Abstract
Objectives
Specialized proresolving lipid mediators have emerged as powerful modulators of inflammation and activators of resolution. Animal models show significant benefits of specialized proresolving lipid mediators on survival and wound healing after major burn trauma. To date, no studies have investigated specialized proresolving lipid mediators and their relation to other lipid mediator pathways in humans after trauma. Here we determine if patients with poor outcomes after trauma have dysregulated lipid mediator pathways.
Design
We studied blood leukocyte expression of 18 genes critical to the synthesis, signaling, and metabolism of specialized proresolving lipid mediators and proinflammatory lipid mediators, and we correlated these expression patterns with clinical outcomes in trauma patients from the Inflammation and the Host Response to Injury study.
Setting
Seven U.S. medical trauma centers.
Subjects
Ninety-six patients enrolled in the Inflammation and Host Response to Injury study, after blunt trauma and unambiguously classified as having uncomplicated or complicated recoveries. Twenty-eight healthy volunteers were enrolled as controls.
Interventions
None.
Measurements and Main Results
Within each patient, the 18 genes of interest were used to calculate scores for distinct families of lipid mediators, including resolvins, lipoxins, prostaglandins, and leukotrienes, as well as leukotriene to resolvin score ratios. Scores were built using a simple weighting scheme, taking into consideration both dependent and independent activities of enzymes and receptors responsible for lipid mediator biosynthesis and function. Individually, ALOX12, PTGS2, PTGES, PTGDS, ALOX5AP, LTA4H, FPR2, PTGER2, LTB4R, HPGD, PTGR1, and CYP4F3 were expressed differentially over 28 days posttrauma between patients with uncomplicated and complicated recoveries (p < 0.05). When all genes were combined into scores, patients with uncomplicated recoveries had differential and higher resolvin scores (p < 0.001) and lower leukotriene scores (p < 0.001). A final combined ratio was calculated for each patient, and posttrauma leukotriene score to resolvin score ratios were significantly lower in patients with uncomplicated clinical courses (p < 0.001).
Conclusions
Trauma patients with uncomplicated recoveries had higher resolvin pathway gene expression and lower gene expression ratios of leukotriene:resolvin pathways. Further validation of these findings with more complex modeling including measures with specialized proresolving lipid mediator lipidomics and/or protein expression, and identifying associated therapeutic targets, may influence the clinical management of trauma patients.
Keywords: gene expression, lipid mediators, outcomes, resolvins, systemic inflammatory response syndrome, trauma
Severe trauma alters the expression of more than 80% of the leukocyte transcriptome during the first 28 days after injury, a response referred to as a “genomic storm” (1). Unlike small injuries that trigger a well-regulated inflammatory cascade to contain infection and stimulate tissue repair, sepsis, major injury, and critical illness trigger exaggerated innate immune responses, leading to multiple organ dysfunction beyond the initial time of injury.
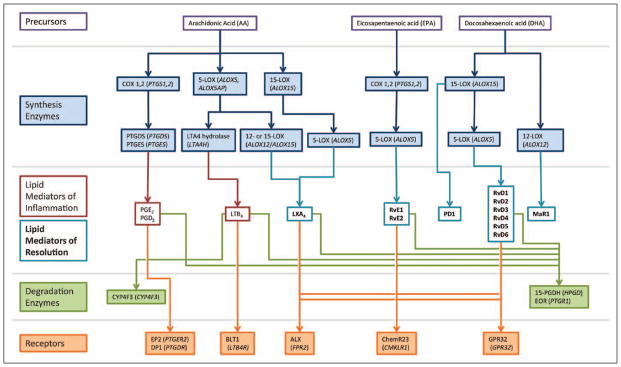
In addition to cytokines and growth factors commonly studied in the context of innate immunity, specialized proresolving lipid mediators (SPM) have emerged as critical for the control of inflammation and its resolution (2). Proinflammatory lipid mediators arise from enzymatic metabolism of arachidonic acid by cyclooxygenases (COX) and 5-lipoxygenase (5-LOX) and include the prostaglandins (PG), which trigger vasodilation, and leukotrienes (LT), which serve in leukocyte adhesion and chemotaxis (Fig. 1). Their counterparts, SPM—including lipoxins, resolvins (Rv), protectins, and maresins—are biosynthesized predominantly via 12- and 15-LOX initiated pathways from arachidonic acid and omega-3 fatty acids (e.g., docosahexaenoic acid [DHA] and eicosapentaenoic acid [EPA]). SPMs thwart inflammation by inhibiting neutrophil chemotaxis and stimulate resolution through the recruitment and differentiation of macrophages to clear cell debris and apoptotic neutrophils from inflamed tissue. The balance between these separate families of lipid mediators contributes to the persistence or resolution of inflammation and therefore has potential implications for wound healing, infection control, and organ failure.
Figure 1.
Schematics of lipid mediator biosynthesis, degradation, and receptor activation. The lipid precursors (arachidonic acid, eicosapentaenoic acid, and docosahexaenoic acid) and active lipid mediators are presented in the open boxes. The enzymes involved in synthesis are presented in blue, with their corresponding genes in parenthesis. Degradation enzymes and receptors are presented in green and orange, respectively, again with corresponding genes in parenthesis. Note that distinct lipid mediators share several of enzymes and receptors. ALX = lipoxin A4 receptor, BLT1 = leukotriene B4 receptor 1, COX = cyclooxygenase, DP = prostaglandin D2 receptor 1, EOR = eicosanoid oxidoreductase, EP2 = prostaglandin E2 receptor 2, LOX = lipoxygenase, LT = leukotriene, LX = lipoxin, MaR = maresin, PD = protectin, PG = prostaglandin, PTGDS = prostaglandin D2 synthase, PTGES = prostaglandin E2 synthase, Rv = resolvin.
