Abstract
In E. coli, errors in newly-replicated DNA, such as the incorporation of a mis-paired base or an accidental insertion or deletion of nucleotides, are corrected by a methyl-directed mismatch repair (MMR) pathway. While the enzymology of MMR has long been established, many fundamental aspects of its mechanisms remain elusive, such as the structures, compositions, and orientations of complexes of MutS, MutL, and MutHas they initiate repair. Using atomic force microscopy, we—for the first time—record the structures and locations of individual complexes of MutS, MutL and MutH bound to DNA molecules during the initial stages of mismatch repair. This technique reveals a number of striking and unexpected structures, such as the growth and disassembly of large multimeric complexes at mismatched sites, complexes of MutS and MutL anchoring latent MutH onto hemi-methylated d(GATC) sites or bound themselves at nicks in the DNA, and complexes directly bridging mismatched and hemimethylated d(GATC) sites by looping the DNA. The observations from these single-molecule studies provide new opportunities to resolve some of the long-standing controversies in the field and underscore the dynamic heterogeneity and versatility of MutSLH complexes in the repair process.
Keywords: Methyl-directed DNA Mismatch Repair, Atomic Force Microscopy, MutS, MutL, MutH, DNA looping
1. Introduction
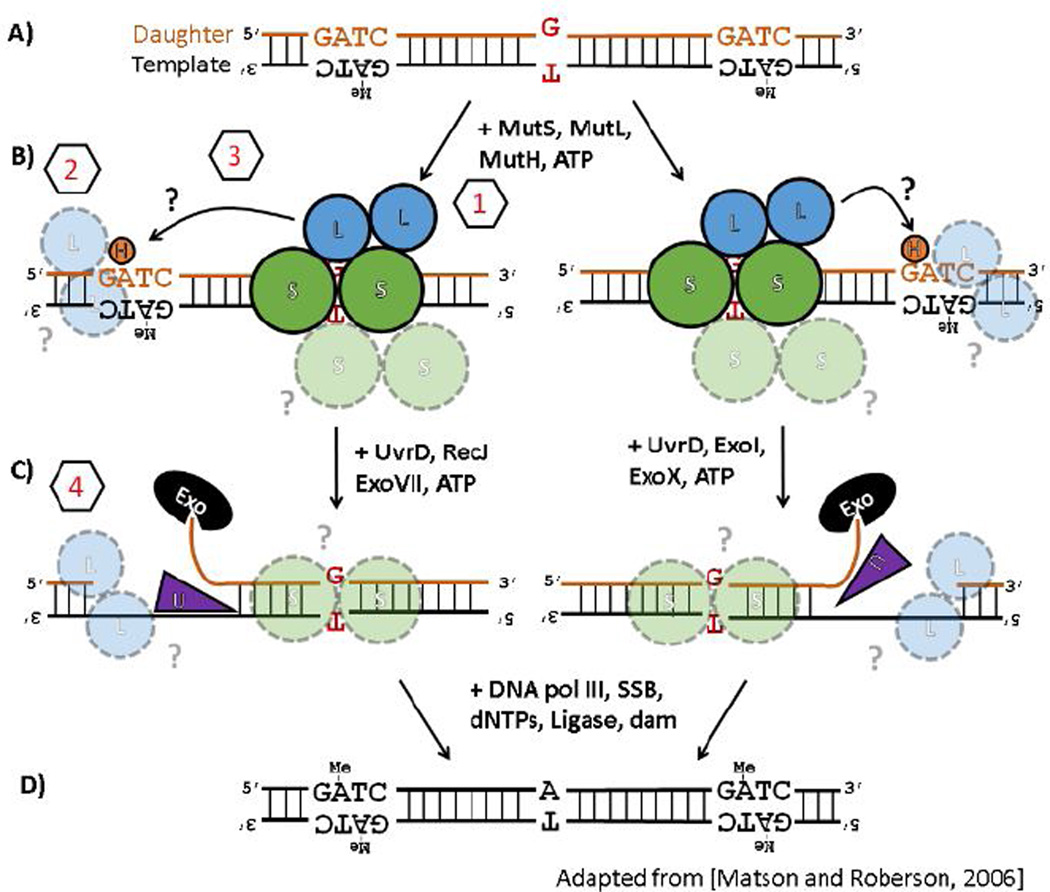
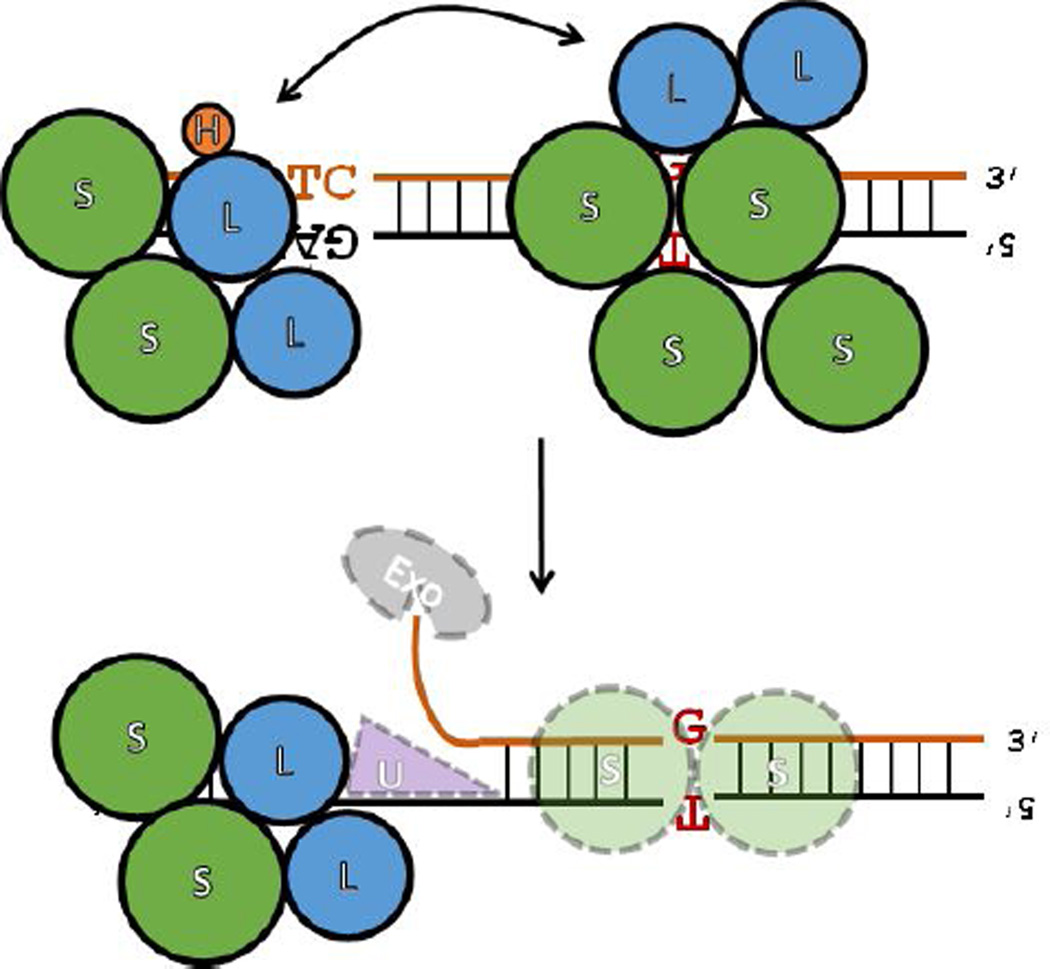
Whenever a DNA molecule is replicated, a mis-paired nucleotide can be introduced or a nucleotide incorrectly inserted or deleted (Figure 1A), errors that can proliferate and damage the genome if not immediately corrected (1–3). In E. coli, these types of errors are identified in newly-replicated DNA when they are bound by a dimer of protein MutS (Figure 1B)(4,5). MutS then associates with protein MutL which, in an ATP-dependent manner (6), in turn activates MutH, a sequence- and methylation-specific nicking endonuclease that is enzymatically latent in the absence of these activating co-factors (MutS, MutL, ATP, and DNA containing a mismatched base-pair or an insertion/deletion bulge). Because DNA in E. coli is methylated a few minutes after replication at d(GATC) sites, the transient hemi-methylation of DNA trailing the replication fork serves to distinguish between the ‘template’ strand being replicated and the erroneous ‘daughter’ strand to be corrected. MutH, after being activated by MutS and MutL, then nicks the hemi-methylated DNA on the unmethylated strand of a nearby d(GATC) site to mark it for correction (Figure 1B). The strand excision machinery (helicase UvrD and the appropriate 5’-to-3’ or 3’-to-5’ exonucleases) is loaded at the nick (Figure 1C) and the unmethylated strand is then digested from the nick back through the site of the error, re-synthesized, ligated and, ultimately, fully methylated by Dam methyltransferase (Figure 1D). Remarkably, errors can still be efficiently repaired even if the nearest strand-discrimination signal is located over 1000 base-pairs (bp) away from the error, regardless of its orientation (5’- or 3’-) relative to the error site and with no apparent directional bias (7,8). Either a single hemi-methylated d(GATC) site or, in the absence of MutH (9,10), a nick in the DNA is absolutely required for an error to be efficiently corrected by this pathway (4,10,11).
Figure 1.
