and L11-12 in PIP3-LRH-1 complex also assume different conformations compared to their corresponding positions in PIP3-SF-1 complex (Fig. 3C; r.m.s.d. between LRH-1 and SF-1 loops L2-3 and L11-12 are 6.1 Å and 1.6 Å, compared to 1.0 Å for the core 210 C alpha atoms; all r.m.s.d. calculations were performed using the UCSF Chimera package; http://www.cgl.ucsf.edu/chimera).
In contrast to PIP3-SF-1 structure, there are no direct interactions between L2-3 and L11-12 residues in the crystalline PIP3-LRH-1 complex. The stabilizing, ligand mediated network of interactions between L2-3 and L11-12 was shown to be essential for configuring loop L11-12 and the following AF-2 helix H12 for optimal interactions of SF-1 receptor with its transcriptional co-regulators (Blind et al., 2014). In the absence of these interactions, differently positioned loop L11-12 leads to an altered configuration of the N-terminal part of helix H12 in PIP3-LRH-1 complex compared to that of SF-1 (Fig. 3C, r.m.s.d. is 1.5 Å for the first four C alpha atoms). Whereas all H12 residues are well ordered in SF-1 structure (in green, Fig. 3A), with the very C-terminal T461 interacting directly with helix H4 and loop L8-9, their counterparts in PIP3-LRH-1 are more flexible, with two C-terminal residues (R540 and A541) disordered and not included in the model. The flexibility of the C-terminus in PIP3-LRH-1 complex prevents its interactions with L8-9, which in the absence of such contacts, adopts a different conformation compared to its counterpart in PIP3-SF-1 (Fig. 3A, r.m.s.d. is 2.7 Å for 9 C alpha atoms). Interestingly, direct interactions between the receptor C-terminus and loop L8-9 are thought to be essential for function of the oxosteroid receptors AR, GR, MR and PR (Billas and Moras, 2013); loop L8-9 is also implicated in allosteric regulation of the retinoic acid receptor alpha (RARα) (Samarut et al., 2011). We note that in the LRH-1 structure bound by its transcriptional co-regulator β-catenin (Yumoto et al., 2012), loop L8-9 is uncharacteristically disordered. In contrast, well ordered L8-9 is an essential part of the receptor’s binding interface with its transcriptional co-regulator Dax-1 in the heterotrimeric mouse LRH-1:(Dax-1)2 complex (Sablin et al., 2008). This comparative analysis suggests that the conformational dynamics built in L8-9 might be essential for allosteric regulation of NR5A receptors by their co-regulators. Differences in configuration of L8-9 observed in PIP3 bound LRH-1 and SF-1 structures suggest an additional, ligand mediated signal transduction pathway that might be relevant for regulation of the NR5A function.
3.3. Structural determinants of DAX-1 NR Box3 binding to LRH-1 LBD
Prior structural analyses of hLRH-1 by our group (Krylova et al., 2005; Sablin et al., 2008) and others (Li et al., 2005a; Ortlund et al., 2005) demonstrate that association of the receptor LBD with specific coregulators or coregulator peptides is imperative for successful protein crystallization and high resolution structural analyses. Several coregulator peptides, including those corresponding to the coactivator NCoA2 (Krylova et al., 2005; Musille et al., 2012) and corepressor SHP (Li et al., 2005a; Ortlund et al., 2005), have been co-crystallized with and imaged bound to the LRH-1 LBD. In addition, two regulatory protein domains corresponding to the armadillo repeats of human β-catenin (Yumoto et al., 2012) and the C-terminal region of mouse Dax-1 (Sablin et al., 2008), were imaged in association with the LRH-1 LBD. However, no LRH-1 structure has been determined with a peptide corresponding to the regulatory N-terminal region of DAX-1 implicated in nuclear translocation of the receptor. Our published work (Benod et al., 2013) demonstrated high affinity interactions between LRH-1 LBD and an N-terminal peptide corresponding to the NR Box 3 motif of DAX-1 (DAX1-3; estimated KD = 90 ± 1 nM). This study reveals the molecular details of these specific regulatory interactions.
