Abstract
We report the results of a series of 1 μs-long explicit-solvent molecular dynamics (MD) simulations performed to compare the free energies of stacking (ΔGstack) of all possible combinations of DNA and RNA nucleoside (NS) pairs and dinucleoside-monophosphates (DNMPs). For both NS-pairs and DNMPs we show that the computed stacking free energies are in reasonable qualitative agreement with experimental measurements, and appear to provide the closest correspondence with experiment yet found among computational studies; in all cases, however, the computed stacking free energies are too favorable relative to experiment. Comparisons of NS-pair systems indicate that stacking interactions are very similar in RNA and DNA systems except when a thymine or uracil base is involved: the presence of a thymine base favors stacking by ~0.3 kcal/mol relative to a uracil base. One exception is found in the self-stacking of cytidines, which are found to be significantly more favorable for the DNA form; an analysis of the rotational orientations sampled during stacking events suggests that this is likely to be due to more favorable sugar-sugar interactions in stacked complexes of deoxycytidines. Comparisons of the DNMP systems indicate that stacking interactions are more favorable in RNA than in DNA except, again, when thymine or uracil bases are involved. Finally, additional simulations performed using a previous generation of the AMBER force field – in which the description of glycosidic bond rotations was less than optimal – produce computed stacking free energies that are in poorer agreement with experimental data. Overall, the simulations provide a comprehensive view of stacking thermodynamics in NS pairs and in DNMPs as predicted by a state-of-the-art MD force field.
Introduction
The extent to which nucleic acid bases stack upon one another is a key determinant of the thermodynamic stability of a wide variety of DNA and RNA based structures.1 In principle, the simplest structural systems in which to study directly the thermodynamics of base stacking interactions are dinucleoside monophosphates (DNMPs), in which two nucleosides are linked by a single phosphate group. A number of early experimental studies were focused on attempts to detect and quantify the extent of stacking in DNMPs, with NMR studies being especially prominent2 but with optical techniques also being used;3 more recently, femtosecond spectroscopy techniques have opened up new possibilities for studying such interactions.4 Another way to study the thermodynamics of base-stacking interactions in relatively simple systems is through analysis of the intermolecular interactions of nucleoside (NS) pairs using techniques such as osmometry5 or through the analysis of changes in diffusion coefficients measured by NMR.6
As a complement to experimental techniques, stacking of nucleic acid bases has been studied by a wide variety of computational methods including calculations based on the Poisson-Boltzmann equation,7 explicit solvent techniques such as Monte Carlo8 and molecular dynamics (MD) simulations,9 quantum mechanical calculations,10 and even combined quantum-mechanical/molecular-mechanical QM/MM approaches.11 The thermodynamics of intramolecular stacking interactions, such as those found in DNMPs, have also been studied computationally by a number of groups using explicit-solvent molecular dynamics (MD) methods. Comprehensive studies of the DNMPs were pioneered by the Nilsson group12 who computed the thermodynamics of stacking in all 16 possible DNMPs for both DNA13 and RNA14 by computing potentials of mean force (PMFs) with umbrella sampling techniques; later work by the same group carried out simulations over a range of temperatures to compute enthalpies of stacking15 and examined the effects of solvents, other than water, on stacking thermodynamics.16 The Okamoto group reported similar PMF calculations17 for all 16 DNA DNMPs using replica exchange techniques in combination with umbrella sampling; quite different results were obtained from those reported by the Nilsson group but these differences were suggested to be most likely due to the use of different parameter sets (force fields) in the simulations.18 Finally and most recently, the Florian group reported a comprehensive equilibrium MD simulation study of the stacking free energies in all possible DNA and RNA DNMPs at 310 K;19 the same paper also reported an extensive analysis of the orientational preferences of the bases in stacked conformations.
Here, we aim to build on these previous studies by simulating all 16 possible DNMPs and all 10 possible NS pairs for both DNA and RNA and by providing direct comparisons of the resulting stacking free energies with experimental data. We use current iterations of the AMBER simulation force fields for both DNA and RNA – both of which have been significantly revised by a number of groups recently20 – and employ simulation times that are many times longer than those used previously (1–3 μs vs. 40 ns19). These simulations allow us to directly compare stacking thermodynamics observed in DNMPs – in which the bases are covalently linked to each other – to those observed in NS pairs – in which the bases are not covalently linked. Quantitative comparisons with experimental data for both NS-pair and DNMP systems suggest that the present simulations, carried out with current force fields, produce results that are somewhat better correlated with experiment than previous results even though, in all cases, the overall strength of stacking appears to be significantly overestimated.
Methods
Initial structures of all nucleic acids simulated here were generated using the Stroud group’s Make-NA server (http://structure.usc.edu/make-na/server.html) and formatted to be recognizable by the MD simulation program GROMACS version 4.6.5.21 Each DNMP and pair of NSs was simulated separately in a 35 × 35 × 35 Å box to which periodic boundary conditions were applied. Each nucleic acid was modeled using the AMBER parm9922 force field supplemented with the bsc0 parameters20a that improve modeling of α and γ torsions, and with improved parameters for the glycosidic torsions for both RNA (χOL3)20d, 23 and DNA (χOL4)20e; for RNA, this set of parameters is now the set recommended for use in AMBER.20e Water was modeled explicitly using the TIP4P-Ew model;24 we selected this model for use in simulations because one of our long-term goals is to study protein-DNA interactions and the combination of the AMBER protein force field with the TIP4P-Ew water model is established as one of the best currently available for modeling peptide systems.25 Na+ and Cl− ions – which were added to 150 mM concentrations in order to crudely mimic physiological concentrations – were modeled using the appropriate set of parameters derived by Joung & Cheatham;26 for simulations of DNMPs, an additional Na+ ion was included to ensure electroneutrality of the system. A view of a typical DNMP system as simulated is shown in Figure S1.
All systems were equilibrated for 1.35 ns, with the temperature being raised incrementally from 50 to 298 K over the course of the first 350 ps. Following this equilibration period all MD simulations were carried out for a production period of 1 μs; in the case of NS-pairs, a single simulation of each system was found to be sufficient; in the case of DNMPs, 3 independent 1 μs simulations of each system were performed in order to improve statistics. Pressure and temperature were maintained at their equilibrium values using the Parrinello-Rahman27 barostat and Nosé28-Hoover29 thermostat, respectively. All covalent bonds were constrained to their equilibrium lengths with LINCS,30 allowing a 2.5 fs timestep to be used. Short-range van der Waals and electrostatic interactions were truncated at 10 Å and longer-range electrostatic interactions were computed using the smooth Particle Mesh Ewald method.31 During the production period of the simulations, all solute coordinates were saved at intervals of 0.1 ps giving a total of 10 million snapshots for each simulation for subsequent analysis.
A number of studies have used alternative geometric criteria to define stacked configurations of nucleic acid bases. The Nilsson group used the distance between the glycosidic nitrogens as a reaction coordinate, with distances below 4.5 or 5 Å, for example, being used to define stacked configurations.13–14 In a combined study using MD simulations together with NMR J-coupling data to examine four RNA DNMPs, the Sychrovský group included additional criteria for the angle between base planes and the angle between vectors oriented along each base plane.32 Finally, the Florian group proposed another very reasonable definition in which the distance between the centers of mass and the angle between the base planes were creatively combined to produce a single reaction coordinate.19 Here, we use a similar but different approach to those used in these previous studies and define stacking using a combination of three criteria: (1) a minimum distance between any pair of heavy atoms in the two bases < 4 Å, (2) a distance between the center of mass of each base of < 5 Å, and (3) a vector angle between the normals to the planes of the two bases between 0 – 45° or between 135 – 180°. The first two of these properties were measured for each snapshot using the standard GROMACS utilities g_mindist and g_dist respectively, the third was measured using a script written in-house. Snapshots simultaneously satisfying all three criteria were considered to be stacked; the appropriateness of this assignment was visually checked for select systems.
Relative free energies of stacking, ΔGstack, were then computed according to the following equations. For DNMPs, ΔGstack = −RT ln (Pstacked / Punstacked), where Pstacked and Punstacked represent, respectively, the populations of the stacked and unstacked conformations. For NS-pairs, we also account for the concentrations of the solutes, which are 0.039 M for the simulation boxes used here; in this case, we use ΔGstack = −RT ln (Pstacked / Punstacked / 0.039 M). Error bars for the computed ΔGstack values of NS-pair systems were obtained by splitting each 1 μs production trajectory into 3 contiguous blocks (0–0.333, 0.333–0.667, 0.667–1.000 μs), computing ΔGstack for each block, and calculating the standard deviation of the three values. Error bars for the computed ΔGstack values of DNMPs were obtained as the standard deviation of the three ΔGstack values obtained for each independent 1 μs production simulation.
In order to compare our computed ΔGstack values with data reported by other groups, either obtained computationally or experimentally, all literature-reported equilibrium constants for stacking, Kstack, were converted to free energies using the standard relation ΔGstack = −RT ln Kstack, with T set to the value used in the original study. Where ΔHstack and ΔSstack terms were instead reported,33 these were used to compute a ΔGstack value appropriate to 298.15 K. The experimental ΔGstack datasets for RNA DNMPs that appear to be the most reliable (see Results) are listed in Table S1.
Finally, in addition to examining stacking interactions we also considered T-shaped configurations of the bases that have been identified as potential intermediates during the stacking-unstacking transition by the Florian group.34 These configurations were defined according to the following criteria: (1) a minimum distance between any pair of heavy atoms in the two bases < 4 Å, and (2) a vector angle between the normals to the planes of the two bases between 45 – 135°. Free energy differences between the T-shaped and stacked configurations, ΔΔGTshape-stack, were then computed according to: ΔGTshape-stack = −RT ln (PTshape / Pstacked), where PTshape and Pstacked represent, respectively, the populations of the T-shaped and stacked configurations.
