Abstract
The Northwestern University Neuroimaging Data Archive (NUNDA), an XNAT-powered data archiving system, aims to facilitate secure data storage; centralized data management; automated, standardized data processing; and simple, intuitive data sharing. NUNDA is a federated data archive, wherein individual project owners regulate access to their data. NUNDA supports multiple methods of data import, enabling data collection in a central repository. Data in NUNDA are available by project to any authorized user, allowing coordinated data management and review across sites. With NUNDA pipelines, users capitalize on existing procedures or standardize custom routines for consistent, automated data processing. NUNDA can be integrated with other research databases to simplify data exploration and discovery. And data on NUNDA can be confidently shared for secure collaboration.
Keywords: Neuroinformatics, data storage, data processing and analysis, data sharing, XNAT
1. Introduction
The Northwestern University Neuroimaging Data Archive (NUNDA, https://nunda.northwestern.edu, RRID: SCR_013664) is an online collaborative environment for managing and sharing neuroimaging and associated data. NUNDA was developed in collaboration with the Neuroinformatics Research Group (NRG) at the Washington University School of Medicine (Marcus et al., 2007a; Marcus et al., 2007b)1. NUNDA aims to facilitate secure data storage; centralized data management; automated, standardized data processing; and intuitive data sharing for the Northwestern University neuroimaging community and collaborators. Within the NUNDA framework, NUNDA project owners retain autonomous control over their data – regulating access, applying project-specific import and anonymization procedures, coding phenotypic data, selecting processing routines, defining quality control guidelines, and setting project-specific notifications. In this sense, NUNDA is best understood as a federation of projects.
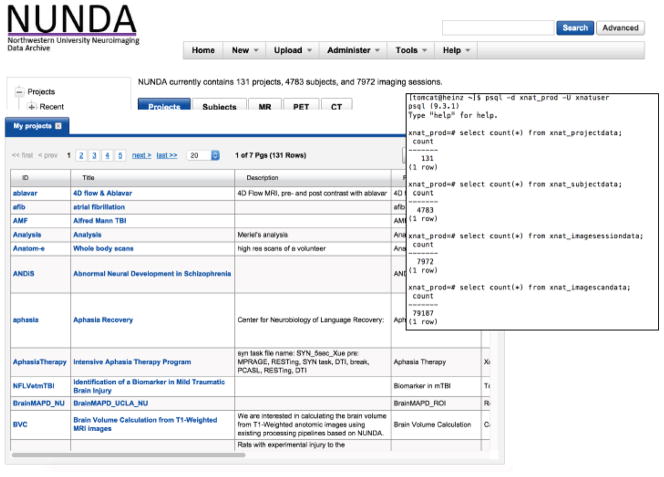
Currently, NUNDA supports 131 projects, 4,783 subjects, 7,972 imaging sessions, and 79,187 scans (Figure 1, Table S1), with new data uploaded daily. Imaging sessions include structural, functional and diffusion magnetic resonance imaging (MRI) scans. Projects range from schizophrenia to dementia, from cancer to cognitive neuroscience, and from humans to primates to rodents. NUNDA is also used as a multisite consortia central collection and repository.
Figure 1. Data stored in NUNDA.
Currently, NUNDA supports 131 projects, 4,783 subjects, 7,972 imaging sessions, and 79,187 scans. New data are uploaded daily. Supplement 1 includes a full breakdown of project, subject, session, and scan counts by project.
NUNDA is powered by the Extensible Neuroimaging Archive Toolkit (XNAT, http://xnat.org, RRID: nif-0000-00531), an open source imaging informatics platform. The XNAT platform allows for easy, portable, secure access to data, and supports a standard research workflow – data acquisition and archiving, data processing and analysis, and data integration. XNAT also offers predefined, configurable security settings, allowing users to designate levels of access for sharing their data. The agility of XNAT enables NUNDA to adapt this workflow to our specific institutional needs at each stage.