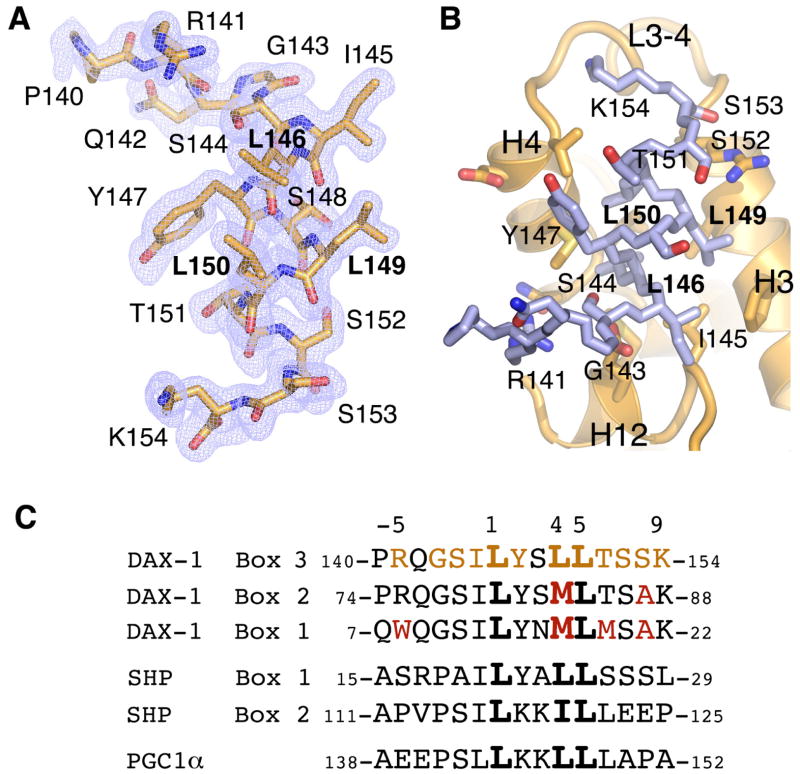
The DAX1-3 peptide (Fig. 4A) in PIP3-LRH-1 structure is docked into the receptor AF-2 site (Fig. 4B), forming an extended network of intermolecular interactions at the buried protein-peptide interface of ~1500 Å2; 12 out of 15 peptide residues (indicated in Fig. 4B) are in direct contact with the receptor LBD at this regulatory site. Although this association is driven by extensive hydrophobic interactions, it is accompanied by formation of seven hydrogen bonds that involve DAX-1 residues R141, S144, I145, L146, L149, and K154, resulting in an estimated free binding energy of ΔG = −9.5 kcal/mol. An electrostatic “clamp” between the peptide main chain and conserved LRH-1 residues R361 and E534 register the NR binding motif of DAX-1 in the receptor AF-2 cleft.
Figure 4. Structural determinants of co-regulator DAX-1 peptide binding to LRH-1.
(A) Electron density map (2Fo−Fc, blue mesh, contoured at 1.5 σ) corresponding to the bound DAX1-3 peptide. (B) DAX1-3 (blue stick model) binds at the receptor AF-2 site. DAX-1 residues forming the receptor-peptide interface are indicated. (C) Alignment of amino acid sequences corresponding to DAX-1 Box 1, 2 and 3, SHP Box 1 and 2, and PGC1α NR binding motifs; the core L(1)XXL(4)L(5) motif is in bold. Interactions of DAX1-3 with LRH-1 extend beyond the NR Box3 motif and involve residues at the positions −5, −3, −2, −1, 1, 2, 4, 5, 6, 7, 8, 9 (in orange). Amino acid substitutions at the corresponding positions in DAX-1 Box1 and Box 2 motifs (in red) as well as in SHP Box 1, Box 2 and PGC-1α peptides result in their lower affinity interactions with LRH-1.
