Abstract
Identifying populations of heart failure (HF) patients is paramount to research efforts aimed at developing strategies to effectively reduce the burden of this disease. The use of electronic medical record (EMR) data for this purpose is challenging given the syndromic nature of HF and the need to distinguish HF with preserved or reduced ejection fraction. Using a gold standard cohort of manually abstracted cases, an EMR-driven phenotype algorithm based on structured and unstructured data was developed to identify all the cases. The resulting algorithm was executed in two cohorts from the Electronic Medical Records and Genomics (eMERGE) Network with a positive predictive value of > 95%. The algorithm was expanded to include three hierarchical definitions of HF (i.e., Definite, Probable, Possible) based on the degree of confidence of the classification to capture HF cases in a whole population whereby increasing the algorithm utility for use in e-Epidemiologic research.
Keywords: Heart failure, Ventricular ejection fraction, Electronic medical records, Natural language processing, Phenotyping
Introduction
Electronic medical record (EMR) systems are increasing in ubiquity, functionality, and comprehensiveness across the United States and thus capitalizing on this data is a practical and cost-effective e-Epidemiology approach. The National Heart Lung and Blood Institute working group on Epidemiology and Population Sciences identified e-Epidemiology as a strategic research priority [1]. Specifically, the recommendation included the active engagement in studies “to establish the validity, reliability, and scalability of electronic tools for data collection.” Given the increasing prevalence and high cost[2–4], an e-Epidemiology approach to study the heart failure (HF) epidemic €would facilitate cost-effective research efforts aimed at developing strategies to effectively reduce the burden and cost[5, 6].
The syndromic nature of HF presents challenges in identifying patients using EMR data given that the diagnosis is clinical [7], at least two distinct types exist [8–11], HF with preserved ejection fraction (HFpEF) and HF with reduced ejection fraction (HFrEF), and previous studies have noted bias using a single modality of EMR data (e.g., diagnoses codes from administrative databases) to identify HF patients [12–16]. However, the Electronic Medical Records and Genomics (eMERGE) Network has demonstrated the applicability and portability of EMR derived phenotype algorithms using different types of clinical data for algorithm execution including billing and diagnoses codes, natural language processing (NLP) of clinical notes and unstructured data, laboratory measurements, patient procedure encounters, and medication data [17]. eMERGE has developed and validated nearly 45 EMR phenotype algorithms, many of which are currently available publically at pheKB.org, to facilitate cost-effective research [18–20]. To date, the predominant focus of eMERGE algorithms has been to accurately identify cases and non-cases of specific medical conditions using multiple types of EMR data and excluding those not meeting strict inclusion or exclusion criteria to facilitate genome-wide association studies [21]. While case/non-case EMR algorithms are powerful tools in research, particularly genome-wide association studies, the ability to characterize real-world clinical patient populations that are a comprised of a mix of primary care patients (i.e., medical home), transient patients, and referral patients resulting in varying patterns of depth and detail in EMR data is more limited. Therefore, the purpose of the current study was to develop and validate an EMR-based algorithm to accurately identify HF patients with characterization of HFpEF and HFrEF. Furthermore, we sought to broaden the EMR algorithm into a tool for e-Epidemiologic research that goes beyond the typical case/non-case identification to characterize HF and HF type of a complete population.
Methods
Development Cohort
Heart Failure in the Community Cohort (HL72435, PI Roger)
Since 2003, the Heart Failure in the Community Cohort, henceforth referred to as the HF Cohort, has prospectively recruited HF patients from Olmsted County, Minnesota, to study the heterogeneity of HF as it relates to outcomes and thus represents a gold standard cohort of manually abstracted cases defined according to Framingham Heart Failure Criteria [22]. NLP of the unstructured EMR text is used to prospectively identify patients presenting with clinical findings compatible with HF [5, 23]. The complete records of potential cases are manually reviewed by trained nurse abstractors to collect clinical data and to verify the diagnosis of HF using the Framingham criteria [24]. The feasibility and reliability of the Framingham criteria to ascertain HF in Olmsted County, Minnesota, have been previously published [22]. Consented HF patients undergo an echocardiogram, blood draw, questionnaires, and hand grip test administered by a registered nurse. Hospitalized patients were contacted during hospitalization, and outpatients at their next clinic appointment. From the HF Cohort, 706 validated HF patients were used in the development of the HF algorithm described herein.
Validation Cohorts
Mayo Genome Consortia (MayoGC)/eMERGE Cohort
MayoGC/eMERGE is a large cohort of Mayo Clinic patients with EMR and genotype data. Eligible patients include those who gave general research (i.e., not disease specific) consent in the contributing studies to share high throughput genotyping data with other investigators. The original design of the cohort has been described previously [25]. In brief, the cohort is a collaborative effort that brings together genomic data on Mayo Clinic patients obtained from research studies and EMR data to facilitate research.
Group Health Cooperative
The Group Health Cooperative/University of Washington eMERGE cohort is a collection of adult patients receiving care at Group Health Cooperative, an integrated delivery health care system in the Pacific Northwest. This cohort includes patients enrolled in the Adult Changes in Thought study and patients recruited for a biorepository by the Northwest Institute for Genetic Medicine. All participants in the cohort provided written and informed consent for their genetic information and EMR data to be used for research purposes. Study participants are at least 50 years of age, have a median of over 23 years of continuous enrollment at Group Health, and have received care in Group Health outpatient clinics documented by a comprehensive Epic© EMR system since 2004. All research involving these participants has been approved by the Group Health Cooperative Human Subjects Review Committee.