SPMs have been well-studied in models of localized inflammation (2–10) and wound healing (11–13) and in animal models of sepsis (14, 15). Few studies, however, have explored their role in the systemic inflammatory response to severe trauma in humans. Supporting the importance of SPM in the immune responses after injury, RvD2 at nanogram doses dramatically increases survival in both a rat model of large burn injury and endotoxin shock and a mouse model of sepsis (14– 16). Elucidating the role of counterregulatory lipid mediator pathways in the inflammatory response after traumatic injury in humans has potential ramifications for organ support and clinical recovery.
We hypothesized that patients with worse outcomes after trauma have dysregulated systemic lipid mediator pathways, particularly, up-regulated proinflammatory lipid mediator pathways, and down-regulated proresolving lipid mediator pathways. To test our hypothesis, we studied the expression of genes critical to the synthesis, signaling, and further metabolism of SPMs and proinflammatory lipid mediators within blood leukocytes. These expression patterns were correlated with clinical outcomes of trauma patients from the Inflammation and the Host Response to Injury study.
MATERIALS AND METHODS
Gene Expression Database for Trauma Patients
Inflammation and the Host Response to Injury is a large study that has collected and analyzed patient blood samples and clinical data with the aim to enhance understanding of serious injury. Patients were 16–55 years old and treated for severe traumatic injuries at one of seven U.S. hospitals. Inclusion criteria for the Inflammation and the Host Response to Injury research database were designed to capture trauma patients at highest risk of physiologic derangement, multiple organ failure, and death. These inclusion criteria included the following: age 16 years or older, suffered blunt trauma, arrival to hospital within 6 hours of injury, shock on presentation (systolic blood pressure less than 90 or base deficit of at least 6), blood transfusion within 12 hours of injury, and body region Abbreviated Injury Scale (AIS) score at least 2 (exclusive of brain) (1, 17). Of note, patients with Glasgow Coma Scale up to 8, with head injury AIS at least 2, and with cervical spine injuries were excluded from the database, to avoid confounding data with the effects of neurogenic shock.
Enrolled patients were managed according to standard procedures adopted and audited across all participating centers to minimize variation in posttrauma treatment, including early goal-directed resuscitation, strict glycemic control, venous thromboembolism prophylaxis, appropriate tidal volume ventilation, ventilator-associated pneumonia management, use of sedation and analgesic, antimicrobial therapy, and restrictive transfusion guidelines (1, 17).
Of 1,637 patients enrolled in the study, 167 consented to blood sampling which was used for full genomic profiling using the Affymetrix platform and buffy coat (1). Patients in this cohort were divided into uncomplicated, complicated, and intermediate groups based on their clinical outcomes (see Definitions of Complicated and Uncomplicated Recovery section). To investigate genomic changes at the extremes of clinical recovery, we analyzed the expression levels of 18 genes in the transcriptome data from the uncomplicated (n = 55) and complicated (n = 41) patients (96 total), for which the genome-wide expression as well as the clinical course of recovery has been carefully mapped (1). In addition, we measured gene expression in 28 healthy controls.
Blood was sampled within 12 hours of injury (day 0) and at 1, 4, 7, 14, 21, and 28 day(s) after injury. Genome-wide expression analysis of whole leukocyte buffy coat was performed using the Affymetrix U133plus2 GeneChip (1). The clinical and laboratory data of the patients are summarized in Table 1. To manage the variability of the actual days of sample collection, we defined nominal days based on time intervals posttrauma, as shown in Table 2. Expression values in Figure 2 were normalized to healthy controls. Nonnormalized expression values are shown in Supplementary Table 2 (Supplemental Digital Content 1, http://links.lww.com/CCM/B436). The study was approved by the institutional review board of each institution. In addition to local institutional oversight, Massachusetts General Hospital reviewed and approved the program’s data center and databases.
TABLE 1.
Patient Characteristics and Clinical Outcomes
| Variables | Uncomplicated Recovery (< 5 D; n = 55) | Complicated Recovery (≥ 14 D, No Recovery by 28 D, or Death; n = 41) |
|---|---|---|
| Demographics | ||
| Age, yr | 32.5±11.2 | 34.2±11.1 |
| Sex, male (n) | 54.5% (30) | 73.1% (30) |
| Race, white (n) | 85.45% (47) | 90.2% (37) |
|
| ||
| Acute care | ||
| Acute Physiology and Chronic Health Evaluation II | 24.4±6.0 | 29.4±5.3a |
| Injury Severity Score | 26.2±13.2 | 35.7±12.6a |
| Total blood administered within the first 12 hr, L | 1.7±1.3 | 3.0±2.7a |
| Total crystalloid within the first 12 hr, L | 10.6±5.7 | 15.2±9.8a |
| Worst base deficit | −9.5±4.1 | −11.3±4.7 |
|
| ||
| Outcomes | ||
| Survival (n) | 100% (55) | 83% (34)a |
| Maximum modified Marshall score | 2.9±1.0 | 8.8±2.6a |
| Hospital length of stay, d | 14.4±12.9 | 35.5±22.9a |
| ICU length of stay, d | 4.8±2.9 | 25.1±14.6a |
|
| ||
| Complications | ||
| Noninfectious complications (n) | 5.5% (3) | 90.2% (37)a |
| Nosocomial infections (n) | 20.0% (11) | 85.4% (35)a |
| Ventilator-associated pneumonia (n) | 1.8% (1) | 63.4% (26)a |
p < 0.01 complicated vs uncomplicated recovery group by Student t test for continuous variables and chi-square or Fisher exact test for categorical variables.