The observed configuration of DAX1-3 - LRH-1 interface provides the molecular basis for the documented preference of LRH-1 for L(+1)XXL(+4)L(+5)-related motifs containing serine, tyrosine and threonine at positions −2, +2, and +6, respectively (Suzuki et al., 2003) (Fig. 4C). Our structural data reveal that: 1) small polar side chain of DAX-1 S144 (position −2) forms a hydrogen bond with conserved LRH-1 residues E534, 2) bulky hydrophobic side chain of Y147 (position +2) fits the pocket formed by LRH-1 residues V371 and M375, and 3) intra-molecular stacking interactions between DAX-1 Y147 and T151 (position +6) further enhance the van der Waals contacts at this site. Our analysis predicts that most substitutions at positions −2 or +6 would either lower or completely abolish the affinity of DAX-1 for LRH-1. This prediction is justified by the observation that mutations S144K and T151Q impair DAX-1 binding to LRH-1 (Suzuki et al., 2003). In contrast to these key residues, substitutions would be tolerated at positions −6, −4 and −3, as solvent exposed DAX-1 residues P140 (position −6) and Q142 (position −4) are not in direct contact with LRH-1. Although the main chain carbonyl of G143 (position −3) is in contact with LRH-1, addition of a side chain at this position is not expected to interfere with this interaction. Consistent with this analysis, mutations P140K, Q142K and G143H did not perturb DAX-1 - LRH-1 association (Suzuki et al., 2003).
4. Conclusions
Our study defines high affinity, specific interactions between LRH-1 and the signaling phosphatidylinositol PIP3, which upon binding stabilizes the receptor LBD. The crystal structure of the PIP3-LRH-1 complex reveals that the phospholipid ligand binds into the receptor hormone pocket, with its hydrophobic tails sequestered inside the pocket, and the polar head group presented on the receptor surface. This ligand binding configuration is consistent with the previously described mode of PIP3 binding to the homologous receptor SF-1 (Blind et al., 2014). In both PIP3 bound receptors, the exposed ligand head group is in direct contact with the NR5A residues from loop L6-7. In different NR5A orthologs, primary sequence variations in this loop control the sensitivity of NR5A receptors to phospholipid binding and ligand-mediated regulation (Krylova et al., 2005; Musille et al., 2013; Sablin et al., 2003). These observations reinforce our earlier hypothesis that signaling phosphatidylinositols play a regulatory role in NR5A function (Blind et al., 2012; Blind et al., 2014).
Supplementary Material
Acknowledgments
We thank members of the Joint Center for Structural Genomics (JCSG) High Throughput Structural Biology pipeline for contributions to this work. Support for this project included Grants R01DK072246 and R01DK099722 (to H.A.I.), National Institute of General Medical Sciences (NIGMS) Fellowship K12GM081266 and Grant 1K01CA172957 (to R.D.B.), Grant R01DK078075, Department of Defense Grant W81XWH-12-1-0396, and Protein Structure Initiative (PSI) Grant U01 GM094614 as part of the PSI-Biology Partnership for Stem Cell Biology (to R.J.F.). The JCSG is supported by NIGMS, PSI Grant U54 GM094586. Use of the Stanford Synchrotron Radiation Lightsource (SSRL) is supported by the US Department of Energy (DOE), Office of Science, and Office of Basic Energy Sciences under Contract DE-AC02-76SF00515. The SSRL Structural Molecular Biology Program is supported by the DOE Office of Biological and Environmental Research and by Grant P41 GM103393.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, McCoy AJ, Moriarty NW, Oeffner R, Read RJ, Richardson DC, Richardson JS, Terwilliger TC, Zwart PH. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta crystallographica Section D, Biological crystallography. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benod C, Carlsson J, Uthayaruban R, Hwang P, Irwin JJ, Doak AK, Shoichet BK, Sablin EP, Fletterick RJ. Structure-based discovery of antagonists of nuclear receptor LRH-1. The Journal of biological chemistry. 2013;288:19830–19844. doi: 10.1074/jbc.M112.411686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Billas I, Moras D. Allosteric controls of nuclear receptor function in the regulation of transcription. Journal of molecular biology. 2013;425:2317–2329. doi: 10.1016/j.jmb.2013.03.017. [DOI] [PubMed] [Google Scholar]
