Abstract
3′-Hydroxyacetanilide or N-acetyl-meta-aminophenol (AMAP) is generally regarded as a non-hepatotoxic analog of acetaminophen (APAP). Previous studies demonstrated absence of toxicity after AMAP in mice, hamsters, primary mouse hepatocytes and several cell lines. In contrast, experiments with liver slices suggested that it may be toxic to human hepatocytes; however, the mechanism of toxicity is unclear. To explore this, we treated primary human hepatocytes (PHH) with AMAP or APAP for up to 48 h and measured several parameters to assess metabolism and injury. Although less toxic than APAP, AMAP dose-dependently triggered cell death in PHH as indicated by alanine aminotransferase (ALT) release and propidium iodide (PI) staining. Similar to APAP, AMAP also significantly depleted glutathione (GSH) in PHH and caused mitochondrial damage as indicated by glutamate dehydrogenase (GDH) release and the JC-1 assay. However, unlike APAP, AMAP treatment did not cause relevant c-jun-N-terminal kinase (JNK) activation in the cytosol or phospho-JNK translocation to mitochondria. To compare, AMAP toxicity was assessed in primary mouse hepatocytes (PMH). No cytotoxicity was observed as indicated by the lack of lactate dehydrogenase release and no PI staining. Furthermore, there was no GSH depletion or mitochondrial dysfunction after AMAP treatment in PMH. Immunoblotting for arylated proteins suggested that AMAP treatment caused extensive mitochondrial protein adducts formation in PHH but not in PMH. In conclusion, AMAP is hepatotoxic in PHH and the mechanism involves formation of mitochondrial protein adducts and mitochondrial dysfunction.
Keywords: 3′-Hydroxyacetanilide (AMAP), acetaminophen, hepatotoxicity, mitochondrial dysfunction, protein adducts
INTRODUCTION
APAP is a safe analgesic and antipyretic drug at therapeutic doses. However, APAP overdose can cause severe liver damage and even liver failure. Currently, it is the most prevalent cause of acute liver failure in the US and UK (Lee, 2012). Mechanistic investigations over the past several decades have revealed key events contributing to toxicity (Jaeschke et al., 2012, 2013). Bioactivation of APAP results in formation of the toxic metabolite N-acetyl-p-benzoquinone imine (NAPQI). At therapeutic doses or mild overdoses, NAPQI can be detoxified by glutathione (GSH) with minimal protein binding (McGill et al., 2013). However, after a more severe overdose, there is extensive hepatic GSH depletion and greater protein adducts formation (McGill et al., 2013). Interestingly, binding to mitochondrial proteins correlates with injury (Tirmenstein and Nelson, 1989; McGill et al., 2012b). Mitochondrial protein binding precedes mitochondrial dysfunction (Meyers et al., 1988), mitochondrial oxidant stress (Jaeschke, 1990; Tirmenstein and Nelson, 1990; Knight et al., 2001) and peroxynitrite formation (Cover et al., 2005). An initial oxidant stress activates and phosphorylates c-jun-N-terminal kinase (JNK), which then translocates to mitochondria (Hanawa et al., 2008) where it amplifies the oxidative stress (Saito et al., 2010) and triggers opening of the mitochondrial permeability transition pore (Kon et al., 2004; Ramachandran et al., 2011; LoGuidice and Boelsterli, 2011), resulting in cell necrosis (Gujral et al., 2002).
3′-Hydroxyacetanilide or N-acetyl-meta-aminophenol (AMAP) is a regioisomer of APAP and also possesses analgesic and antipyretic properties (Baker et al., 1963). However, unlike APAP, studies in mice (Tirmenstein and Nelson, 1989), hamsters (Roberts et al., 1990), primary mouse hepatocytes (Holme et al., 1991) and TAMH cells (Pierce et al., 2002) have suggested that AMAP is not hepatotoxic. Several groups have proposed possible mechanisms to explain the discrepancy in toxicity, and most of the evidence suggests a critical role for mitochondria. It was demonstrated that, although AMAP and APAP caused similar total cellular protein binding, there was significantly less mitochondrial protein binding after AMAP (Tirmenstein and Nelson, 1989; Myers et al., 1995). Similarly, it was shown that the reactive metabolite of APAP extensively bound to mitochondrial proteins, while metabolites of AMAP preferentially bound to microsomal and cytosolic proteins (Tirmenstein and Nelson, 1989; Streeter et al., 1984; Matthews et al., 1997; Qiu et al., 2001). APAP also triggered robust nitrotyrosine protein adduct formation in mitochondria (Cover et al., 2005), while there was no appreciable mitochondrial protein nitration after AMAP (Abdelmegeed et al., 2013). Consistent with those data, there is less cytosolic and mitochondrial GSH depletion after AMAP compared to APAP (Nelson, 1980; Tirmenstein and Nelson, 1989; Rashed et al., 1990; Hanawa et al., 2008). Furthermore, the lack of JNK activation and translocation to mitochondria in mice after AMAP suggested that AMAP did not particularly target mitochondria (Hanawa et al., 2008). Together these data indicated that mitochondrial dysfunction and mitochondrial protein binding are critical in determining toxicity. A widely accepted hypothesis to explain the difference in mitochondrial effects is that the reactive metabolite(s) of AMAP is more active and not stable enough to escape the site of formation in the endoplasmic reticulum (ER), leading to greater microsomal protein binding, while NAPQI can escape the ER and attack other organelles, including mitochondria (Rashed et al., 1990).
A recent study revisited AMAP toxicity in precision-cut liver slices and found that AMAP was as toxic as APAP based on ATP depletion in rat and human liver slices (Hadi et al., 2013). However, the mechanism of AMAP toxicity in human liver cells was not investigated. These findings prompted us to evaluate the differences in the mechanisms of action between AMAP and APAP in PHH, and to compare across species. A better understanding of AMAP toxicity in PHH could provide new insights into APAP-induced hepatotoxicity.
MATERIALS AND METHODS
Isolation and treatment of primary human hepatocytes (PHH)
All human tissues were acquired with informed consent from each patient according to ethical and institutional guidelines. The study was approved by the University of Kansas Medical Center Institutional Review Board. Table 1 provides a summary of medical information of donors. No donor tissue was obtained from executed prisoners or other institutionalized persons. All liver specimens were acquired in accordance with a Human Subject Committee approved protocol from patients undergoing a hepatic resection procedure or from donor livers. Hepatocytes were isolated as described in detail (Xie et al., 2014). Around 3 h after being seeded, cells were washed with sterile phosphate-buffered saline (PBS) and treated with AMAP or APAP. Cells were harvested at different time points according to assays.
Table 1.
Liver donor medical information
| Donor | Source | Age | Gender | Diagnosis |
|---|---|---|---|---|
|
| ||||
| 1 | Resection | 41 | Female | Metastatic colon cancer |
| 2 | Donor | 25 | Female | Head trauma |
| 3 | Donor | 64 | Female | Anoxia |
| 4 | Donor | 38 | Male | Head trauma |
| 5 | Donor | 29 | Female | CVA/Stroke |
| 6 | Donor | 25 | Male | Cardiac arrest |
| 7 | Donor | 13 | Male | Asthma attack |
| 8 | Donor | 50 | Male | Head trauma |
| 9 | Donor | N/A | Male | Anoxia |
| 10 | Donor | 19 | Male | Head trauma |
| 11 | Donor | 24 | Female | Cardiac arrest |
| 12 | Donor | 16 | Male | Head trauma |
CVA; cerebrovascular accident; N/A; not available
Isolation and treatment of primary mouse hepatocytes (PMH)
Primary hepatocytes were isolated as described in detail (Bajt et al., 2004). Cell viability of each isolation was generally more than 90%, and hepatocyte purity was >95%. Following isolation, primary hepatocytes were plated with a concentration of 6×105 cells per well and allowed 3 h to attach. The cells were then washed with PBS and exposed to AMAP or APAP. Cells were harvested at the indicated time points.