Results
Adequacy of sampling
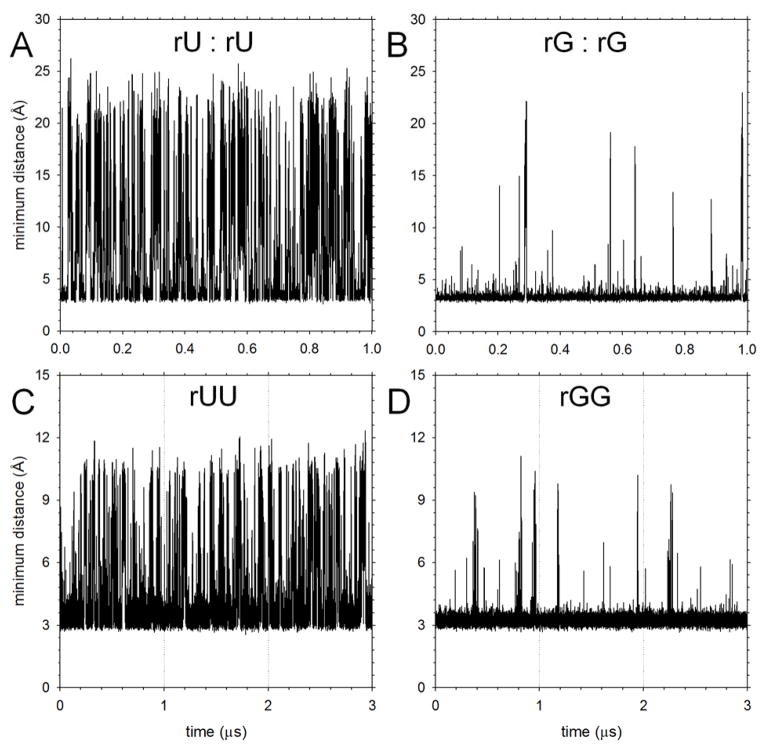
As outlined in Methods, we computed stacking free energies, ΔGstack, for each system by directly measuring the populations of stacked and unstacked conformations during the MD simulations. Stacked conformations are identified on the basis of three criteria (see Methods), one of which involves the minimum distance measured between any of the heavy atoms of the two bases. To provide an indication of the extent of sampling achieved during the 1 μs MD simulations, Figure 1 shows how this distance between bases varies with simulation time for four representative systems; complete sets of plots for all systems are shown in Figures S2–S5. Figures 1A and 1B show, respectively, the sampling in simulations of the RNA NS pairs rU+rU and rG+rG; these systems are, respectively, the most weakly and most strongly stacked of any of the modeled NS pairs. Figures 1C and 1D show, respectively, the sampling in simulations of the RNA DNMPs rUU and rGG, which again, we find to be among the most weakly and strongly stacked of the DNMPs; for these cases, dashed vertical lines are used to indicate that the data shown represent the results of three independent 1 μs simulations (see Methods) that have been concatenated together. For NS-pair systems, the weakest and strongest interacting pairs are found to be stacked 33 and 81% of the time respectively; for the DNMPs, the corresponding numbers are 55 and 97%.
Figure 1. Sampling of stacked conformations versus simulation time.
A. Plot showing the minimum distance between heavy atoms of the two bases in a MD simulation of two rU nucleosides as a function of simulation time. B. Same as A but showing results for simulation of two rG nucleosides. C. Same as A but showing results for simulation of the dinucleoside monophosphate rUU; the data shown are concatenated from 3 independent MD simulations of 1 μs duration. D. Same as C but showing results for rGG.
For weakly stacked systems (Figures 1A and 1C) the 1 μs MD simulations sample many stacking and unstacking events, the former being identifiable by minimum distances in the range of 3–4 Å. The distances sampled during unstacked periods of the NS-pair simulations are typically greater than those seen with the DNMPs since in the case of NS pairs the bases are not covalently linked to each other: once unstacked, therefore, they are free to explore the entire simulation box. For strongly stacked systems (Figures 1B and 1D), transitions to fully unstacked states are much less frequent but occur with sufficient frequency that relatively low standard deviations are obtained for all of the computed stacking free energies. In particular, the standard deviations of the ΔGstack values of the 32 DNMP simulations average 0.16 kcal/mol, with the largest individual value being a standard deviation of 0.34 kcal/mol obtained for the dAT DNMP; for the 20 NS-pair systems, the standard deviations of the ΔGstack values average 0.05 kcal/mol. Additional replicate simulations performed for five NS-pair systems chosen at random produced ΔGstack values in excellent agreement with the original simulations, further suggesting that the values reported here are likely to be reliable (Figure S6).
The full set of computed ΔGstack values for DNMPs and NS-pair systems are listed in Tables 1 and 2, respectively. We note in passing that although we define stacked conformations here using a combination of three geometric criteria (see Methods), we obtain very similar trends using less restrictive definitions in which the requirement that the ring centers be separated by < 5 Å is dropped (see Figure S7A for DNMPs) or in which the only criterion for stacking is that the minimum distance between any pair of heavy atoms of the two bases is < 4 Å (see Figure S7B). Free energies computed with the latter, distance-only criterion implicitly include conformations that are effectively stacked but which do not quite satisfy simultaneously all three of the geometric criteria; they also include non-stacked conformations in which the two bases are in physical contact (e.g. hydrogen bonding arrangements). None of the qualitative trends described in what follows are changed when this less restrictive criterion is used instead.
Table 1.
Computed stacking free energies for all DNA and RNA DNMPs at 298.15 K.
| DNA DNMP | ΔGstack (kcal/mol) | RNA DNMP | ΔGstack (kcal/mol) |
|---|---|---|---|
| AA | −0.89 ± 0.10 | AA | −1.64 ± 0.10 |
| AC | −0.72 ± 0.09 | AC | −1.25 ± 0.24 |
| AG | −1.26 ± 0.32 | AG | −1.72 ± 0.30 |
| AT | −1.50 ± 0.34 | AU | −1.36 ± 0.11 |
| CA | −0.27 ± 0.19 | CA | −0.60 ± 0.03 |
| CC | −0.00 ± 0.10 | CC | −0.37 ± 0.17 |
| CG | −0.40 ± 0.10 | CG | −0.83 ± 0.15 |
| CT | −0.68 ± 0.06 | CU | −0.50 ± 0.15 |
| GA | −1.26 ± 0.32 | GA | −2.13 ± 0.08 |
| GC | −1.52 ± 0.11 | GC | −1.78 ± 0.26 |
| GG | −1.65 ± 0.29 | GG | −1.87 ± 0.18 |
| GT | −1.59 ± 0.13 | GU | −1.52 ± 0.25 |
| TA | −0.29 ± 0.08 | UA | −0.51 ± 0.12 |
| TC | −0.23 ± 0.12 | UC | −0.38 ± 0.05 |
| TG | −0.37 ± 0.16 | UG | −0.53 ± 0.05 |
| TT | −0.57 ± 0.20 | UU | −0.06 ± 0.08 |
Table 2.
Computed stacking free energies for all DNA and RNA NS-pairs at 298.15 K.
| DNA NS pair | ΔGstack (kcal/mol) | RNA NS pair | ΔGstack (kcal/mol) |
|---|---|---|---|
| A + A | −2.41 ± 0.07 | A + A | −2.41 ± 0.05 |
| A + C | −2.06 ± 0.08 | A + C | −2.17 ± 0.03 |
| A + G | −2.54 ± 0.05 | A + G | −2.61 ± 0.02 |
| A + T | −2.25 ± 0.07 | A + U | −2.10 ± 0.09 |
| C + C | −1.93 ± 0.06 | C + C | −1.76 ± 0.04 |
| C + G | −2.24 ± 0.06 | C + G | −2.30 ± 0.06 |
| C + T | −2.01 ± 0.11 | C + U | −1.75 ± 0.04 |
| G + G | −2.66 ± 0.06 | G + G | −2.77 ± 0.06 |
| G + T | −2.46 ± 0.04 | G + U | −2.25 ± 0.03 |
| T + T | −1.82 ± 0.05 | U + U | −1.51 ± 0.00 |
Comparison with experiment & previous computational results
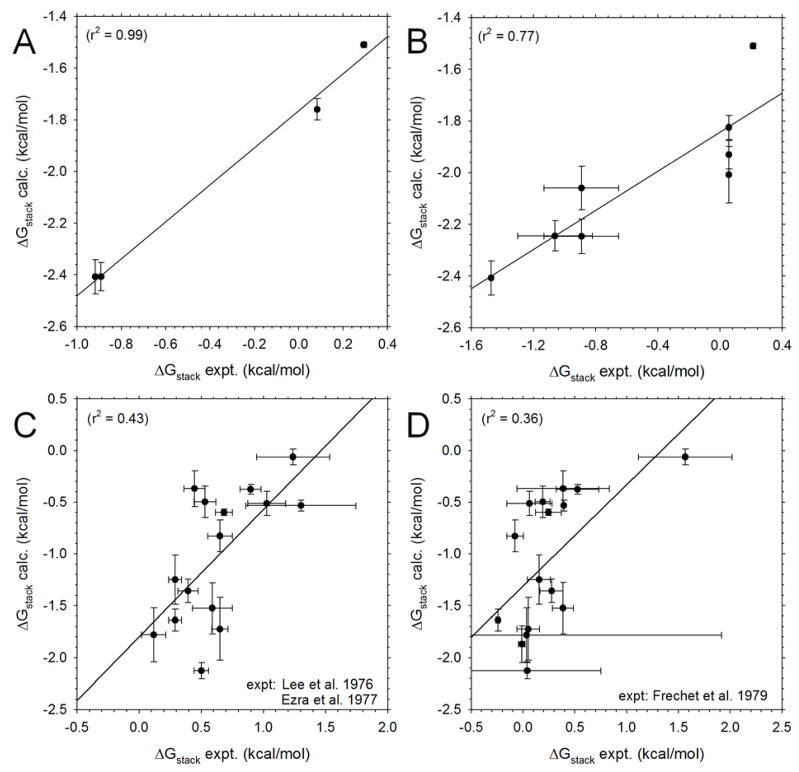
Before examining the full set of predictions in detail, we consider the extent to which our computed ΔGstack values agree with prior experimental estimates. As noted by the Florian group,19 it is a challenge to find appropriate experimental data to fully validate the computed thermodynamics of stacking interactions and since there are often significant differences between values reported by different groups14 or obtained by different methods we have restricted ourselves to comparing with data obtained by a single group using a single methodology. For the NS pairs, vapor pressure osmometry measurements were used by the Ts’o group to determine equilibrium constants for stacking interactions of rC and rU,5a with a subsequent study also providing numbers for rA and dA.5b We plot their data versus our results for the same systems in Figure 2A. The Pearson correlation coefficient is 1.00 indicating very good qualitative agreement, but it is clear that the computed ΔGstack values are significantly more favorable than the experimental estimates. Since it is not hugely surprising to find that the simulations correctly reproduce the relative stacking free energies of purine (Pu+Pu) and pyrimidine (Py+Py) self-associations, it is more appropriate to focus on the more impressive result that the simulations also successfully capture the relative strengths of self-association of rC and rU: the experimental data for rC and rU (Kstack values of 0.87 and 0.61 M−1 respectively) correspond to a ΔΔGstack of 0.21 kcal/mol, which can be compared with our value of 0.25 kcal/mol. The very small difference between the experimental data5b for dA (dimerization Kstack = 4.7 M−1) and rA (Kstack = 4.5 M−1) is also in good agreement with what is found in our simulations (Figure 2A).