- Blind RD, Suzawa M, Ingraham HA. Direct modification and activation of a nuclear receptor-PIP(2) complex by the inositol lipid kinase IPMK. Science signaling. 2012;5:ra44. doi: 10.1126/scisignal.2003111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blind RD, Sablin EP, Kuchenbecker KM, Chiu HJ, Deacon AM, Das D, Fletterick RJ, Ingraham HA. The signaling phospholipid PIP3 creates a new interaction surface on the nuclear receptor SF-1. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:15054–15059. doi: 10.1073/pnas.1416740111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta crystallographica Section D, Biological crystallography. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diederichs K, Karplus PA. Improved R-factors for diffraction data analysis in macromolecular crystallography. Nature structural biology. 1997;4:269–275. doi: 10.1038/nsb0497-269. [DOI] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta crystallographica Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- French S, Wilson K. On the treatment of negative intensity observations. Acta Crystallographica Section A. 1978;34:517–525. [Google Scholar]
- Ingraham HA. Metabolism: A lipid for fat disorders. Nature. 2011;474:455–456. doi: 10.1038/474455a. [DOI] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. Journal of molecular biology. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Krylova IN, Sablin EP, Moore J, Xu RX, Waitt GM, MacKay JA, Juzumiene D, Bynum JM, Madauss K, Montana V, Lebedeva L, Suzawa M, Williams JD, Williams SP, Guy RK, Thornton JW, Fletterick RJ, Willson TM, Ingraham HA. Structural analyses reveal phosphatidyl inositols as ligands for the NR5 orphan receptors SF-1 and LRH-1. Cell. 2005;120:343–355. doi: 10.1016/j.cell.2005.01.024. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lee JM, Lee YK, Mamrosh JL, Busby SA, Griffin PR, Pathak MC, Ortlund EA, Moore DD. A nuclear-receptor-dependent phosphatidylcholine pathway with antidiabetic effects. Nature. 2011;474:506–510. doi: 10.1038/nature10111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie AGW, Powell HR. Processing diffraction data with Mosflm. In: Read RJ, Sussman J, editors. Evolving methods for macromolecular crystallography : the structural path to the understanding of the mechanism of action of CBRN agents. NATO science series Series II, Mathematics, physics, and chemistry. Springer Verlag; Dordrecht: 2007. pp. 41–51. [Google Scholar]
- Li Y, Choi M, Suino K, Kovach A, Daugherty J, Kliewer SA, Xu HE. Structural and biochemical basis for selective repression of the orphan nuclear receptor liver receptor homolog 1 by small heterodimer partner. Proceedings of the National Academy of Sciences of the United States of America. 2005a;102:9505–9510. doi: 10.1073/pnas.0501204102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Choi M, Cavey G, Daugherty J, Suino K, Kovach A, Bingham NC, Kliewer SA, Xu HE. Crystallographic identification and functional characterization of phospholipids as ligands for the orphan nuclear receptor steroidogenic factor-1. Molecular cell. 2005b;17:491–502. doi: 10.1016/j.molcel.2005.02.002. [DOI] [PubMed] [Google Scholar]
- Lin BC, Suzawa M, Blind RD, Tobias SC, Bulun SE, Scanlan TS, Ingraham HA. Stimulating the GPR30 estrogen receptor with a novel tamoxifen analogue activates SF-1 and promotes endometrial cell proliferation. Cancer research. 2009;69:5415–5423. doi: 10.1158/0008-5472.CAN-08-1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McPhillips TM, McPhillips SE, Chiu HJ, Cohen AE, Deacon AM, Ellis PJ, Garman E, Gonzalez A, Sauter NK, Phizackerley RP, Soltis SM, Kuhn P. Blu-Ice and the Distributed Control System: software for data acquisition and instrument control at macromolecular crystallography beamlines. Journal of synchrotron radiation. 2002;9:401–406. doi: 10.1107/s0909049502015170. [DOI] [PubMed] [Google Scholar]