Biochemical assays
ALT activities were measured using an ALT reagent kit (Pointe Scientific, MI). LDH and GDH levels were determined as described (Xie et al., 2013; Du et al., 2013). A modified Tietze assay was used to measure glutathione levels as described (Jaeschke and Mitchell, 1990). The JC-1 Mitochondrial Membrane Potential Kit (Cell Technology, Mountain View, CA) was used to measure the mitochondrial membrane potential as described in detail (Bajt et al., 2004).
Propidium iodide (PI) staining
PHH and PMH were treated with AMAP or APAP 3 h after seeding. The medium was removed after 48 h (PHH) or 15h (PMH) and cells were stained with 500 nM of PI (Invitrogen) for 5 min. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) and images were obtained with a fluorescence microscope.
Isolation of subcellular fractions and western blotting
Mitochondria and cytosolic fractions were isolated using differential centrifugation as described (Xie et al., 2014). Western blotting was performed as described (Bajt et al., 2000). A rabbit anti-JNK antibody and a rabbit anti-phospho-JNK antibody (Cell Signaling Technology, Danvers, MA) were used for JNK and phospho-JNK detection. A rabbit primary antibody was used to detect APAP or AMAP protein adducts. The antibody was raised against acetamidobenzoic acid and has been demonstrated to detect both APAP and AMAP protein adducts (Matthews et al., 1997; Salminen et al., 1998).
Statistics
All results are expressed as mean ± standard error (SE). Comparisons between multiple groups were performed with one-way ANOVA followed by post hoc Student-Newman-Keul’s test. If the data were not normally distributed, the Kruskal-Wallis Test was used (nonparametric ANOVA) followed by Dunn’s Multiple Comparisons Test. P < 0.05 was considered significant.
RESULTS
AMAP induces cell death and GSH depletion in primary human hepatocytes
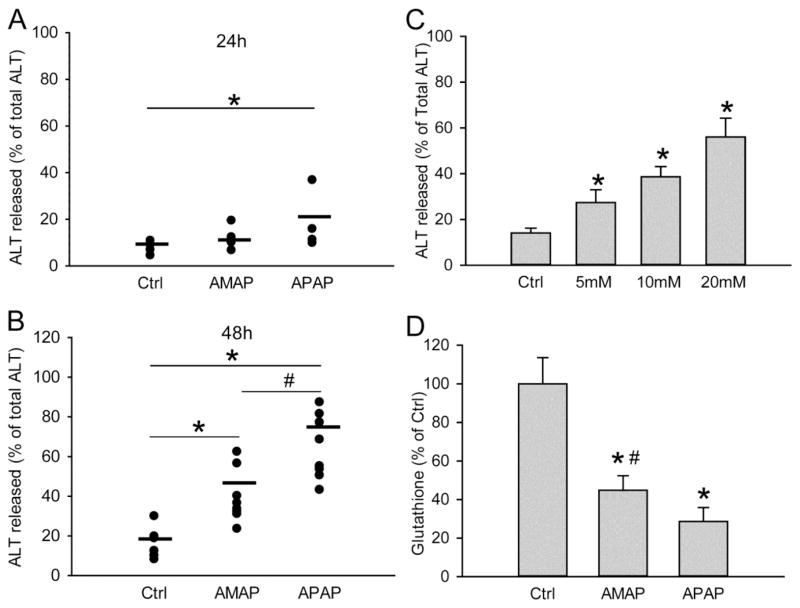
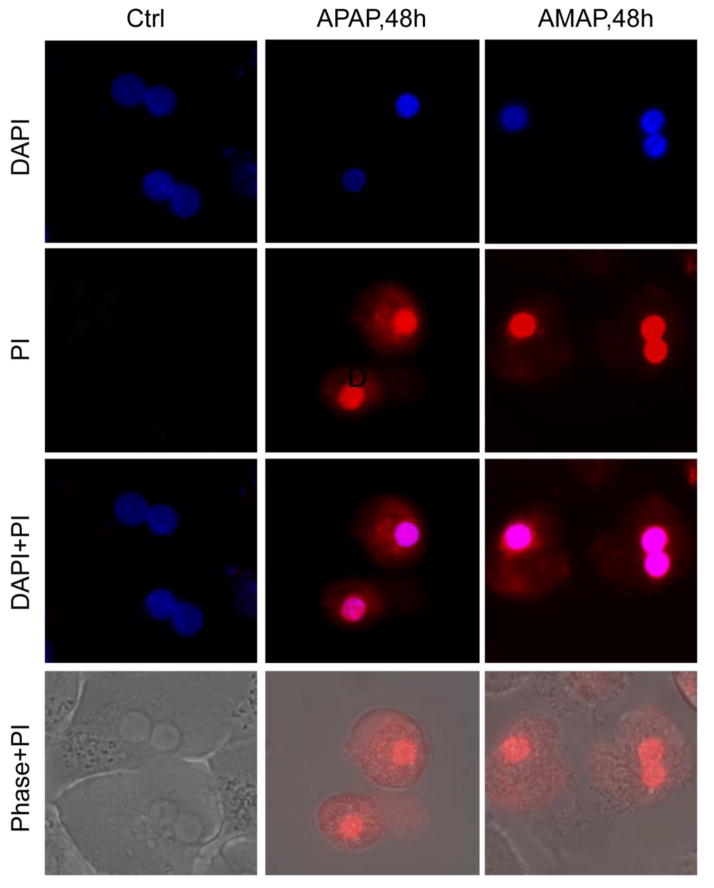
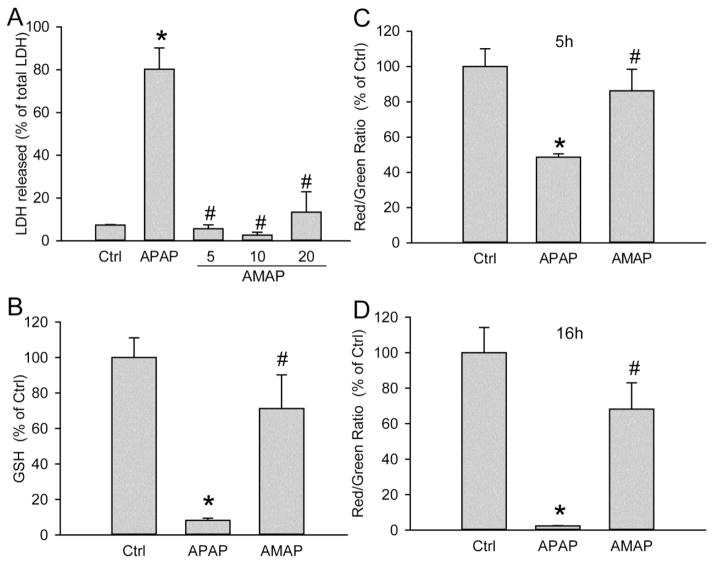
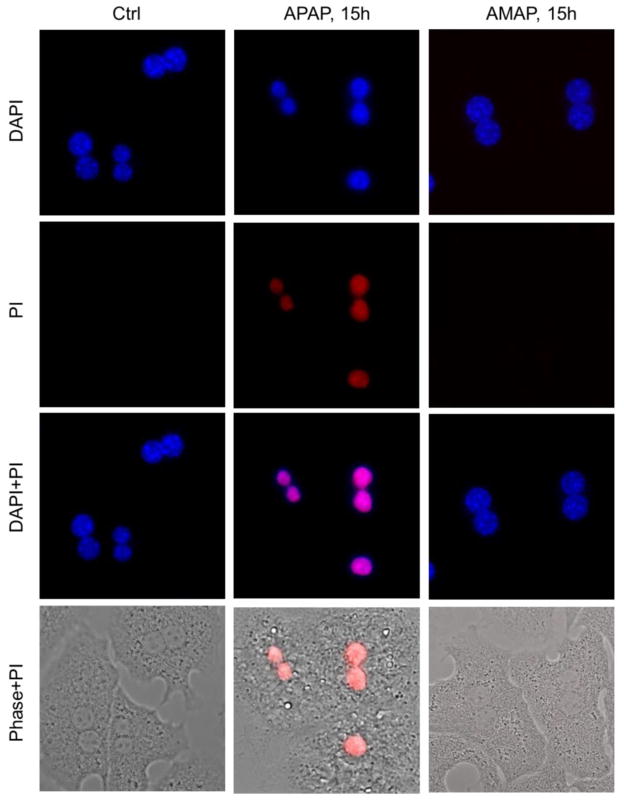
Freshly isolated primary human hepatocytes (PHH) were treated with 10 mM AMAP or APAP for 48 h. APAP triggered significant cell death as indicated by the percentage of alanine aminotransferase (ALT) released into the cell culture medium at 24 h and 48 h after treatment (Fig. 1A,B). AMAP did not cause significant cytotoxicity until 48 h (Fig. 1B). Consistent with that, toxicities of both AMAP and APAP at 48 h were confirmed by the cellular uptake of red fluorescent PI stain (Fig. 2). PI cannot enter live cells but upon membrane permeabilization during necrosis, the stain enters the cells and binds to nuclear DNA, which enhances the red fluorescence. An overlay of PI with the nuclear stain DAPI or the phase contrast images confirms the nuclear localization of the PI staining indicative of necrotic cell death in PHH after exposure to APAP and AMAP (Fig. 2). At both 24 h and 48 h, AMAP was less toxic than APAP (Fig. 1A,B). The hepatotoxicity of AMAP was dose-dependent (Fig. 1C). Exposure to 10 mM AMAP triggered substantial GSH depletion at 24 h, although this effect was less dramatic than after APAP (Fig. 1D). Overall, although AMAP has a reduced propensity to cause necrotic cell death compared to APAP, our data clearly show that it is toxic in PHH.