Values represent the mean ± SD or % and number (n) from total n as appropriate.
TABLE 2.
Nominal Days and Patient Follow-Up
| Posttrauma Day | Nominal Day | No. of Patients | |
|---|---|---|---|
| Uncomplicated | Complicated | ||
| 0 | 0 | 55 | 41 |
| 1, 2 | 1 | 51 | 40 |
| 3, 4, 5 | 4 | 41 | 37 |
| 6, 7, 8, 9, 10 | 7 | 33 | 35 |
| 11, 12, 13, 14, 15, 16, 17 | 14 | 10 | 30 |
| 18, 19, 20, 21, 22, 23, 24 | 21 | 5 | 26 |
| 25, 26, 27, 28, 29, 30, 31 | 28 | 2 | 20 |
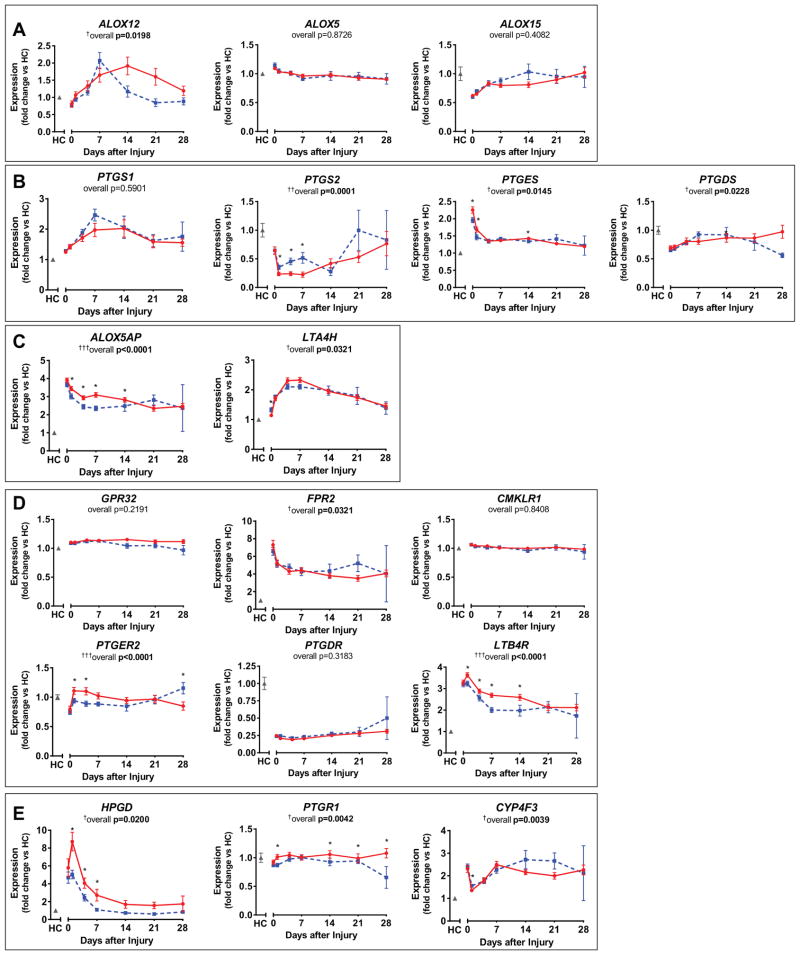
Figure 2.
Individual lipid mediator pathway genes showed minor differences between patients with uncomplicated (blue dashed line) and complicated (red solid line) recovery from trauma, including resolvin synthesis enzymes ALOX12, ALOX5, and ALOX15 (A); prostaglandin synthesis enzymes PTGS1 (cyclooxygenase, COX-1), PTGS2 (COX-2), PTGES, and PTGDS (B); leukotriene synthesis enzymes ALOX5AP and LTA4H (C); lipid mediator receptors FPR2 (ALX/FPR2), GPR32, CMKLR1 (ChemR23), PTGER2, PTGDR, and LTB4R (D); and degradation enzymes HPGD (15-prostaglandin dehydrogenase, 15-PGDH), PTGR1 (eicosanoid oxidoreductase, EOR), and CYP4F3 (E). Expression is normalized to healthy controls (HC; gray), nonnormalized values are listed in Supplementary Table 2 (Supplemental Digital Content 1, http://links.lww.com/CCM/B436). Plotted are mean and SEM. Overall (longitudinal) F test for complicated versus uncomplicated groups given under each gene title: †p < 0.05, ††p < 0.001, †††p < 0.0001. Significant differences between complicated and uncomplicated groups at specific days denoted by *p < 0.05.
Definitions of Complicated and Uncomplicated Recovery
We compared gene expression patterns among patients at the extremes of clinical recovery as has been done previously (1), by categorizing patients according to their length of recovery, according to the following criteria:
Uncomplicated (n = 55): Recovery within less than 5 days; maximum modified Marshall score less than 6.
Complicated (n = 41): Recovery within at least 14 days, no recovery by 28 days, or death; maximum modified Marshall score more than 6.
Patients who did not meet criteria for either uncomplicated or complicated recovery were classified as intermediate recovery and were not included in this analysis (n = 71). This exclusion allowed for a comparison of two unambiguously defined cohorts of patients.