Figure 2. Comparison of computed stacking free energies with experiment.
A. Comparison of ΔGstack values computed for four NS pairs (rU, rC, rA and dA) with values calculated from osmometry data reported by Ts’o et al.5a, 5b B. Same as A but comparing the following NS pairs (dA+dA, dC+dC, dT+dT, rU+rU, dA+dC, dA+dT, dC+dT, dG+dC) with values reported by Solie & Schellman.5c C. Comparison of ΔGstack values computed for 15 RNA DNMPs with corresponding values derived from 1H NMR data measured by Sarma et al.2b, 2c and compiled by Davies2d; no experimental data were reported for rGG. Error bars for experimental data obtained from reported uncertainties in stacking populations.2d D. Same as C but showing a comparison with ΔGstack values derived from hypochromism measurements by Frechet et al.33a
A considerably broader set of data were described by Solie and Schelmann5c in a report using thermal osmometry, ultracentrifugation and hypochromicity measurements. Experiments of the first type were conducted on a number of single component nucleoside systems (dA, dT, dC and rU) as well as on a number of mixed systems (dA+dT, dG+dC, dA+dC, dA+rU, dT+dC); the mixed systems were interpreted in terms of a single effective association constant which, as noted by the authors, will be some composite of the three possible types of associations in each system (e.g. for dA+dT the data will, in reality, reflect the combined effects of dA:dA, dT:dT and dA:dT interactions). A comparison of our data with those of Solie and Schellman for all systems studied by them is shown in Figure 2B. The Pearson correlation coefficient, Rcorr, in this case is 0.88, and improves further when only the single-component systems are considered (Rcorr = 0.92); again, however, it is noticeable that the computed ΔGstack values are significantly more favorable than the experimental estimates.
For the RNA DNMPs, we have three different sources of experimental data, and two different sets of computational data to compare with; the correlation coefficients obtained for all combinations of data sets are listed in Table 3. Figure 2C compares our computed ΔGstack values with those derived from 1H NMR J-coupling constants for 15 of the 16 RNA DNMPs reported by the groups of Danyluk and Sarma2b, 2c and reviewed by Davies.2d Figure 2D compares our computed ΔGstack values with those derived from temperature-dependent hypochromism measurements reported by Frechet et al.33a Both figures make clear that the level of agreement between the computations and experiments is only modest, with Rcorr being 0.66 and 0.60 for Figures 2C and 2D respectively (in the latter case, Rcorr drops to 0.55 when the outlier rUU datapoint at the top-right of Figure 2D is removed). This level of agreement, however, is comparable to that obtained when these two sets of experimental data are compared with each other: in that case, Rcorr = 0.60 (see the intersection of the “Danyluk” and “Frechet” entries in Table 3). Yet again, we find that the computed values are significantly more favorable than the corresponding experimental estimates (Figures 2C and 2D).
Table 3. Correlation coefficients for computed and experimental stacking free energies for RNA DNMPs.
The three columns headed “This work”, “Nilsson”, and “Florian” represent computational datasets; the three columns headed “Danyluk”, “Frechet” and “Tinoco” represent experimental datasets.
| This work | computation Nilssona | Florianb | Danylukc | experiment Frechetd | Tinocoe | |
|---|---|---|---|---|---|---|
| This work | 1.00 | 0.52 | 0.84 | 0.66 | 0.60 | 0.25 |
| Nilsson | 0.52 | 1.00 | 0.45 | 0.27 | 0.45 | 0.16 |
| Florian | 0.84 | 0.45 | 1.00 | 0.24 | 0.38 | 0.13 |
| Danyluk | 0.66 | 0.27 | 0.24 | 1.00 | 0.60 | 0.20 |
| Frechet | 0.60 | 0.45 | 0.38 | 0.60 | 1.00 | 0.20 |
| Tinoco | 0.25 | 0.16 | 0.13 | 0.20 | 0.20 | 1.00 |
Encouragingly, the agreement with experiment achieved here is better than is obtained using previous computational estimates. Comparison of the computed values reported almost twenty years ago by the Nilsson group with the 1H NMR-derived and hypochromism-derived datasets, gives Rcorr = 0.27 and 0.45 respectively (Figures S8A and S8B). Comparison of the computed results reported more recently by the Florian group with the same two experimental datasets gives Rcorr = 0.24 and 0.38, respectively (Figures S8C and S8D). Amongst the simulation datasets the level of correspondence is highest between our results and those of the Florian group (Rcorr = 0.84): this is to be expected given that both used variants of the AMBER force fields.
We note that there is a third set of experimental data to which comparison can be made: this is the optical rotation dispersion (ORD) data set reported by Davis & Tinoco 46 years ago.33b We obtain very poor agreement between those data and our simulation results (Rcorr = 0.25; Table 3). But we observe still poorer agreement when the ORD-derived measurements are compared with the two other experimental datasets and the two computational datasets (see comparison of “Tinoco” entry with all the other datasets considered here (see Table 3). For this reason we have not considered the ORD-derived dataset further.
Finally, we have considered whether better correspondence between the various data sets might be obtained if ΔGstack values are grouped according to the purine/pyrimidine composition of the DNMPs (i.e. Pu-Pu, Pu-Py, Py-Pu, Py-Py). The data grouped and averaged in this way for all data sets are shown in Table S2 and the resulting table of correlation coefficients is shown in Table 4. Among the computational data sets, the correlation coefficients are improved considerably by grouping the data in this way, as is the correspondence between the computational results reported here and the 1H NMR-derived and hypochromism-derived datasets. Notably, however, the agreement between these two sets of experimental data is worsened by grouping the data according to composition; this further indicates that there are likely to be limits to what can be concluded about the quality of current force fields from comparison with these experimental data.
Table 4. Correlation coefficients for computed and experimental stacking free energies for RNA DNMPs when grouped by purine/pyrimidine composition.
Same as Table 3 but showing results obtained when the ΔGstack values are grouped and averaged by base composition (see Table S2 for the averaged ΔGstack values).
| This work | computation Nilssona | Florianb | Danylukc | experiment Frechetd | Tinocoe | |
|---|---|---|---|---|---|---|
| This work | 1.00 | 0.66 | 0.97 | 0.84 | 0.79 | 0.48 |
| Nilsson | 0.66 | 1.00 | 0.63 | 0.39 | 0.89 | −0.10 |
| Florian | 0.97 | 0.63 | 1.00 | 0.71 | 0.84 | 0.64 |
| Danyluk | 0.84 | 0.39 | 0.71 | 1.00 | 0.38 | 0.20 |
| Frechet | 0.79 | 0.89 | 0.84 | 0.38 | 1.00 | 0.35 |
| Tinoco | 0.48 | −0.10 | 0.64 | 0.20 | 0.35 | 1.00 |
Comparisons of computed stacking free energies
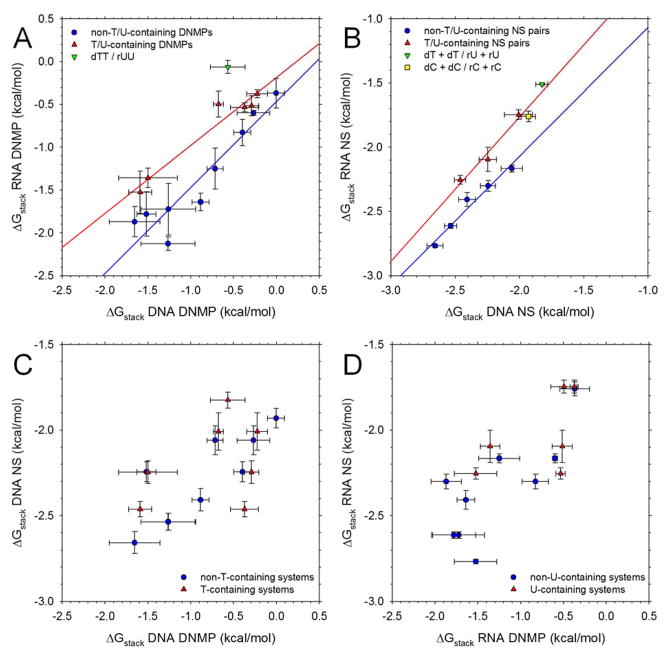
We now proceed with a more detailed examination of the computed results reported here. Figure 3 compares the computed stacking free energies of different types of systems in various combinations. Figure 3A compares the ΔGstack values of RNA DNMPs with those of the corresponding DNA DNMPs. For reasons that will become clear below, we plot the results for DNMPs that contain T or U bases separately from those that do not. For the non-T/U-containing systems, the ΔGstack values of the RNA DNMPs are in general significantly more favorable than those of the corresponding DNA DNMPs: linear regression of these data (blue line) gives a slope of 1.00, an intercept of −0.47 kcal/mol, and an Rcorr of 0.94. In contrast, for the systems that contain a single T or U base, the ΔGstack values of the RNA and DNA systems are more similar: linear regression (red line) gives a slope of 0.79, an intercept of −0.18 kcal/mol, and an Rcorr of 0.97. In general, therefore, we can conclude that with the present iteration of the AMBER force field, RNA DNMPs stack significantly more favorably than their corresponding DNAs, but that this difference can be cancelled out if the corresponding sequences contain T or U bases. In fact, for the DNMPs rUU and dTT the effect is reversed: the ΔGstack value for dTT is 0.5 kcal/mol more favorable than that of rUU (see green downward triangle in Figure 3A).