(A–D) Enzymology of the E. coli methyl-directed DNA mismatch repair pathway. See text for details. The pathway is shown to emphasize that it is bidirectional, i.e. can efficiently repair errors whether the nearest strand-discrimination signal (a hemi-methylated d(GATC) site) is located either 5’- or 3’-relative to the error, and that errors can still be corrected when the distance between the error and the nearest d(GATC) site is quite far (> 1000 bp). Figure adapted from Ref. (12) to highlight segments of the pathway where persistent questions regarding the mechanistic details remain (see Sections 3.1–3.4 for details): (1) the initial encounter with mis-matched base-pair, (2) the identification a hemi-methylated d(GATC) site, (3) the search for and nicking of a hemi-methylated d(GATC) site after the mismatch has been identified, and (4) the directional loading of the strand excision machinery toward the error.
While the enzymology of methyl-directed mismatch repair (MMR) has long been established (10), many fundamental aspects of its mechanism remain elusive. This may be a result of the heterogeneous and dynamic nature of the protein complexes hypothesized to form during the initial stages of MMR, which are difficult to capture in traditional biochemical experiments (13). In particular, it is still subject of debate how, in the initial stages of MMR the proteins MutS, MutL, and MutH are able to rapidly identify a (potentially distant) nick orhemi-methylated d(GATC) site after a replication error is found and, furthermore, how the strand excision machinery can be directed from this distant site back toward the error (14). Here we use atomic force microscopy (AFM) to visualize, at the single-molecule level, the initiation of methyl-directed mismatch repair reconstituted at physiological protein concentrations (15). Previous AFM studies of MMR proteins focused primarily on capturing the interactions of MutS alone with DNA (16–18), without investigating interactions with MutL and MutH on DNA. We, for the first time, succeeded in visualizing complexes of all three MMR-initializing enzymes, simultaneously, on heteroduplex DNA. Using engineered DNA substrates to simulate different conditions under which mismatch repair occurs, AFM allows us to discern various structures and complexes as the pathway proceeds. This technique reveals marked structural heterogeneity within the population of DNA and MMR proteins undergoing the initiation of the MMR reaction, and by making use of a novel statistical analysis of complexes’ size and location on the DNA we are able to identify a number of striking and unexpected structures: the growth and disassembly of large multimeric complexes at mismatched sites; complexes of MutS and MutL anchoring latent MutH onto hemi-methylated d(GATC) sites or bound themselves at nicks in the DNA; and these complexes directly bridging mismatched and hemi-methylated d(GATC) sites by looping the DNA. The observations from these single-molecule studies serve as snapshots of the repair process as it progresses and provide new opportunities to reexamine some of the long-standing controversies in the field.
2. Materials and Methods
2.1 Materials / Protein Preparation
Tris-HCl (pH 7.6) buffer was obtained from Rockland Immunochemicals. L-glutamic acid monopotassium salt monohydrate, dithiothreitol (DTT), adenosine 5’-triphosphate (ATP) disodium salt hydrate, and magnesium chloride were obtained from Sigma Aldrich Co., LLC. Only ultra-pure (> 17 MΩ) water was used. Plasmids for expression of MutS, MutL, and MutH containing an N-terminal, six histidine tag (his-MutS, his-MutL, his-MutH) were provided as a generous gift by Prof. Malcolm Winkler (Indiana University Bloomington). (19) All three proteins were expressed in E. coli and purified on a TALON metal affinity resin column (Clontech, Inc.). For MutS and MutH, the his-tag was removed via thrombin cleavage kit (Novagen, Inc.). The his-tag was not removed from MutL, which has a thrombin sensitive internal site, but his-MutL was demonstrated to be able to activate MutH effectively during mismatch repair. (19) Protein functionality was verified by assessing the MutS/MutL/mismatch-stimulated MutH nicking activity of d(GATC) sites via denaturing gel electrophoresis stained by SYBR Gold (Figure S1). Nicking activity by 224 nM MutH alone in Buffer A (used for experiments described below, 20 mM Tris-HCl (pH 7.6), 100 mM potassium glutamate, 5 mM magnesium chloride, 0.4 mM DTT, and, when indicated in the text, 0.5 mM ATP) with 20 nM DNA containing a single hemi-methylated d(GATC) site was not detected after 1 hour (not shown), and ~<1% of DNA imaged by AFM appeared to have been truncated.
2.2 Preparation of DNA Substrates with Different Methylation Patterns
A 690 bp DNA construct containing a single d(GATC) site and multiple restriction cut-sites was synthesized (GeneScript, Inc.) into a pUC57 plasmid (Supporting Information). DNA containing no additional d(GATC) sites was copied out of lambda DNA (New England Biolabs) via polymerase chain reaction (PCR) and sub-cloned into the pUC57 plasmid to generate substrates that were 876 bp in length. DNA primers were obtained from IDT (Integrated DNA Technologies, Inc.). The substrates were prepared by PCR from the pUC57 plasmid using primers of which one was biotinylated (Integrated DNA Technologies); all substrates were labeled at the biotinylated end with monovalent streptavidin (20) (gift of Modrich laboratory, Duke University Medical Center) prior to incubation with proteins MutSLH and imaging.
A single mismatched G-T site was incorporated into the substrates following the nicking-ligation method described in Ref. (21): briefly, a series of Nt. BbvCI nicking sites were serially incorporated near the middle of the synthetic DNA sequence and after nicking by Nt. BbvCI, DNA with an short, exposed single-stranded DNA region is purified. The substrate is annealed slowly from 95 °C with a designed oligo which is complementary to the bare region except for a single mismatched G-T site and is then sealed with T4 ligase. The substrates were then, as appropriate, fully-methylated using Dam methyltransferase (New England Biolabs) or hemimethylated using TaqI methyltransferase (New England Biolabs), which can methylate d(TCGA) sites that were engineered to asymmetrically overlap the d(GATC) sites (i.e., TCGATCTT) (22). Proper methylation was verified by analysis of the digestion patterns via agarose gel electrophoresis after incubation individually with methylation-sensitive restriction enzymes TaqαI or DpnI (New England Biolabs) (Figure S2). Nicked substrates were prepared by nicking with BsrDI at an engineered site near the single d(GATC) site.
2.3 Atomic Force Microscopy (AFM)
Atomic force microscopy (AFM) was performed in air using a Bruker Nanoscope V Multimode with RTSEP (Bruker) probes (nominal spring constant 40 N/m, resonance frequency, 300 kHz). Proteins were centrifuged briefly prior to incubation with DNA. DNA (2.5 nM) and proteins at physiological concentrations (102 nM MutS dimers, 66 nM MutL dimers, and 80 nM MutH monomers (15), using new estimates of E. coli cellular volumes (23)) were mixed in a solution of Buffer A for 10 minutes, deposited for 5 seconds on freshly cleaved mica (Ted Pella, Inc.) that had been treated with 1-(3-aminopropyl)silatrane (prepared as previously described (24) to give the mica surface a positive charge), rinsed with ultra-pure (> 17 MΩ) water, and dried in air. In general at least two but often three to four preparations for each experimental condition were imaged and in general at least three images recorded at 2048 × 2048 pixel resolution over 5.5 micron square areas at 1 line/s for each sample.
2.4 Image processing and analysis
Acquired images were flattened and levelled using an open-source image analysis software for scanning probe microscopy, Gwyddion (http://gwyddion.net/), and analyzed using Gwyddion and MATLAB (Mathworks, Inc.). DNA molecules were each selected, and the presence of bound protein complexes and the occurrence of looping at which a complex was identified at the base of the loop were individually determined. Protein complexes and streptavidin were individually selected, and their size determined as the contiguous region taller than the mean height of the bound DNA. The volume of the protein complexes were scaled relative to the volumes of the streptavidin on the same DNA molecule or, in cases when streptavidin was absent on that individual DNA molecule, the volume of the protein was scaled to the median volume of all streptavidin molecules from that experimental set. Fractional occupancy of bound proteins or protein complexes on DNA was estimated by dividing the number of DNA observed with a monovalent streptavidin end-label in addition to a bound protein (complex) for a given experiment by the total number of DNA observed with streptavidin end-labels. Fraction of looped DNA molecules was estimated by dividing the number of streptavidin-labeled DNA molecules observed with a protein bound at the base of a loop by the total number of streptavidin-labeled DNA molecules (complexes) for a given experiment.
The locations of the protein complexes relative to the streptavidin were determined by tracing by hand DNA molecules with (i) bound complexes, (ii) a single, unambiguously attached streptavidin, and (iii) an unambiguous contour. Loop position and length were considered as the segment of traced DNA between two contacts of the same protein complex. To identify significant enrichment of different species of protein complexes at specific locations on the DNA, the volume-location data of traced complexes was first clustered by their local densities in the volume-location plane. This array was then subjected to a repeated (10,000) permutations of the locational data. Those species (with the same volumes) were considered to be significantly enriched at that location if they were found to have higher local densities in the volume-location plane than >90% or >95% of that species in the permuted data sets.
3. Results
3.1 Quantitative volumetric analysis of MutSLH complexes on DNA
After decades of study, the composition and stoichiometry of MutSLH complexes over the course of mismatch repair remains the subject of debate. MutS forms primarily dimeric and tetrameric complexes in solution (25), however the respective functional roles of these MutS complexes in MMR has not been established definitively (tetrameric MutS was found to play a role inhibiting homeologous recombination (26,27)). On one hand, mutational disruption of the ability of MutS to tetramerize resulted in only a modest increase of E. coli mutation rates in vivo (28), but on the other hand tetrameric MutS was identified at the base of looped DNA originating from mismatched sites (17). The looping of DNA by MutS tetramers was proposed to be a potential mechanism to search for and physically couple mismatched (MM) sites to d(GATC) sites (14), although direct evidence of these bridged structures had never been observed (see Sections 3.2.2 – 3.2.4). MutL is primarily a dimer in solution (29), and while its stoichiometry in mismatch repair has also not been established (30)it has been termed the ‘molecular matchmaker’ of mismatch repair as it has been shown to interact directly with MutS (6), (monomeric) MutH (31), and other enzymes involved in DNA repair (in particular UvrD (12)). Fluorescent microscopy of MutS and MutL at unrepaired mismatch sites in vivo suggested there is approximately three times as many MutL proteins at these sites than MutS (30), although whether this relationship still holds during actual repair of the mismatched sites remains unclear.