Mayo Clinic Primary Care Practice (PCIM)
PCIM is an adult internal medicine practice caring for patients over the age of 16 living within the local area. This patient population is self-insured; thus, PCIM has developed strategies including case management of chronic illnesses and EMR clinical decision support [26] to assist primary care providers with preventative services. Mayo Clinic has standardized care process models [27] throughout Mayo Clinic for chronic diseases including both HF with preserved and reduced EF. These process models and treatment recommendations differ for preserved and reduced EF.
Mayo Clinic Biobank
This Biobank is an institutional resource for biological specimens, patient-provided risk factor data, and clinical data that has been described in detail elsewhere [28]. In brief, adult patients from the Mayo Clinic/Mayo Clinic Health System sites in Rochester, Minnesota; LaCrosse, Wisconsin; and Jacksonville, Florida are invited to participate. For this study, only participants from the Rochester, MN site (n = 30,461) were included in the analyses. These participants were actively recruited from the Department of Medicine Divisions of Community Internal Medicine (18%), Executive Health (4%), Family Medicine (23%), General Internal Medicine (27%), Obstetrics and Gynecology (3%), Orthopedics (9%), or Preventive Medicine (10%). Community volunteers (6%) interested in participating were also included. Data were available from the EMR and from patient-provided information on current health, family health history, and various important factors known to confer risk for disease. Specifically, participants self-reported whether they had a personal and/or family history of HF as well as age of HF onset for those with a positive HF history.
Algorithm Development
Case/Non-case algorithm
International Classification of Diseases, 9th Revision (ICD-9) code 428 was used based on previous previously reported yields [22]. In addition, we searched for positive mentions of HF from structured problem lists or problem list sections in clinical notes. For structured problem lists which are typically coded with SNOMED-CT, we applied recursive traversal of the descendants of the SNOMEDCT code 84114007 (HF) to indicate if the subject had a positive mention of HF. An NLP system, MedTagger, was used to help determine HF diagnosis from problem list sections of clinical notes [29]. In MedTagger (publically distributed www.ohnlp.org), besides a rule-based concept extraction engine which extracts concept mentions defined using regular expression, it also consists of i) a sectionizer adapted from SecTag to detect sections and ii) a rule-based context annotator adapted from ConText [30] assigning each concept mention a status modifier (i.e., positive, negative, and probable). Note that clinical notes in Mayo Clinic EMR are Clinical Document Architecture 1.0 compliant where sections have been codified. For non-Clinical Document Architecture compliant documents (Group Health EMR), the sectionizer was used to detect Diagnosis and other sections (i.e., Chief Complaints or Impressions as the Secondary Problem List section). To determine the date of first documented HF, the cross product of all ICD-9 and problem list dates were considered. Echocardiography measurements of left ventricular ejection fraction (EF) were extracted from structured database (Mayo Clinic EMR) and by deploying NLP to search the radiology reports for EF measurements (Group Health EMR). Multiple EF measurements from the same examination were averaged. HFrEF was defined as an average EF <50% and HFpEF ≥50% [31].
e-Epidemiology tool
Among the participants with “unknown” (i.e., not meeting the case or non-case definition), 100 patients were randomly selected for medical record abstraction. This information was used to broaden the algorithm by creating definitions for definite, probable, and possible HF as well as refinements to the non-case definition. These three hierarchical definitions of HF (i.e., definite, probable, possible) have decreasing stringency in terms of level of evidence. This classification strategy was adopted as it is used extensively to classify disease in cardiovascular epidemiologic research [32]. Definite HF requires the presence of ICD-9 and NLP within a relatively narrow time window. Probable HF requires five or more unique dates of either ICD-9 or NLP with a more liberal time window. In contrast, possible HF has minimal evidence of HF albeit the presence of an ICD-9 code, NLP hit, or low EF measurement, thus prohibiting them from being classified as non-cases. Patients are further classified by HF type within definite and probable groups, and are considered unknown to HF type if no qualifying EF is available. Non-cases were defined as the absence of any of these elements (i.e. ICD9, NLP) and a normal EF (i.e., ≥50%) if measured.
Validation and Statistical Analysis
The validation of the case/non-case algorithm was completed in two phases in the Mayo Clinic cohorts. First, 50 cases and 50 non-cases identified by the algorithm were randomly selected in the MayoGC/eMERGE Cohort. Trained nurse abstractors, blinded to disease, reviewed medical records to determine HF using Framingham Heart Failure Criteria [22], and HF date and type (i.e., HFrEF or HFpEF) for those who were identified as having HF. External validation was performed at Group Health using a trained medical chart abstractor who reviewed the charts of random samples of patients identified by the automated phenotype algorithm at Group Health. To determine the accuracy of the algorithm, positive and negative predictive values (PPV and NPV, respectively) were calculated as well as sensitivity and specificity correcting for verification bias [33, 34]. Estimated prevalence of HF corresponding to the sex averaged rates for 60–79 year olds for MayoGC/eMERGE and the 80+ year olds for Group Health were used based on published reports [35]. The expanded algorithm was validated by randomly selecting a total of 300 patients, 100 definite, 100 probable, 50 possible, and 50 non-cases equally divided between PCIM and Biobank cohorts. Trained abstractors reviewed the records to determine case/non-case, case type, and incident date. For the latter, the incident date was considered validated if the abstracted data and algorithm date occurred within 1 year of each other.