Survival, hospital and ICU lengths of stay, infectious and noninfectious complications, and maximum modified multiple organ dysfunction (Marshall) scores (18) were followed for all patients and compared between the uncomplicated and complicated recovery groups.
Genes of Interest
We analyzed blood leukocyte gene expression levels for 18 genes coding for enzymes involved in the synthesis and degradation of lipid mediators, as well as for their leukocyte receptors. These 18 genes included all currently identified genes of importance in lipid mediator pathways (Fig. 1; and Supplementary Table 1, Supplemental Digital Content 1, http://links.lww.com/CCM/B436). Enzymes and receptors in any given lipid mediator pathway have synergistic activities; thus, we created scores to evaluate groups of genes controlling separate lipid mediator pathways. These scores do not attempt to capture actual measures of pathway activity but rather serve as crude estimators of pathway activity useful for comparative analyses. The interactions between these genes are scored in the form of logical operators, where activities of coded entities (i.e., genes of specific enzymes or receptors) use AND (i.e., multiplication) clauses for dependent and OR (i.e., addition) clauses for independent activities. For example, to define a resolvin score, we first included genes pivotal in the synthesis of resolvins, ALOX15 and ALOX5 (Fig. 1). These enzymes work together in a dependent fashion (AND) for resolvin pathway activation, with further dependence (AND) on known G-protein-coupled receptors of resolvins, FPR2, GPR32, and CMKLR1, which themselves function independently (OR) of each other in resolvin signaling. To attribute the degradation of resolvins to specific enzymes, we included the expression of HPDG and LTB4DH, genes coding for two enzymes that work in parallel, independent (OR) of each other. The final score was calculated by dividing the activities of synthesis and signaling entities by the activities of degradation entities: resolvin score = (ALOX5 × ALOX15 × (FPR2 + GPR32 + CMKLR1))/(HPGD + PTGR1). These same principles were applied in the creation of scores for lipoxin, prostaglandin, and leukotriene pathways, as they appear below.
Statistical Methods
For clinical data, univariate analysis was performed to compare characteristics between groups using the Student t test for continuous variables and chi-square or Fisher exact test for categorical variables. Comparisons of gene expression between patients with complicated and uncomplicated recoveries were assessed with repeated-measures analysis of variance with a compound symmetric within patient covariance matrix and empirical (sandwich estimator) SES. Gene scores were divided by 107 prior to analysis in order to obtain numerical stability in SAS PROC MIXED. An overall F test for any difference between groups over time and a test for a between-group difference at each day were included in the analysis.
RESULTS
Patient Outcomes
Patients with uncomplicated and complicated recoveries did not differ significantly in age or sex (Table 1). The healthy controls matched the age, sex, and race distributions of patients (~54% men, aged 28.7 ± 9.4 yr, ~89% Caucasian). When comparing clinical variables between cohorts, patients in the complicated recovery group had statistically significantly higher Acute Physiology and Chronic Health Evaluation (APACHE) II scores, Injury Severity Scores, volume of crystalloid resuscitation, and blood transfusions in the first 12 hours. There was no difference in base deficit between groups. Patients with complicated recoveries had higher mortality rates, as well as longer hospital and ICU lengths of stay and higher modified multiple organ dysfunction (Marshall) scores. Complicated trauma patients also had statistically significantly higher rates of ventilator-associated pneumonia and other nosocomial infections, as well as a greater occurrence rate of noninfectious complications (Table 1).
Control Versus Trauma Patient Data
In the majority of cases, traumatic injury induced a statistically significant change in lipid mediator pathway gene expression when compared with healthy controls. Patients had significantly lower expression levels of ALOX12, ALOX15, PTGS2, PTGDS, PTGER2, and PTGDR immediately after injury (day 0) compared with controls (data not shown), indicating a potential initial down-regulation of prostaglandin synthesis (Fig. 1). Levels of ALOX5, PTGS1, PTGES, ALOX5AP, LTA4H, GPR32, FPR2, CMKLR1, LTB4R, HPGD, and CYP4F3, meanwhile, were significantly increased compared with controls at immediately postinjury (day 0; data not shown). The only gene in for which there was not a statistically significant change early after trauma was PTGR1. As expected, our patient cohort had neutrophil and total WBC counts higher than normal ranges for healthy individuals (data not shown).
Association Between Individual Gene Expression and Clinical Outcomes
Resolvin Biosynthesis Enzymes
Proresolving lipid mediators are biosynthesized by the sequential actions of 15-LOX (ALOX15), 12-LOX (ALOX12), and 5-LOX (ALOX5) on DHA and arachidonic acid (Fig. 1). Of these genes, only the overall course of leukocyte ALOX12 expression was different between patients with uncomplicated and complicated recoveries (overall F test p = 0.0198), although expression on any single day was not significantly different between the groups (Fig. 2A).
Prostaglandin Biosynthesis Enzymes
Prostanoids PGE2 and PGD2 are both proinflammatory and proresolving in self-limited inflammatory responses. They stimulate vascular leakage that allows leukocytes to transmigrate into tissues but also act as critical triggers of lipid mediator class switching (19). PGE2 and PGD2 are synthesized by the sequential actions of COX-1 (PTGS1) or COX-2 (PTGS2) followed by PGE2 synthase (PTGES) for PGE2 or PGD2 synthase (PTGDS) for PGD2 (Fig. 1). COX-1 and COX-2 are also involved in the first step of E-series resolvin synthesis. The overall course of expression of PTGS2, PTGES, and PTGDS were altered between patients with uncomplicated and complicated recoveries (Fig. 2B). PTGS2 expression was higher in leukocytes of patients with uncomplicated recoveries and closer to that of healthy controls in the first week postinjury, compared with patients with complicated recoveries. PTGES expression, on the other hand, was slightly higher initially following injury (days 0 and 1) in patients who experienced complicated recoveries. Total WBC, neutrophil, and monocyte counts were not significantly different between our patient cohorts over this same time period (data not shown).