Figure 3. Comparison of stacking free energies for different nucleic acid types.
A. Plot comparing ΔGstack values of RNA DNMPs with those of the corresponding DNA DNMPs; error bars indicate the standard deviation of the ΔGstack values computed from 3 independent 1 μs trajectories; blue and red lines indicate linear regressions. Data are separated into groups with separate regression lines drawn for those DNMPs that do not contain any T or U bases (blue) and those that do contain T or U bases (red); the special case of dTT/rUU (green symbol) is not included in any regression (see text). B. Same as A but comparing ΔGstack values of RNA nucleoside (NS) pairs with those of the corresponding DNA NS pairs; error bars indicate the standard deviation computed from separating each 1 μs trajectory into three blocks. C. Same as A but comparing ΔGstack values of DNA nucleoside (NS) pairs with those of the corresponding DNA DNMPs. D. Same as A but comparing ΔGstack values of RNA nucleoside (NS) pairs with those of the corresponding RNA DNMPs.
Figure 3B compares the ΔGstack values of RNA NS pairs with those of the corresponding DNA NS pairs; again we plot the results for NS pairs that contain T or U bases separately from those that do not. Datapoints for those pairs that do not involve T or U bases are plotted in blue; linear regression of these data (excluding the C+C pair; see below) gives a slope of 1.00, an intercept of −0.06 kcal/mol, and an Rcorr of 0.98 (blue line). There is, therefore, a near-perfect agreement between the ΔGstack values calculated for corresponding DNA and RNA systems that contain only A, C and G bases; given the very small structural differences between the two sets of systems this is to be expected if the simulation data are appropriately converged. Datapoints for NS pairs that contain a single T or U base are plotted in red; linear regression of these data gives a slope of 1.12, an intercept of 0.48 kcal/mol, and an Rcorr of 0.98 (red line). In the case of NS pairs that contain a single T or a U, therefore, there is again a clear offset in the data, with the computed ΔGstack values for the DNA systems being significantly more favorable than those of the corresponding RNA systems. In fact, for the 3 T/U-containing datapoints the ΔGstack values of the DNA systems are on average 0.21 +/−0.06 kcal/mol more favorable than those of the corresponding RNA system. For the 5 datapoints that do not involve T/U NS pairs (used for blue line), the ΔGstack values of the DNA systems differ by on average only −0.07 +/− 0.04 kcal/mol (i.e. they are slightly less favorable for DNA than for RNA). These results indicate, therefore, that for NS pairs, the thymine methyl group makes a favorable contribution to the stacking free energy of ~0.3 kcal/mol. An interesting explanation for the apparently anomalous behavior of the C+C system is proposed below.
Figure 3C compares the ΔGstack values of DNA NS pairs with those of DNA DNMPs; Figure 3D compares those of RNA NS pairs with those of RNA DNMPs. Again, for consistency with Figures 3A and 3B we have used separate coloring schemes for those systems that contain T/U bases and those systems that do not. In these cases, however, there is no longer such a clean separation of the two datasets; the most notable aspect of both Figures is that the ΔGstack values of the NS pairs are somewhat more favorable than those of the corresponding DNMPs.
Sequence dependence of computed stacking free energies in DNMPs
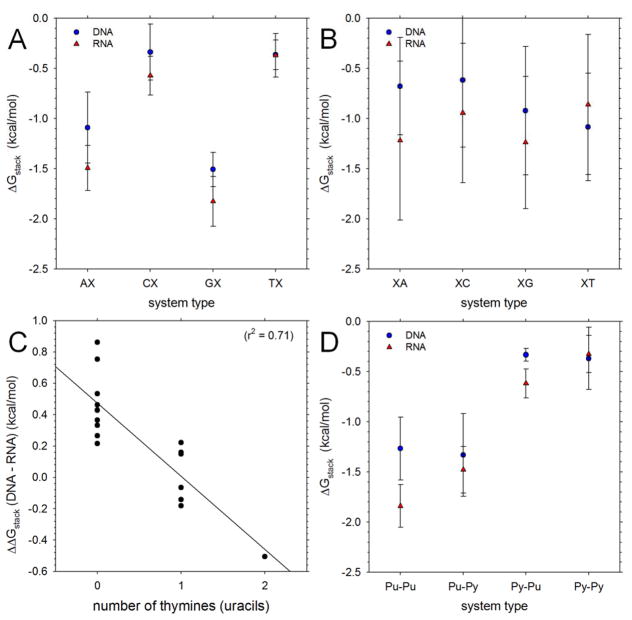
For the DNMP systems it is of interest to combine data from separate simulations in order to obtain an averaged view of the ΔGstack values associated with each type of base at both the 5′ and 3′ positions. To this end, Figure 4A shows the average ΔGstack values for each type of base at the 5′ position. For both DNA and RNA it can be seen that the most favorable ΔGstack values are associated with either an adenine or a guanine (i.e. a purine) at the 5′ position of the DNMP. As expected from the data described earlier, however, it is also clear that, with the exception of 5′-T/U-containing DNMPs (far right of Figure 4A), the ΔGstack values are on average significantly more favorable for RNA systems (red triangles) than for the corresponding DNA systems (blue circles). Figure 4B shows the average ΔGstack values for each type of base at the 3′ position. Here, the trends with respect to the identity of the 3′ base are less dramatic, but for RNA DNMPs we again see that purines are associated with generally more favorable ΔGstack values than pyrimidines; for DNA DNMPs, on the other hand, we see that the average ΔGstack value of DNMPs containing a 3′-T is more favorable than that of any other type of 3′-base.
Figure 4. Computed stacking free energies averaged for given sequence types. A.
Plot ′ showing average ΔGstack values of DNA and RNA DNMPs for each type of base at the 5′ position; error bars indicate the standard deviation of all sequences contributing to the averages: error bars for entry ‘AX’ for example, represent the standard deviation of ΔGstack values of AA, AC, AG and AT DNMPs. Note that with the regard to the labeling of the x-axis, T is substituted by U in RNA simulations. B. Same as A but showing average ΔGstack values of DNA and RNA DNMPs for each type of base at the 3′ position. C. Plotting showing difference of ΔGstack values of DNA and RNA DNMPs versus number of thymines (uracils) in the DNMP. D. Same as A but showing average ΔGstack values of DNA and RNA DNMPs for each purine/pyrimidine composition.
The observation that in both Figures 4A and 4B the results involving T or U bases are distinct from those of the other bases suggests that the differences in ΔGstack values between corresponding DNA and RNA DNMPs are likely to be a function of the number of T/U bases in the sequence. Figure 4C shows that this is indeed the case: when the differences in ΔGstack values of the DNA and RNA DNMPs are plotted versus the number of T or Us in each DNMP a strong anti-correlation is observed (Rcorr = 0.84). For DNMPs that contain no T/Us the RNA ΔGstack values are on average 0.47 +/− 0.22 kcal/mol more favorable than those of the corresponding DNA DNMP. For those DNMPs that contain 1 T/U, this difference is effectively negated: the differences between the RNA and DNA ΔGstack values average 0.02 +/− 0.17 kcal/mol. Finally, for the DNMPs that contain 2 T/U bases (i.e. dTdT and rUrU), the effect is completely reversed: the RNA ΔGstack value is 0.51 kcal/mol less favorable than that of the corresponding DNA.
Finally, Figure 4D shows the ΔGstack values averaged according to the purine/pyrimidine nature of the base at each position (see earlier). In this case, it is apparent that for RNA there is a clear trend that the ΔGstack values increase (i.e. become less favorable) in the order Pu-Pu < Pu-Py < Py-Pu < Py-Py. This trend is disrupted in the case of DNA, being replaced with Pu-Pu ~ Pu-Py < Py-Pu ~ Py-Py; again, however, when systems containing T or U bases are omitted from consideration, a clearer picture emerges (Figure S9). Interestingly, regardless of whether T/U bases are included or not, with both RNA and DNA, the ΔGstack values of Pu-Py DNMPs are typically much more favorable than those of the corresponding Py-Pu DNMPs: for RNA, they are 0.86 +/− 0.15 kcal/mol more favorable, while for DNA, they are 1.00 +/− 0.37 kcal/mol more favorable. For comparison, the 1H data also indicate that the ΔGstack values of Pu-Py DNMPs are more favorable than those of Py-Pu DNMPs2b–d (by 0.57 +/− 0.14 kcal/mol), while the hyperchromicity data of Frechet et al.33a suggest that they are less favorable by 0.06 +/− 0.13 kcal/mol.
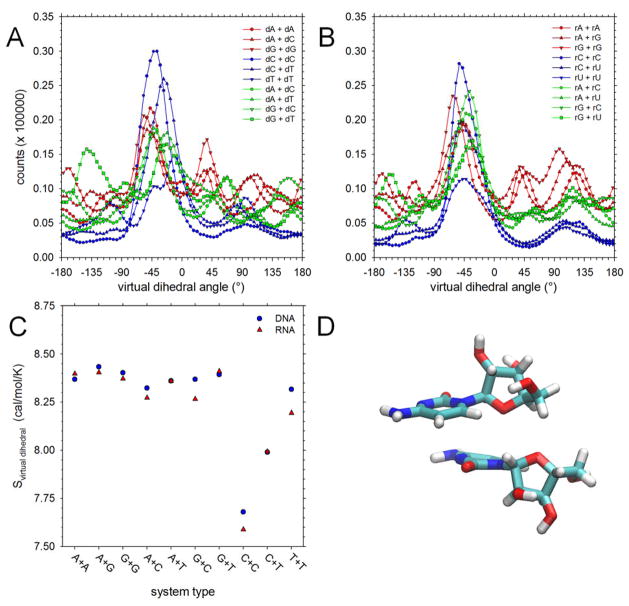
Relative base orientations in stacked NS pairs
In contrast to DNMPs, the association of NS pairs is not subject to geometric constraints imposed by the covalent linkage provided by the sugar-phosphate backbone. It is therefore possible to imagine that the stacked configurations of NS pairs might freely adopt a wide range of rotational orientations of the two bases. To explore this question we calculated, for every stacked snapshot of the bases, the value of a virtual dihedral angle connecting the center of each base via their C1′ atoms. Histograms of these virtual dihedral angles are shown in Figure 5 for all of the simulated systems: Figures 5A and 5B show, respectively, the histograms sampled by the DNA and RNA NS pairs; Figures S10A and S10B show, respectively, the corresponding results for the DNA and RNA DNMPs. The results for the latter are neither surprising nor especially informative: in a DNMP, the intramolecular linkage between the two bases provided by the backbone places significant constraints on the relative orientations that the two bases can adopt in a stacked conformation. It is no surprise, therefore, to find that the histogram of virtual dihedral angles is restricted to a (comparatively) narrow range spanning ~−90° to ~+120°.