Atomic force microscopy (AFM) can be used to approximate the composition of MutSLH complexes during mismatch repair, since the molecular weight of a protein complex is linearly correlated with its apparent volume in topographical AFM images (32,33). This technique has been previously exploited, e.g., to identify oligomeric states of MutS bound to mismatched DNA (17), and has resolution on the scale of 30 – 50 kDa. As described in more detail in Section 3.2, we performed AFM imaging on various combinations of MutS, MutL, and MutH bound to DNA molecules designed to simulate the different states of the DNA during MMR. By performing the volumetric analysis on the bound protein complexes we can begin to identify their approximate compositions as mismatch proceeds. Since, prior to imaging, DNA molecules were biotinylated at one end and bound with monovalent streptavidin (60 kDa)(20), we scale the apparent volume of the complexes (Vc) to that of the bound monovalent streptavidin (VSA)to account for differences in AFM tip geometry across large numbers of experiments; the streptavidin should remain consistent under each condition and can serve as a reference. Note that for all the experiments described here, we use 2.5 nM DNA and proteins at their approximate physiological concentrations (102 nM MutS dimers, 66 nM MutL dimers, and 80 nM MutH monomers (15), using new estimates of E. coli cellular volumes (23)), where appropriate.
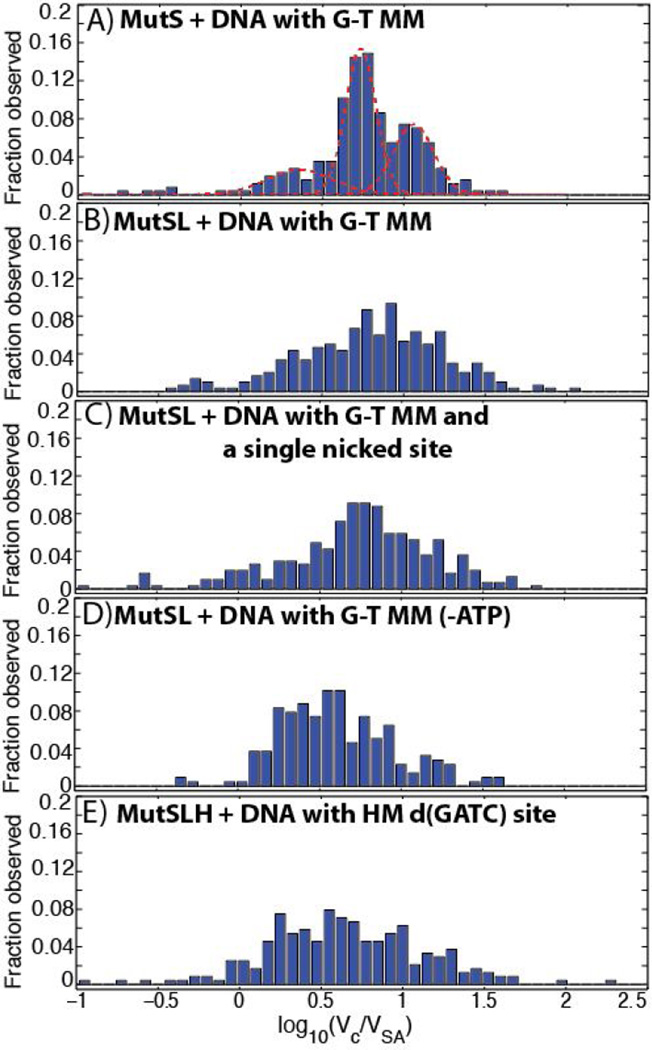
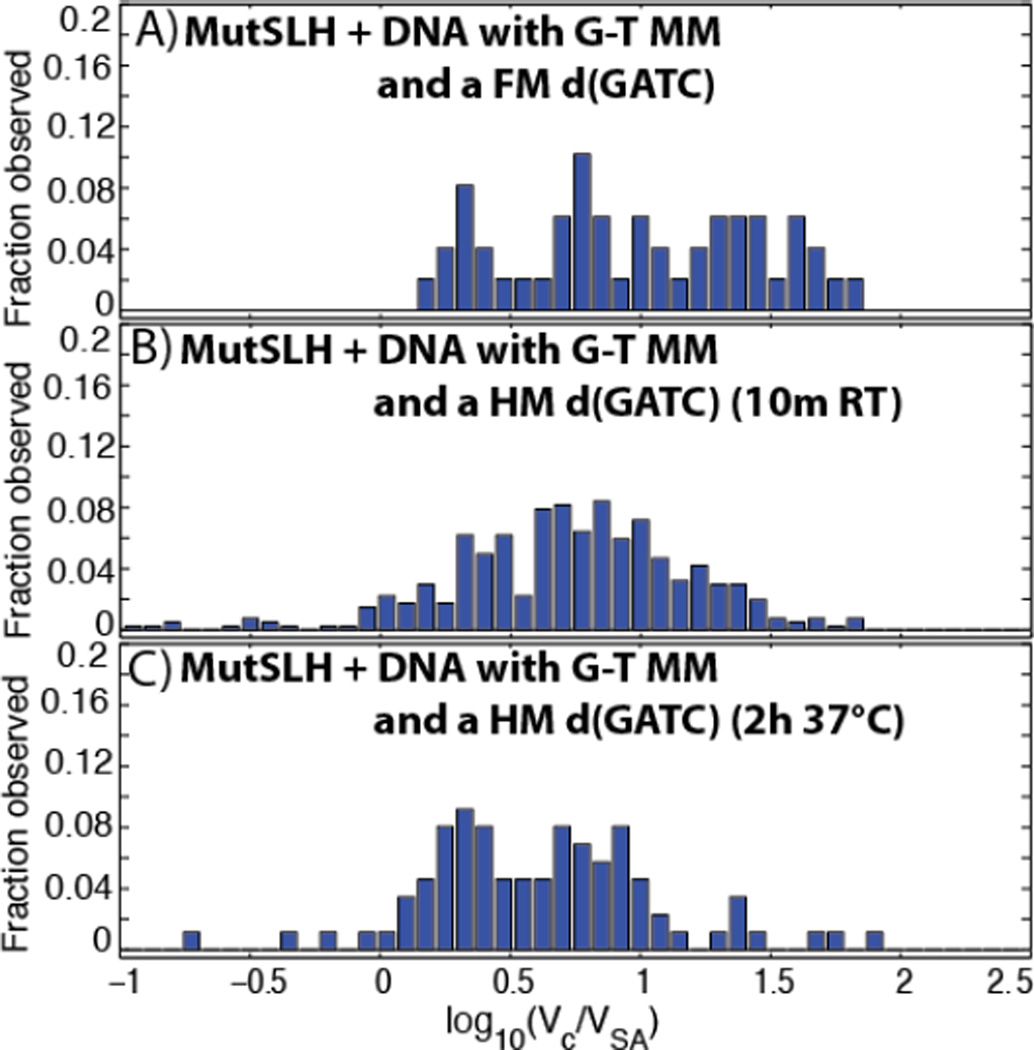
When MutS(92 kDa monomer) was imaged on DNA with a mismatch in the presence of 0.5 mM ATP, we observe three distinct peaks in the histogram of scaled volumes (Figure 2A) at log10(Vc/VSA) = 0.3514 (95% confidence: 0.272 – 0.4308),0.7287 (0.7213 – 0.736), and 1.058 (1.041 – 1.075), likely corresponding to MutS monomers, dimers, and tetramers, respectively, as previously observed (17,34). The addition of MutL (66 kDa monomer) and MutH (28 kDa monomer) dramatically increases the complexity of the observed distribution of volumes, and these data cannot be well or uniquely fit by Gaussian mixture models. In addition to larger sized complexes, there appear to be relatively consistent ‘peaks’ at log10(Vc/VSA) = 0.9 and 1.1in samples containing both MutS and MutL—likely MutL dimers bound to MutS dimers and tetramers, respectively (see below)—although the resolution of this technique is not sufficient to differentiate individual species of complexes unambiguously. Furthermore, the small size of MutH places it at the limit of AFM imaging so that its presence cannot be consistently and unambiguously determined. These challenges limit the utility of volumetric analysis alone in identifying the composition and stoichiometry of MutSLH complexes bound to DNA.
Figure 2.
Histogram of the volumes of DNA-bound MutSLH complexes (Vc) scaled to those of the streptavidin label (VSA). See Section 3.2 for description of experimental conditions. Unless noted, all proteins incubated with DNA in 0.5 mM ATP. (A) MutS on 876 bp DNA molecule containing a single G-T mismatch (MM) at its center, with Gaussian fits (see Section 3.2.1 and Figure 3A). Number of proteins measured n= 256. (B) MutS and MutL incubated with DNA molecule containing a single G-T mismatch at its center(see Section 3.2.1 and Figure 3B).n = 300.(C) MutS and MutL incubated with DNA molecule containing a single G-T mismatch at its center and a single-strand nick located ~270 bp away (see Section 3.2.4 and Figure 10B). n = 308. (D) MutS and MutL incubated in the absence of ATP with DNA molecule containing a single G-T mismatch at its center.n = 217. (E) MutS, MutL, and MutH incubated with DNA molecule containing a single hemi-methylated d(GATC) site (see Section 3.2.2 and Figure 6).n = 405.