Figure 1. AMAP induces cell death and GSH depletion in human hepatocytes.
Primary human hepatocytes (PHH) were treated with 10 mM of AMAP or APAP over a period of 48 h, and the percentage of alanine aminotransferase (ALT) release was measured at 24 h (A) and 48 h (B) to evaluate toxicity. (C) Dose-response of AMAP toxicity at 48 h after treatment. (D) Glutathione depletion after 10 mM AMAP or APAP exposure for 24 h. Data represent mean ± SE from experiments using cells from 3 to 10 donors. *P< 0.05 (compared with controls); #P< 0.05 (compared with APAP group).
Figure 2. AMAP and APAP induce necrosis in human hepatocytes.
Primary human hepatocytes (PHH) were treated with 10 mM of AMAP or APAP over a period of 48 h and necrotic cell death as indicated by nuclear PI staining was assessed. DAPI, 4′,6-Diamidino-2-Phenylindole (nuclear stain); PI, propidium iodide (nuclear stain of necrotic cells)
AMAP triggers mitochondrial dysfunction in PHH without JNK activation or P-JNK translocation to mitochondria
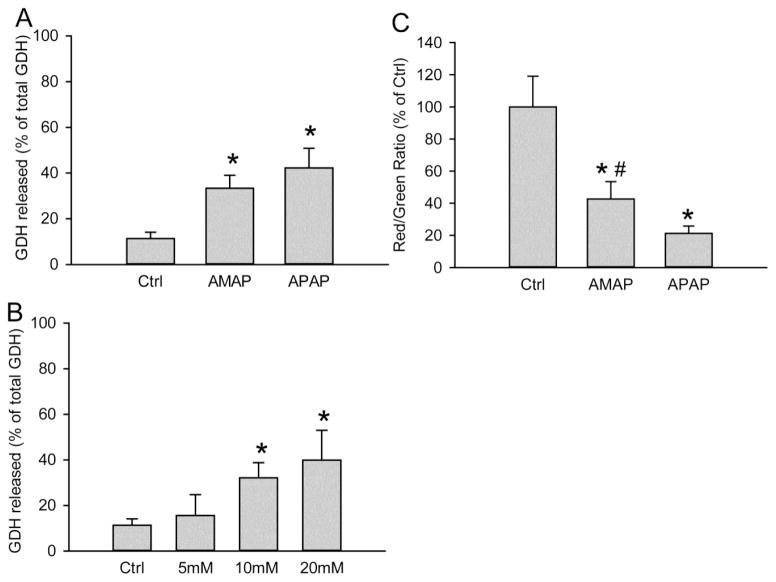
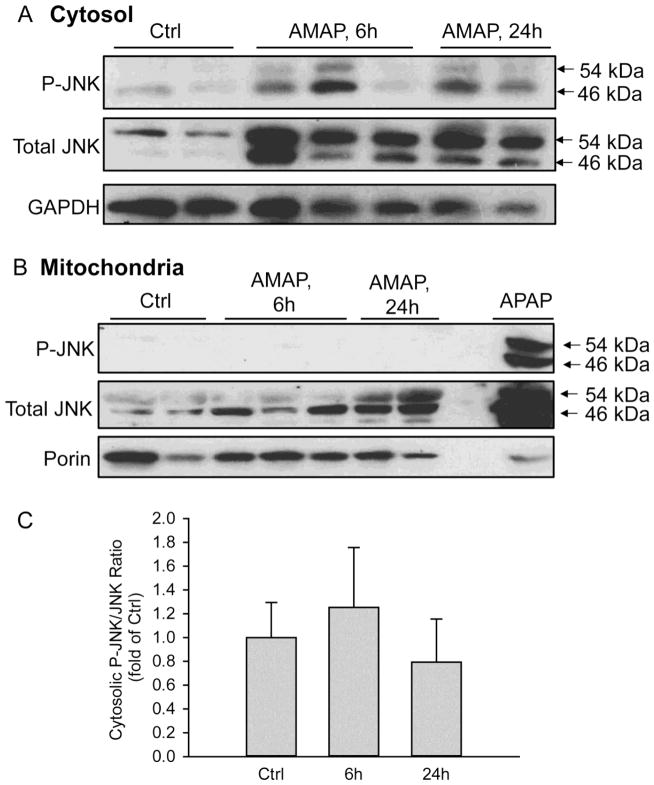
Given the importance of mitochondria in the toxicity after APAP in mice and humans, we hypothesized that AMAP caused mitochondrial dysfunction in PHH. To test this hypothesis, release of glutamate dehydrogenase (GDH), an enzyme localized in the mitochondrial matrix that is released into the serum upon mitochondrial damage (McGill et al., 2012a), was evaluated. Significant release of GDH was observed in both AMAP- and APAP-treated hepatocytes at 48 hours after treatment, and the effect of AMAP was dose-dependent (Fig. 3A,B). Consistent with that, the JC-1 assay revealed the significant loss of mitochondrial membrane potential after AMAP by a decline of the red-to-green fluorescence ratio, although the decrease was less extensive than after exposure to APAP (Fig. 3C). Importantly, compared with APAP, the less severe mitochondrial dysfunction after AMAP correlated with the lower ALT release and GSH depletion after AMAP compared to APAP (Fig. 1). JNK activation and translocation, which are critical amplifying events in APAP-induced hepatotoxicity, were also assessed after AMAP treatment (Fig. 4). At 6 and 24 h after AMAP exposure, an increase in total JNK compared to untreated cells was observed in the cytosol, which was paralleled by a modest increased expression of P-JNK (Fig. 4A). However, the ratio of P-JNK-to-total JNK did not change significantly before or after AMAP (Fig. 4C). Furthermore, there was no P-JNK translocation to mitochondria and only a minor JNK translocation at 24 h (Fig. 4B). Compared to APAP, JNK activation and P-JNK translocation to the mitochondria after AMAP is negligible (Fig. 4B). Taken together, AMAP causes significant mitochondrial dysfunction, but JNK activation and phospho-JNK translocation likely do not contribute to the cell death in PHH.