Figure 5. Orientational preferences of NS bases involved in stacking interactions.
A. Histograms of values of the virtual dihedral angle describing relative orientations of bases in stacked configurations of DNA NS pairs. B. Same as A but showing values for RNA NS pairs. C. Configurational entropies, Svirtual dihedral, computed using the histograms shown in A and B for DNA and RNA NS pairs. Note that with the regard to the labeling of the x-axis, T is substituted by U in RNA simulations. D. Representative snapshot of the rC + rC system in a configuration corresponding to the peak shown panel B.
The results for the NS pairs, however, are more interesting (Figures 5A and 5B). In particular, while the histograms for NS pairs in which both nucleosides are purines (red), or in which one nucleoside is a purine and the other pyrimidine (green) show quite broad distributions spanning ~−180° to ~+180°, the histograms for NS pairs in which both nucleosides are pyrimidines (blue) show a strong preference for virtual dihedral angles of ~−45°. This trend is especially pronounced for the C+C system. It is possible to convert the histograms into effective configurational entropies using the standard relation S = –R Σ pi ln(pi), where pi represents the probability of finding a given value of the virtual dihedral angle (binned in 5° intervals), R is the gas constant and the summation extends over all of the bins from −180° to 180°. The resulting entropies, which we term Svirtual dihedral, are compared for all NS pair systems in Figure 5C, from which it can be seen that C+C and C+T(U) systems produce entropies that are clearly much lower than those of other systems. In passing we note that the fact that very similar Svirtual dihedral values are obtained for the DNA and RNA systems provides a further indication of the convergence of the simulation data.
The origins of this surprising orientational preference are difficult to determine unambiguously. Figure 5D shows a representative snapshot of the rC+rC system when in the most populated conformation. From this, it would appear that favorable nonbonded interactions between the ribose/deoxyribose rings are likely to play a significant role in stabilizing the geometry, although this would not explain the stronger preferences exhibited by the C+C and C+T(U) systems relative to all other systems. Instead, as suggested by a reviewer, it may well be that favorable electrostatic interactions between the exocyclic amino group of one cytosine and the carbonyl group of the other cytosine (or thymidine/uracil) help to dictate the orientational preference. Such interactions have been shown experimentally to make significant contributions to the favorable stacking of a 3′ dangling U onto a 3′ terminal C:G basepair;35 a potentially pivotal role for such interactions would also be consistent with the observation that the Svirtual dihedral values for the rU+rU or dT+dT systems are not unusual (see Figure 5C), as the bases in these systems lack exocylic amino groups.
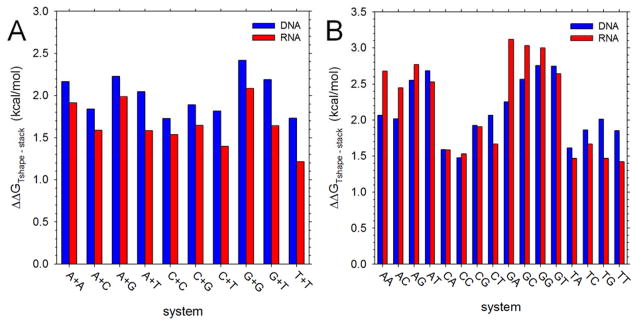
Relative free energies of T-shaped geometries
Finally, following the work of the Florian group34 who identified them as intermediate structures along the stacking-unstacking transition, we determined the extent to which T-shaped base-base geometries are sampled during the simulations (see Methods). Figure 6A shows the ΔΔGTshape-stack values, i.e. the free energies of T-shaped geometries relative to those of stacked geometries, for all NS pairs; Figure 6B shows the same for DNMPs. For NS pairs (Figure 6A), the T-shaped geometries are computed to be less stable than the more conventionally stacked geometries by anywhere from 1 to 2.5 kcal/mol. Interestingly, however, in all cases the relative free energies of T-shaped geometries are significantly more favorable for RNA NS pairs than for the corresponding DNA NS pairs: for NS pairs that contain no T or U bases, the difference amounts to an average of 0.25 +/−0.05 kcal/mol, whereas for NS pairs that contain 1 T or U base, the average difference is even greater: 0.48 +/−0.05 kcal/mol. The finding that ΔΔGTshape-stack is generally more favorable for RNA NS pairs than for DNA NS pairs presumably reflects stabilization of the T-shaped geometry in RNA due to hydrogen bonding between the 2′-OH group of the ribose ring of one NS with base heteroatoms of the second NS. The finding that the difference between ΔΔGTshape-stack values of RNA and DNA NS pairs is greater when T- or U-containing bases are involved, on the other hand, likely reflects instead stabilization of the stacked geometry in DNA due to the presence of the thymine methyl group noted earlier.
Figure 6. Relative free energies of T-shaped and stacked geometries.
A. Plot showing the difference in free energies of T-shaped and stacked geometries for all NS pairs; note that with the regard to the labeling of the x-axis, T is substituted by U in RNA simulations. B. Same as A but showing results for DNMPs; again note that T is substituted by U in RNA simulations.
For DNMPs (Figure 6B), the T-shaped geometries are again found to be much less stable than the stacked geometries – by from 1.5 to 3 kcal/mol – but, with the notable exception of T or U-containing DNMPs, the relative free energies of T-shaped geometries are more favorable for the DNA DNMPs than for the corresponding RNA DNMPs. This suggests that stabilization of T-shaped geometries by hydrogen bonding interactions with the ribose ring is not a major factor in the more conformationally constrained DNMPs. If so, then one might expect ΔΔGTshape-stack to be primarily determined by the stability of the stacked conformation; consistent with this, Figure S11 shows that there is a strong anticorrelation between the ΔΔGTshape-stack and the ΔGstack values of both DNA and RNA DNMPs.
Discussion
Molecular dynamics simulations continue to be a valuable method for gaining insights into the dynamics and thermodynamics of nucleic acid systems.36 A large number of studies have recently examined the ability of the AMBER force field,22 in particular, to accurately describe conformational features20a of a variety of nucleic acid systems and important adjustments to the force field to improve the agreement with experiment have been proposed20c, 20e, 23, 37; other studies have directly addressed the impact of these and other force field parameters on the thermodynamics of single-, double- and quadruple-stranded nucleic acid systems.38 The present work is effectively a complement to these other studies as it focuses on the simplest possible systems in which base stacking events are expected to dominate both intra- and intermolecular interactions. Importantly, in addition to highlighting similarities and differences between stacking in DNA and RNA systems, the comparisons with experiment reported here provide an important test of AMBER parameters that are now widely used to model nucleic acid systems; the generally low standard deviations and the close correspondence between results of replicate simulations (e.g. Figure S6) argue that the numbers reported here are reliable representations of the underlying force field. This is also (to our knowledge) the first time that a direct quantitative comparison of computed and experimental stacking free energies of DNMPs has been reported.
With regard to the NS pairs, our computed values are in good qualitative agreement with the experimental data reported by the groups of Ts’o5a, 5b (Figure 2A) and Schellman5c (Figure 2B), even though the computed numbers are consistently too negative. We have already noted that the relative computed ΔGstack values of the two pyrimidines, rC and rU (ΔΔGstack of 0.25 kcal/mol) are nicely in line with the Ts’o group’s data (ΔΔGstack of 0.21 kcal/mol), but they are also consistent with Kstack values derived from diffusion coefficient measurements6 (respective Kstack values for rC and rU of 4.83 and 2.48 M−1, which translate to a ΔΔGstack of 0.39 kcal/mol). Our relative ΔGstack values for dT, rC and rU (0.00, 0.07 and 0.32 kcal/mol, respectively) are also in good agreement with values derived from thermal osmometry data reported by Plesiewicz et al.5e (ΔΔGstack values of 0.0, 0.03 and 0.26 kcal/mol respectively).
Less good, however, is our correspondence with another source of data. Using 1H NMR, Mitchell and Sigel39 reported Kstack values of 15 and <0.5 M−1 for rA and rU, respectively, which correspond to a ΔΔGstack of 2.02 kcal/mol; qualitatively, this is in agreement with our result but at a quantitative level it is significantly greater than our value of 0.90 kcal/mol. Subsequent work by the same group40 added data for rC (Kstack = 1.4 M−1) and reported an estimate for rG of 8 M−1 based on comparisons of data for inosine and GTP: direct measurement of a value for rG was prevented by its low solubility. While the reported values for rA, rC and rU are in qualitative agreement with ours, the estimated value for rG is very clearly at variance with our data: the computed ΔGstack of rG with itself is the most favorable of any of our computed ΔGstack values (Table 2). Solie & Schellman reported5c similar solubility-related difficulties in measuring Kstack for dG, but interestingly, they suggested on the basis of its anomalous behavior that its stacking interaction was likely to be the strongest of any of the NS pairs: this interpretation would be more in line with the results of our simulations.