2. Collaboration through NUNDA
In addition to supporting the Northwestern University research community as a resource for archiving and processing neuroimaging data, NUNDA is a platform for collaboration with researchers who are not affiliated with Northwestern University. NUNDA contains several multisite projects wherein collaboration and data sharing are part of the study design. Examples include the Aphasia Recovery project, an NIH-funded 5-site study examining language learning/re-learning in aphasia, and the Illinois Stroke Intervention Registry and Trials Network, a cooperative registry and clinical/imaging trial network of 6 academic institutions in Illinois studying endovascular stroke intervention.
NUNDA also lends itself to post-hoc collaboration. Many NUNDA projects began as single-site studies only to invite outside-institution researchers to access their data as new collaborations formed. For example, the schizophrenia project (“Neuromorphometry by Computer Algorithm NUSRG”) has collaborators from Duke University and the University of Minnesota; the primary progressive aphasia project (“Language in Primary Progressive Aphasia”) has collaborators from the Banner Institute; and the infant imaging project (“NUBridge: Prenatal Stress and Early Brain Development (12 & 24 months)”) has shared its data with Massachusetts General Hospital.
Another common form of data sharing and collaboration through NUNDA involves housing data collected at other institutions for access by collaborators at Northwestern. In these cases, new projects are created on NUNDA and data are uploaded into these projects from remote sites. Once on NUNDA, the data are easily retrieved by collaborators at Northwestern. Examples include the Tourette syndrome project with Washington University in St. Louis (Williams et al., 2013), the study on language development with the University of Chicago, and the study on childhood onset schizophrenia with the National Institute on Mental Health (Johnson et al., 2013). Work is also underway to share the entire collection of schizophrenia studies on NUNDA through SchizConnect.2
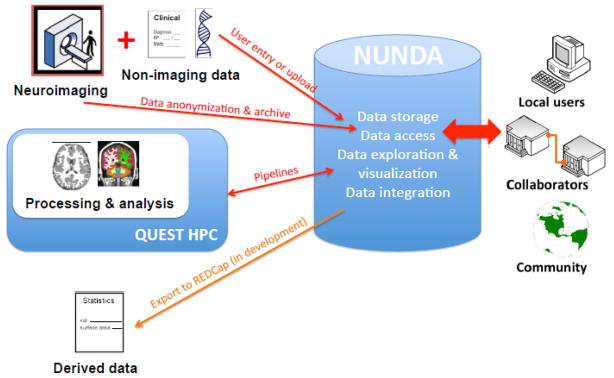
3. The Research Workflow
The NUNDA research workflow, illustrated in Figure 1, adapted from (Marcus et al., 2007b), begins with data acquisition from subjects in IRB-approved protocols at the scanner and the transfer of this data into the NUNDA archive. Annotations are added as needed to identify the subject number, study, scanner, scanning parameters, and any other information needed for later dataset retrieval. The imaging data are then made available for automated image processing routines, and derived images and quantitative measures are stored within archive datasets for integration with demographic, clinical, and neurobiological measures. Some or all data within a project can be shared simply and securely by project with other NUNDA users.
3.1 Data Access & Data Sharing
NUNDA sign-ups are publically available at the web portal. Prospective NUNDA users must receive administrator approval to activate an account. With an active account, any user can create a project (i.e., become the project’s owner) or request access to existing projects.
Within NUNDA, individual project owners regulate access to their data. When creating a project, the owner can choose to make the project “Private”, “Protected”, or “Public”. For “Private” projects, the project title and description are visible to the project owner and invited users only, and only these users can access the data. For “Protected” projects, the project title and description are visible to all NUNDA users, who can access data upon owner approval. For “Public” projects, data are accessible to all NUNDA users. It should be noted that NUNDA does not manage Data Use Agreements; this is the responsibility of the individual project owner.