Results
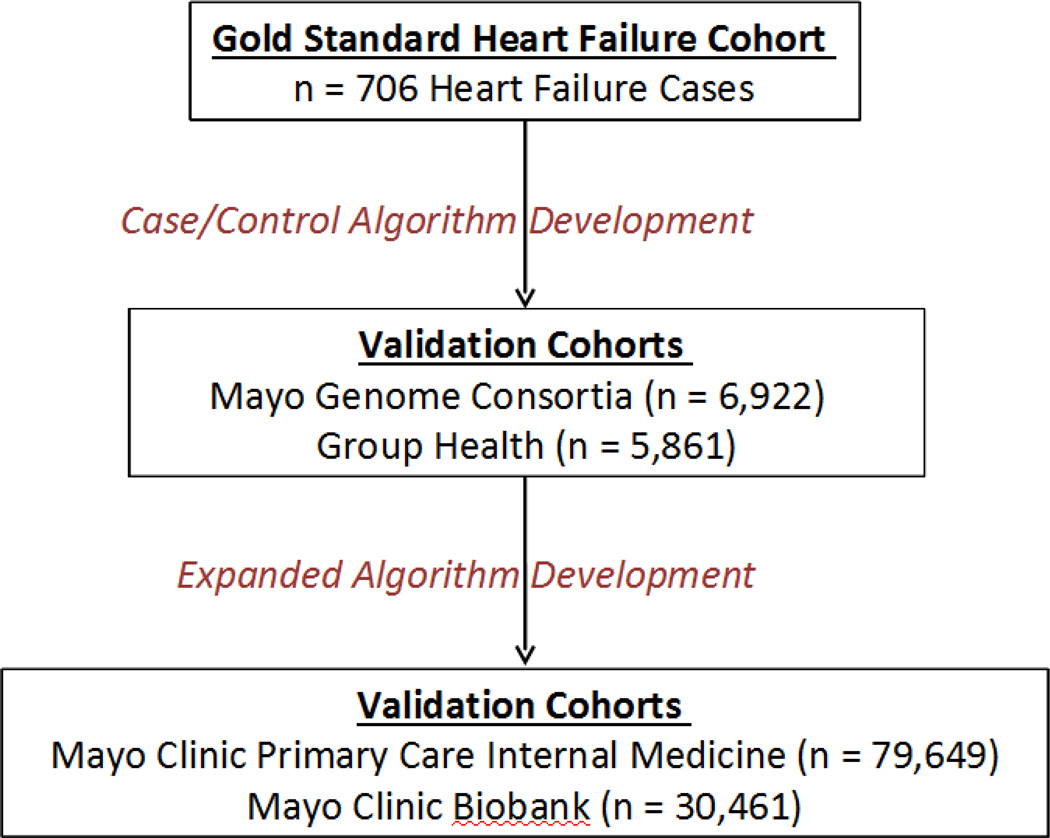
Figure 1 illustrates the cohorts used in the development and validation of the case/non-case algorithm and the expanded algorithm.
Figure 1.
Algorithm development
An ICD-9 code for HF (428.X) was present in 93% of the cases. NLP analyses of the clinical notes identified six common terms/acronyms present in 89% of cases from the HF Cohort: multi-organ failure, cardiac failure, heart failure, CHF, LVF, and ventricular failure in the primary and secondary diagnosis sections. Using the combination of ICD-9 code and positive NLP hit, 99% of the cases were identified. Abstraction of the seven cases without an ICD-9 or an NLP hit revealed that all patients were in critical condition in the ICU at the time they met the Framingham HF criteria. Thus only a symptom-based algorithm would have been able to identify these patients. Since the HF Cohort study protocol included echocardiography, all cases were able to be classified as either preserved or reduced EF HF type.
Algorithm validation
The algorithm was run in 6,922 participants in the MayoGC/eMERGE (mean age 65 ± 12 years) and 5,861 at Group Health (mean age 90 ± 12 years). The algorithm performed with a positive predictive value of 0.94, a negative predictive value of 0.98 (MayoGC/eMERGE) and a positive predictive value of 0.80 and a negative predictive value of 1.0. (Group Health, Table 1). Sensitivity was 0.71 and 1.0 for MayoGC/eMERGE and Group Health, respectively. Specificity was similar across the two sites (Table 1).
Table 1.
Validation results per prevalence of the disease in each population
|
*Estimated prevalence of heart failure |
Positive predictive value |
Negative predictive value |
Sensitivity | Specificity | |
|---|---|---|---|---|---|
| Mayo Genome Consortia (MayoGC)/eMERGE | 6.2% | 0.94 | 0.98 | 0.71 | 0.99 |
| Group Health | 10.1% | 0.80 | 1.0 | 1.0 | 0.97 |
Estimated prevalence based on Go, A.S., Mozaffarian, D., Roger, V.L., et al. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129, e28–e292 corresponding to the sex averaged rates for 60–79 year olds for MayoGC/eMERGE and the 80+ year olds for Group Health.
e-Epidemiology tool validation
The complete algorithm is provided in Web material and available online at pheKB.org. Validation of HF cases, case type, and index date was similar for the two cohorts and did not differ substantially between definite and probable definitions (Table 2). Likewise, the non-case definition which requires the complete absence of evidence of HF had good performance in both cohorts. Proportion of those with HF based on abstraction for those classified as possible HF cases differed between PCIM (48%) and Biobank (16%) populations but was poor in both cohorts despite the high prevalence of HF ICD9 codes (62% and 45% for PCIM and Biobank respectively).
Table 2.
Proportion of agreement comparing the electronic medical record algorithm to manual chart abstraction
| Case statusa | Case typeb | Incident datec | ||||
|---|---|---|---|---|---|---|
| Heart failure definitions | PCIM (%) |
Biobank (%) |
PCIM (%) | Biobank (%) | PCIM (%) |
Biobank (%) |
| Definite | 100 | 96 | 96 | 98 | 80 | 83 |
| Probable | 98 | 100 | 100 | 96 | 84 | 76 |
| Possible | 48 | 16 | n/a | n/a | n/a | n/a |
| Non-Case | 100 | 100 | n/a | n/a | n/a | n/a |
PCIM Mayo Clinic Primary Care Practice
Validation included 300 cases randomly selected within the following strata; 100 definite, 100 probable, 50 possible, and 50 non-cases equally divided between PCIM and Biobank cohorts.