However, often times we can identify individual components of these complexes from their AFM topographical images directly which are structurally dissimilar enough to distinguish (see Figures 3,5,7, 10, and Supporting Information); for example, in Figure 3B, a small dimer of MutL can be distinguished from its bound MutS complex. These features will be described with their respective experimental observations in Section 3.2, although discrete features become more difficult to interpret as the size of the complexes increase under different experimental conditions. While current work is presently underway in our laboratory to develop molecular labels so that MutS, MutL, and MutH can be uniquely identified via AFM, at present we can only use the distribution of volumes to provide a sense of the variety and sizes of different species bound to DNA as mismatch repair is being initiated.
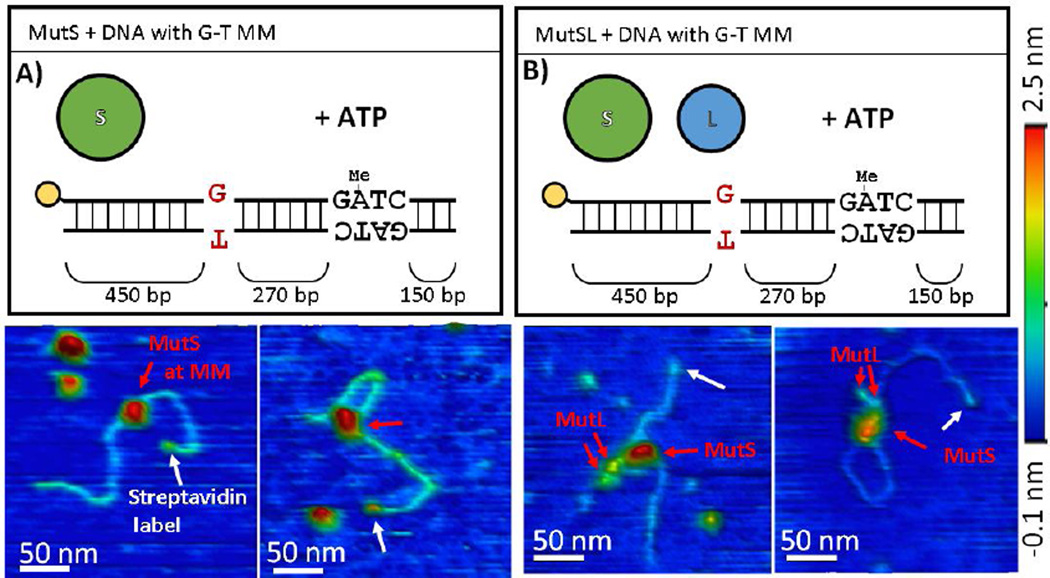
Figure 3.
AFM images of (A) MutS and (B) MutSL bound to DNA containing a mismatch. Top) Schematic representations of the DNA substrate and proteins used for the experiment. Below) 3D topographical rendering of AFM images (color scale −0.1 nm (blue) to 2.5 nm (red)). White arrows point to location of monovalent streptavidin label (yellow in schematic) with the mismatch located approximately in the middle of the DNA molecule. Red arrows to assigned MutS and MutL oligomers in complex.
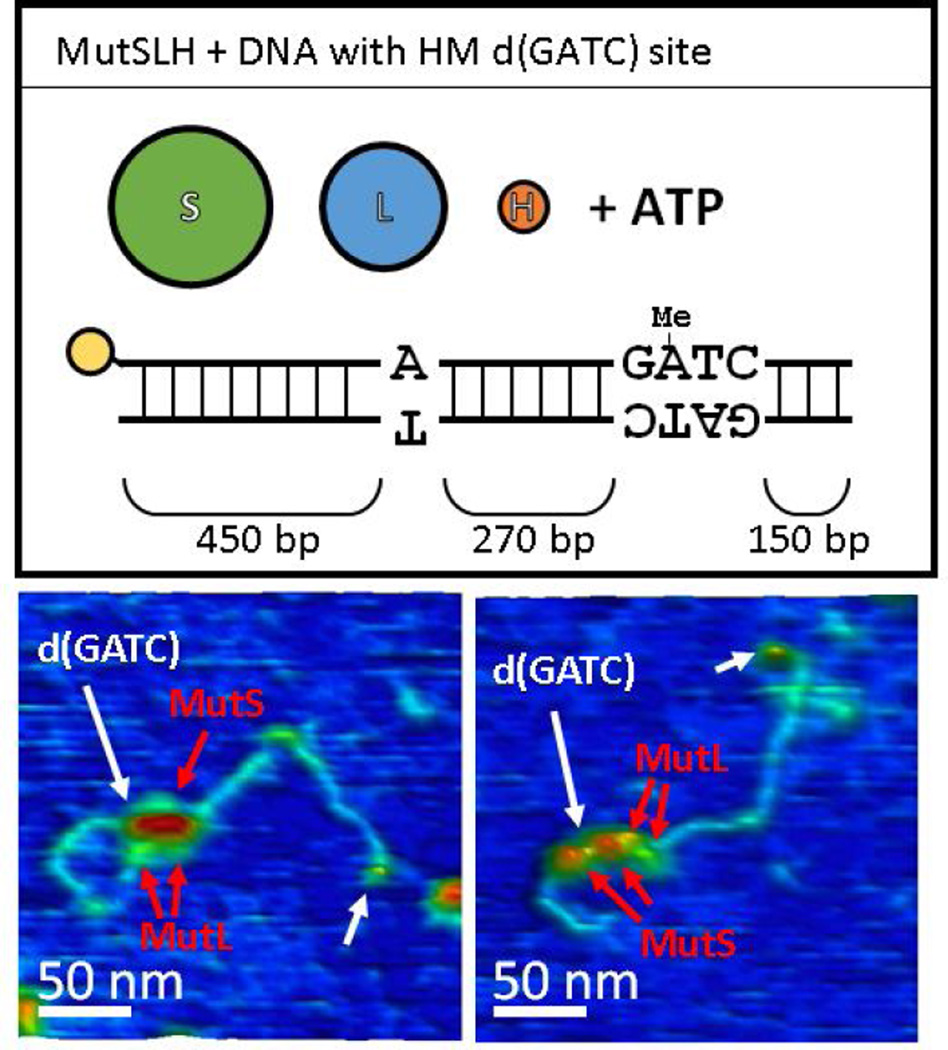
Figure 5.
AFM images of MutSLH complexes bound to DNA (without a mismatch) at hemimethylated d(GATC) sites. Top) Schematic representation of the DNA substrate and proteins used for the experiment. Below) 3D topographical rendering of AFM images (color scale −0.1 nm (blue) to 2.5 nm (red) as in Figure 3). White arrows point to location of monovalent streptavidin label (yellow in schematic), with the d(GATC) site approximately 3/4 the length of the DNA away from this label. While MutS and MutL (red arrows) can be assigned based on their sizes as components of the complexes at d(GATC) sites, MutH is too small to be clearly resolved under these conditions.
Figure 7.
AFM images of MutSLH incubated with DNA containing a mismatch that was (A) fully-methylated (and hence lacks a strand discrimination signal) or (B) hemi-methylated at the d(GATC) site before the experiment. Top) Schematic representation of the DNA substrate and proteins used for the experiment. Below) 3D topographical rendering of AFM images (color scale −0.1 nm (blue) to 2.5 nm (red) as in Figure 3). White arrows point to location of monovalent streptavidin label (yellow in schematic).
Figure 10.
AFM images of MutSLH after significant incubation (A) with hemi-methylated DNA containing a mismatch or (B) with DNA which had been nicked by BsrDI at a site near the d(GATC) before the experiment. Top) Schematic representation of the DNA substrate and proteins used for the experiment. Below) 3D topographical rendering of AFM images (color scale −0.1 nm (blue) to 2.5 nm (red) as in Figure 3). White arrows point to location of monovalent streptavidin label (yellow in schematic).
3.2 Imaging the structure and distribution of MutSLH complexes on model DNA substrates provides snapshots of the initiation of mismatch repair
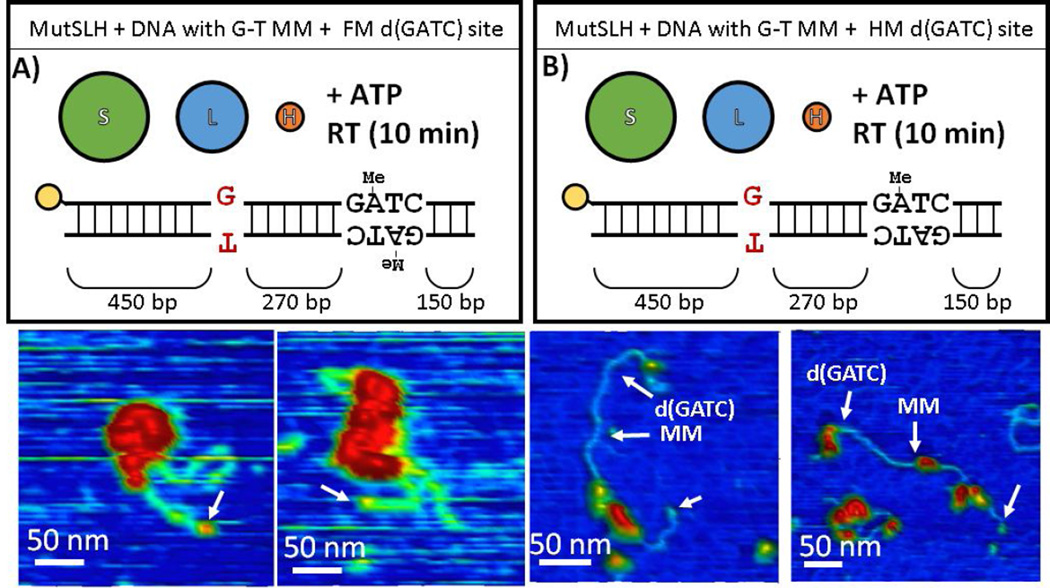
In order to simulate various conditions at the initiation of mismatch repair, we engineered a DNA molecule which we can modularly manipulate to possess a single G-T mismatch and/or various patterns of methylation on its sole d(GATC) site located 270 bp away. The experiments, as well as the fraction of DNA with bound complexes and DNA observed to be looped by proteins, are summarized in Table 1. To ease their interpretation, we categorized these experimental conditions by the stages of MMR they can help to elucidate (Figure 1): (A) the initial encounter of a mismatch (Section 3.2.1); (B) the identification a hemi-methylated d(GATC) site (Section 3.2.2); (C) the search for and nicking of a hemi-methylated d(GATC) site after the mismatch has been identified (Section 3.2.3); and (D) the directional loading of the strand excision machinery toward the error (Section 3.2.4).