Leukotriene Biosynthesis Enzymes
The potent chemoattractant and proinflammatory mediator LTB4 is synthesized from arachidonic acid by 5-LOX (ALOX5) acting with 5-LOX activating protein (ALOX5AP), followed by LTA4 hydrolase (LTA4H) (Fig. 1). Expression of both ALOX5AP and LTA4H were significantly different over time between the two patient cohorts (p < 0.05) (Fig. 2C). ALOX5AP expression was lower at days 1–14 posttrauma in patients with uncomplicated compared with complicated clinical outcomes, with the largest difference at day 7 (4,964 ± 249 vs 7,008 ± 272; p < 0.0001).
Lipid Mediator Receptors
D-series resolvin receptor GPR32 (GPR32) (specifically, for RvD1, RvD3, and RvD5) and RvE1 receptor ChemR23 (CMKLR1) gene expression were not different overall or at any specific day posttrauma between the two patient cohorts (Fig. 2D). In contrast, lipoxin receptor ALX/FPR2 (FPR2) gene expression was significantly different posttrauma between the two groups (overall F test p = 0.03); however, there were no differences at any particular day (Fig. 2D). Regarding prostaglandin and leukotriene receptors, both the LTB4 receptor LTB4R (LTB4R) and the PGE2 receptor EP2 (PTGER2) gene expression were highly significantly different between patients cohorts (Fig. 2D). Both LTB4R and PTGER2 levels were lower in leukocytes of patients with uncomplicated outcomes on several days (Fig. 2D). LTB4R levels reached the largest differences at day 7 between uncomplicated and complicated patients (603 ± 35 vs 836 ± 31; p < 0.0001). PTGER2 expression in patients with uncomplicated recoveries was significantly lower by day 4 compared with patients with complicated recoveries (429 ± 21 vs 510 ± 28; p = 0.02). Comparisons at days 21 and 28 should be interpreted with caution, as many patients were lost to follow up. Only five and two uncomplicated recovery patients, respectively, participated in analyses at 21 and 28 days (Table 2).
Lipid Mediator Further Metabolism/Inactivation Enzymes
Enzymes that convert lipid mediators to their mostly less-active further metabolites include 15-prostaglandin dehydrogenase (HPGD) and eicosanoid oxidoreductase (PTGR1), which often act sequentially. The CYP450 enzyme, CYP4F3 (CYP4F3), is primarily involved in degradation of LT. Gene expression of all three of these further metabolism enzymes was significantly different postinjury between the two cohorts (Fig. 2E). Both HPGD and PRGR1 expression was lower in patients with uncomplicated reveries at several sample days between days 1 and 28, indicating a potential lower rate of conversion of lipid mediators. The greatest differential in expression of a single gene, between patient cohorts, was observed in the case of HPGD at day 1, which was expressed at lower levels in patients with uncomplicated recoveries (301 ± 28 vs 516 ± 61; p = 0.001). However, the overall course of HPGD expression was only weakly associated with outcome (p = 0.02) (Fig. 2E).
Association Between Pathway Scores and Clinical Outcomes
Given that lipid mediator levels and bioactivity are the result of multiple components working in concert, we next developed equations that reflected the multigene nature of each lipid mediator pathway. We hypothesized that scores considering the formation, signaling, and degradation of lipid mediators would more accurately reflect inflammatory changes and their influence on clinical outcome than would expression measurements of isolated genes.
For D-series resolvins, the following equation was developed:
This algorithm takes into consideration the sequential and dependent nature of 5-LOX and 15-LOX enzymes in the production of resolvins, as well as their known receptors which function independently of each other, and enzymes that can independently convert resolvins to inactive further metabolites (20).
Using the same principles, the following equations were developed for lipoxins, PG, and LT:
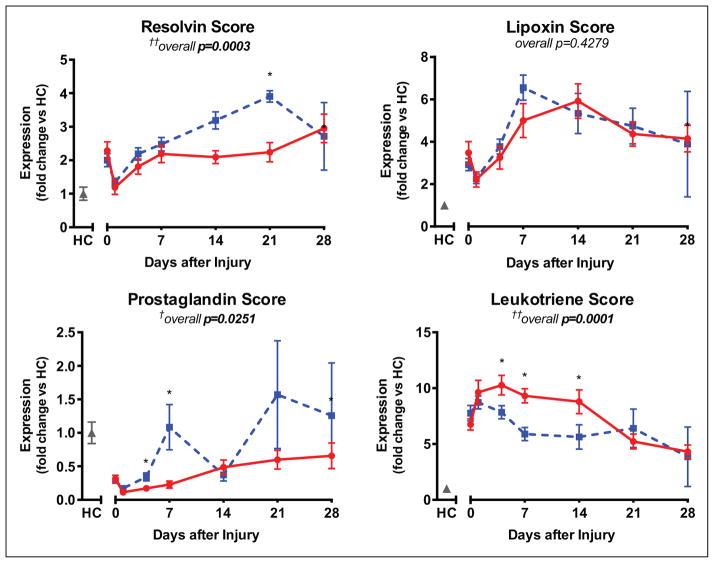
Patients with worse clinical outcomes had overall significantly lower resolvin scores over the time course of recovery (p = 0.0003). For individual days, there were significantly higher resolvin scores at day 21 posttrauma in uncomplicated patients compared with patients with complicated recoveries (7.23 ± 0.41 × 10−3 vs 5.08 ± 0.55 × 10−3; p = 0.002) (Fig. 3). Similarly, the prostaglandin score was different between the two cohorts posttrauma (p = 0.03), with higher scores at days 4 and 7 posttrauma among uncomplicated patients (1.23 ± 0.20 vs 0.622 ± 0.149, p = 0.01 at day 4; 3.80 ± 1.16 vs 0.80 ± 0.17, p = 0.01 at day 7) (Fig. 3). The lipoxin score, meanwhile, was not significantly associated with outcomes posttrauma (Fig. 3). Finally, leukotriene scores over the course of recovery posttrauma were significantly associated with outcome (p = 0.0001). Leukotriene scores were lower between days 4 and 14 postinjury for patients with uncomplicated recoveries, with the most significant difference at day 7 (487 ± 45 vs 744 ± 51; p = 0.0002) (Fig. 3).