Figure 3. AMAP triggers mitochondrial dysfunction in PHH.
(A) Percentage of glutamate dehydrogenase (GDH) release into the culture medium at 48 h after 10 mM AMAP or APAP. (B) Dose-response of GDH release at 48 h after AMAP (5–20 mM). (C) Loss of mitochondria membrane potential after 24 h exposure to 10 mM AMAP or APAP as indicated by the decrease of the red/green fluorescence ratio using the JC-1assay. Data represent mean ± SE from experiments using cells from 8 donors. *P< 0.05 (compared with controls); #P< 0.05 (compared with APAP group).
Figure 4. No JNK activation or mitochondrial translocation after AMAP in PHH.
JNK activation in the cytosol (A) and P-JNK translocation to the mitochondria (B) were evaluated by western blotting. Mitochondrial fraction from APAP-treated PHH was used as positive control. (C) Densitometry of cytosolic JNK activation. Data represent mean ± SE from densitometry using cells from 3 donors.
No injury or significant mitochondrial dysfunction after AMAP in primary mouse hepatocytes (PMH) under the conditions studied
For comparison, primary mouse hepatocytes were treated with 5 mM APAP or various concentrations of AMAP. Consistent with previous findings, AMAP was not significantly toxic even at 20 mM as demonstrated by the lack of lactate dehydrogenase (LDH) release. However, 5 mM APAP resulted in extensive cell death (80% release of total LDH into culture medium) (Fig. 5A). The necrotic cell death after APAP was confirmed by cellular uptake of the red PI stain using immunofluorescent staining (Fig. 6). Consistent with the lack of LDH release, AMAP-treated cells did not show any cellular PI uptake. Overlay of PI with the nuclear stain DAPI or phase contrast images confirmed the nuclear localization of the PI stain in APAP but not AMAP-treated cells (Fig. 6). A similar trend was observed with GSH content, as exposure of AMAP had no significant effect on GSH levels while APAP caused 92% depletion of total GSH (Fig. 5B). In contrast to PHH, no mitochondrial dysfunction was observed in PMH at either 5 h or 16 h after AMAP (Fig. 5C, D). Based on these data we conclude that AMAP does not induce cell death or mitochondrial dysfunction in PMH under the conditions studied.
Figure 5. No significant mitochondrial dysfunction or cell death after AMAP in primary mouse hepatocytes (PMH).
(A) Lactate dehydrogenase was measured as an indicator of cell death after exposure to 5–20 mM AMAP or 5 mM APAP for 15 h. (B) GSH depletion at 5 h after 5 mM AMAP or APAP. Loss of mitochondrial membrane potential at 5 h (C) and 16 h (D) after AMAP and APAP were assessed by decrease of red/green fluorescence ratio using the JC-1assay. Data represent mean ± SE from experiments using primary mouse hepatocytes from 4 different isolations. *P< 0.05 (compared with controls); #P<0.05 (compared with the APAP group).
Figure 6. APAP but not AMAP caused cell necrosis in mouse hepatocytes.
Primary mouse hepatocytes (PMH) were treated with 5 mM of AMAP or APAP over a period of 15 h and necrotic cell death as indicated by nuclear PI staining was assessed. DAPI, 4′,6-Diamidino-2-Phenylindole (nuclear stain); PI, propidium iodide (nuclear stain of necrotic cells)
AMAP induces a significant increase of mitochondrial protein binding in PHH but not in PMH under the conditions studied
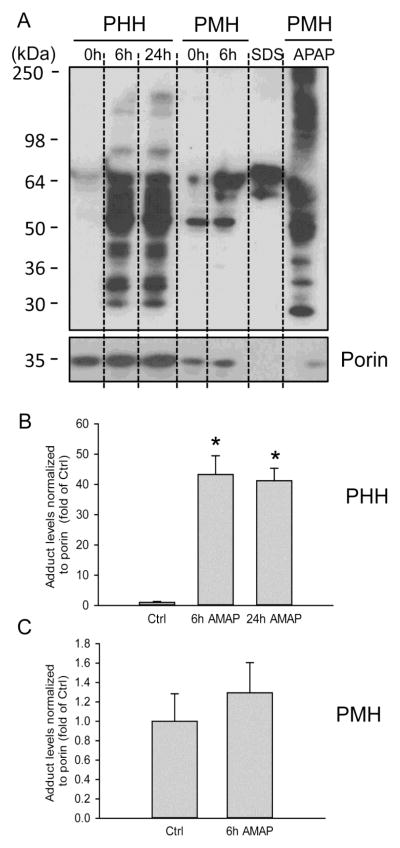
As mitochondrial protein binding is thought to be a key event in the initiation of APAP toxicity in mice, covalent binding in mitochondria from PHH was measured by immunoblotting for arylated proteins. An antibody against acetamidobenzoic acid was used that can detect both APAP and AMAP protein adducts (Matthews et al., 1997; Salminen et al., 1998). In PHH, extensive mitochondrial protein adducts formation was detected at 6 and 24 h after AMAP exposure (Fig. 7A,B). Note that the adduct bands consist of various proteins with molecular weights ranging from 30–250 kDa. A comparison to adducts detection in AMAP-treated PMH clearly indicates lower overall mitochondrial protein adducts formation in PMH than in PHH (Fig. 7A,B,C). On the other hand, a substantial number of mitochondrial protein adducts were observed in PMH after APAP treatment (Fig. 7A). The unexpected bands in the SDS control lane (Fig. 7A) were caused by SDS as this band was absent when only buffer was used (Supplemental Figure). Thus, despite less protein loading on the gel (as seen by the lower porin detection), APAP appeared to trigger much higher mitochondrial protein adducts formation than AMAP in PMH. Nevertheless, these data suggest that AMAP exposure results in significantly more mitochondrial covalent protein binding in PHH, which may be involved in AMAP-induced mitochondrial damage and hepatotoxicity in these cells.
Figure 7. AMAP induces covalent protein binding in PHH but not PMH.