-
g
Fraction of end-labeled DNA molecules with bound protein(s) that are observed with a loop that has a protein complex located at its base.
-
h
95% binomial confidence interval.
-
i
Proteins incubated with DNA for 10 minutes at room temperature prior to imaging.
-
j
Proteins incubated with DNA for 2 hours at 37°C prior to imaging.
Table 1.
Summary of experimental conditions and observed number of DNA molecules which possessed a bound protein / complex or loop with protein / protein complex at its base.
| Proteina | MMb | d(GATC)c | ATPd | Figures | nDe | Bound complex?f |
Looped?g |
|---|---|---|---|---|---|---|---|
| 3.2.1 Encountering a mismatched base-pair | |||||||
| S | + | HM | + | 3A, 4A-D, S3 | 206 | 56.3%(±6.8%)h | 10.7% (±4.2%) |
| SL | + | HM | + | 3B, 4E-H, S6, S7A | 196 | 76.0% (±6.0%) | 20.9% (±5.7%) |
| SL | + | HM | − | S7B | 120 | 75.8% (±7.7%) | 24.2% (±7.7%) |
| 3.2.2 Identifying a hemi-methylated d(GATC) site | |||||||
| SLH | − | HM | + | 5, 6, S8, S9A | 184 | 76.6% (±6.1%) | 19.0% (±5.7%) |
| SL | − | HM | + | S10 | 209 | 49.3% (±6.8%) | 13.9% (±4.7%) |
| LH | + | HM | + | S11A | 42 | 26.2% (±13.3%) | 4.8% (±6.4%) |
| 3.2.3 Searching for and nicking a hemi-methylated d(GATC) site | |||||||
| SLH | + | FM | + | 7A, S12, S13, S14 | 24 | 95.8% (±8.0%) | 50.0% (±20.0%) |
| SLH (10m)i | + | HM | + | 7B, 9, S9B, S15 | 276 | 65.6% (±5.6%) | 9.4% (±3.5%) |
| 3.2.4 MutSL(H) complexes on nicked sites of DNA containing a mispair | |||||||
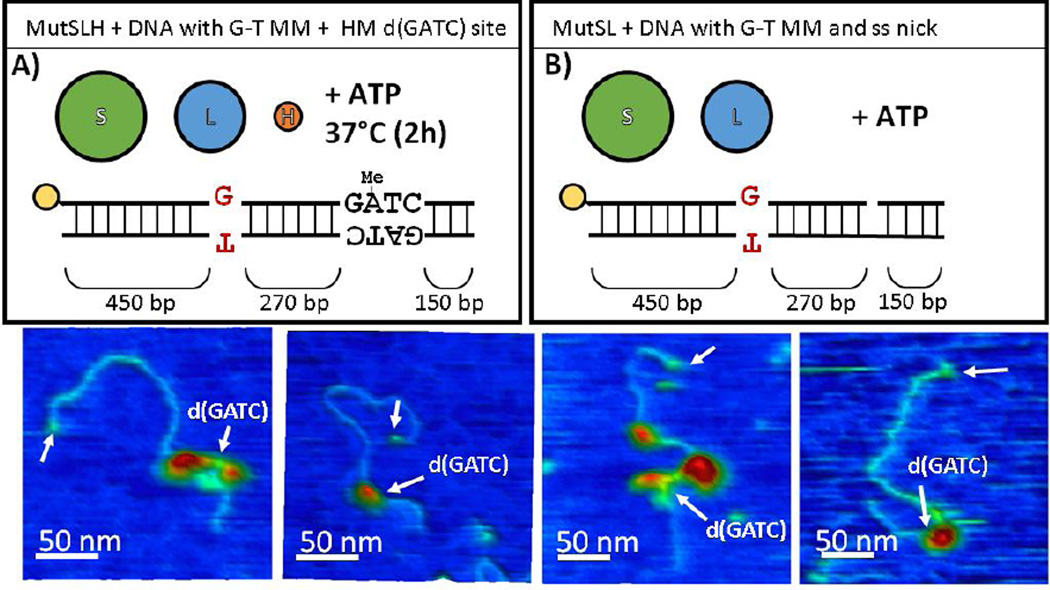
| SLH (2h)j | + | HM | + | 10A, 11A-D, S16, S17 | 202 | 20.8% (±5.6%) | 5.0% (±3.0%) |
| SL | + | Ni | + | 10B, 11E-H, S18, S19 | 257 | 66.9% (±5.8%) | 9.0% (±3.5%) |
| L | + | Ni | + | S20 | 20 | 25.0% (±19.0%) | 6.0% (±9.6%) |
Proteins incubated with DNA prior to imaging. S = MutS, L = MutL, H = MutH.
Whether the DNA molecule contained a mismatched G-T site (+) or not (−).
Methylation of the sole d(GATC) site in the molecule. HM = hemi-methylated, FM = fully methylated, Ni = DNA nicked at nearby BsrDI site prior to experiment.
Whether proteins were incubated with DNA in the presence (+) or absence (+) of 0.5 mM ATP.
Total number of end-labeled DNA molecules observed for each experimental condition.
Fraction of end-labeled DNA molecules observed to possess a bound protein complex.
For these experiments, the size of observed complexes were mapped to their positions on the DNA molecules. As noted above, we find remarkable heterogeneity in both the sizes of the complexes and their distribution on the DNA. We therefore performed a bootstrapping analysis (see Methods) on this data to identify species of complexes which were significantly enriched at specific sites above this ‘background’ level of binding (see Figures 4, 6, 9, 11, and S14). This technique allows us to identify important features and structures which are otherwise lost when examining the one-dimensional histograms of complex size and binding site distributions which are traditionally analyzed independently in AFM studies of DNA-protein interactions.
Figure 4.
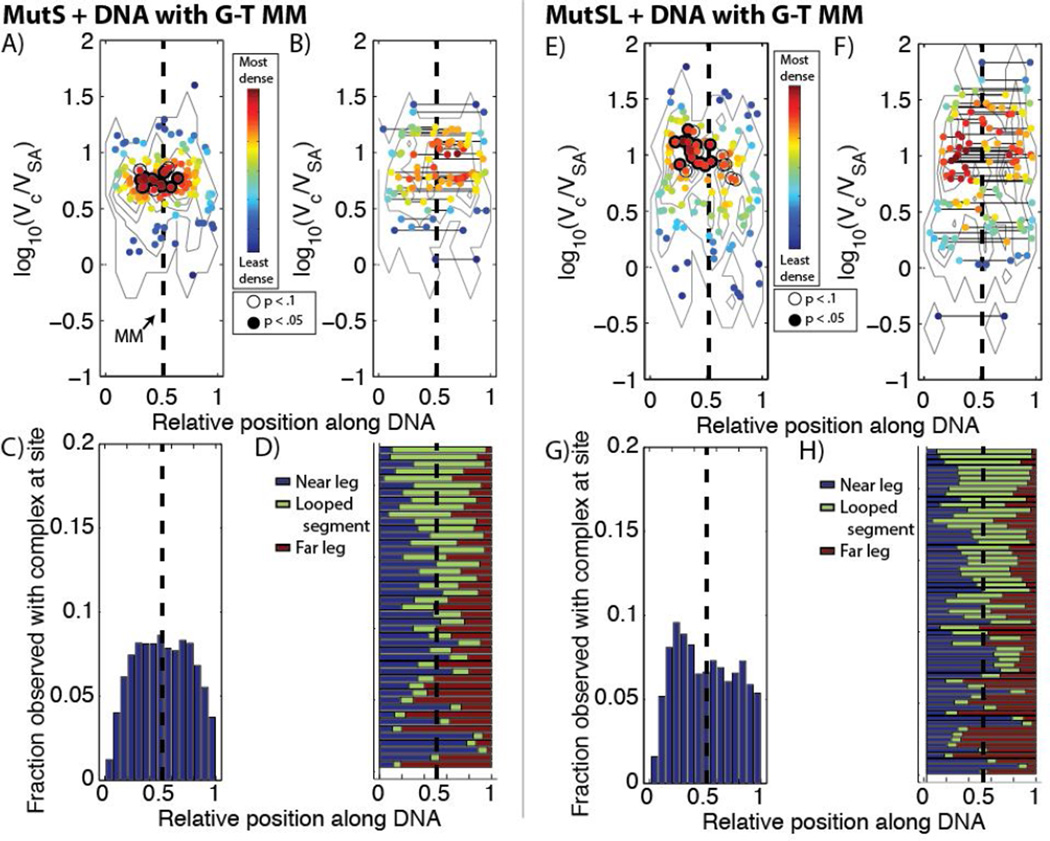
Distribution of different complexes of (A–D) MutS and (E–H) MutS and MutL bound to DNA containing a single G-T mismatch (MM, dashed line). (A,E) Scatter plots of observed complex size vs. relative position on unlooped DNA molecules, colored by their local densities in the size-position plane. Points marked by an open circle were more enriched than >90% of the species with that volume when the positional data was permuted 10,000 times (see Methods), and points marked in a filled black circle were more enriched than >95% of permuted species. The contours are derived from the surface of the two-dimensional empirical probability distributions (a two-dimensional histogram) of volumes and locations to guide the eye. (B,F) Scatter plot of observed complex size vs. relative position on the DNA molecules which were looped, with solid line connecting complexes at the two sites they were observed. Coloring and significance are labeled as described in (A) and (E). (C,G) Histogram of binding occupancy along DNA by all (looped and unlooped) observed complexes. (D, H) Normalized contour lengths of looped DNA, with blue highlighting the length of the DNA nearest the streptavidin label, green the length of the looped segment, and red the length of the DNA outside the loop at the end away from the streptavidin.