For user requests to access “Private” or “Protected” projects, the project owner assigns the user a predefined role within the project, which specifies access privileges to the data. Currently on NUNDA, these roles are 1) Owners, who can read, insert, modify, and delete anything associated with the project and can add additional users to the project and modify the data types associated with the project; 2) Members, who can read, insert, and modify subjects and experiments in the project, but cannot modify the project users and data types; and 3) Collaborators, who cannot insert or modify data, but have read-only access to all of the data within the project.
NUNDA also offers the ability to share subjects or imaging sessions between its projects. Scan data and related metadata acquired in the context of a particular study can thus be leveraged for use in other studies. To access, process, or modify shared data, users may still need permission from the original project owner (depending on the project’s accessibility setting). However, by enabling the reuse of existing data, this sharing utility can greatly reduce the overhead involved in study initiation.
While NUNDA currently does not offer a means of making data available to unregistered users, we do maintain a list of the protected and public NUNDA projects at https://nunda.northwestern.edu/nunda/app/template/ProjectInfo.vm. Interested users can reach out the project’s principal investigator about accessing data through NUNDA. Also, there exist a number of XNAT installations (for example, XNAT Central - https://central.xnat.org, RRID: nif-0000-04375)3 that do offer open access projects wherein data are available to anyone. Methods for exporting data from XNAT installations like NUNDA into open-access installations like XNAT Central are under development, in collaboration with the XNAT team.
3.2 Data Acquisition, Anonymization & Archive
NUNDA supports data acquisition in several ways: 1) all research scans collected at Northwestern University’s Center for Translational Imaging (CTI), 2) clinical scans collected at the Northwestern Memorial Hospital (NMH) identified for research purposes on a study-by-study basis, 3) research scans collected at other institutions participating in multisite studies, and 4) ad-hoc uploads initiated from the web portal. In most cases, imaging data can be uploaded to NUNDA from remote sites using DICOM-send (C-STORE, http://medical.nema.org/Dicom/2011/11_07pu.pdf). This standardizes data acquisition from DICOM-compliant systems such as scanners, picture archiving and communication system (PACS), Vendor Neutral Archives (VNA), or de-identification systems. In some cases, the XNAT web-based upload tools are used for data uploading.
In order to handle unexpected events during data uploading and archiving, NUNDA employs a cache space for temporary storage of data in transition. If any errors occur, the data can be retrieved from the cache space for troubleshooting or another archiving attempt. This feature is especially important for large data uploads, as locating data at the source and re-uploading it to NUNDA would take considerable time.
Currently on NUNDA, project-specific anonymization is applied when data is archived, moved, or renamed. XNAT anonymization scripts (https://wiki.xnat.org/display/XNAT16/Anonymization), which can be edited from the NUNDA web portal, control this procedure. Their functionalities include clearing specific DICOM header tag values, changing header tag values, clearing private tags, and setting header tag values based on inputs (project, subject, session, and visit).
For clinical imaging data stored within the hospital PACS, researchers request the PACS team to send selected studies to a computer workstation within the hospital firewall where anonymization routines are to be executed. A qualified Imaging Informatics Professional (IIP)/compliance officer supervises this process by identifying the specific DICOM header tags that need to be cleared or masked. Once the IIP verifies the anonymization process, the transfer runs in a semi-automated mode wherein a staff member initiates the transfer, and data is automatically anonymized and forwarded to NUNDA via DICOM-send.
Data arriving at NUNDA are automatically placed either in a temporary pre-archive location or the permanent archive, depending on each project’s specific settings. Data in the pre-archive location can be reviewed, annotated with study information, and then moved into permanent archive, all via the NUNDA web portal.
Phenotypic data, such as clinical, cognitive, and demographic data, is available on NUNDA in two ways. Basic demographic data – age, gender, handedness, SES, employment, education, race, ethnicity, weight, height, recruitment source – and clinical research group (coded by the project owner and thus project-specific) can be added for each subject directly through the NUNDA web portal. This information is stored in the NUNDA database, and it is searchable and downloadable. Because clinical group is project-specific, we do not have an overall breakdown of clinical populations within NUNDA.