(A) Mitochondrial fractions were isolated from PHH and PMH which were treated with AMAP for 6 h or 24h. Mitochondrial fraction from APAP-treated PMH was used as a positive control, and a blank (bl) lane loaded with only SDS loading buffer was used as a negative control. 0 h lanes represent mitochondria samples of control PHH or control PMH without any treatment. Porin was used as a loading control. (B) (C) Densitometry of adduct levels normalized to porin for PHH and PMH. The results of each 0 h lane were set to 1 and the data for AMAP-treated cells are expressed as fold increase over baseline. Data represent mean ± SE from experiments using primary human or mouse hepatocytes from 3 different isolations per group. *P< 0.05 (compared with corresponding controls)
DISCUSSION
The aim of this study was to investigate the mechanisms of AMAP hepatotoxicity in freshly isolated PHH and PMH. In contrast to results in mice and PMH, we found that AMAP is toxic to PHH. Comparison between PHH and PMH suggested the importance of mitochondrial protein adducts formation and mitochondrial dysfunction in the toxicity.
Mitochondrial protein adducts formation in AMAP-induced hepatotoxicity
AMAP has long been considered a non-hepatotoxic analog of APAP. The main reason for the difference in toxicity in mice appears to be protein adducts formation. When directly compared to APAP, AMAP triggers comparable total protein adducts formation but induces significantly less mitochondrial adducts (Myers et al., 1995; Tirmenstein and Nelson, 1989; Qiu et al., 2001). Therefore, it was concluded that mitochondrial protein adduct levels, rather than total cellular adduct levels, correlate with toxicity. The more recent data showing that AMAP can cause toxicity (ATP depletion) in human liver slices cast some doubt on this conclusion (Hadi et al., 2013). However, our data suggest that mitochondrial protein binding in fact correlates with toxicity in PHH, and the significant disparity in mitochondrial adduct levels readily explains the species difference in our study. Currently two approaches are commonly used for assessment of protein adducts formation after AMAP: detection of radiolabeled adducts and detection with antibodies. A comparison of the two methods indicate similar performance of adduct detection after AMAP either quantitatively or qualitatively. Importantly, both approaches suggest low levels of mitochondrial adducts in PMH (Myers et al., 1995). As antibody detection performs equally as well as radiolabeling, which is generally the gold standard method to detect protein adducts, the limited detection of mitochondrial protein adducts observed here in PMH after AMAP is most likely due to actually low levels of adducts. Admittedly, we cannot guarantee that this antibody is able to detect all mitochondrial adducts in mouse samples. However, since the antibody appears to detect large numbers of mitochondrial AMAP adducts in PHH, one would have to assume a dramatically different efficacy in detecting AMAP adducts in human versus mouse samples. This appears unlikely. Nevertheless, alternative detection methods are necessary for further verification.
Although the mitochondrial adducts profile in PHH after AMAP treatment appears to be similar to that in PMH after APAP (Fig. 7), a more detailed analysis of the adducted proteins is needed to determine if similar proteins were adducted. Proteomic investigation in mice demonstrated that proteins modified by AMAP constitute a subgroup of proteins modified by APAP in that species (Fountoulakis et al., 2000). Other studies narrowed down the list of adducted proteins and indicated that microsomal proteins, especially Cyp2e1, are the main targets of AMAP (Myers et al., 1995; Matthews et al., 1997; Halmes et al., 1998). However, that is beyond the scope of the current study. The fact that there appear to be more mitochondrial protein adducts after APAP correlates with the more severe mitochondrial dysfunction and higher cell death after APAP compared to AMAP under the conditions studied. In addition, previous data from a comparison of APAP toxicity in rats and mice suggest that the extent of mitochondrial protein binding correlates with liver injury (McGill et al., 2012b). Collectively, our data together with previous findings consistently support the idea that mitochondrial protein adduct levels correlate with hepatotoxicity.
Mitochondrial dysfunction in AMAP-induced hepatotoxicity
The critical role of mitochondria in APAP-induced hepatotoxicity has been well established (Jaeschke et al., 2012, 2013). Specifically, mitochondria are involved in APAP toxicity by covalent protein binding (Tirmenstein and Nelson, 1989; McGill et al., 2012b), mitochondrial oxidative stress (Jaeschke, 1990) and peroxynitrite formation (Cover et al., 2005), JNK activation and P-JNK translocation to mitochondria (Hanawa et al., 2008), oxidant stress amplification (Saito et al., 2010), mitochondrial Bax pore formation (Bajt et al., 2008), the mitochondrial membrane permeability transition pore opening (Kon et al., 2004) and mitochondrial endonuclease translocation to the nucleus (Bajt et.al., 2006). Similar mechanisms were observed in the metabolically competent human hepatoma cell line HepaRG (McGill et al., 2011) and in primary human hepatocytes (Xie et al., 2014). For AMAP, previous reports have shown a limited relevance of mitochondria. AMAP leads to microsomal protein binding rather than the mitochondrial protein binding observed after APAP (Tirmenstein and Nelson 1989; Rashed et al., 1990; Myers et.al., 1995). Compared with APAP, AMAP depletes total GSH and mitochondrial GSH to a lesser extent (Nelson, 1980; Tirmenstein and Nelson 1989; Rashed et al., 1990; Hanawa et al., 2008), despite the fact that a number of glutathione conjugates were identified in vitro (Rashed and Nelson, 1989). In contrast to APAP, AMAP did not disrupt mitochondrial calcium homeostasis by impairing the capacity to sequester calcium (Tirmenstein and Nelson, 1989). Thus, consistent evidence in mice indicates that AMAP does not appear to target mitochondria in that species. However, pharmacological intervention using BSO to deplete mitochondrial GSH made mice more vulnerable to AMAP (Tirmenstein and Nelson, 1991). Under these conditions, there was more mitochondrial protein binding correlating with AMAP toxicity in mice (Tirmenstein and Nelson, 1991). Overall, this led to the hypothesis that metabolites of AMAP may be too reactive to reach mitochondria under physiological conditions in mice (Rashed et al., 1990; Myers et al., 1995). Our data are consistent with the hypothesis that AMAP triggered mitochondrial dysfunction as a result of mitochondrial protein binding in PHH and the resulting mitochondrial damage likely led to cell death. This suggests more of a species difference in AMAP toxicity rather than a totally different mechanism. In support of this hypothesis, we have previously shown that the species difference in the time course of APAP toxicity between mice and humans is caused by the much more delayed protein adducts formation, especially in mitochondria, leading to delayed oxidant stress and JNK activation (Xie et al., 2014). In addition, the apparently nontoxic APAP analog, 2,6-dimethyl APAP, (2,6-DMA) was cytotoxic in PMH from phenobarbital-pretreated mice (Birge et al. 1989). Although the authors did not specifically assess mitochondrial protein adducts, the cytotoxicity of 2,6-DMA in PMH correlated with enhanced protein binding (Birge et al., 1989). These examples suggest that the cytotoxicity of AMAP or 2,6-DMA in PMH is mainly dependent on the experimental conditions. However, whether these difference in AMAP toxicity is only caused by a different metabolite profile and spectrum of adducts formed in human compared to mouse cells or if there are other factors involved remains to be investigated.
The metabolism profile of AMAP is different between mice and humans. In mice, around 40% of AMAP is eliminated either unaltered or conjugated with glucuronic acid or sulfate (Rashed et al., 1990). The remaining AMAP is metabolized by P450 enzymes to 3 major proximate metabolites which are 2,5-diOH-acetanilide, 3-OH-APAP and 3-OMe-APAP. These hydroquinones can be conjugated with glucuronic acid or sulfate, or they could be further converted to reactive quinones, which bind to GSH or attack cellular proteins (Rashed et al., 1990; Kenna, 2013). In precision-cut liver slices, it was reported that sulfation is the predominant pathway for AMAP in mouse livers, while glucuronidation prevails in human liver slices (Hadi et al., 2013). Besides, levels of three hydroquinones, which are precursors of the toxic quinones differ significantly in mice and humans (Hadi et al., 2013). It is also possible that different cytochrome P450s are involved in AMAP metabolism in different models. Future investigations regarding the species differences are necessary to elucidate the different mechanisms of actions.