Heart failure type at the time of initial diagnosis; reduced ejection fraction (<50) or preserved ejection fraction (≥50) for validated cases.
Incident dates within 1 year of each other were considered to be in agreement for validated cases.
Characteristics of the PCIM are summarized in Table 3 by HF case/non-case and type. Of the 79,649 patients, the algorithm identified 3,318 definite and probable HF cases. Of these, case type was identified in 79% of those classified as definite and 65% of probable cases. By definition, all definite cases had ICD-9 code and NLP evidence, however, for probable cases, 99% had ICD-9 code evidence but only about 7% had NLP evidence. In either category, those of unknown HF type were less likely to have an EF measurement in their EMR. Of the PCIM population, 4.6% were categorized as possible HF. ICD-9 HF codes and EF measurements were common but NLP hits were less frequent.
Table 3.
Characteristics of Mayo Clinic Primary Care Internal Medicine Practice
| Characteristics | Definite HF cases with reduced EF |
Definite HF cases with preserved EF |
Definite HF cases (unknown type) |
Probable HF cases with reduced EF |
Probable HF cases with preserved EF |
Probable HF case (unknown type) |
Possible HF cases |
Non-cases |
|---|---|---|---|---|---|---|---|---|
| n | 760 | 754 | 391 | 422 | 497 | 494 | 3,668 | 72,663 |
| Sex, % female | 39 | 61 | 53 | 35 | 62 | 57 | 50 | 56 |
| Race, % white | 93 | 92 | 94 | 96 | 96 | 96 | 93 | 80 |
| Diabetes, % yes | 45 | 48 | 48 | 42 | 46 | 43 | 34 | 12 |
| Hypertension, % yes | 92 | 95 | 96 | 93 | 97 | 95 | 86 | 31 |
| ICD-9 Code 428, % yes | 100 | 100 | 100 | 99 | 99 | 99 | 62 | 0 |
| ICD-9 Code 428 unique dates, mean ± SD (range)* | 19 ± 20 (1–165) | 15 ± 17 (1–159) | 16 ± 19 (1–189) | 20 ± 19 (1–144) | 15 ± 14 (1–97) | 15 ± 13 (1–91) | 2.0 ± 1.2 (1–10) | n/a |
| NLP HF term, % yes | 100 | 100 | 100 | 7.6 | 7.7 | 5.1 | 12 | 0 |
| NLP HF term unique dates, mean ± SD (range)* | 12 ± 17 (1–200) | 6.6 ± 8.6 (1–67) | 8.8 ± 15 (1–154) | 2.4 ± 2.6 (1–11) | 2.0 ± 2.2 (1–12) | 2.4 ± 2.1 (1–8) | 1.4 ± 0.9 (1–8) | n/a |
| EF measured, % yes | 100 | 100 | 80 | 100 | 99 | 70 | 87 | 21 |
| EF measurement unique dates, mean ± SD (range)* | 15 ± 13 (1–132) | 11 ± 9.5 (1–131) | 7.3 ± 7.0 (1–63) | 12 ± 11 (1–87) | 9.1 ± 7.1 (1–54) | 5.7 ± 5.3 (1–48) | 7.1 ± 6.3 (1–52) | 4.1 ± 3.7 (1–54) |
| EF, mean ± SD | 42 ± 16 | 59 ± 9.6 | 51 ± 15 | 40 ± 15 | 59 ± 10 | 51 ± 15 | 54 ± 12 | 61 ± 6.2 |
| History of myocardial infarction, % yes | 88 | 76 | 80 | 91 | 82 | 82 | 65 | 12 |
| Medication history | ||||||||
| Angiotensin converting enzyme use, % yes | 79 | 69 | 73 | 83 | 70 | 67 | 55 | 14 |
| Angiotensin receptor blocker use, % yes | 20 | 26 | 17 | 21 | 23 | 16 | 12 | 3 |
| Beta blocker use, % yes | 80 | 73 | 69 | 76 | 69 | 60 | 58 | 16 |
| Calcium channel blocker use, % yes | 39 | 48 | 39 | 39 | 53 | 42 | 31 | 7 |
EF ejection fraction, HF heart failure, ICD-9 International Classification of Diseases, 9th Revision, NLP natural language processing, SD standard deviation
For those with non-missing data
Of the 30,461 Biobank participants, the algorithm identified 606 definite or probable HF cases (Table 4). The Biobank was similar to PCIM, however, ICD-9 codes were somewhat less common (92–95%) and NLP evidence was more common (7–21%) compared to PCIM. Furthermore, case type was available for a greater number of patients in the Biobank, 90% of definite and 85% of probable cases.
Table 4.