Project owners and members can also upload spreadsheets with phenotypic data as resources within their projects, and these will have a unique URI and will be available for download by approved users. Data uploaded in this way is not searchable; however, NUNDA can be extended to incorporate additional phenotypic parameters in the database upon request.
3.3 Data Processing & Analysis
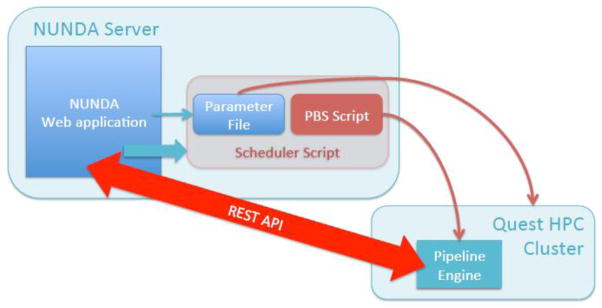
NUNDA supports a variety of processing routines, or pipelines, all of which use the XNAT Pipeline Engine running on Northwestern University’s High Performance Computing Cluster, Quest (Figure 3). Pipelines are XML specification documents that detail workflow steps. For NUNDA, these steps include downloading images from NUNDA onto Quest using the XNAT REST API, running processing scripts, generating quality control snapshot images, updating the database with derived measures via the XNAT REST API, uploading processed images from Quest back to NUNDA using the XNAT REST API, and delivering e-mail notifications. The Pipeline Engine monitors the progress of the pipeline, updating the pipeline status on the web portal as each step completes. Users can view the status of all their pipelines from a custom NUNDA “View My Pipelines” page.
Figure 3. The NUNDA-QUEST Connection.
NUNDA pipelines run on Quest, Northwestern University’s high performance computing cluster (http://www.it.northwestern.edu/research/user-services/quest/index.html). A scheduler script on the NUNDA server copies the NUNDA-generated parameter file onto Quest and submits the pipeline job to the Quest PBS. Once running, the pipeline uses the XNAT REST API to download raw data from NUNDA onto Quest and to upload processed data uploaded from Quest back to NUNDA. The XNAT Pipeline Engine, installed on Quest, monitors the progress of the pipeline and updates the pipeline status on web portal as each step completes.
NUNDA pipelines include standard image processing routines, such as the FreeSurfer fully automatic structural imaging processing stream, and custom routines adapted from the local workflows of our users. Table 1 lists the currently available NUNDA pipelines. Users can request a custom pipeline through a web form (http://niacal.northwestern.edu/nunda_pipeline_requests/new) in which they specify inputs, outputs, and software dependencies. The NUNDA software development team reviews these requests and works with the users to implement their routines on NUNDA. Examples of NUNDA custom pipelines include quality assurance and robust processing routines for fMRI, DTI, and T1 scans (Song et al., 2014); perfusion image vascular territory processing; preprocessing routines for EPI, DTI, T1, and ASL images; subcortical segmentation (Khan et al., 2008); and first- and second- level analysis with SPM/FEAT. Pipelines may be executed with different priorities depending on whether the project owner has access to Quest dedicated nodes (http://www.it.northwestern.edu/research/user-services/quest/allocation-guidelines.html#types).
Table 1. Pipelines currently available on NUNDA.