Figure 6.
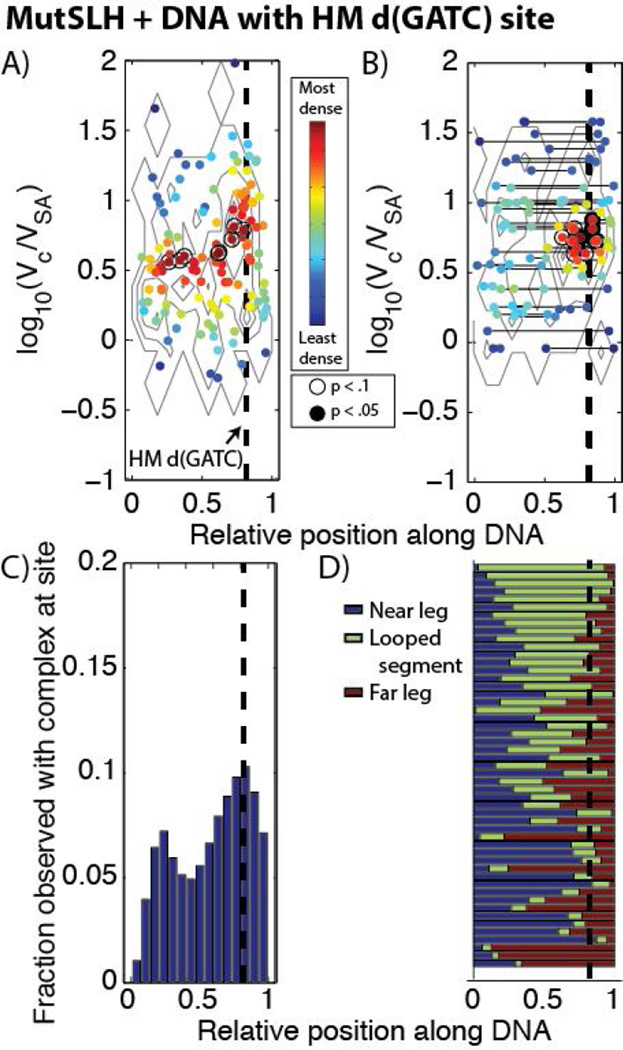
Distribution of different complexes of MutS, MutL, and MutH bound to DNA containing a single hemi-methylated d(GATC) site (dashed line) with no mismatches. See Figure 4 for details regarding the labels and coloring schemes for Figures 6A–D. (A) In the scatter plot of observed complex size vs. relative position on un-looped DNA molecules, there is a significantly enriched population log10(Vc/VSA) ~0.8−1.3 located at or near the hemi-methylated d(GATC) site. (B) In the scatter plot for looped DNA molecules, there is a significant enrichment of complexes of similar sizes located at the hemi-methylated site. C) In the binding histogram, we observe a subtle enrichment of total occupancy of protein complexes located near the site of the d(GATC). (D) Normalized contour lengths of looped DNA show that a large population of looped DNA molecules, particularly those with larger loops, have loops which originate at or near the hemi-methylated d(GATC) site (compare, e.g., Figure 4D, 4H).
Figure 9.
Distribution of different complexes of MutS, MutL, and MutH bound to DNA containing a G-T mismatch and single hemi-methylated d(GATC) site (dashed lines). See Figure 4 for details regarding the labels and coloring schemes for Figures 9A–D. (A,C) While in the binding histogram (C) the population appears approximately uniformly distributed on the DNA, in the scatter plot (A) there is a significantly enriched population (log10(Vc/VSA) ~0.8−1.1)just upstream of the hemi-methylated d(GATC) site.(B, D) In the scatter plot (B) for looped DNA molecules, we observe complexes (log10(Vc/VSA) ~0.7−1.5) looping DNA which (D) preferentially bridge at or near the mismatch to the d(GATC) site. Compare with Figures 4D, H.
Figure 11.
Distribution of different complexes of (A-D) MutS, MutL, and MutH incubated for 2 hours at 37°C to DNA containing a single G-T mismatch and hemi-methylated d(GATC) site (dashed lines) and (E-H) MutS and MutL incubated with DNA containing a G-T mismatch and a nicked site. See Figure 4 for details regarding the labels and coloring schemes for Figures 11AH. (A–D) While the occupancy of proteins on DNA and fraction looped drops (see Table 1) after the 2 hour incubation, we see an enrichment of complexes at the mismatch and d(GATC) site. (E–H) On nicked DNA, there is a (E, G) significant enrichment of complexes (log10(Vc/VSA) ~ 0.8 − 1.1) located slightly upstream of the mismatch, and (F,H) there is an increased tendency for loops to bridge the mismatched site and the nick.
3.2.1 Encountering a mismatched base-pair-random looping of and diffusion on mismatched DNA by MutSL complexes
It is well-documented that MutS binds to a variety of replication errors as a dimer (13,35), but what occurs immediately after the identification of these sites has been obfuscated by conflicting experimental results, generally interpreted by three different models(reviewed in (14,36)). A series of recent single molecule fluorescence studies (37–40)of MutS (without MutL or MutH) at low concentrations (1 – 5 nM) show that ADP-bound dimers initially diffuse slowly and transiently (~1 s) along the DNA molecules and, upon encountering a mismatched base-pair, bind to it for several seconds. This binding is followed by an ADP/ATP exchange and conformational change to a ‘sliding clamp’that is highly stabilized on DNA (diffuses on DNA for > 5 min)(5,41,42). This sliding clamp then releases from the mismatch and rapidly (and randomly) diffuses away. It has been suggested that the rapidly diffusing MutS dimers may be involved in the subsequent search for d(GATC) sites (6,13), however how the relative location of the mismatch is retained has not been established. Conversely, the results of in vivo fluorescence microscopy suggested that MutL might polymerize on the DNA after MutS identifies a mismatch, presumably remaining at the site of the error, although the observations occurred on mismatch sites which were not repaired over the course of a cell cycle (30). Additionally, as mentioned above, a third set of experiments using atomic force microscopy of MutS at physiological concentrations (100 nM dimer) (17) and electron microscopy studies of MutS and MutL on mismatched DNA (43)have identified DNA molecules bent into looping structures with MutS (or MutS and MutL) located at the base of the loop near the mismatch. The loops were found to grow rapidly in presence of ATP (43) and the MutS at these sites was found to be mostly in tetrameric form (as a dimer of dimers) (17). The looping of DNA is suggestive of a mechanism where one dimer of MutS remains at or near the mismatch site while the other bound dimer translocates toward d(GATC) sites (either using actively translocation, through random diffusion on the DNA, or by random collisions through space), providing intrinsic memory of the mismatch’s location as the sites are brought into contact.
We imaged hemi-methylated DNA containing a single mismatch after incubation with MutS (95 kDa monomer) in the presence of ATP (Figure 3A and S3). As previously observed on mismatched DNA incubated with MutS (17), we note the presence of discrete particulate features (~2 – 2.5 nm in height) on ~60% of DNA molecules (Table 1), which we assign to be MutS oligomers using the volumetric analysis described above. The MutS on the DNA is approximately 18% monomer, 50% dimer, and 32% tetramer (Figures 2A). Mapped to their positions on the DNA molecule (Figures 4A and 4C), the population of dimers and tetramers appear approximately symmetrically distributed on the DNA molecule around the site of the mismatch, which may either represent nonspecific binding events or captured MutS that have slid away from the mismatch site along the DNA. We do note that imaging was performed after a 10 minute incubation with ATP, and from single-molecule fluorescence studies of MutS on DNA containing mismatches diffusion of MutS away from the mismatch is expected in the presence of ATP (5).
A population (~10%, Table 1) of the DNA appear to have loops with MutS located at the base and near the site of the mismatch (Figure 4B and 4D). The proportion of the MutS population assigned to be tetrameric is enriched over the population average over dimers (Figure S5), consistent with earlier studies which showed the majority of looped DNA possessed a tetrameric MutS complex at the base of the loops. DNA looping is centered around the site of the mismatch and appears random and symmetrically distributed, consistent with previous studies (17,43). We do note that some smaller features (MutS monomers or unbound streptavidin) have been classified as having ‘looped’ the DNA. ‘Looping’ by these small proteins or complexes is observed in every experiment; however, the loops tend to be small and they are uniformly distributed across the entire length of the DNA molecules, which is suggestive that these loops may be an artifact of deposition on the surface for imaging (Figure S4). These small loops comprise ~30% of loops observed (see, e.g., Figure 4D) and are noted in the text.