With regard to DNMPs, we have shown that our computed ΔGstack values for RNA systems correlate moderately well with two different sources of experimental data.2b–d, 33a We should perhaps be careful not to over-interpret this result: as noted above, the level of correlation between the different sets of experimental data is comparatively low (Table 3) and for several DNMPs the disagreement between the experimental estimates is very significant (Table S1). That said, it remains interesting that our values compare somewhat better than the two sets of computed values reported previously; it is to be noted that we did not adjust our criteria for defining stacking in order to optimize agreement with experiment (see Methods). In one respect this result is encouraging, but in another it is simply to be expected: force fields in general have developed considerably in the years since the Nilsson group’s seminal studies12–16 were performed and a number of important updates20a, 20e, 23 have since been made to the AMBER force field used in the Florian group’s studies.19 Differences between our data and the Florian group’s data may also be at least partly due to the different temperatures at which the studies were conducted (298 versus 310 K respectively): the Nilsson group has previously shown that stacking PMFs can be sensitive to temperature,15 an issue that has also been examined in detail by Chen & Garcia (see below).41 We have not explored the effects of temperature here but we have attempted to gauge the potential impact of the most recent of the changes to the AMBER force field – i.e. the reparameterization of the glycosidic bond dihedrals – by repeating our calculations of the DNMPs with only the AMBER parm9922 and bsc020a parameters retained. Interestingly, we obtain a significantly poorer agreement with experiment using the ‘old’ glycosidic dihedral terms (Rcorr = 0.47 and 0.44 for the 1H NMR-derived2b, 2c and the hypochromism-derived33a data, respectively; Figure S12). This suggests that updates to the glycosidic terms – which were made to RNA, for example, in order to improve modeling of syn/anti/high-anti equilibria and to penalize the formation of ‘ladder-like’ structures in simulations of RNA tetraloops23 – may also have improved the force field’s description of the relative stacking thermodynamics of different DNMP systems. As such, it also provides an illustration of how reparameterization of one aspect of a force field can have consequences for the force field’s description of other thermodynamic quantities.
In addition to comparisons with experiment, comparisons of the ΔGstack values that we obtained for corresponding DNA and RNA systems produce a number of insights. The data for the NS pairs, in particular, are interesting for two reasons. First, the clear offset of the datapoints for systems containing T or U bases in Figure 3B indicates that the current iteration of the AMBER force fields predict base stacking interactions in free nucleosides to be stabilized by ~0.3 kcal/mol by the substitution of a T relative to a U. This effect – which is reflected also in the data for DNMPs (Figures 3A, 4A, 4B and 4C) and which was also noticed in the seminal calculations of the Nilsson group14 – is supported by a variety of experimental evidence. A general effect of base methylation increasing self-association of nucleosides (purines) was noticed in the early studies of the T’so group5b and was also echoed in the osmotic experimental data of Plesiewicz et al.5e referred to above; the latter work also showed that progressive addition of alkyl groups to uracil rings promoted stacking significantly albeit in a non-monotonic manner. Further support for the idea that the addition of a methyl group can increase self-association of nucleosides comes from a comparative microcalorimetry study of cytidine and 5-methylcytidine,42 with Kstack values reported in that work (0.74 and 1.1 M−1 respectively) corresponding to a ΔΔGstack 0.23 kcal/mol. Still another source of similar data comes from 1D NMR experiments (at 35 °C) which, for example, report Kstack values of 0.091 and 0.165 M−1 for pyrimidine and 5-methylpyrimidine respectively, again corresponding to a ΔΔGstack of 0.35 kcal/mol.43 Finally, the same effect is apparent in larger nucleic acid systems: in thermal denaturation studies of a variety of double and triple helical systems, the 5-methyl group of T was shown to always provide a stabilizing effect.44
The second, more intriguing finding that emerges from a comparison of the DNA and RNA NS pairs is the anomalous behavior of the C+C system apparent in Figure 3B. The explanation for this behavior appears to lie in the orientational specificity of the stacking interaction for this system that is indicated in Figures 5A and 5B: relative to all other systems, the C+C system produces the strongest preference for some relative orientations of the two bases in stacked geometries. Since these preferred structures have the two sugars in close contact we think that this provides a straightforward explanation for why there might be a more significant difference between the ΔGstack values of the DNA and RNA versions of this NS pair. The finding that the ΔGstack value of the DNA version is slightly more favorable than that of the corresponding RNA system might be consistent with additional hydrophobic interactions between the rings attending the loss of the 2′-OH group.
A third result that emerges from the simulations presented here comes from the DNMPs: this is that stacking in the RNA DNMPs is computed to be significantly more favorable than that in DNA DNMPs. For the following reasons, however, it is unclear to us how much weight to attach to this result. First, it is to be noted that this is not a consistent feature of all computational studies that compared DNA and RNA DNMPs: while the Florian group19 did find that RNA DNMPs are on average 0.19 +/− 0.49 kcal/mol more stable than the corresponding DNAs (0.20 +/− 0.49 kcal/mol when sequences containing a T/U base are omitted), Norberg & Nilsson explicitly noted13 that in the nine DNMPs that did not involve a T/U base, three stacked better in the DNA form, three stacked better in the RNA form and three stacked similarly in both forms. It is also worth noting that while with the ‘new’ glycosidic terms DNMPs that do not contain T/U bases stack more favorably in their RNA form by 0.50 +/− 0.31 kcal/mol, in simulations that use the ‘old’ glycosidic terms the situation is reversed: on average the DNA DNMPs are more stable by 0.41 +/− 0.26 kcal/mol. If, as seems likely, the different modifications made to the glycosidic terms of RNA23 and DNA20e have also changed their relative stacking thermodynamics it further reinforces the point made above that changes to correct one aspect of a force field’s behavior can have significant consequences for other properties.
A second reason for not wishing to overstate the finding that the computed ΔGstack values of RNA DNMPs are more favorable than those of DNA DNMPs is that it appears to be at odds with thermal denaturation data reported and analyzed by the Inoue group:45 for both AA and GG it was reported that the DNA form was significantly more stable than the RNA form. The experimental stacking populations of DNMPs that we compared with here – those due to the Danyluk & Sarma groups2b, 2c – were obtained by comparing J3′4′ coupling constants in RNA DNMPs with those of corresponding 3′ and 5′ mononucleotides. Unfortunately, while corresponding J3′4′ coupling constants were also subsequently reported by the same group for the DNA DNMPs,46 it appears that the significant assumptions47 used to convert these to stacking populations for RNAs do not hold for DNAs,2d which means that a direct comparison of DNA and RNA stacking populations using the 1H NMR data cannot be carried out.
Finally, our simulation results are to be considered in the light of important recent work from Chen and Garcia describing significant adjustments to the AMBER Lennard-Jones parameters in order to allow high-resolution structures of three different RNA tetraloops to be accurately predicted by replica-exchange MD simulations.41 During the course of that work the authors investigated self-interactions of the rG nucleoside and found that in simulations of 32 rG molecules (performed below the solubility limit) stacking-driven aggregation rapidly occurred. This overestimation of stacking interactions was mitigated by scaling of the standard AMBER Lennard-Jones parameters for base-base interactions: appropriately scaled parameters allowed a Kstack value to be obtained that was essentially identical to that reported experimentally by Solie and Schellman using the same method of analysis.5c The reparameterization process was extended by scaling the base-water interactions in order to reproduce entropies of stacking for rAA, and by reparameterizing, as others have done20b, 20c, 20e, 23, the glycosidic dihedral parameters to reproduce relative populations of syn and anti conformers.
Although we have not explored the Chen & Garcia RNA parameters here for reasons of wishing to directly compare the computed free energies of stacking interactions in DNAs and RNAs (different parameterizations of the glycosidic terms, in particular, may be required for DNA20e), their finding that the standard AMBER parameters produce excessively favorable stacking interactions of the rG nucleoside is entirely consistent with the results presented here. Given that this result is also additionally supported by high-level QM calculations,48 the emphasis placed by Chen & Garcia on reparameterizing stacking interactions to better match osmometry and thermal denaturation data appears to be an appropriate direction in which to pursue further development of the AMBER nucleic acid force fields. It would, for example, be especially interesting to determine whether the use of osmometry data to explicitly parameterize nucleosides other than rG would improve the force field’s ability to predict stacking interaction trends seen in RNA DNMPs (Figure 2C).
Supplementary Material
Acknowledgments
The authors wish to thank the anonymous reviewers for a number of suggestions that strengthened the manuscript. This work was supported by NIH R01 GM099865 and R01 GM087290 awarded to A.H.E.