NUNDA pipelines include standard image processing routines, such as the FreeSurfer fully automatic structural imaging processing stream, and custom routines adapted from the local workflows of our users. Users can request a custom pipeline through a web form http://niacal.northwestern.edu/nunda_pipeline_requests/new).
| Pipeline Type | Pipeline Name | Pipeline Description |
|---|---|---|
| Structural | StdStructuralBuild | DICOM to analyze conversion, T1 processing and averaging, and T2 geometric distortion correction |
| Structural | Freesurfer | Run FreeSurfer’s recon-all |
| Structural | FSLDDMM | Freesurfer-Initiated Large-Deformation Diffeomorphic Metric Mapping for Subcortical Segmentation |
| Structural | TemplateInjection | Re-index surfaces so that all have common indexing, perform denoising |
| Structural | ReillyPreProcessing | DICOM to nifti conversion and brain extraction of T1 |
| Structural | QA_Anat | Quality assurance for T1 scans |
| Functional | QA_EPI_Human/QA_EPI_Phantom | fBIRN QA of EPI scan types |
| Functional | GenericBoldPreProcessing | Pre-process BOLD scan types, includes QC |
| Functional | RobustfMRIPreProcessing | Pre-process fMRI data |
| Functional | SPMAnalysis | Second-level analysis of EPI data (requires RobustfMRIPreProcessing outputs) |
| Diffusion | QA_DTI | QA of DTI scan types |
| Diffusion | StdDTIPreprocessing | Pre-process DTI scan types |
| Perfusion | StdASLProcessing | Pre-process ASL scan types |
| Perfusion | Automatic Vascular Territory Processing | Analyze MRI perfusion images using vascular territory ROIs |
After pipeline execution, users return to the NUNDA web portal to review results, fine-tune parameters for re-execution, or download images for further processing. Some pipelines, such as the FreeSurfer structural imaging processing stream, can resume processing after data has been edited locally and uploaded back to NUNDA. Other pipelines automatically update image quality flags (e.g., “questionable” scan), which direct downstream algorithms that utilize these images.
NUNDA offers several customized options for launching pipelines. In addition to the default XNAT single-session launch, users can launch a pipeline on some or all sessions within a project using a single, project-level launch form. In it, the user can configure parameters and then batch-select sessions for launch based on “Group” or “Acquisition Site” or manually by session label. Sessions on which the pipeline has already run will be skipped, though the user can request a forced re-run. Project owners can further configure pipelines to automatically run on their data after archiving. Although XNAT provides this functionality by default, it doesn’t allow for any session-specific manipulation of parameters set at the project level. As such, NUNDA has custom launch scripts that run automatically as cron tasks. These scripts interact directly with the NUNDA database, scanning for newly added data, determining appropriate parameters, and launching pipelines as needed.
3.4 Data Integration
NUNDA has the capability to be integrated with other research databases such as Research Electronic Data Capture (REDCap, http://project-redcap.org, RRID: nif-0000-33254) (Harris et al., 2009). REDCap is a secure, web-based application designed to support data capture for research studies. It provides 1) an intuitive interface for validated data entry, 2) audit trails for tracking data manipulation and export procedures, 3) automated export procedures for seamless data downloads to common statistical packages, and 4) procedures for importing data from external sources. The integration of NUNDA and REDCap enhances the functionality of the research workflow by integrating two very different research databases in order to create and maintain dynamically growing research datasets.
We are currently working on integrating NUNDA and Northwestern University’s instance of REDCap. After a NUNDA pipeline completes, we will upload the non-imaging data to REDCap. This process will proceed as follows: 1) NUNDA pipeline output will be stored on the file system in a designated location, 2) a “spider” (http://xnat.vanderbilt.edu/index.php/Short_Course_on_Managing_Imaging_Studies_with_XNAT_and_RedCAP) will check this designated location for new output, 3) upon discovery of new output, the spider will use the REDCap API to upload the pipeline output to a REDCap project and notify research personnel.
3.5 Data Security
The XNAT platform offers superior data security. NUNDA administrators monitor user activity and can track all logins and access to data via XNAT’s internal audit system. The NUNDA web application is protected by secure socket layer (SSL) technology to ensure secure communication. The built-in XNAT upload tool, also protected by SSL, offers secure image upload to NUNDA. NUNDA database snapshots are created daily, and a comprehensive database snapshot is captured monthly. All snapshots are backed up on a tape-based backup system, so reconstruction of the database from any time point is possible. NUNDA archived data is backed up with several sets of incremental, differential and full backups.