When heteroduplex DNA was imaged after incubation with both MutS and MutL (66 kDa) under ATP (Figures 3B, 4 E–H, and S7) this distribution of protein complexes across the DNA remains approximately the same as before, but now is composed mostly of larger complexes (Figures 2B and 4E–F). As mentioned above, the size of these complexes is consistent with MutL dimers binding to MutS dimers and tetramers (log10(Vc/VSA) = 0.9 and 1.1), in addition to larger observed complexes (log10(Vc/VSA) ~1.2 − 1.5). MutS and MutL were previously observed at the base of looped DNA (43), and here we find an enrichment in the populations of looped DNA molecules upon the addition of MutL (Figures 4E and S7) (Table 1). Oftentimes, we are able to resolve two smaller, dimeric protrusions positioned close to the taller MutS complex which we assign to be MutL dimers (~1–1.5 nm in height) (Figures 3B and S7, red arrows). The structure of many of these complexes appears similar to that predicted by chemical trapping and computational modeling of MutSL complexes (31,44). The looping is again symmetrically and randomly distributed about the mismatch (Figure 4H).
If the experiment is repeated in the absence of ATP, the fraction of DNA molecules bound by MutSL complexes and fraction of looped molecules are observed to increase slightly (Table 1). However there is a strong downward shift in the sizes of observed protein complexes and the fraction of smaller complexes, likely comprised of MutS monomers, increases sharply. These results are consistent with previously reports of the effects of ATP on MutS/MutSL complex formation (6,17,45); the increase in DNA occupancy is attributed to the observation that MutS can diffuse rapidly from the mismatch in the presence of ATP and, hence can diffuse off of the DNA molecule. Both with and without ATP (Figure S7), we do note that the MutSL complexes appear distributed on the DNA away from the location of the mismatch, which is consistent with the idea that MutL may diffuse along with MutS multimers.
3.2.2 Identifying a hemi-methylatedd(GATC) site-MutS and MutL anchor latent MutH at hemimethylated d(GATC) sites
The status of the latent MutH prior to activation is also not well understood. On one hand, it is often implicitly assumed that MutH remains bound to hemi-methylated d(GATC) sites awaiting activation by MutSL (12), although its dissociation constant (0.9 µM) remains far greater than its physiological concentration (80 nM) (46,47). On the other hand, it has been suggested that MutH scans DNA for hemi-methylated d(GATC) sites in complex with MutSL during a diffusive search (31). However, in either case it should be noted thatin vivo MutH must compete for hemimethylated d(GATC) sites with SeqA (48), which packages and bundles newly-replicated DNA into large-scale focal structures that persist for several minutes and which are thought to provide some genome-wide structural and topological order during replication. SeqA foci have been observed trailing the replication fork closely (49), and while the interactions between MutH and SeqA at d(GATC) sites are not well-characterized, the observed persistence of these SeqA structures in vivo implies that SeqA can likely displace MutH. How MutH may access these sites under these conditions remains unclear.
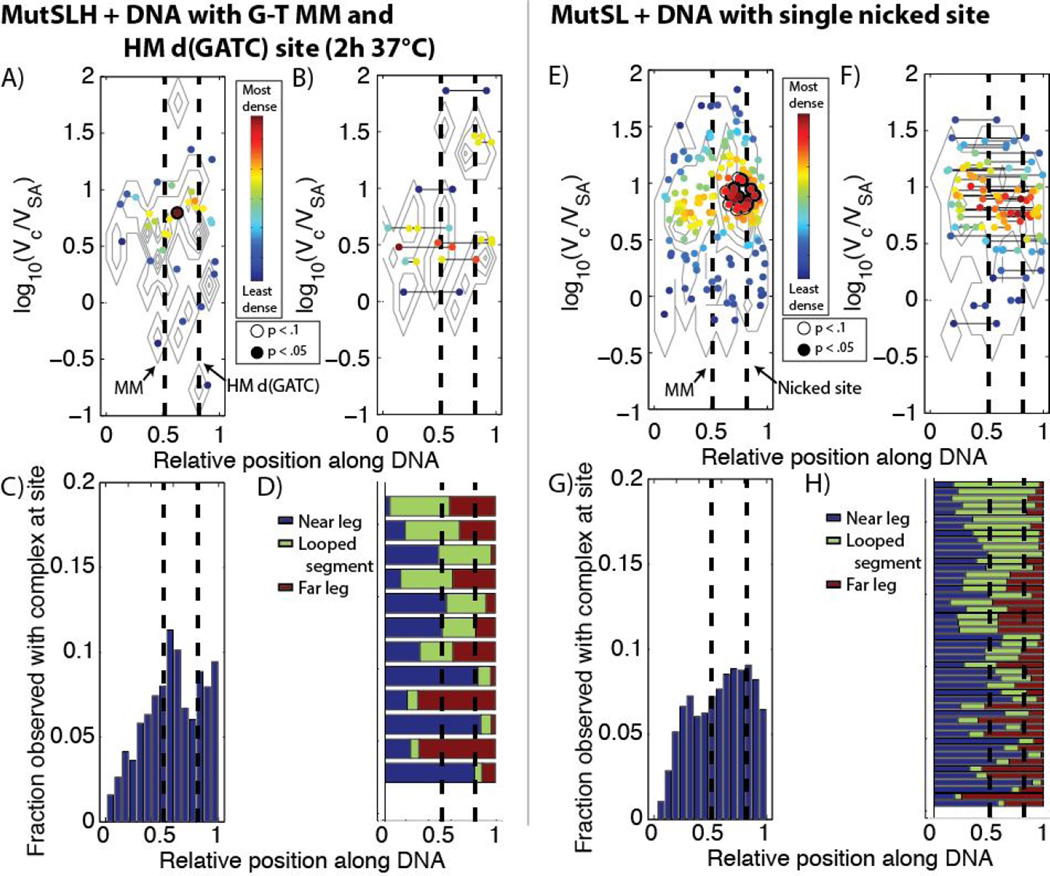
We imaged hemi-methylated DNA which did not contain a mismatch with MutS, MutL, and MutH under ATP (Figures 5, 6, S8, and S9A). Surprisingly, the occupancy on DNA was similar to that of MutSL on DNA containing a mismatch, and there was a slight increase in the fraction of looped DNA molecules as well (Table 1). While MutH cannot often be directly resolved by AFM in these complexes, there is an apparent upward shift in the population of larger molecular complexes over that of MutSL from Section 3.2.1 (compare Figures 2B and 2E). We observe a population of smaller protein features (sizes ranging from log10(Vc/VSA) ~ 0.5 – 0.7) distributed approximately uniformly on the DNA (Figure 6A), likely MutS dimers (see below), however, a significant population of larger features (log10(Vc/VSA) ~0.8−1.3), likely composed of MutSLH complexes, are observed precisely at the location of the hemi-methylated d(GATC) site (Figures 6A (brackets), C). Furthermore, of the population of looped molecules, a substantial fraction in that same size range appear to have large loops terminating (or originating from) the hemimethylated site (21 DNA molecules out of 52 traced, with 18 having short loops assigned to be non-specific, Figures 6D) and there is significant enrichment of looped complexes bound at one end at these sites (Figure 6B).
We compared these images of MutSLH on hemi-methylated homoduplex DNA with those of just MutS and MutL (without MutH, Figure S10), just MutH and MutL (without MutS, Figure S11A), or MutH alone (Figure S11B). In the absence of MutH, MutS and MutL are observed bound to homoduplex DNA at significantly lower rates (from 76% to ~45% fractional occupancy). The size of the complexes are generally smaller than on DNA containing a mispair (Figure S10A), suggesting that the larger MutSL complexes do not form in the absence of a mismatch; this finding is consistent with earlier reports (50). The distribution of complexes across the DNA does not appear enriched at the hemi-methylated d(GATC) site in the absense of MutH (Figure S10B). This result would suggest that MutH, though not clearly resolvable via AFM under these conditions, is responsible for targeting the complexes to these sites in the absence of a mismatch. Imaging hemi-methylated DNA incubated with both MutL and MutH (without MutS) there is further drop in the occupancy of DNA with protein complexes compared with those with MutS, MutL, and MutH (26%), but in those which possess complexes we do observe small species of sizes log10(Vc/VSA) ~ 0.4 – 0.7 (Figure S11A), where a dimer of MutL is sometimes apparent. Imaging the hemi-methylated DNA with MutH alone (Figure S11B) we observe negligible interaction between the DNA and the protein, consistent with its high rate of its dissociation from DNA. Taken together, these results suggest that MutS anchors MutL and MutH at these hemi-methylated d(GATC) sites even in the absence of a detected mismatched DNA.
3.2.3 Searching for and nicking a hemi-methylatedd(GATC) site-growth and deconstruction of large MutSLH complexes suggests a search mechanism
To investigate the outward search for d(GATC) sites originating from the mismatch, we performed AFM first on fully-methylated DNA containing a mismatch in the presence of MutS, MutL, and MutH (hence with no viable strand discrimination signal), then on hemi-methylated DNA with a mismatch (incubated at room temperature for 10 minutes prior to imaging), both in the presence of ATP.
On fully-methylated DNA (Figures 7A, S12, S13, and S14), the fraction of occupied DNA molecules rises considerably with the addition of MutH, even in the absence of a strand discrimination signal (Table 1). A population of quite large (log10(Vc/VSA) ~ 1.2 − 2) complexes is observed on approximately the vast majority of the DNA molecules (Figure 8A), of which we cannot unambiguously determine the components and stoichiometry at present, as well a population with complexes (log10(Vc/VSA) ~ 0.7 − 1.2) which appear away from the mismatch site (Figures S12 and S13). Looping in both classes of DNA molecule is also increased significantly relative to those incubated with MutS and MutL alone (Table 1).
Figure 8.