Figure 3.
Patients without complications (blue dashed line) have a higher resolvin and lower leukotriene scores than patients with complications (red solid line). Expression is normalized to healthy controls (HC; gray). Overall (longitudinal) F test for uncomplicated versus complicated groups given under each score title: †p < 0.05, ††p < 0.001. Significant differences between groups at specific days denoted by *p < 0.05.
Combined Lipid Mediator Score
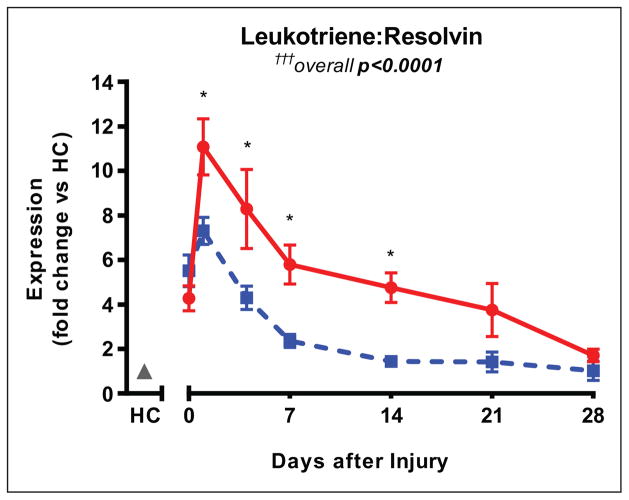
Considering the opposing effects of LT (proinflammatory) and resolvins (proresolving) during inflammation, we next questioned if a combined score comparing LT to resolvins would be most highly associated with clinical course posttrauma. To calculate this value, we divided the leukotriene score by the resolvin score for each patient at each time point. As with the pathway scores themselves, this combined score is a crude estimate of pathway activity that enables relative comparison of the two patient cohorts, rather than a measurement of pathway activity itself. This combined score takes into account that higher levels of proinflammatory lipid mediators and lower levels of proresolving lipid mediators often occur simultaneously in dysregulated inflammation. Posttrauma, the combined ratio of leukotriene to resolvin score was highly associated with clinical outcome (overall p < 0.0001) (Fig. 4). The leukotriene:resolvin score ratio was lower starting day 1 in patients with uncomplicated recoveries (3.39 ± 0.28 × 105 vs 5.08 ± 0.57 × 105; p = 0.008) and the difference increased by day 7 (1.26 ± 0.17 × 105 vs 2.66 ± 0.37 × 105; p = 0.0007) (Fig. 4). Scores were comparable by day 28, and in both uncomplicated and complicated patients, this ratio had decreased close to the values seen in healthy controls. These results indicate that a high patient score of leukotriene pathway to resolvin pathway gene expression— indicative of imbalance of inflammatory mediator activity weighted toward a proinflammatory state—is most powerfully associated with complicated recovery posttrauma.
Figure 4.
Leukotriene:resolvin gene expression ratio is lower in patients with uncomplicated recoveries (blue dashed line) compared with those with uncomplicated recoveries (red solid line). Changes in the ratio within blood leukocytes begin within 24 hr after trauma and persist for an extended period, up to 14 d in uncomplicated patients and more than 3 weeks in complicated patients. Expression is normalized to healthy controls (HC; gray). Overall (longitudinal) F test for uncomplicated versus complicated groups is †††p < 0.0001. Significant differences between groups at specific days denoted by *p < 0.05.
DISCUSSION
In this study, we demonstrate that lipid mediator pathway gene expression in blood leukocytes correlates with clinical outcomes after trauma. We found that the ratio of expression between endogenous proinflammatory (LT) and proresolving (resolvins) lipid mediator pathway genes was significantly higher in patients with prolonged, complicated recovery after blunt trauma. These results suggest that patients with complicated recoveries have dysregulated lipid mediator signaling, which can be detected as early as 24 hours postinjury. It is also possible that severe traumatic injury itself may contribute to this dysregulation of inflammatory mediators, suggested by the lower baseline Injury Severity Scores and APACHE II scores in the uncomplicated recovery group compared with the complicated recovery group. Changes in individual lipid mediator pathway genes were associated with clinical recovery, although to a lesser extent than for any of the conglomerate scores.
The use of Boolean logic and various weighting schemes has been established in the analysis of genomic data in the context of existing knowledge about biochemical pathways (21–23). Here, we applied these concepts in the context of lipid mediator pathways of both inflammation initiators (LT, prostaglandin) and proresolving (resolving, protectin) pathways. Our pathway scores capture crude estimates of pathway activity, which provide values useful for relative comparisons between cohorts.
