Abstract
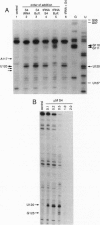
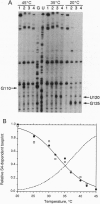
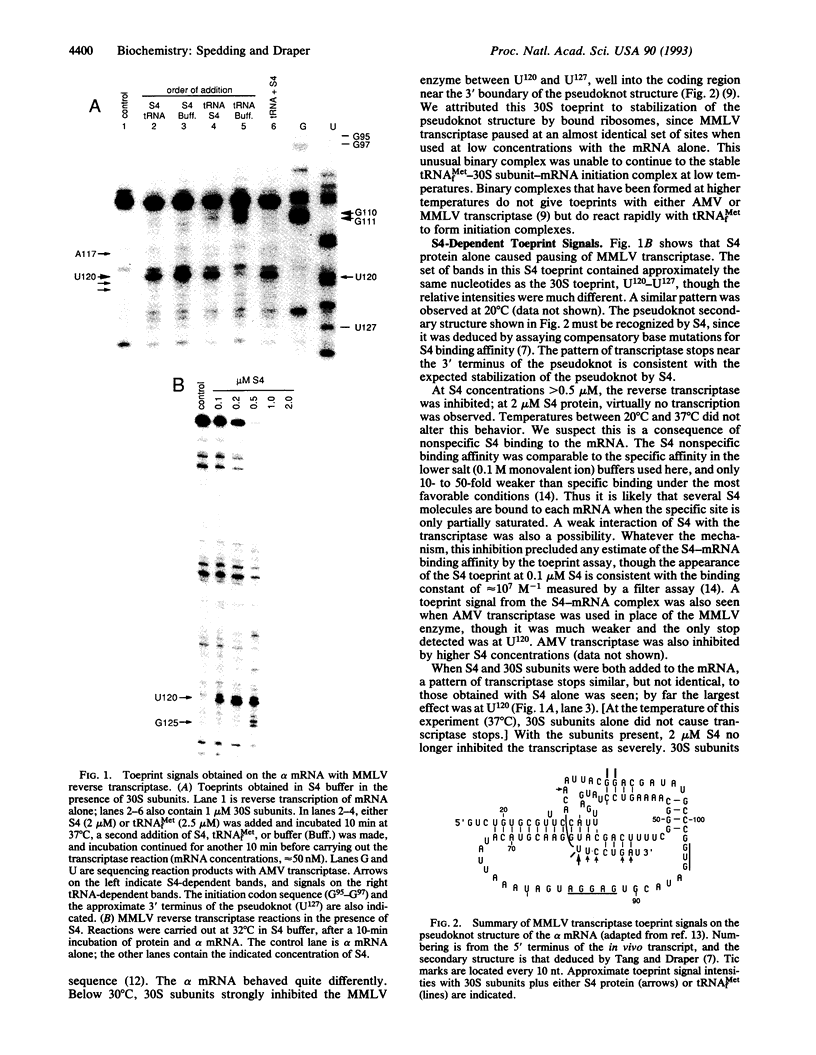
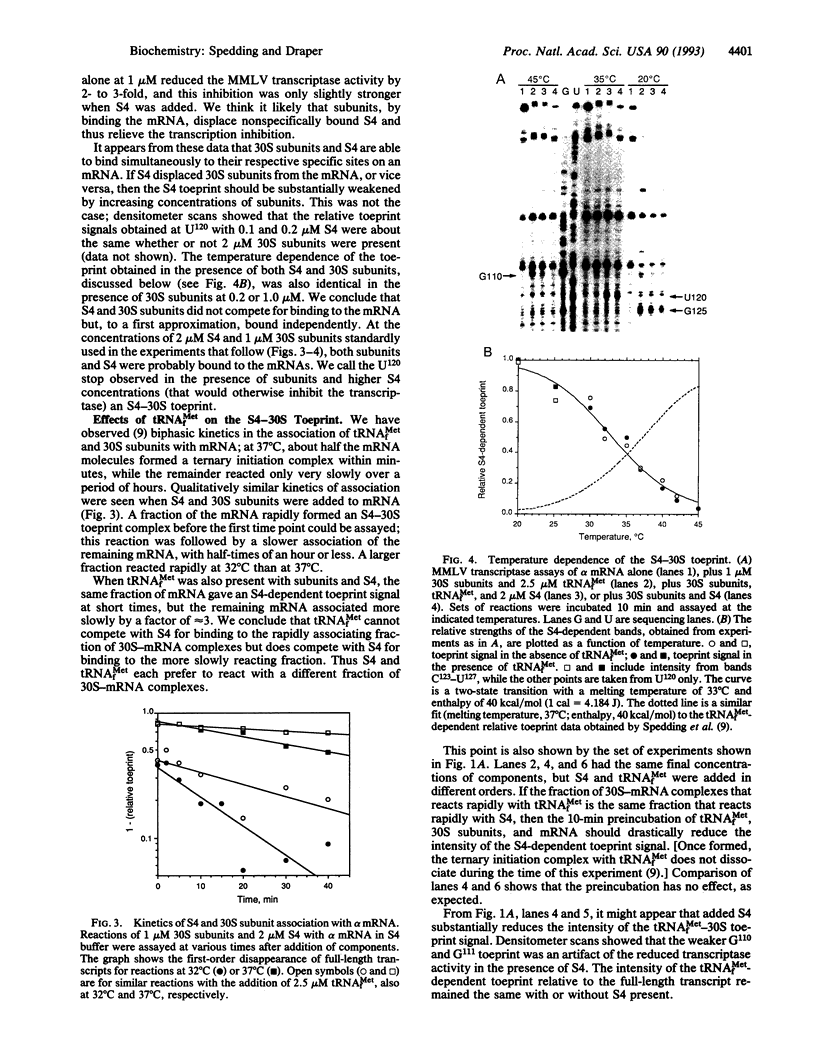
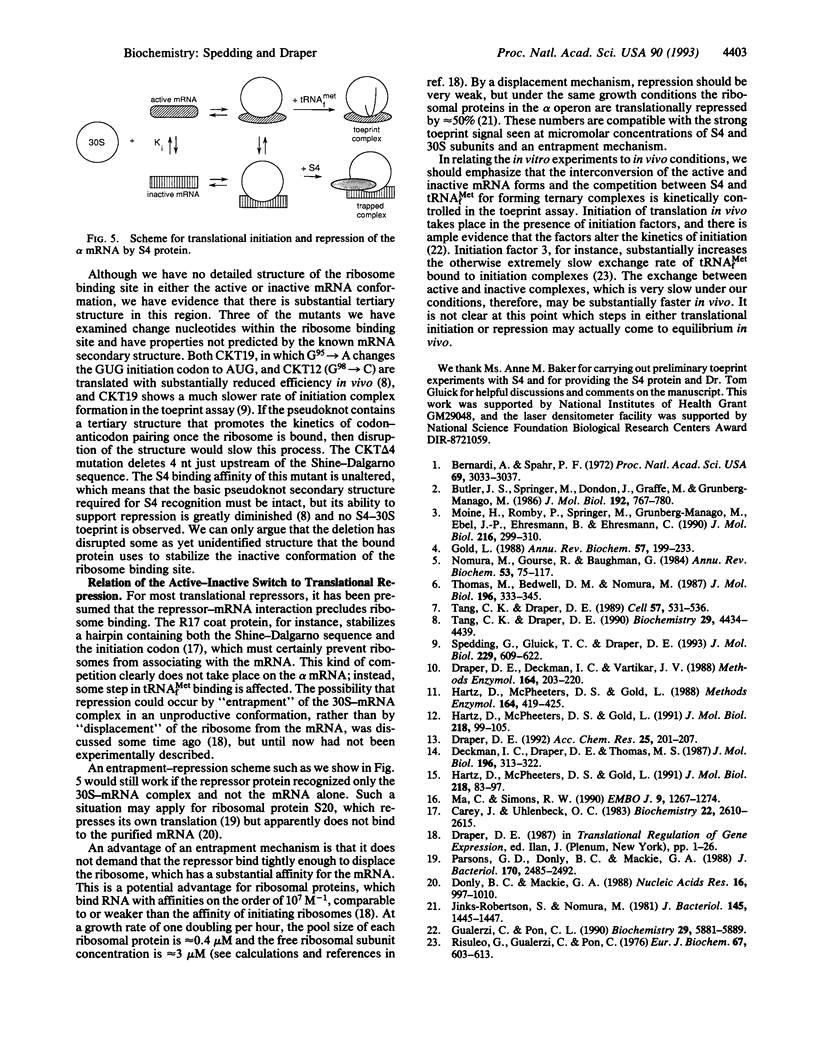
The ribosomal protein S4 is a translational repressor that binds to a complex mRNA pseudoknot structure containing the ribosome binding site for the first gene of the alpha operon. Either 30S subunits or S4 protein bound to the mRNA causes Moloney murine leukemia virus reverse transcriptase to pause near the 3' terminus of the pseudoknot. There is no competition between subunits and S4 for mRNA binding. The kinetics of forming S4-30S-mRNA complexes are biphasic, and the fraction of mRNA molecules reacting more rapidly decreases as the temperature is increased from 30 degrees C to 40 degrees C. The complex cannot be detected with mRNA mutants that cannot be repressed. We have previously shown similar kinetic behavior for the formation of tRNA(fMet) initiation complexes with tRNA(fMet), 30S subunits, and mRNA, except that the fraction reacting rapidly increases when the temperature is increased over the same 30-40 degrees C range. Thus the two sets of experiments show that there are two forms of 30S-mRNA complexes that differ in their abilities to bind S4 and tRNA(fMet). The results support an allosteric model for translational repression in which S4 traps the mRNA in a conformation able to bind 30S subunits but unable to form an initiation complex with tRNA(fMet).