JNK activation and AMAP-induced hepatotoxicity
Interestingly, no P-JNK translocation to the mitochondria was observed in PHH after AMAP treatment. After APAP overdose, JNK activation and mitochondrial P-JNK translocation are critical for the mechanism of liver injury in mice (Hanawa et al., 2008). In addition, knock-down or inhibition of many kinases, including apoptosis signal-regulating kinase 1 (Nakagawa et al., 2008; Xie et al., 2015), mixed-lineage kinase 3 (Sharma et al., 2012) and glycogen synthase kinase-3beta (Shinohara et al., 2010) has revealed that they are involved in activation of JNK and translocation of P-JNK to mitochondria, which are critical amplifying events for the initial oxidative stress. For AMAP, previous studies demonstrated the absence of JNK activation in other models after AMAP treatment. In TGF-alpha transgenic mouse hepatocytes (TAMH), which exhibited susceptibility to APAP and resistance to AMAP, AMAP induced less JNK and c-Jun activation compared to APAP (Stamper et al., 2010). In fact, JNK siRNA had no effect on cell death after AMAP while it was able to attenuate the cytotoxicity after APAP (Stamper et al., 2010). Similarly, in mice there is no JNK activation or mitochondrial translocation after AMAP (Hanawa et al., 2008). However, there was no toxicity after AMAP in these models. In contrast, our data indicate AMAP hepatotoxicity in PHH without relevant JNK activation and mitochondrial P-JNK translocation suggesting cell death in the absence of JNK activation. Interestingly, even in APAP-induced liver injury, JNK activation is not always necessary for the cell death mechanisms. In HepaRG cells, APAP triggers mitochondrial dysfunction and cell death without JNK activation (McGill et al., 2011; Xie et al., 2014). In PHH hepatocytes, APAP causes JNK activation but JNK inhibitors are only moderately protective (Xie et al., 2014). Similarly, PKC inhibitors attenuate APAP-induced liver injury in mice by preventing phosphorylation of AMPK and by activating autophagy in a JNK-independent way (Saberi et al., 2014). Together these data suggest that although JNK is important for most models of APAP hepatotoxicity, especially in mice, it does not contribute to cell death after AMAP in PHH.
In summary, our results demonstrated the hepatotoxicity of AMAP in PHH. The cell death was preceded by GSH depletion and loss of mitochondrial membrane potential. JNK activation was likely not involved in AMAP toxicity. In contrast, no GSH depletion, mitochondrial dysfunction or cell death was observed in PMH after AMAP. Comparison between PHH and PMH revealed that AMAP induces a significant increase of mitochondrial protein binding in PHH but not in PMH under the conditions studied. Our data emphasize the importance of mitochondrial dysfunction and sustained mitochondrial protein binding in determining hepatotoxicity in human hepatocytes after AMAP treatment, and also indicate that species differences in protein adduct formation should be considered when evaluating the toxic potential of a compound.
Supplementary Material
HIGHLIGHTS.
AMAP induces cell death in primary human hepatocytes (PHH).
AMAP does not cause cell death in primary mouse hepatocytes (PMH).
AMAP leads to mitochondria dysfunction in PHH but not PMH.
Protein adduct formation and dysfunction in mitochondria correlate with toxicity.
Acknowledgments
The authors thank Dr. Lance Pohl (NIH) for generously providing the antibody for detection of APAP and AMAP protein adducts. This antibody was originally generated by Drs. Neil Pumford and Jack Hinson (University of Arkansas for Medical Sciences). This work was supported in part by grants from the National Institutes of Health R01 DK102142 and R01 AA12916), and from the National Center for Research Resources (5P20RR021940-07) and the National Institute of General Medical Sciences (8 P20 GM103549-07) of the National Institutes of Health. Additional support came from the “Training Program in Environmental Toxicology” T32 ES007079-26A2 (to M.R.M.) from the National Institute of Environmental Health Sciences.
Abbreviations
- ALT
alanine aminotransferase
- AMAP
N-acetyl-meta-aminophenol
- APAP
acetaminophen
- BSO
buthionine sulfoximine
- DAPI
4′,6-diamidino-2-phenylindole
- GDH
glutamate dehydrogenase
- GSH
glutathione
- GSSG
glutathione disulfide
- JNK
c-jun-N-terminal kinase
- LDH
lactate dehydrogenase
- NAPQI
N-acetyl-p-benzoquinone imine
- PBS
phosphate-buffered saline
- PHH
primary human hepatocytes
- PI
propidium iodide
- PMH
primary mouse hepatocytes
- TAMH
TGF-alpha transgenic mouse hepatocytes
Footnotes
CONFLICT OF INTEREST DISCLOSURE
The authors declare no competing financial interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abdelmegeed MA, Jang S, Banerjee A, Hardwick JP, Song BJ. Robust protein nitration contributes to acetaminophen-induced mitochondrial dysfunction and acute liver injury. Free Radic Biol Med. 2012;60:211–22. doi: 10.1016/j.freeradbiomed.2013.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajt ML, Lawson JA, Vonderfecht SL, Gujral JS, Jaeschke H. Protection against Fas receptor-mediated apoptosis in hepatocytes and nonparenchymal cells by a caspase-8 inhibitor in vivo: evidence for a postmitochondrial processing of caspase-8. Toxicol Sci. 2000;58:109–17. doi: 10.1093/toxsci/58.1.109. [DOI] [PubMed] [Google Scholar]
- Bajt ML, Cover C, Lemasters JJ, Jaeschke H. Nuclear translocation of endonuclease G and apoptosis-inducing factor during acetaminophen-induced liver cell injury. Toxicol Sci. 2006;94:217–25. doi: 10.1093/toxsci/kfl077. [DOI] [PubMed] [Google Scholar]
- Bajt ML, Farhood A, Lemasters JJ, Jaeschke H. Mitochondrial bax translocation accelerates DNA fragmentation and cell necrosis in a murine model of acetaminophen hepatotoxicity. J Pharmacol Exp Ther. 2008;324:8–14. doi: 10.1124/jpet.107.129445. [DOI] [PubMed] [Google Scholar]
- Bajt ML, Knight TR, Lemasters JJ, Jaeschke H. Acetaminophen-induced oxidant stress and cell injury in cultured mouse hepatocytes: protection by N-acetyl cysteine. Toxicol Sci. 2004;80:343–9. doi: 10.1093/toxsci/kfh151. [DOI] [PubMed] [Google Scholar]
- Baker JA, Hayden J, Marshall PG, Palmer CH, Whittet TD. Some antipyretics related to aspirin and phenacetin. J Pharm Pharmacol Suppl. 1963:97–100. doi: 10.1111/j.2042-7158.1963.tb11193.x. [DOI] [PubMed] [Google Scholar]