The data from this study hint at a potential therapeutic role for SPM in protecting against posttraumatic multiple organ failure. Prior studies in other areas of critical illness support this theory. For example, randomized controlled trials suggest that administration of IV fish oil—which contains DHA and EPA, precursors to SPM—may decrease mortality and ventilator days in critically ill patients (24, 25). Evidence also suggests that statins and nonsteroidal anti-inflammatory drugs exert their benefits in part through increased production of SPM such as LXA4 (26). Immunomodulating therapies that restore the balance of lipid mediators, favoring proresolving SPM, may similarly exert a protective effect in trauma patients, limiting organ damage from a prolonged inflammatory state. Furthermore, in recent studies in animals, we found that survival improved significantly after administration of resolvins to burn-injured and septic rats (16).
Although our analyses reveal associations between time to recovery and lipid mediator pathway scores within the first week after trauma, scores from later assessment days should be interpreted with caution. Many patients in the uncomplicated recovery group were lost to follow up by days 21 and 28, reducing the statistical power for those observations, which may account for the lack of differences between cohorts at later time points. It remains an open question whether lipid mediator profiles return to homeostasis in all patients or not, a point that may be important to study in the context of long-term complications after trauma. A further drawback to our analyses is the absences of a positive control arm, that is, patients with systemic inflammatory response syndrome but no traumatic injury (e.g., septic shock). The fact that complicated patients had higher APACHE II scores raises the question of whether or not critical illness from nontraumatic causes is also associated with lipid mediator dysregulation and whether such scores as the APACHE II are predictive of lipid mediator dysfunction. Our analyses included gene expression of 18 currently known important enzymes and receptors in lipid mediator pathways, and these analyses could be significantly shifted by taking into consideration as-yet-unidentified genes of importance. Finally, future studies on purified cell populations of blood neutrophils and monocytes would help incorporate cell-cell interactions of lipid mediator biosynthesis into metabolism equations.
Gene expression analyses, particularly of multiple associated pathway genes, offer insight into pathway kinetics that cannot be ascertained through direct measurements. However, despite this strength, gene expression cannot precisely capture the interplay of lipid mediators in vivo; enzymes exert cumulative effects on substrates over time, and receptors may undergo amplification and recycling. Therefore, incorporating measurements of serum lipid mediator, enzyme activity, and protein expression levels, particularly receptor surface expression, would complement and further validate the present findings. Lipid mediators are present in blood and tissue at picomolar to nanomolar concentrations, usually have short half-lives, and require sophisticated technologies, for example, liquid chromatography mass spectrometry, for quantification, currently limiting their use in large clinical studies. The results attained via this study provide preliminary data that substantiate the need for further investigation in the face of these technical challenges. In the clinical arena, IV n-3 essential fatty acids (EPA and DHA) supplementation may offer a compelling avenue of research for trauma patients with significant physiologic derangement.
CONCLUSIONS
We found that trauma patients with complicated courses and worse clinical outcomes have higher expression ratios of leukotriene pathway genes to resolvin pathway genes. Future studies are warranted to confirm correlations between lipid mediator levels and clinical responses in trauma, in particular validating the current findings on the lipid mediator lipidomic level. If further laboratory work validates our findings, such research, complemented with clinical studies, could potential identify a therapeutic role of SPMs in regulating the excessive posttrauma inflammatory response, to the ultimate benefit of patients.
Supplementary Material
Acknowledgments
Dr. Orr received support from the Canadian Institutes of Health Research PDF (Post-Doctoral Fellowship—salary support for all research activities). Dr. Butler received support for article research from the National Institutes of Health (NIH). Her institution received grant support from the NIH. Dr. Hayden’s institution received grant support from the NIH (5P50GM021700-33). Dr. Tompkins received support for article research from the NIH. His institution received grant support from the NIH. Dr. Serhan served as a board member for the Inflammation Research Foundation (Scientific Advisory Board member), has stock (scientific founder Resolvyx Pharma—stock of unknown value), and received support for article research from the NIH (GM095467) and Canadian Medical Research Council. His institution received grant support from the NIH, has a patent with Resolvyx, and received royalties. Dr. Irimia received support for article research from the NIH (GM092804). His institution received grant support from the NIH.
Footnotes
Supplemental digital content is available for this article. Direct URL citations appear in the printed text and are provided in the HTML and PDF versions of this article on the journal’s website (http://journals.lww.com/ccmjournal).