Footnotes
Snapshot of a typical MD system; plots of base-base distance versus time for all simulated systems; comparisons of stacking free energies for replicate simulations; comparisons of stacking free energies with different stacking criteria; comparisons of previously reported computed stacking free energies with experiment; computed stacking free energies averaged by sequence composition omitting sequences that contain T or U bases; orientational preferences of DNMP bases when in stacked conformations; comparison of stacking free energies with the difference in free energy of stacked and T-shaped conformations; comparisons of stacking free energies computed without updated glycosidic terms with experiment; table listing previous experimental estimates of stacking free energies in RNA DNMPs; computed and experimental stacking free energies grouped according to purine/pyrimidine composition. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Saenger W. Principles of Nucleic Acid Structure. Springer-Verlag; New York: 1984. [Google Scholar]
- 2.(a) Chan SI, Nelson JH. Proton magnetic resonance studies of ribose dinucleoside monophosphates in aqueous solution. I. Nature of the base-stacking interaction in adenylyl(3′->5′)adenosine. J Am Chem Soc. 1969;91:168–183. doi: 10.1021/ja01029a033. [DOI] [PubMed] [Google Scholar]; (b) Lee CH, Ezra FS, Kondo NS, Sarma RH, Danyluk SS. Conformational properties of dinucleoside monophosphates in solution: dipurines and dipyrimidines. Biochemistry. 1976;15:3627–3639. doi: 10.1021/bi00661a034. [DOI] [PubMed] [Google Scholar]; (c) Ezra FS, Lee CH, Kondo NS, Danyluk SS, Sarma RH. Conformational properties of purine-pyrimidine and pyrimidine-purine dinucleoside monophosphates. Biochemistry. 1977;16:1977–1987. doi: 10.1021/bi00628a035. [DOI] [PubMed] [Google Scholar]; (d) Davies DB. Conformations of nucleosides and nucleotides. Prog Nucl Magn Reson Spectrosc. 1978;12:135–225. [Google Scholar]
- 3.Warshaw MM, Tinoco I., Jr Optical properties of sixteen dinucleoside phosphates. J Mol Biol. 1966;20:29–38. doi: 10.1016/0022-2836(66)90115-x. [DOI] [PubMed] [Google Scholar]
- 4.(a) Chen J, Kohler B. Base stacking in adenosine dimers revealed by femtosecond transient absorption spectroscopy. J Am Chem Soc. 2014;136:6362–72. doi: 10.1021/ja501342b. [DOI] [PubMed] [Google Scholar]; (b) Takaya T, Su C, de La Harpe K, Crespo-Hernández CE, Kohler B. UV excitation of single DNA and RNA strands produces high yields of exciplex states between two stacked bases. Proc Natl Acad Sci USA. 2008;105:10285–10290. doi: 10.1073/pnas.0802079105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.(a) Ts’o POP, Melvin IS, Olson AC. Interaction and Association of Bases and Nucleosides in Aqueous Solutions. J Am Chem Soc. 1963;85:1289–1296. doi: 10.1021/ja00950a045. [DOI] [PubMed] [Google Scholar]; (b) Broom AD, Schweizer MP, Ts’o POP. Interaction and association of bases and nucleosides in aqueous solutions. V. Studies of the association of purine nucleosides by vapor pressure osmometry and by proton magnetic resonance. J Am Chem Soc. 1967;89:3612–3622. [Google Scholar]; (c) Solie TN, Schellman JA. The interaction of nucleosides in aqueous solution. J Mol Biol. 1968;33:61–77. doi: 10.1016/0022-2836(68)90281-7. [DOI] [PubMed] [Google Scholar]; (d) Nakano NI, Igarashi SJ. Molecular interactions of pyrimidines, purines, and some other heteroaromatic compounds in aqueous media. Biochemistry. 1970;9:577–583. doi: 10.1021/bi00805a019. [DOI] [PubMed] [Google Scholar]; (e) Plesiewicz E, Stepień E, Bolewska K, Wierzchowski KL. Osmometric studies on self-association of pyrimidines in aqueous solutions: evidence for involvement of hydrophobic interactions. Biophys Chem. 1976;4:131–141. doi: 10.1016/0301-4622(76)85003-x. [DOI] [PubMed] [Google Scholar]
- 6.Stokkeland I, Stilbs P. A multicomponent self-diffusion NMR study of aggregation of nucleotides, nucleosides, nucleic acid bases and some derivatives in aqueous solution with divalent metal ions added. Biophys Chem. 1985;22:65–75. doi: 10.1016/0301-4622(85)80026-0. [DOI] [PubMed] [Google Scholar]
- 7.(a) Friedman RA, Honig B. A free energy analysis of nucleic acid base stacking in aqueous solution. Biophys J. 1995;69:1528–1535. doi: 10.1016/S0006-3495(95)80023-8. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Luo R, Gilson HSR, Potter MJ, Gilson MK. The Physical Basis of Nucleic Acid Base Stacking in Water. Biophys J. 2001;80:140–148. doi: 10.1016/S0006-3495(01)76001-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.(a) Pohorille A, Burt SK, MacElroy RD. Monte Carlo simulation of the influence of solvent on nucleic acid base associations. J Am Chem Soc. 1984;106:402–409. doi: 10.1021/ja00314a025. [DOI] [PubMed] [Google Scholar]; (b) Danilov VI, Dailidonis VV, Van Mourik T, Früchtl HA. A study of nucleic acid base-stacking by the Monte Carlo method: Extended cluster approach. Central Eur J Chem. 2011;9:720–727. [Google Scholar]
- 9.(a) Cieplak P, Kollman PA. Calculation of the free energy of association of nucleic acid bases in vacuo and water solution. J Am Chem Soc. 1988;110:3734–3739. [Google Scholar]; (b) Dang LX, Kollman PA. Molecular dynamics simulations study of the free energy of association of 9-methyladenine and 1-methylthymine bases in water. J Am Chem Soc. 1990;112:503–507. [Google Scholar]
- 10.(a) Sponer J, Sponer JE, Mladek A, Jurecka P, Banas P, Otyepka M. Nature and magnitude of aromatic base stacking in DNA and RNA: Quantum chemistry, molecular mechanics, and experiment. Biopolymers. 2013;99:978–88. doi: 10.1002/bip.22322. [DOI] [PubMed] [Google Scholar]; (b) Gu J, Wang J, Leszczynski J. Stacking and H-bonding patterns of dGpdC and dGpdCpdG: Performance of the M05-2X and M06-2X Minnesota density functionals for the single strand DNA. Chem Phys Lett. 2011;512:108–112. [Google Scholar]; (c) Sponer J, Sponer JE, Mladek A, Banas P, Jurecka P, Otyepka M. How to understand quantum chemical computations on DNA and RNA systems? A practical guide for non-specialists. Methods. 2013;64:3–11. doi: 10.1016/j.ymeth.2013.05.025. [DOI] [PubMed] [Google Scholar]
- 11.Florián J, Šponer J, Warshel A. Thermodynamic parameters for stacking and hydrogen bonding of nucleic acid bases in aqueous solution: Ab initio/langevin dipoles study. J Phys Chem B. 1999;103:884–892. [Google Scholar]
- 12.Norberg J, Nilsson L. Stacking-unstacking of the dinucleoside monophosphate guanylyl-3′,5′-uridine studied with molecular dynamics. Biophys J. 1994;67:812–824. doi: 10.1016/S0006-3495(94)80541-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Norberg J, Nilsson L. Potential of mean force calculations of the stacking-unstacking process in single-stranded deoxyribodinucleoside monophosphates. Biophys J. 1995;69:2277–2285. doi: 10.1016/S0006-3495(95)80098-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Norberg J, Nilsson L. Stacking Free Energy Profiles for All 16 Natural Ribodinucleoside Monophosphates in Aqueous Solution. J Am Chem Soc. 1995;117:10832–10840. [Google Scholar]
- 15.Norberg J, Nilsson L. Temperature dependence of the stacking propensity of adenylyl-3′,5′-adenosine. J Phys Chem. 1995;99:13056–13058. [Google Scholar]
- 16.Norberg J, Nilsson L. Solvent Influence on Base Stacking. Biophys J. 1998;74:394–402. doi: 10.1016/S0006-3495(98)77796-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murata K, Sugita Y, Okamoto Y. Free energy calculations for DNA base stacking by replica-exchange umbrella sampling. Chem Phys Lett. 2004;385:1–7. [Google Scholar]
- 18.Norberg J, Nilsson L. Comment on ‘Free energy calculations for DNA base stacking by replica-exchange umbrella sampling’ by Katsumi Murata, Yuji Sugita, Yuko Okamoto. Chem Phys Lett. 2004;393:282–283. [Google Scholar]
- 19.Jafilan S, Klein L, Hyun C, Florián J. Intramolecular Base Stacking of Dinucleoside Monophosphate Anions in Aqueous Solution. J Phys Chem B. 2012;116:3613–3618. doi: 10.1021/jp209986y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.(a) Pérez A, Marchán I, Svozil D, Sponer J, Cheatham TE, III, Laughton CA, Orozco M. Refinement of the AMBER Force Field for Nucleic Acids: Improving the Description of α/γ Conformers. Biophys J. 2007;92:3817–3829. doi: 10.1529/biophysj.106.097782. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ode H, Matsuo Y, Neya S, Hoshino T. Force field parameters for rotation around χ torsion axis in nucleic acids. J Comput Chem. 2008;29:2531–2542. doi: 10.1002/jcc.21006. [DOI] [PubMed] [Google Scholar]; (c) Yildirim I, Stern HA, Kennedy SD, Tubbs JD, Turner DH. Reparameterization of RNA χ Torsion Parameters for the AMBER Force Field and Comparison to NMR Spectra for Cytidine and Uridine. J Chem Theory Comput. 2010;6:1520–1531. doi: 10.1021/ct900604a. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Zgarbova M, Otyepka M, Sponer J, Mladek A, Banas P, Cheatham TE, 3rd, Jurecka P. Refinement of the Cornell et al. Nucleic Acids Force Field Based on Reference Quantum Chemical Calculations of Glycosidic Torsion Profiles. J Chem Theory Comput. 2011;7:2886–2902. doi: 10.1021/ct200162x. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Krepl M, Zgarbova M, Stadlbauer P, Otyepka M, Banas P, Koca J, Cheatham TE, 3rd, Jurecka P, Sponer J. Reference simulations of noncanonical nucleic acids with different chi variants of the AMBER force field: quadruplex DNA, quadruplex RNA and Z-DNA. J Chem Theory Comput. 2012;8:2506–2520. doi: 10.1021/ct300275s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.(a) Van Der Spoel D, Lindahl E, Hess B, Groenhof G, Mark AE, Berendsen HJC. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26:1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]; (b) Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: Algorithms for Highly Efficient, Load-Balanced, and Scalable Molecular Simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- 22.Cheatham TE, Cieplak P, Kollman PA. A Modified Version of the Cornell et al. Force Field with Improved Sugar Pucker Phases and Helical Repeat. J Biomol Struct Dyn. 1999;16:845–862. doi: 10.1080/07391102.1999.10508297. [DOI] [PubMed] [Google Scholar]