Characteristics of Mayo Clinic Biobank Population
| Characteristics | Definite HF cases with reduced EF |
Definite HF cases with preserved EF |
Definite HF cases (unknown type) |
Probable HF cases with reduced EF |
Probable HF cases with preserved EF |
Probable HF case (unknown type) |
Possible HF cases |
Non- cases |
|---|---|---|---|---|---|---|---|---|
| n | 207 | 173 | 40 | 75 | 84 | 27 | 1429 | 28,426 |
| Sex, % Female | 28 | 49 | 33 | 41 | 46 | 48 | 41 | 59 |
| Race, % white | 98 | 98 | 98 | 97 | 99 | 100 | 98 | 96 |
| Diabetes, % yes | 41 | 45 | 63 | 32 | 46 | 44 | 31 | 14 |
| Hypertension, % yes | 89 | 94 | 95 | 80 | 93 | 100 | 77 | 42 |
| ICD-9 Code 428, % yes | 100 | 100 | 100 | 92 | 95 | 100 | 45 | 0 |
| ICD-9 Code 428 unique dates, mean ± SD (range)* | 11 ± 11 (1–65) | 9.6 ± 9.6 (1–61) | 11 ± 9.1 (1–36) | 14 ± 12 (1–54) | 10 ± 7.4 (1–44) | 11 ± 7.5 (1–30) | 1.8 ± 1.3 (1–25) | n/a |
| NLP HF term, % yes | 100 | 100 | 100 | 21 | 13 | 7.4 | 15 | 0 |
| NLP HF term unique dates, mean ± SD (range)* | 6.2 ± 8.7 (1–69) | 4.7 ± 6.6 (1–47) | 4.0 ± 4.7 (1–21) | 4.6 ± 4.7 (1–17) | 2.8 ± 2.4 (1–7) | 1.5 ± 0.7 (1–2) | 1.4 ± 0.9 (1–8) | n/a |
| EF measured, % yes | 100 | 100 | 83 | 100 | 100 | 93 | 94 | 32 |
| EF measurement unique dates, mean ± SD (range)* | 17 ± 13 (1–96) | 13 ± 9.0 (2–42) | 9.3 ± 6.4 (2–23) | 20 ± 16 (1–91) | 14 ± 12 (2–82) | 12 ± 10 (2–43) | 9.2 ± 8.1 (1–66) | 4.4 ± 4.2 (1–68) |
| EF, mean ± SD | 43 ± 14 | 58 ± 10 | 46 ± 17 | 43 ± 13 | 58 ± 8.8 | 56 ± 7.1 | 53 ± 11 | 61 ± 6.1 |
| History of Myocardial Infarction, % yes | 88 | 81 | 93 | 79 | 83 | 89 | 62 | 19 |
| Medication History | ||||||||
| Angiotensin converting enzyme use, % yes | 88 | 74 | 78 | 80 | 77 | 70 | 52 | 20 |
| Angiotensin receptor blocker use, % yes | 27 | 23 | 18 | 23 | 30 | 33 | 16 | 5.5 |
| Beta blocker use, % yes | 73 | 60 | 75 | 68 | 61 | 63 | 44 | 19 |
| Calcium channel blocker use, % yes | 30 | 47 | 23 | 35 | 44 | 44 | 25 | 9.4 |
| Self-reported family history of HF, % Yes | 36 | 40 | 36 | 47 | 32 | 36 | 34 | 26 |
| Self-reported history of HF, n (%) | 47 | 44 | 50 | 57 | 36 | 50 | 11 | 0.8 |
EF ejection fraction, HF heart failure, ICD-9 International Classification of Diseases, 9th Revision, NLP natural language processing, SD standard deviation
For those with non-missing data
Discussion
The rapid expansion of electronic health information necessitates studies to establish valid and reliable e-Epidemiologic tools. We developed a multi-modal EMR HF algorithm that combines structured and unstructured EMR data to accurately classify HF in clinical populations. We further expanded the utility of the algorithm by incorporating hierarchical categories enabling the classification of a complete population and providing multiple methods to extract EF measurement. The latter is essential for the algorithm to accurately distinguish HF with preserved or reduced EF, a critical feature to characterize the burden of HF in populations.
We have developed a cost-effective and robust EMR HF algorithm and have demonstrated its effectiveness in characterizing HF patients across institutions with different EMR systems (i.e., Mayo Clinic and Group Health) and in two different clinic-based populations (i.e., primary care and Biobank). Further, we demonstrate that combining structured (e.g., ICD-9 codes) and unstructured (i.e., NLP) data improves the accuracy of identifying HF patients compared to administrative data alone [36]. Importantly the algorithm provides several methods for capturing EF measurements to use in the classification of HF as some institutions store this information in structured databases and other sites require NLP to either extract the EF value or free text responses (i.e., normal EF). This algorithm will enable population management strategies by identifying patients in practice to build registries to facilitate quality measures.
Furthermore, understanding the performance of the algorithm in a Biobank population is crucial as participants in Biobanks may have greater diversity in terms of EMR depth and completeness as compared to a primary care population. Biobanks are commonly comprised of community volunteers who may or may not receive regular care at the institution supporting the Biobank and thus missing or incomplete data within the EMR may hinder the ability to accurately classify case/non-case for both EMR-based algorithms as well as manual abstraction and results in subsets of patients with indeterminate disease. For example, patients may not be billed if a HF episode happened in the past or it occurred at another institution and they are currently asymptomatic. Likewise, patients may be inappropriately coded with HF when in fact it was another diagnosis.
While the distinction of definite, probable, and possible HF is not clinically meaningful for a physician treating a patient, the ability to characterize who, in the given population, falls outside the case/non-case definition is essential for an effective e-Epidemiology tool to facilitating population research and understanding the potential biases present.
In conclusion, the magnitude of the HF epidemic necessitates the development of cost-effective methods to study and identify strategies to alleviate the substantial global burden and cost of HF. Further, the differentiation of HF by type (i.e., HFrEF and HFpEF) has clinical implications that may improve quality metrics for health care institutions. Using a combination of structured and unstructured EMR data, we have developed and validated a transportable e-Epidemiologic tool to facilitate population research.