4. Working with NUNDA Data
Each project, subject, and session in NUNDA can be accessed through a unique URI, restricted to approved users. NUNDA does not offer DOIs at this time. All NUNDA data, including raw imaging data, analyzed/derived data from pipeline processing or direct upload, and phenotypic data (clinical, cognitive, demographic), are available for download by approved users. Functional magnetic resonance imaging stimulus data are not stored on NUNDA. Imaging data are downloaded in native format, meaning that DICOM will be downloaded as DICOM, nifti will be downloaded as nifti, etc. The phenotypic data stored within the NUNDA database can be exported in xml or csv format, while any clinical, cognitive, or demographic data uploaded separately can be downloaded in its native format (csv for csv, xls for xls, etc).
NUNDA data can be downloaded from the web portal or through the REST API. The latter enables NUNDA access from the command line, so users can write scripts for automated downloading. There is currently no browser-independent method for resuming an interrupted download.
Quality control procedures are performed at the project owners’ discretion. NUNDA offers a “usability” parameter for each scan, which can be modified directly from the NUNDA web portal at any point during the project lifecycle. Certain QA pipelines also update this “usability” parameter. When a scan is marked as “unusable”, processing pipelines will ignore it by default, and users can opt to remove the scan completely. Some derived data, such as FreeSurfer output, also contains a quality field. Again, it is the responsibility of the individual project owner to implement his/her own quality control procedures; NUNDA simply offers a means of tracking data quality.
Notifications within NUNDA currently exist for the archive of new imaging data and the completion of processing pipelines. Project owners can configure a list of notification addresses for these events within their project. Currently, NUNDA does not support automatic notifications when data is withdrawn or revised; however, we are in the process of developing this option.
5. Discussion
By offering a secure, robust archive for data storage, management, processing, and sharing, NUNDA has become an essential component in an integrated research program. It provides a resource for research projects to collect and share their data, with many of these datasets serving as invaluable sources for pilot studies.
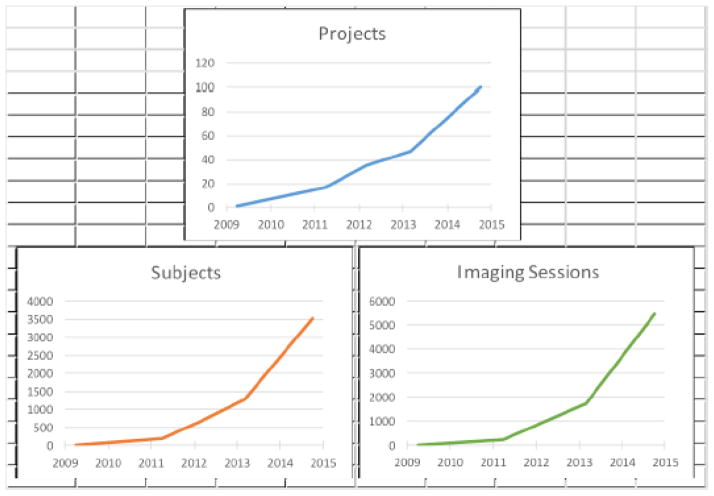
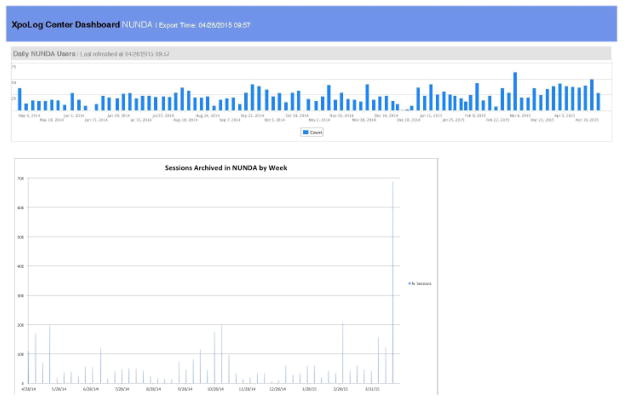
NUNDA has been steadily growing since its inception in 2009 (Figure 4), supporting the research, education and training mission of our institution and its collaborators. In the past year, approximately 75 imaging sessions have been archived each week, with an average of 20 users accessing NUNDA daily (Figure 5). Grants and institutional support enable NUNDA’s continued management and maintenance, and the flexibility of the XNAT platform enables NUNDA to continuously expand and adapt to the growing needs of its users. An essential resource for neuroimaging research, NUNDA is a collaborative environment for cross-fertilization at the institutional level as well as the research community at large.