- 23.Banáš P, Hollas D, Zgarbová M, Jurečka P, Orozco M, Cheatham TE, Sponer Ji, Otyepka M. Performance of Molecular Mechanics Force Fields for RNA Simulations: Stability of UUCG and GNRA Hairpins. J Chem Theory Comput. 2010;6:3836–3849. doi: 10.1021/ct100481h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. J Chem Phys. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 25.(a) Beauchamp KA, Lin YS, Das R, Pande VS. Are Protein Force Fields Getting Better? A Systematic Benchmark on 524 Diverse NMR Measurements. J Chem Theory Comput. 2012;8:1409–1414. doi: 10.1021/ct2007814. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Li S, Andrews CT, Frembgen-Kesner T, Miller MS, Siemonsma SL, Collingsworth TD, Rockafellow IT, Ngo NA, Campbell BA, Brown RF, Guo C, Schrodt M, Liu YT, Elcock AH. Molecular Dynamics Simulations of 441 Two-Residue Peptides in Aqueous Solution: Conformational Preferences and Neighboring Residue Effects with the Amber ff99SB-ildn-NMR Force Field. J Chem Theory Comput. 2015;11:1315–1329. doi: 10.1021/ct5010966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joung IS, Cheatham TE. Determination of Alkali and Halide Monovalent Ion Parameters for Use in Explicitly Solvated Biomolecular Simulations. J Phys Chem B. 2008;112:9020–9041. doi: 10.1021/jp8001614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parrinello M, Rahman A. Polymorphic transitions in single crystals: A new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 28.Nosé S. A unified formulation of the constant temperature molecular dynamics methods. J Chem Phys. 1984;81:511–519. [Google Scholar]
- 29.Hoover WG. Canonical dynamics: Equilibrium phase-space distributions. Phys Rev A. 1985;31:1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 30.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 31.Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen LG. A smooth particle mesh Ewald method. J Chem Phys. 1995;103:8577. [Google Scholar]
- 32.Vokáčová Z, Buděšínský M, Rosenberg I, Schneider B, Šponer J, Sychrovský V. Structure and Dynamics of the ApA, ApC, CpA, and CpC RNA Dinucleoside Monophosphates Resolved with NMR Scalar Spin–Spin Couplings. J Phys Chem B. 2009;113:1182–1191. doi: 10.1021/jp809762b. [DOI] [PubMed] [Google Scholar]
- 33.(a) Frechet D, Ehrlich R, Remy P, Gabarro-Arpa J. Thermal perturbation differential spectra of ribonucleic acids. II. Nearest neighbour interactions. Nucleic Acids Res. 1979;7:1981–2001. doi: 10.1093/nar/7.7.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Davis RC, Tinoco I. Temperature-dependent properties of dinucleoside phosphates. Biopolymers. 1968;6:223–242. doi: 10.1002/bip.1968.360060206. [DOI] [PubMed] [Google Scholar]
- 34.(a) Bren U, Martínek V, Florián J. Free Energy Simulations of Uncatalyzed DNA Replication Fidelity: Structure and Stability of T·G and dTTP·G Terminal DNA Mismatches Flanked by a Single Dangling Nucleotide. J Phys Chem B. 2006;110:10557–10566. doi: 10.1021/jp060292b. [DOI] [PubMed] [Google Scholar]; (b) Bren U, Lah J, Bren M, Martínek V, Florián J. DNA Duplex Stability: The Role of Preorganized Electrostatics. J Phys Chem B. 2010;114:2876–2885. doi: 10.1021/jp9064246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burkard ME, Kierzek R, Turner DH. Thermodynamics of unpaired terminal nucleotides on short RNA helixes correlates with stacking at helix termini in larger RNAs1. J Mol Biol. 1999;290:967–982. doi: 10.1006/jmbi.1999.2906. [DOI] [PubMed] [Google Scholar]
- 36.(a) Cheatham TE, 3rd, Case DA. Twenty-five years of nucleic acid simulations. Biopolymers. 2013;99:969–77. doi: 10.1002/bip.22331. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Šponer J, Banáš P, Jurečka P, Zgarbová M, Kührová P, Havrila M, Krepl M, Stadlbauer P, Otyepka M. Molecular Dynamics Simulations of Nucleic Acids. From Tetranucleotides to the Ribosome. J Phys Chem Lett. 2014;5:1771–1782. doi: 10.1021/jz500557y. [DOI] [PubMed] [Google Scholar]; (c) Turner DH. Fundamental interactions in RNA: Questions answered and remaining. Biopolymers. 2013;99:1097–104. doi: 10.1002/bip.22294. [DOI] [PubMed] [Google Scholar]
- 37.(a) Zgarbova M, Luque FJ, Sponer J, Cheatham TE, 3rd, Otyepka M, Jurecka P. Toward Improved Description of DNA Backbone: Revisiting Epsilon and Zeta Torsion Force Field Parameters. J Chem Theory Comput. 2013;9:2339–2354. doi: 10.1021/ct400154j. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Yildirim I, Stern HA, Tubbs JD, Kennedy SD, Turner DH. Benchmarking AMBER force fields for RNA: comparisons to NMR spectra for single-stranded r(GACC) are improved by revised chi torsions. J Phys Chem B. 2011;115:9261–70. doi: 10.1021/jp2016006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.(a) Yildirim I, Kennedy SD, Stern HA, Hart JM, Kierzek R, Turner DH. Revision of AMBER Torsional Parameters for RNA Improves Free Energy Predictions for Tetramer Duplexes with GC and iGiC Base Pairs. J Chem Theory Comput. 2012;8:172–181. doi: 10.1021/ct200557r. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Spasic A, Serafini J, Mathews DH. The Amber ff99 Force Field Predicts Relative Free Energy Changes for RNA Helix Formation. J Chem Theory Comput. 2012;8:2497–2505. doi: 10.1021/ct300240k. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Stadlbauer P, Krepl M, Cheatham TE, 3rd, Koca J, Sponer J. Structural dynamics of possible late-stage intermediates in folding of quadruplex DNA studied by molecular simulations. Nucleic Acids Res. 2013;41:7128–43. doi: 10.1093/nar/gkt412. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Tubbs JD, Condon DE, Kennedy SD, Hauser M, Bevilacqua PC, Turner DH. The nuclear magnetic resonance of CCCC RNA reveals a right-handed helix, and revised parameters for AMBER force field torsions improve structural predictions from molecular dynamics. Biochemistry. 2013;52:996–1010. doi: 10.1021/bi3010347. [DOI] [PMC free article] [PubMed] [Google Scholar]; (e) Zgarbová M, Otyepka M, Šponer J, Lankaš F, Jurečka P. Base Pair Fraying in Molecular Dynamics Simulations of DNA and RNA. J Chem Theory Comput. 2014:140703110835007. doi: 10.1021/ct500120v. [DOI] [PubMed] [Google Scholar]; (f) Aytenfisu AH, Spasic A, Seetin MG, Serafini J, Mathews DH. Modified Amber Force Field Correctly Models the Conformational Preference for Tandem GA pairs in RNA. J Chem Theory Comput. 2014;10:1292–1301. doi: 10.1021/ct400861g. [DOI] [PMC free article] [PubMed] [Google Scholar]; (g) Besseova I, Banas P, Kuhrova P, Kosinova P, Otyepka M, Sponer J. Simulations of A-RNA duplexes. The effect of sequence, solute force field, water model, and salt concentration. J Phys Chem B. 2012;116:9899–916. doi: 10.1021/jp3014817. [DOI] [PubMed] [Google Scholar]; (h) Deb I, Sarzynska J, Nilsson L, Lahiri A. Conformational preferences of modified uridines: comparison of AMBER derived force fields. J Chem Inf Model. 2014;54:1129–42. doi: 10.1021/ci400582a. [DOI] [PubMed] [Google Scholar]
- 39.Mitchell PR, Sigel H. A Proton Nuclear-Magnetic-Resonance Study of Self-Stacking in Purine and Pyrimidine Nucleosides and Nucleotides. Eur J Biochem. 1978;88:149–154. doi: 10.1111/j.1432-1033.1978.tb12432.x. [DOI] [PubMed] [Google Scholar]
- 40.Scheller KH, Hofstetter F, Mitchell PR, Prijs B, Sigel H. Macrochelate formation in monomeric metal ion complexes of nucleoside 5′-triphosphates and the promotion of stacking by metal ions. Comparison of the self-association of purine and pyrimidine 5′-triphosphates using proton nuclear magnetic resonance. J Am Chem Soc. 1981;103:247–260. [Google Scholar]
- 41.Chen AA, García AE. High-resolution reversible folding of hyperstable RNA tetraloops using molecular dynamics simulations. Proc Natl Acad Sci USA. 2013;110:16820–16825. doi: 10.1073/pnas.1309392110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Soccorsi L, Cotta-Ramusino M, Cignitti M, Boccacci M. Effect of C(5) methylation on self- and heteroassociation of cytidine in aqueous solution. Bioelectrochem Bioenerg. 1986;16:449–454. [Google Scholar]
- 43.Aradi F. Study of the self-association of methylated pyrimidines in aqueous solutions by 1H NMR chemical shifts. Magn Reson Chem. 1990;28:246–249. [Google Scholar]
- 44.Wang S, Kool ET. Origins of the Large Differences in Stability of DNA and RNA Helixes: C-5 Methyl and 2′-Hydroxyl Effects. Biochemistry. 1995;34:4125–4132. doi: 10.1021/bi00012a031. [DOI] [PubMed] [Google Scholar]
- 45.(a) Sakurai M, Tazawa I, Inoue Y. Simple model to account for the deoxy- versus ribodimer stacking quotient data: Estimation of apparent and intrinsic equilibrium quotients for intramolecular stacking association of purine deoxy- and ribodinucleoside monophosphates. J Mol Biol. 1983;163:683–686. doi: 10.1016/0022-2836(83)90119-5. [DOI] [PubMed] [Google Scholar]; (b) Ogasawara N, Inoue Y. Titration and temperature-dependent properties of homodinucleoside monophosphates. Evaluation of stacking equilibrium quotients for neutral and half-ionized ApA, CpC, GpG, and UpU. J Am Chem Soc. 1976;98:7054–7060. doi: 10.1021/ja00438a053. [DOI] [PubMed] [Google Scholar]
- 46.Cheng DM, Sarma RH. Intimate details of the conformational characteristics of deoxyribodinucleoside monophosphates in aqueous solution. J Am Chem Soc. 1977;99:7333–7348. doi: 10.1021/ja00464a038. [DOI] [PubMed] [Google Scholar]
- 47.Altona C, Hartel AJ, Olsthoorn CSM, de Leeuw HPM, Haasnoot CAG. The Quantitative Separation of Stacking and Self-Association Phenomena in a Dinucleoside Monophosphate by Means of NMR Concentration-Temperature Profiles: 6-N-(Dimethyl)Adenylyl- (3′,5′)-Uridine. In: Pullman B, editor. Nuclear Magnetic Resonance Spectroscopy in Molecular Biology. Vol. 11. Springer; Netherlands: 1978. pp. 87–101. [Google Scholar]
- 48.Banáš P, Mládek A, Otyepka M, Zgarbová M, Jurečka P, Svozil D, Lankaš F, Šponer J. Can We Accurately Describe the Structure of Adenine Tracts in B-DNA? Reference Quantum-Chemical Computations Reveal Overstabilization of Stacking by Molecular Mechanics. J Chem Theory Comput. 2012;8:2448–2460. doi: 10.1021/ct3001238. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.