Supplementary Material
Acknowledgments
FUNDING:
The Mayo Clinic Biobank and the Mayo Genome Consortia is funded by the Mayo Clinic Center for Individualized Medicine. Additional funding for this work came from National Institutes of Health grants R01HL72435 (Heart Failure in the Community Cohort), R01AG034676 (The Rochester Epidemiology Project), R01GM102282 (Natural Language Processing for Clinical and Translational Research), the Electronic Medical Record and Genomics (eMERGE) Network U01 HG06379 (Mayo Clinic), U01HG006375 (Group Health/University of Washington); U01HG006382 (Geisinger Health System); U01HG006389 (Essentia Health & Marshfield Clinic Research Foundation); U01HG006388 (Northwestern University); HG004438 (Center for Inherited Disease Research, Johns Hopkins University); HG004424 (Broad Institute of Harvard & MIT); U01HG006378, U01HG006385 (Vanderbilt University); U01HG006380 (The Mt. Sinai Hospital); U01HG006828 (Cincinnati Children’s Hospital Medical Center/Harvard); U01HG006830 (Children’s Hospital of Philadelphia), NIA grant U01AG006781-25, Life Sciences Discovery Fund Grant #2065508, and additional support was provided by a State of Washington Life Sciences Discovery Fund award to the Northwest Institute of Genetic Medicine.
Abbreviations
- EF
ejection fraction
- eMERGE
Electronic Medical Records and Genomics
- EMR
Electronic medical record
- HF
Heart failure
- HFpEF
HF with preserved ejection fraction
- HFrEF
HF with reduced ejection fraction
- ICD-9
International Classification of Diseases, 9th Revision
- NLP
Natural language processing
- PCIM
Mayo Clinic Primary Care Practice
Footnotes
The online version contains supplementary material which is available to authorized users.
Conflict of Interest All authors have reported that they have no relationships to disclose.
Human subjects/informed consent statement All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000. All research procedures were approved by the Institutional Review Committee of the Mayo Clinic and the participants from each participating study provided written and informed consent for general research. The inclusion of Group Health participants for studies was approved by Group Health Cooperative Human Subjects Review Committee, Seattle, Washington.
No animal studies were carried out by the authors for this article.
Contributor Information
Suzette J. Bielinski, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Jyotishman Pathak, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
David S. Carrell, Group Health Research Institute, Seattle, WA 98101, USA
Paul Y. Takahashi, Department of Medicine, Division of Primary Care Internal Medicine, Mayo Clinic, Rochester, MN 55905, USA
Janet E. Olson, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Nicholas B. Larson, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Hongfang Liu, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Sunghwan Sohn, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Quinn S. Wells, Department Medicine, Vanderbilt University, Nashville, TN 37232, USA
Joshua C. Denny, Departments of Biomedical Informatics and Medicine, Vanderbilt University, Nashville TN 37232, USA
Laura J. Rasmussen-Torvik, Department of Preventive Medicine, Northwestern University Feinberg School of Medicine, Chicago, IL 60611, USA.
Jennifer Allen Pacheco, Center for Genetic Medicine, Northwestern University, Feinberg School of Medicine, Chicago, IL 60611, USA
Kathryn L. Jackson, Center for Healthcare Studies, Northwestern University Feinberg School of Medicine, Chicago, Illinois 60611, USA
Timothy G. Lesnick, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Rachel E. Gullerud, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Paul A. Decker, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Naveen L. Pereira, Division of Cardiovascular Diseases, Mayo Clinic Rochester, MN 55905, USA
Euijung Ryu, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Richard A. Dart, Center for Human Genetics, Marshfield Clinic Research Foundation, Marshfield, WI 54449, USA
Peggy Peissig, Biomedical Informatics Research Center, Marshfield Clinic Research Foundation, Marshfield, WI 54449, USA
James G. Linneman, Biomedical Informatics Research Center, Marshfield Clinic Research Foundation, Marshfield, WI 54449, USA
Gail P. Jarvik, Departments of Medicine (Medical Genetics) and Genome Sciences, University of Washington, Seattle, WA 98195, USA
Eric B. Larson, Group Health Research Institute, Seattle, WA 98101, USA
Jonathan A. Bock, The Sigfried and Janet Weis Center for Research, Geisinger Health System, Danville, PA 17822, USA
Gerard C. Tromp, The Sigfried and Janet Weis Center for Research, Geisinger Health System, Danville, PA 17822, USA.
Mariza de Andrade, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA
Véronique L. Roger, Department of Health Sciences Research, Mayo Clinic, Rochester, MN 55905, USA; Division of Cardiovascular Diseases, Mayo Clinic Rochester, MN 55905, USA.
References
- 1.Roger VL, Boerwinkle E, Crapo JD, Douglas PS, Epstein JA, Granger CB, Greenland P, Kohane I, Psaty BM. Strategic transformation of population studies: recommendations of the working group on epidemiology and population sciences from the National Heart, Lung, and Blood Advisory Council and Board of External Experts. American Journal of Epidemiology. 2015;181(6):363–368. doi: 10.1093/aje/kwv011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roger VL, Go AS, Lloyd-Jones DM, Benjamin EJ, Berry JD, Borden WB, Bravata DM, Dai S, Ford ES, Fox CS, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, et al. Heart disease and stroke statistics--2012 update: a report from the American Heart Association. Circulation. 2012;125(1):e2–e220. doi: 10.1161/CIR.0b013e31823ac046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Heidenreich PA, Trogdon JG, Khavjou OA, Butler J, Dracup K, Ezekowitz MD, Finkelstein EA, Hong Y, Johnston SC, Khera A, Lloyd-Jones DM, Nelson SA, Nichol G, Orenstein D, Wilson PW, et al. Forecasting the future of cardiovascular disease in the United States: a policy statement from the American Heart Association. Circulation. 2011;123(8):933–944. doi: 10.1161/CIR.0b013e31820a55f5. [DOI] [PubMed] [Google Scholar]
- 4.Heidenreich PA, Albert NM, Allen LA, Bluemke DA, Butler J, Fonarow GC, Ikonomidis JS, Khavjou O, Konstam MA, Maddox TM, Nichol G, Pham M, Pina IL, Trogdon JG. Forecasting the impact of heart failure in the United States: a policy statement from the American Heart Association. Circulation Heart failure. 2013;6(3):606–619. doi: 10.1161/HHF.0b013e318291329a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlay SM, Shah ND, Shi Q, Morlan B, VanHouten H, Long KH, Roger VL. Lifetime costs of medical care after heart failure diagnosis. Circulation. 2011;4(1):68–75. doi: 10.1161/CIRCOUTCOMES.110.957225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Medicare & Medicaid Services. Readmissions Reduction Program. [Accessed January 30, 2015]; [cited; Available from: http://www.cms.gov/Medicare/Medicare-Fee-for-Service-Payment/AcuteInpatientPPS/Readmissions-Reduction-Program.html.