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernardi A., Spahr P. F. Nucleotide sequence at the binding site for coat protein on RNA of bacteriophage R17. Proc Natl Acad Sci U S A. 1972 Oct;69(10):3033–3037. doi: 10.1073/pnas.69.10.3033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. S., Springer M., Dondon J., Graffe M., Grunberg-Manago M. Escherichia coli protein synthesis initiation factor IF3 controls its own gene expression at the translational level in vivo. J Mol Biol. 1986 Dec 20;192(4):767–780. doi: 10.1016/0022-2836(86)90027-6. [DOI] [PubMed] [Google Scholar]
- Carey J., Uhlenbeck O. C. Kinetic and thermodynamic characterization of the R17 coat protein-ribonucleic acid interaction. Biochemistry. 1983 May 24;22(11):2610–2615. doi: 10.1021/bi00280a003. [DOI] [PubMed] [Google Scholar]
- Deckman I. C., Draper D. E., Thomas M. S. S4-alpha mRNA translation repression complex. I. Thermodynamics of formation. J Mol Biol. 1987 Jul 20;196(2):313–322. doi: 10.1016/0022-2836(87)90692-9. [DOI] [PubMed] [Google Scholar]
- Donly B. C., Mackie G. A. Affinities of ribosomal protein S20 and C-terminal deletion mutants for 16S rRNA and S20 mRNA. Nucleic Acids Res. 1988 Feb 11;16(3):997–1010. doi: 10.1093/nar/16.3.997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draper D. E., Deckman I. C., Vartikar J. V. Physical studies of ribosomal protein-RNA interactions. Methods Enzymol. 1988;164:203–220. doi: 10.1016/s0076-6879(88)64044-4. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Gualerzi C. O., Pon C. L. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990 Jun 26;29(25):5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Gold L. Influence of mRNA determinants on translation initiation in Escherichia coli. J Mol Biol. 1991 Mar 5;218(1):83–97. doi: 10.1016/0022-2836(91)90875-7. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Green L., Gold L. Detection of Escherichia coli ribosome binding at translation initiation sites in the absence of tRNA. J Mol Biol. 1991 Mar 5;218(1):99–105. doi: 10.1016/0022-2836(91)90876-8. [DOI] [PubMed] [Google Scholar]
- Hartz D., McPheeters D. S., Traut R., Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S., Nomura M. Regulation of ribosomal protein synthesis in an Escherichia coli mutant missing ribosomal protein L1. J Bacteriol. 1981 Mar;145(3):1445–1447. doi: 10.1128/jb.145.3.1445-1447.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Simons R. W. The IS10 antisense RNA blocks ribosome binding at the transposase translation initiation site. EMBO J. 1990 Apr;9(4):1267–1274. doi: 10.1002/j.1460-2075.1990.tb08235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moine H., Romby P., Springer M., Grunberg-Manago M., Ebel J. P., Ehresmann B., Ehresmann C. Escherichia coli threonyl-tRNA synthetase and tRNA(Thr) modulate the binding of the ribosome to the translational initiation site of the thrS mRNA. J Mol Biol. 1990 Nov 20;216(2):299–310. doi: 10.1016/S0022-2836(05)80321-3. [DOI] [PubMed] [Google Scholar]
- Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu Rev Biochem. 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- Parsons G. D., Donly B. C., Mackie G. A. Mutations in the leader sequence and initiation codon of the gene for ribosomal protein S20 (rpsT) affect both translational efficiency and autoregulation. J Bacteriol. 1988 Jun;170(6):2485–2492. doi: 10.1128/jb.170.6.2485-2492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risuleo G., Gualerzi C., Pon C. Specificity and properties of the destabilization, induced by initiation factor IF-3, of ternary complexes of the 30-S ribosomal subunit, aminoacyl-tRNA and polynucleotides. Eur J Biochem. 1976 Aug 16;67(2):603–613. doi: 10.1111/j.1432-1033.1976.tb10726.x. [DOI] [PubMed] [Google Scholar]
- Spedding G., Gluick T. C., Draper D. E. Ribosome initiation complex formation with the pseudoknotted alpha operon messenger RNA. J Mol Biol. 1993 Feb 5;229(3):609–622. doi: 10.1006/jmbi.1993.1067. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Evidence for allosteric coupling between the ribosome and repressor binding sites of a translationally regulated mRNA. Biochemistry. 1990 May 8;29(18):4434–4439. doi: 10.1021/bi00470a025. [DOI] [PubMed] [Google Scholar]
- Tang C. K., Draper D. E. Unusual mRNA pseudoknot structure is recognized by a protein translational repressor. Cell. 1989 May 19;57(4):531–536. doi: 10.1016/0092-8674(89)90123-2. [DOI] [PubMed] [Google Scholar]
- Thomas M. S., Bedwell D. M., Nomura M. Regulation of alpha operon gene expression in Escherichia coli. A novel form of translational coupling. J Mol Biol. 1987 Jul 20;196(2):333–345. doi: 10.1016/0022-2836(87)90694-2. [DOI] [PubMed] [Google Scholar]