Figure 4. Growth of data stored in NUNDA since inception in 2009.
NUNDA has been steadily growing since its inception in 2009. Currently, NUNDA supports 131 projects, 4,783 subjects, and 7,972 imaging sessions.
Figure 5. NUNDA Analytics.
In the past year, approximately 75 imaging sessions have been archived each week, and an average of 20 users access NUNDA each day.
Supplementary Material
Figure 2. The NUNDA research workflow, adapted from (Marcus et al., 2007a).
The NUNDA research workflow begins with data acquisition and archiving, which brings data into NUNDA. Once a part of the NUNDA archive, data can be processed and analyzed, shared with collaborators, integrated with non-imaging data, and downloaded for local use.
Acknowledgments
This work was supported in part by NIH grants 1R01 MH084803, 1U01 MH097435-01A1, P50 DC012283-01A1, and a grant from the Northwestern Memorial Hospital.
Footnotes
See also paper on the original XNAT implementation of Central Neuroimaging Data Archive (CNDA) at Washington University in St. Louis in this special issue.
See also paper on SchizConnect in this special issue.
See also paper on XNAT Central in this special issue.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)--a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42:377–381. doi: 10.1016/j.jbi.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SL, Wang L, Alpert KI, Greenstein D, Clasen L, Lalonde F, Miller R, Rapoport J, Gogtay N. Hippocampal shape abnormalities of patients with childhood-onset schizophrenia and their unaffected siblings. J Am Acad Child Adolesc Psychiatry. 2013;52:527–536. e522. doi: 10.1016/j.jaac.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan AR, Wang L, Beg MF. FreeSurfer-initiated fully-automated subcortical brain segmentation in MRI using Large Deformation Diffeomorphic Metric Mapping. Neuroimage. 2008;41:735–746. doi: 10.1016/j.neuroimage.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Archie KA, Olsen TR, Ramaratnam M. The open-source neuroimaging research enterprise. J Digit Imaging. 2007a;20(Suppl 1):130–138. doi: 10.1007/s10278-007-9066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus DS, Olsen TR, Ramaratnam M, Buckner RL. The Extensible Neuroimaging Archive Toolkit: an informatics platform for managing, exploring, and sharing neuroimaging data. Neuroinformatics. 2007b;5:11–34. doi: 10.1385/ni:5:1:11. [DOI] [PubMed] [Google Scholar]
- Song X, Wang X, Alpert K, Chen Y, Huang L, Wang L, Parrish T. Human Brain Mapping. Hamburg, Germany: 2014. Rapid automatic comprehensive quality assurance metrics evaluation for neuroimaging Studies. [Google Scholar]
- Williams AC, McNeely ME, Greene DJ, Church JA, Warren SL, Hartlein JM, Schlaggar BL, Black KJ, Wang L. A pilot study of basal ganglia and thalamus structure by high dimensional mapping in children with Tourette syndrome. 2013;1:F1000Research 2. doi: 10.12688/f1000research.2-207.v1. ref status: approved 1, http://f1000r.es/1yu. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.