- 7.Schellenbaum GD, Rea TD, Heckbert SR, Smith NL, Lumley T, Roger VL, Kitzman DW, Taylor HA, Levy D, Psaty BM. Survival associated with two sets of diagnostic criteria for congestive heart failure. American Journal of Epidemiology. 2004;160(7):628–635. doi: 10.1093/aje/kwh268. [DOI] [PubMed] [Google Scholar]
- 8.Redfield MM, Jacobsen SJ, Burnett JC, Jr, Mahoney DW, Bailey KR, Rodeheffer RJ. Burden of systolic and diastolic ventricular dysfunction in the community: appreciating the scope of the heart failure epidemic. JAMA. 2003;289(2):194–202. doi: 10.1001/jama.289.2.194. [DOI] [PubMed] [Google Scholar]
- 9.Bursi F, Weston SA, Redfield MM, Jacobsen SJ, Pakhomov S, Nkomo VT, Meverden RA, Roger VL. Systolic and diastolic heart failure in the community. JAMA. 2006;296(18):2209–2216. doi: 10.1001/jama.296.18.2209. [DOI] [PubMed] [Google Scholar]
- 10.Owan TE, Hodge DO, Herges RM, Jacobsen SJ, Roger VL, Redfield MM. Trends in prevalence and outcome of heart failure with preserved ejection fraction. New England Journal of Medicine. 2006;355(3):251–259. doi: 10.1056/NEJMoa052256. [DOI] [PubMed] [Google Scholar]
- 11.Gerber Y, Jacobsen SJ, Frye RL, Weston SA, Killian JM, Roger VL. Secular trends in deaths from cardiovascular diseases: a 25-year community study. Circulation. 2006;113(19):2285–2292. doi: 10.1161/CIRCULATIONAHA.105.590463. [DOI] [PubMed] [Google Scholar]
- 12.Schellenbaum GD, Heckbert SR, Smith NL, Rea TD, Lumley T, Kitzman DW, Roger VL, Taylor HA, Psaty BM. Congestive heart failure incidence and prognosis: case identification using central adjudication versus hospital discharge diagnoses. Annals of Epidemiology. 2006;16(2):115–122. doi: 10.1016/j.annepidem.2005.02.012. [DOI] [PubMed] [Google Scholar]
- 13.Pakhomov S, Weston SA, Jacobsen SJ, Chute CG, Meverden R, Roger VL. Electronic medical records for clinical research: application to the identification of heart failure. American Journal of Managed Care. 2007;13(6 Part 1):281–288. [PubMed] [Google Scholar]
- 14.Heliovaara M, Aromaa A, Klaukka T, Knekt P, Joukamaa M, Impivaara O. Reliability and validity of interview data on chronic diseases. The Mini-Finland Health Survey. Journal of Clinical Epidemiology. 1993;46(2):181–191. doi: 10.1016/0895-4356(93)90056-7. [DOI] [PubMed] [Google Scholar]
- 15.Ermenc B. Minimizing mistakes in clinical diagnosis. Journal of Forensic Sciences. 1999;44(4):810–813. [PubMed] [Google Scholar]
- 16.Psaty BM, Boineau R, Kuller LH, Luepker RV. The potential costs of upcoding for heart failure in the United States. American Journal of Cardiology. 1999;84(1):108–109. doi: 10.1016/s0002-9149(99)00205-2. [DOI] [PubMed] [Google Scholar]
- 17.Kho AN, Pacheco JA, Peissig PL, Rasmussen L, Newton KM, Weston N, Crane PK, Pathak J, Chute CG, Bielinski SJ, Kullo IJ, Li R, Manolio TA, Chisholm RL, Denny JC. Electronic medical records for genetic research: results of the eMERGE consortium. Science translational medicine. 2011;3(79):79re71. doi: 10.1126/scitranslmed.3001807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kho AN, Hayes MG, Rasmussen-Torvik L, Pacheco JA, Thompson WK, Armstrong LL, Denny JC, Peissig PL, Miller AW, Wei WQ, Bielinski SJ, Chute CG, Leibson CL, Jarvik GP, Crosslin DR, et al. Use of diverse electronic medical record systems to identify genetic risk for type 2 diabetes within a genome-wide association study. Journal of the American Medical Informatics Association. 2012;19(2):212–218. doi: 10.1136/amiajnl-2011-000439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peissig PL, Rasmussen LV, Berg RL, Linneman JG, McCarty CA, Waudby C, Chen L, Denny JC, Wilke RA, Pathak J, Carrell D, Kho AN, Starren JB. Importance of multi-modal approaches to effectively identify cataract cases from electronic health records. Journal of the American Medical Informatics Association: JAMIA. 2012;19(2):225–234. doi: 10.1136/amiajnl-2011-000456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denny JC, Ritchie MD, Crawford DC, Schildcrout JS, Ramirez AH, Pulley JM, Basford MA, Masys DR, Haines JL, Roden DM. Identification of genomic predictors of atrioventricular conduction: using electronic medical records as a tool for genome science. Circulation. 2010;122(20):2016–2021. doi: 10.1161/CIRCULATIONAHA.110.948828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Denny JC, Crawford DC, Ritchie MD, Bielinski SJ, Basford MA, Bradford Y, Chai HS, Bastarache L, Zuvich R, Peissig P, Carrell D, Ramirez AH, Pathak J, Wilke RA, Rasmussen L, et al. Variants near FOXE1 are associated with hypothyroidism and other thyroid conditions: using electronic medical records for genome- and phenome-wide studies. American Journal of Human Genetics. 2011;89(4):529–542. doi: 10.1016/j.ajhg.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roger VL, Weston SA, Redfield MM, Hellermann-Homan JP, Killian J, Yawn BP, Jacobsen SJ. Trends in heart failure incidence and survival in a community-based population. JAMA. 2004;292(3):344–350. doi: 10.1001/jama.292.3.344. [DOI] [PubMed] [Google Scholar]