- Mullaney BC, Blind RD, Lemieux GA, Perez CL, Elle IC, Faergeman NJ, Van Gilst MR, Ingraham HA, Ashrafi K. Regulation of C. elegans fat uptake and storage by acyl-CoA synthase-3 is dependent on NR5A family nuclear hormone receptor nhr-25. Cell metabolism. 2010;12:398–410. doi: 10.1016/j.cmet.2010.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musille PM, Pathak M, Lauer JL, Griffin PR, Ortlund EA. Divergent sequence tunes ligand sensitivity in phospholipid-regulated hormone receptors. The Journal of biological chemistry. 2013;288:20702–20712. doi: 10.1074/jbc.M113.472837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musille PM, Pathak MC, Lauer JL, Hudson WH, Griffin PR, Ortlund EA. Antidiabetic phospholipid-nuclear receptor complex reveals the mechanism for phospholipid-driven gene regulation. Nature structural and molecular biology. 2012;19:532–537. doi: 10.1038/nsmb.2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura M, Naito S, Yokoi T. Tissue-specific mRNA expression profiles of human nuclear receptor subfamilies. Drug metabolism and pharmacokinetics. 2004;19:135–149. doi: 10.2133/dmpk.19.135. [DOI] [PubMed] [Google Scholar]
- Ortlund EA, Lee Y, Solomon IH, Hager JM, Safi R, Choi Y, Guan Z, Tripathy A, Raetz CR, McDonnell DP, Moore DD, Redinbo MR. Modulation of human nuclear receptor LRH-1 activity by phospholipids and SHP. Nature structural and molecular biology. 2005;12:357–363. doi: 10.1038/nsmb910. [DOI] [PubMed] [Google Scholar]
- Ruggiero C, Doghman M, Lalli E. How genomic studies have improved our understanding of the mechanisms of transcriptional regulation by NR5A nuclear receptors. Molecular and cellular endocrinology. 2014 doi: 10.1016/j.mce.2014.10.022. In press. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Krylova IN, Fletterick RJ, Ingraham HA. Structural basis for ligand-independent activation of the orphan nuclear receptor LRH-1. Molecular cell. 2003;11:1575–1585. doi: 10.1016/s1097-2765(03)00236-3. [DOI] [PubMed] [Google Scholar]
- Sablin EP, Woods A, Krylova IN, Hwang P, Ingraham HA, Fletterick RJ. The structure of corepressor Dax-1 bound to its target nuclear receptor LRH-1. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:18390–18395. doi: 10.1073/pnas.0808936105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sablin EP, Blind RD, Krylova IN, Ingraham JG, Cai F, Williams JD, Fletterick RJ, Ingraham HA. Structure of SF-1 bound by different phospholipids: evidence for regulatory ligands. Molecular endocrinology. 2009;23:25–34. doi: 10.1210/me.2007-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarut E, Amal I, Markov GV, Stote R, Dejaegere A, Laudet V, Rochette-Egly C. Evolution of nuclear retinoic acid receptor alpha (RARalpha) phosphorylation sites. Serine gain provides fine-tuned regulation. Molecular biology and evolution. 2011;28:2125–2137. doi: 10.1093/molbev/msr035. [DOI] [PubMed] [Google Scholar]
- Suzuki T, Kasahara M, Yoshioka H, Morohashi K, Umesono K. LXXLL-related motifs in Dax-1 have target specificity for the orphan nuclear receptors Ad4BP/SF-1 and LRH-1. Molecular and cellular biology. 2003;23:238–249. doi: 10.1128/MCB.23.1.238-249.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terwilliger TC. Maximum-likelihood density modification. Acta crystallographica Section D, Biological crystallography. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Zhang C, Marimuthu A, Krupka HI, Tabrizizad M, Shelloe R, Mehra U, Eng K, Nguyen H, Settachatgul C, Powell B, Milburn MV, West BL. The crystal structures of human steroidogenic factor-1 and liver receptor homologue-1. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:7505–7510. doi: 10.1073/pnas.0409482102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yumoto F, Nguyen P, Sablin EP, Baxter JD, Webb P, Fletterick RJ. Structural basis of coactivation of liver receptor homolog-1 by beta-catenin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:143–148. doi: 10.1073/pnas.1117036108. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.