- Birge RB, Bartolone JB, McCann DJ, Mangold JB, Cohen SD, Khairallah EA. Selective protein arylation by acetaminophen and 2,6-dimethylacetaminophen in cultured hepatocytes from phenobarbital-induced and uninduced mice. Relationship to cytotoxicity. Biochem Pharmacol. 1989;38:4429–38. doi: 10.1016/0006-2952(89)90653-9. [DOI] [PubMed] [Google Scholar]
- Cover C, Mansouri A, Knight TR, Bajt ML, Lemasters JJ, Pessayre D, Jaeschke H. Peroxynitrite-induced mitochondrial and endonuclease-mediated nuclear DNA damage in acetaminophen hepatotoxicity. J Pharmacol Exp Ther. 2005;315:879–87. doi: 10.1124/jpet.105.088898. [DOI] [PubMed] [Google Scholar]
- Du K, Williams CD, McGill MR, Xie Y, Farhood A, Vinken M, Jaeschke H. The gap junction inhibitor 2-aminoethoxy-diphenyl-borate protects against acetaminophen hepatotoxicity by inhibiting cytochrome P450 enzymes and c-jun N-terminal kinase activation. Toxicol Appl Pharmacol. 2013;273:484–91. doi: 10.1016/j.taap.2013.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fountoulakis M, Berndt P, Boelsterli UA, Crameri F, Winter M, Albertini S, Suter L. Two-dimensional database of mouse liver proteins: changes in hepatic protein levels following treatment with acetaminophen or its nontoxic regioisomer 3-acetamidophenol. Electrophoresis. 2000;21:2148–61. doi: 10.1002/1522-2683(20000601)21:11<2148::AID-ELPS2148>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- Gujral JS, Knight TR, Farhood A, Bajt ML, Jaeschke H. Mode of cell death after acetaminophen overdose in mice: apoptosis or oncotic necrosis? Toxicol Sci. 2002;67:322–8. doi: 10.1093/toxsci/67.2.322. [DOI] [PubMed] [Google Scholar]
- Hadi M, Dragovic S, van Swelm R, Herpers B, van de Water B, Russel FG, Commandeur JN, Groothuis GM. AMAP, the alleged non-toxic isomer of acetaminophen, is toxic in rat and human liver. Arch Toxicol. 2013;87:155–65. doi: 10.1007/s00204-012-0924-1. [DOI] [PubMed] [Google Scholar]
- Halmes NC, Samokyszyn VM, Hinton TW, Hinson JA, Pumford NR. The acetaminophen regioisomer 3′-hydroxyacetanilide inhibits and covalently binds to cytochrome P450 2E1. Toxicol Lett. 1998;94:65–71. doi: 10.1016/s0378-4274(97)00100-8. [DOI] [PubMed] [Google Scholar]
- Hanawa N, Shinohara M, Saberi B, Gaarde WA, Han D, Kaplowitz N. Role of JNK translocation to mitochondria leading to inhibition of mitochondria bioenergetics in acetaminophen-induced liver injury. J Biol Chem. 2008;283:13565–77. doi: 10.1074/jbc.M708916200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holme JA, Hongslo JK, Bjørge C, Nelson SD. Comparative cytotoxic effects of acetaminophen (N-acetyl-p-aminophenol), a non-hepatotoxic regioisomer acetyl-m-aminophenol and their postulated reactive hydroquinone and quinone metabolites in monolayer cultures of mouse hepatocytes. Biochem Pharmacol. 1991;42:1137–42. doi: 10.1016/0006-2952(91)90299-k. [DOI] [PubMed] [Google Scholar]
- Jaeschke H. Glutathione disulfide formation and oxidant stress during acetaminophen-induced hepatotoxicity in mice in vivo: the protective effect of allopurinol. J Pharmacol Exp Ther. 1990;255:935–41. [PubMed] [Google Scholar]
- Jaeschke H, Mitchell JR. Use of isolated perfused organs in hypoxia and ischemia/reperfusion oxidant stress. Methods Enzymol. 1990;186:752–759. doi: 10.1016/0076-6879(90)86175-u. [DOI] [PubMed] [Google Scholar]
- Jaeschke H, McGill MR, Ramachandran A. Oxidant stress, mitochondria, and cell death mechanisms in drug-induced liver injury: lessons learned from acetaminophen hepatotoxicity. Drug Metab Rev. 2012;44:88–106. doi: 10.3109/03602532.2011.602688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaeschke H, Williams CD, McGill MR, Xie Y, Ramachandran A. Models of drug-induced liver injury for evaluation of phytotherapeutics and other natural products. Food Chem Toxicol. 2013;55:279–89. doi: 10.1016/j.fct.2012.12.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenna JG. A new twist to an old tale: novel insights into the differential toxicities of acetaminophen and its regioisomer N-acetyl-meta-aminophenol (AMAP) Arch Toxicol. 2013;87:15–8. doi: 10.1007/s00204-012-0945-9. [DOI] [PubMed] [Google Scholar]
- Knight TR, Kurtz A, Bajt ML, Hinson JA, Jaeschke H. Vascular and hepatocellular peroxynitrite formation during acetaminophen toxicity: role of mitochondrial oxidant stress. Toxicol Sci. 2001;62:212–20. doi: 10.1093/toxsci/62.2.212. [DOI] [PubMed] [Google Scholar]
- Kon K, Kim JS, Jaeschke H, Lemasters JJ. Mitochondrial permeability transition in acetaminophen-induced necrosis and apoptosis of cultured mouse hepatocytes. Hepatology. 2004;40:1170–9. doi: 10.1002/hep.20437. [DOI] [PubMed] [Google Scholar]
- Lee WM. Acute liver failure. Semin Respir Crit Care Med. 2012;33:36–45. doi: 10.1055/s-0032-1301733. [DOI] [PubMed] [Google Scholar]
- LoGuidice A, Boelsterli UA. Acetaminophen overdose-induced liver injury in mice is mediated by peroxynitrite independently of the cyclophilin D-regulated permeability transition. Hepatology. 2011;54:969–78. doi: 10.1002/hep.24464. [DOI] [PubMed] [Google Scholar]
- McGill MR, Lebofsky M, Norris HR, Slawson MH, Bajt ML, Xie Y, Williams CD, Wilkins DG, Rollins DE, Jaeschke H. Plasma and liver acetaminophen-protein adduct levels in mice after acetaminophen treatment: dose-response, mechanisms, and clinical implications. Toxicol Appl Pharmacol. 2013;269:240–9. doi: 10.1016/j.taap.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Sharpe MR, Williams CD, Taha M, Curry SC, Jaeschke H. The mechanism underlying acetaminophen-induced hepatotoxicity in humans and mice involves mitochondrial damage and nuclear DNA fragmentation. J Clin Invest. 2012a;122:1574–83. doi: 10.1172/JCI59755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Yan HM, Ramachandran A, Murray GJ, Rollins DE, Jaeschke H. HepaRG cells: a human model to study mechanisms of acetaminophen hepatotoxicity. Hepatology. 2011;53:974–82. doi: 10.1002/hep.24132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGill MR, Williams CD, Xie Y, Ramachandran A, Jaeschke H. Acetaminophen-induced liver injury in rats and mice: comparison of protein adducts, mitochondrial dysfunction, and oxidative stress in the mechanism of toxicity. Toxicol Appl Pharmacol. 2012b;264:387–94. doi: 10.1016/j.taap.2012.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews AM, Hinson JA, Roberts DW, Pumford NR. Comparison of covalent binding of acetaminophen and the regioisomer 3′-hydroxyacetanilide to mouse liver protein. Toxicol Lett. 1997;90:77–82. doi: 10.1016/s0378-4274(96)03831-3. [DOI] [PubMed] [Google Scholar]