For information regarding this article, dirimia@mgh.harvard.edu
References
- 1.Xiao W, Mindrinos MN, Seok J, et al. Inflammation and Host Response to Injury Large-Scale Collaborative Research Program: A genomic storm in critically injured humans. J Exp Med. 2011;208:2581–2590. doi: 10.1084/jem.20111354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Serhan CN. Pro-resolving lipid mediators are leads for resolution physiology. Nature. 2014;510:92–101. doi: 10.1038/nature13479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Serhan CN, Chiang N. Resolution phase lipid mediators of inflammation: Agonists of resolution. Curr Opin Pharmacol. 2013;13:632–640. doi: 10.1016/j.coph.2013.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwab JM, Chiang N, Arita M, et al. Resolvin E1 and protectin D1 activate inflammation-resolution programmes. Nature. 2007;447:869–874. doi: 10.1038/nature05877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Norling LV, Spite M, Yang R, et al. Cutting edge: Humanized nano-proresolving medicines mimic inflammation-resolution and enhance wound healing. J Immunol. 2011;186:5543–5547. doi: 10.4049/jimmunol.1003865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miyahara T, Runge S, Chatterjee A, et al. D-series resolvin attenuates vascular smooth muscle cell activation and neointimal hyperplasia following vascular injury. FASEB J. 2013;27:2220–2232. doi: 10.1096/fj.12-225615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy BD, Serhan CN. Resolution of acute inflammation in the lung. Annu Rev Physiol. 2014;76:467–492. doi: 10.1146/annurev-physiol-021113-170408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spite M, Clària J, Serhan CN. Resolvins, specialized proresolving lipid mediators, and their potential roles in metabolic diseases. Cell Metab. 2014;19:21–36. doi: 10.1016/j.cmet.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Serhan CN, Jain A, Marleau S, et al. Reduced inflammation and tissue damage in transgenic rabbits overexpressing 15-lipoxygenase and endogenous anti-inflammatory lipid mediators. J Immunol. 2003;171:6856–6865. doi: 10.4049/jimmunol.171.12.6856. [DOI] [PubMed] [Google Scholar]
- 10.Hsiao HM, Sapinoro RE, Thatcher TH, et al. A novel anti-inflammatory and pro-resolving role for resolvin D1 in acute cigarette smoke-induced lung inflammation. PLoS One. 2013;8:e58258. doi: 10.1371/journal.pone.0058258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hasturk H, Kantarci A, Goguet-Surmenian E, et al. Resolvin E1 regulates inflammation at the cellular and tissue level and restores tissue homeostasis in vivo. J Immunol. 2007;179:7021–7029. doi: 10.4049/jimmunol.179.10.7021. [DOI] [PubMed] [Google Scholar]
- 12.Wang SB, Hu KM, Seamon KJ, et al. Estrogen negatively regulates epithelial wound healing and protective lipid mediator circuits in the cornea. FASEB J. 2012;26:1506–1516. doi: 10.1096/fj.11-198036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang Y, Zhang MJ, Hellmann J, et al. Proresolution therapy for the treatment of delayed healing of diabetic wounds. Diabetes. 2013;62:618–627. doi: 10.2337/db12-0684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spite M, Norling LV, Summers L, et al. Resolvin D2 is a potent regulator of leukocytes and controls microbial sepsis. Nature. 2009;461:1287–1291. doi: 10.1038/nature08541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Walker J, Dichter E, Lacorte G, et al. Lipoxin a4 increases survival by decreasing systemic inflammation and bacterial load in sepsis. Shock. 2011;36:410–416. doi: 10.1097/SHK.0b013e31822798c1. [DOI] [PubMed] [Google Scholar]
- 16.Kurihara T, Jones CN, Yu YM, et al. Resolvin D2 restores neutrophil directionality and improves survival after burns. FASEB J. 2013;27:2270–2281. doi: 10.1096/fj.12-219519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cuschieri J, Bulger E, Schaeffer V, et al. Inflammation and the Host Response to Injury Collaborative Research Program: Early elevation in random plasma IL-6 after severe injury is associated with development of organ failure. Shock. 2010;34:346–351. doi: 10.1097/SHK.0b013e3181d8e687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marshall JC, Cook DJ, Christou NV, et al. Multiple organ dysfunction score: A reliable descriptor of a complex clinical outcome. Crit Care Med. 1995;23:1638–1652. doi: 10.1097/00003246-199510000-00007. [DOI] [PubMed] [Google Scholar]
- 19.Levy BD, Clish CB, Schmidt B, et al. Lipid mediator class switching during acute inflammation: Signals in resolution. Nat Immunol. 2001;2:612–619. doi: 10.1038/89759. [DOI] [PubMed] [Google Scholar]
- 20.Sun YP, Oh SF, Uddin J, et al. Resolvin D1 and its aspirin-triggered 17R epimer. Stereochemical assignments, anti-inflammatory properties, and enzymatic inactivation. J Biol Chem. 2007;282:9323–9334. doi: 10.1074/jbc.M609212200. [DOI] [PubMed] [Google Scholar]
- 21.Patil KR, Nielsen J. Uncovering transcriptional regulation of metabolism by using metabolic network topology. Proc Natl Acad Sci U S A. 2005;102:2685–2689. doi: 10.1073/pnas.0406811102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baron R, Lioubashevski O, Katz E, et al. Elementary arithmetic operations by enzymes: A model for metabolic pathway based computing. Angew Chem Int Ed Engl. 2006;45:1572–1576. doi: 10.1002/anie.200503314. [DOI] [PubMed] [Google Scholar]
- 23.Ha SS, Kim I, Wang Y, et al. Applications of different weighting schemes to improve pathway-based analysis. Comp Funct Genomics. 2011;2011:463645. doi: 10.1155/2011/463645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Manzanares W, Dhaliwal R, Jurewitsch B, et al. Parenteral fish oil lipid emulsions in the critically ill: A systematic review and meta-analysis. JPEN J Parenter Enteral Nutr. 2014;38:20–28. doi: 10.1177/0148607113486006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fullerton JN, O’Brien AJ, Gilroy DW. Lipid mediators in immune dysfunction after severe inflammation. Trends Immunol. 2014;35:12–21. doi: 10.1016/j.it.2013.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Birnbaum Y, Ye Y, Lin Y, et al. Augmentation of myocardial production of 15-epi-lipoxin-a4 by pioglitazone and atorvastatin in the rat. Circulation. 2006;114:929–935. doi: 10.1161/CIRCULATIONAHA.106.629907. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.