- 23.Pakhomov SV, Buntrock J, Chute CG. Prospective recruitment of patients with congestive heart failure using an ad-hoc binary classifier. Journal of Biomedical Informatics. 2005;38(2):145–153. doi: 10.1016/j.jbi.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 24.Ho KK, Pinsky JL, Kannel WB, Levy D. The epidemiology of heart failure: the Framingham Study. Journal of the American College of Cardiology. 1993;22(4) Suppl A:6A–13A. doi: 10.1016/0735-1097(93)90455-a. [DOI] [PubMed] [Google Scholar]
- 25.Bielinski SJ, Chai HS, Pathak J, Talwalkar JA, Limburg PJ, Gullerud RE, Sicotte H, Klee EW, Ross JL, Kocher JP, Kullo IJ, Heit JA, Petersen GM, de Andrade M, Chute CG. Mayo Genome Consortia: a genotype-phenotype resource for genome-wide association studies with an application to the analysis of circulating bilirubin levels. Mayo Clinic Proceedings. 2011;86(7):606–614. doi: 10.4065/mcp.2011.0178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhry R, Tulledge-Scheitel SM, Parks DA, Angstman KB, Decker LK, Stroebel RJ. Use of a Web-based clinical decision support system to improve abdominal aortic aneurysm screening in a primary care practice. Journal of Evaluation in Clinical Practice. 2012;18(3):666–670. doi: 10.1111/j.1365-2753.2011.01661.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cook DA, Sorensen KJ, Nishimura RA, Ommen SR, Lloyd FJ. A comprehensive information technology system to support physician learning at the point of care. Academic Medicine. 2015;90(1):33–39. doi: 10.1097/ACM.0000000000000551. [DOI] [PubMed] [Google Scholar]
- 28.Olson JE, Ryu E, Johnson KJ, Koenig BA, J MK, Morrisette JA, Liebow M, Takahashi PY, Fredericksen ZS, Sharma RG, Anderson KS, Hathcock MA, Carnahan JA, Pathak J, Lindor NM, et al. The Mayo Clinic Biobank: a building block for individualized medicine. Mayo Clinic Proceedings. 2013;88(9):952–962. doi: 10.1016/j.mayocp.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu H, Bielinski SJ, Sohn S, Murphy S, Wagholikar KB, Jonnalagadda SR, Ravikumar KE, Wu ST, Kullo IJ, Chute CG. An information extraction framework for cohort identification using electronic health records. AMIA Joint Summits on Translational Science proceedings AMIA Summit on Translational Science. 2013;2013(149–153) [PMC free article] [PubMed] [Google Scholar]
- 30.Harkema H, Dowling JN, Thornblade T, Chapman WW. ConText: an algorithm for determining negation, experiencer, and temporal status from clinical reports. Journal of biomedical informatics. 2009;42(5):839–851. doi: 10.1016/j.jbi.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Borlaug BA, Redfield MM. Diastolic and systolic heart failure are distinct phenotypes within the heart failure spectrum. Circulation. 2011;123(18):2006–2013. doi: 10.1161/CIRCULATIONAHA.110.954388. discussion 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luepker RV, Apple FS, Christenson RH, Crow RS, Fortmann SP, Goff D, Goldberg RJ, Hand MM, Jaffe AS, Julian DG, Levy D, Manolio T, Mendis S, Mensah G, Pajak A, et al. Case definitions for acute coronary heart disease in epidemiology and clinical research studies: a statement from the AHA Council on Epidemiology and Prevention; AHA Statistics Committee; World Heart Federation Council on Epidemiology and Prevention; the European Society of Cardiology Working Group on Epidemiology and Prevention; Centers for Disease Control and Prevention; and the National Heart, Lung, and Blood Institute. Circulation. 2003;108(20):2543–2549. doi: 10.1161/01.CIR.0000100560.46946.EA. [DOI] [PubMed] [Google Scholar]
- 33.Begg CB, Greenes RA. Assessment of diagnostic tests when disease verification is subject to selection bias. Biometrics. 1983;39(1):207–215. [PubMed] [Google Scholar]
- 34.Genç Y, Tüccar E. Effect of vertification bias on sensitivity and specificity of diagnostic tests. Journal of Ankara Medical School. 2003;25(3):107–112. [Google Scholar]
- 35.Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Blaha MJ, Dai S, Ford ES, Fox CS, Franco S, Fullerton HJ, Gillespie C, Hailpern SM, Heit JA, Howard VJ, et al. Heart disease and stroke statistics--2014 update: a report from the American Heart Association. Circulation. 2014;129(3):e28–e292. doi: 10.1161/01.cir.0000441139.02102.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mähönen M, Jula A, Harald K, Antikainen R, Tuomilehto J, Zeller T, Blankenberg S, Salomaa V. The validity of heart failure diagnoses obtained from administrative registers. European Journal of Preventive Cardiology. 2013;20(2):254–259. doi: 10.1177/2047487312438979. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.