- Meyers LL, Beierschmitt WP, Khairallah EA, Cohen SD. Acetaminophen-induced inhibition of hepatic mitochondrial respiration in mice. Toxicol Appl Pharmacol. 1988;36:1193–6. doi: 10.1016/0041-008x(88)90040-3. [DOI] [PubMed] [Google Scholar]
- Myers TG, Dietz EC, Anderson NL, Khairallah EA, Cohen SD, Nelson SD. A comparative study of mouse liver proteins arylated by reactive metabolites of acetaminophen and its nonhepatotoxic regioisomer, 3′-hydroxyacetanilide. Chem Res Toxicol. 1995;8:403–13. doi: 10.1021/tx00045a012. [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Maeda S, Hikiba Y, Ohmae T, Shibata W, Yanai A, Sakamoto K, Ogura K, Noguchi T, Karin M, Ichijo H, Omata M. Deletion of apoptosis signal-regulating kinase 1 attenuates acetaminophen-induced liver injury by inhibiting c-Jun N-terminal kinase activation. Gastroenterology. 2008;135:1311–21. doi: 10.1053/j.gastro.2008.07.006. [DOI] [PubMed] [Google Scholar]
- Nelson EB. The pharmacology and toxicology of meta-substituted acetanilide I: acute toxicity of 3-hydroxyacetanilide in mice. Res Commun Chem Pathol Pharmacol. 1980;28:447–56. [PubMed] [Google Scholar]
- Pierce RH, Franklin CC, Campbell JS, Tonge RP, Chen W, Fausto N, Nelson SD, Bruschi SA. Cell culture model for acetaminophen-induced hepatocyte death in vivo. Biochem Pharmacol. 2002;64:413–24. doi: 10.1016/s0006-2952(02)01180-2. [DOI] [PubMed] [Google Scholar]
- Qiu Y, Benet LZ, Burlingame AL. Identification of hepatic protein targets of the reactive metabolites of the non-hepatotoxic regioisomer of acetaminophen, 3′-hydroxyacetanilide, in the mouse in vivo using two-dimensional gel electrophoresis and mass spectrometry. Adv Exp Med Biol. 2001;500:663–73. doi: 10.1007/978-1-4615-0667-6_99. [DOI] [PubMed] [Google Scholar]
- Ramachandran A, Lebofsky M, Baines CP, Lemasters JJ, Jaeschke H. Cyclophilin D deficiency protects against acetaminophen-induced oxidant stress and liver injury. Free Radic Res. 2011;45:156–64. doi: 10.3109/10715762.2010.520319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rashed MS, Myers TG, Nelson SD. Hepatic protein arylation, glutathione depletion, and metabolite profiles of acetaminophen and a non-hepatotoxic regioisomer, 3′-hydroxyacetanilide, in the mouse. Drug Metab Dispos. 1990;18:765–70. [PubMed] [Google Scholar]
- Rashed MS, Nelson SD. Characterization of glutathione conjugates of reactive metabolites of 3′-hydroxyacetanilide, a nonhepatotoxic positional isomer of acetaminophen. Chem Res Toxicol. 1989;2:41–5. doi: 10.1021/tx00007a007. [DOI] [PubMed] [Google Scholar]
- Roberts SA, Price VF, Jollow DJ. Acetaminophen structure-toxicity studies: in vivo covalent binding of a nonhepatotoxic analog, 3-hydroxyacetanilide. Toxicol Appl Pharmacol. 1990;105:195–208. doi: 10.1016/0041-008x(90)90181-s. [DOI] [PubMed] [Google Scholar]
- Saberi B, Ybanez MD, Johnson HS, Gaarde WA, Han D, Kaplowitz N. Protein kinase C (PKC) participates in acetaminophen hepatotoxicity through c-jun-N-terminal kinase (JNK)-dependent and -independent signaling pathways. Hepatology. 2014;59:1543–54. doi: 10.1002/hep.26625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito C, Lemasters JJ, Jaeschke H. c-Jun N-terminal kinase modulates oxidant stress and peroxynitrite formation independent of inducible nitric oxide synthase in acetaminophen hepatotoxicity. Toxicol Appl Pharmacol. 2010;246:8–17. doi: 10.1016/j.taap.2010.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salminen WF, Jr, Roberts SM, Pumford NR, Hinson JA. Immunochemical comparison of 3′-hydroxyacetanilide and acetaminophen binding in mouse liver. Drug Metab Dispos. 1998;26:267–71. [PubMed] [Google Scholar]
- Sharma M, Gadang V, Jaeschke A. Critical role for mixed-lineage kinase 3 in acetaminophen-induced hepatotoxicity. Mol Pharmacol. 2012;82:1001–7. doi: 10.1124/mol.112.079863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara M, Ybanez MD, Win S, Than TA, Jain S, Gaarde WA, Han D, Kaplowitz N. Silencing glycogen synthase kinase-3beta inhibits acetaminophen hepatotoxicity and attenuates JNK activation and loss of glutamate cysteine ligase and myeloid cell leukemia sequence 1. J Biol Chem. 2010;285:8244–55. doi: 10.1074/jbc.M109.054999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stamper BD, Bammler TK, Beyer RP, Farin FM, Nelson SD. Differential regulation of mitogen-activated protein kinase pathways by acetaminophen and its nonhepatotoxic regioisomer 3′-hydroxyacetanilide in TAMH cells. Toxicol Sci. 2010;116:164–73. doi: 10.1093/toxsci/kfq100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Streeter AJ, Bjorge SM, Axworthy DB, Nelson SD, Baillie TA. The microsomal metabolism and site of covalent binding to protein of 3′-hydroxyacetanilide, a nonhepatotoxic positional isomer of acetaminophen. Drug Metab Dispos. 1984;12:565–76. [PubMed] [Google Scholar]
- Tirmenstein MA, Nelson SD. Subcellular binding and effects on calcium homeostasis produced by acetaminophen and a nonhepatotoxic regioisomer, 3′-hydroxyacetanilide, in mouse liver. J Biol Chem. 1989;264:9814–9. [PubMed] [Google Scholar]
- Tirmenstein MA, Nelson SD. Acetaminophen-induced oxidation of protein thiols. Contribution of impaired thiol-metabolizing enzymes and the breakdown of adenine nucleotides. J Biol Chem. 1990;265:3059–65. [PubMed] [Google Scholar]
- Tirmenstein MA, Nelson SD. Hepatotoxicity after 3′-hydroxyacetanilide administration to buthionine sulfoximine pretreated mice. Chem Res Toxicol. 1991;4:214–7. doi: 10.1021/tx00020a014. [DOI] [PubMed] [Google Scholar]
- Xie Y, Williams CD, McGill MR, Lebofsky M, Ramachandran A, Jaeschke H. Purinergic receptor antagonist A438079 protects against acetaminophen-induced liver injury by inhibiting p450 isoenzymes, not by inflammasome activation. Toxicol Sci. 2013;131:325–35. doi: 10.1093/toxsci/kfs283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, McGill MR, Dorko K, Kumer SC, Schmitt TM, Forster J, Jaeschke H. Mechanisms of acetaminophen-induced cell death in primary human hepatocytes. Toxicol Appl Pharmacol. 2014;279:266–74. doi: 10.1016/j.taap.2014.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y, Ramachandran A, Breckenridge DG, Liles JT, Lebofsky M, Farhood A, Jaeschke H. Inhibitor of apoptosis signal-regulating kinase 1 protects against acetaminophen-induced liver injury. Toxicol Appl Pharmacol. 2015;286:1–9. doi: 10.1016/j.taap.2015.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.