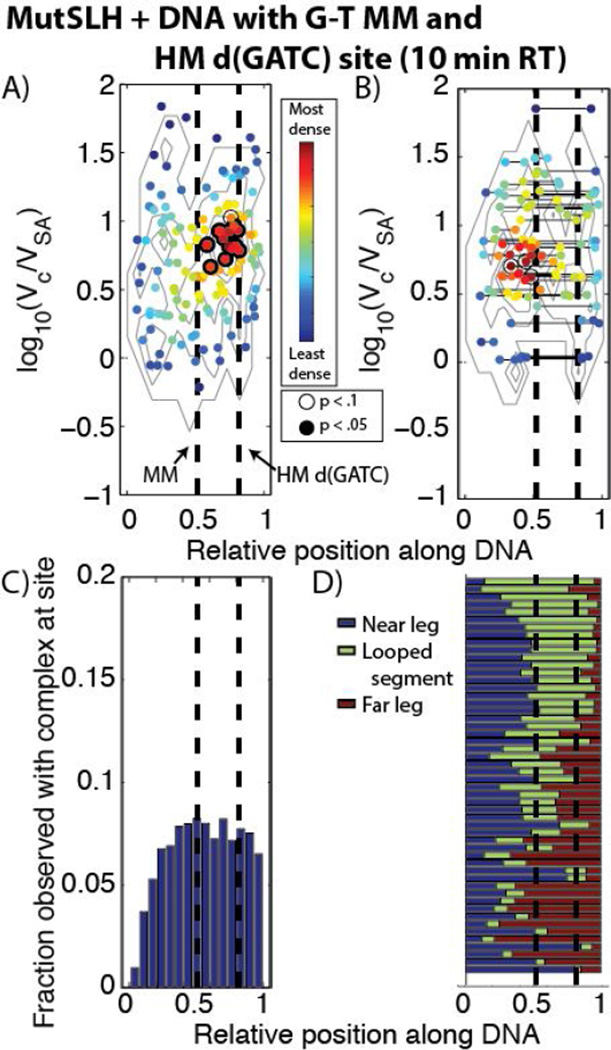
Histogram of the volumes of DNA-bound MutSLH complexes (Vc) scaled to those of the streptavidin label (VSA). (A) MutSLH on DNA containing a single G-T mismatch at its center and a fully-methylated (FM) d(GATC) site (see Section 3.2.3 and Figure 7A). Number of proteins measured:n= 49. (B) MutSLH on DNA containing a single G-T mismatch at its center and a hemi-methylated (HM) d(GATC) site after 10 minutes of incubation at room temperature (see Section 3.2.3 and Figure 7B). n= 405.(C) MutSLH on DNA containing a single G-T mismatch at its center and HM d(GATC) site after incubation for 2 hours at 37°C (see Section 3.2.4 and Figure 10A).n= 87.
However, when the DNA is hemi-methylated (Figures 7B, 9, S9, and S15)—and now possessive of a stand-discrimination signal— the number of very large, aggregated complexes observed drops considerably (Figure 8B), suggesting either that the loops dissociate and complexes break up upon identifying a hemi-methylated d(GATC) after a mismatched site is identified, or that these very large complexes rarely form on the hemi-methylated substrate. We find that of those looped molecules (with sizes ranging log10(Vc/VSA) ~ 0.7 − 1.5), the loops preferentially bridge the segment of DNA between the mismatch site and the hemi-methylated d(GATC) site: 24 out of 52 traced, looped molecules appear to bridge at or near the mismatch and d(GATC) sites;16 have short loops uniformly distributed on the DNA assigned to be non-specific; and the remained appear centered either at the mismatch or at the hemi-methylated d(GATC) site (Figure 9D). Of those unlooped DNA molecules, a significant population of the complexes (log10(Vc/VSA) ~ 0.8 − 1.1), appear slightly upstream of the hemi-methylated site (Figure 9A). We note that at present we cannot determine via AFM whether the sites have been nicked at this stage.
3.2.4 MutSL complexes on nicked sites of DNA containing a mispair- a potential mechanism to direct the excision machinery
MutL is known to stimulate the loading rate and helicase activity of UvrD onto nicked DNA,(12)and as UvrD travels3’-to-5’ on DNA it must be loaded on the correct strand of DNA toward the error. It is presently unknown how MutSLH complexes are able to retain the relative position of the error at the d(GATC) site to orient UvrD correctly. We investigated the structure of MutSLH complexes by imaging DNA with a G-T mismatch and a single hemi-methylated d(GATC) site incubated at 37°C for two hours (Figures 10A, 11A–D, S16, and S17), where the nicking reaction by MutH should be essentially complete, as well as by imaging MutSL on mismatched DNA which we had nicked prior to experiments at a BsrDI site near the d(GATC) site (Figure 11B, 12E–H, S18, and S19).
Figure 12.
Proposed dual-search / proofreading model of the initiation of mismatch repair in light of the presented AFM imaging data. (Top) MutS can directly identify mismatches, and from these sites search for strand discrimination signals with MutL and MutH through diffusion or direct looping of the DNA. At the same time, MutS, presumably closely trailing the replisome, also anchors MutLH at d(GATC) sites or MutL at nicked DNA (at the unligated edges of Okazaki fragments), and through direct looping or diffusion, scans (proofreads) the local DNA for mismatched base-pairs. The contact between the two sites may occur as a result of ‘mutual’ search from both mismatches and strand discrimination signals. Failure to quickly identify a strand discrimination signal by MutS complexes at mismatch sites results in the recruitment and loading onto DNA of additional MMR complexes. (Bottom) After both a strand discrimination signal and a mismatch are mutually identified, the loops dissociate and MutS anchors MutL in at the nicked sites asymmetrically, in a manner that retains directional orientation relative to the error.
For the DNA with a G-T mismatch and a single hemi-methylated d(GATC) incubated with MutSLH for 2 hours, we note a drop in the population of occupied DNA to 20%, as well as a significant decrease in the population of looped DNA observed to ~5% (Table 1). Those with protein complexes are preferentially bound at either the mismatch site or slightly upstream of the d(GATC) site (Figure 11C). Additionally, a further downward shift in the observed volume of most of the complexes is observed (Figure 8C), suggesting a further dissociation of the larger complexes at the late states of MMR initiation.
When MutS and MutL were incubated with pre-nicked DNA containing a G-T mismatch, the size distribution of complexes is similar to that observed of MutSL on DNA with a hemi-methylated d(GATC) site (Figures 2C, S18, and S19) but we observe a significant enrichment of complexes (log10(Vc/VSA) ~ 0.8 − 1.1) located slightly upstream the nicked site (Figure 11E and 11G). Similar to the case of MutSLH on hemi-methylated DNA with a mismatch, we find that looped DNA molecules tend to bridge at or near the mismatch and the nicked site; 17 of 47 traced, looped DNA appeared to bridge the sites (10 appeared non-specifically looped, and the remainder were largely centered on the mismatch or the nick, Figure 11H). Imaging of MutL alone incubated with this DNA substrate exhibits very low affinity for nicked DNA (Table 1) and we rarely observed MutL dimers on DNA at that site after incubation with MutL alone (Figure S20).
4. Discussion
4.1 MutS as a scaffold for MutL and MutH at strand discrimination signals and to direct excision
The results of AFM imaging presented here reveal that MutS, in addition to its well-known role in identifying and binding to mismatched base-pairs in DNA, also serves as a ‘scaffold’ anchoring MutL and latent MutH onto DNA. It is also important to note that the targeting of these complexes to hemi-methylated d(GATC) sites and MutL to nicked sites after a mismatch has been identified is abrogated in the absence of MutH or MutL, respectively, suggestive of a more passive role rather than in direct search for these features. This role for MutS, as a scaffold for MutL and MutH, is consistent with the in vitro observation that while MutL and MutS increase nicking activity of MutH approximately twenty-fold, MutL alone increases MutH nicking activity approximately ten-fold over basal MutH nicking rates (47,51)—the modest increase in nicking activity following the addition of MutS may be simply a result of MutS sequestering MutLH onto DNA containing a hemi-methylated d(GATC) site.
On unlooped DNA molecules containing mismatches, we find that MutSL(H) complexes preferentially bind slightly upstream of either the hemi-methylated d(GATC) or nicked site, toward the G-T mismatch. Because of the limited time resolution of our AFM, it is difficult to discern the dynamics of how complexes assemble at these sites. Recent fluorescence microscopy experiments suggested that MutS binds asymmetrically to DNA at mismatches (37), and this asymmetry in binding by MutS has been proposed to be the mechanism by which the location of the error is retained at d(GATC) sites (37). Hence, these asymmetric complexes may assemble at these sites after diffusion by the MutS away from the mismatch in a manner that retains the directionality of the mismatch. Alternatively, these results may suggest that when the strand discrimination signal is discovered after a mismatch has been identified, any looped structures may fall apart and leave complexes asymmetrically anchored at the strand discrimination signal toward the mismatch. The complexes we observe at nicked sites appear to contain both MutS and MutL, as MutL has very low affinity for the nicked sites on its own (Table 1), and while MutL interacts directly with UvrD (52) and stimulates helicase activity (12), this activity is greatly enhanced by the additional presence of MutS (53). Hence, while MutL may be responsible for loading UvrD onto the DNA, the relative location of the error may also be imparted by MutS in addition to anchoring MutL to the DNA(see Section 4.3). Further studies using different orientations between the mismatch and d(GATC) sites will be necessary to confirm this hypothesis.
4.2 Remarkable heterogeneity of protein complexes size and location on DNA suggests versatility in DNA search mechanisms
We also note again the remarkable heterogeneity in the structures and locations of complexes bound to DNA under nearly all experimental conditions (for example, in the scatter plots of Figures 4, 6, 9, and 11). Even when distinct enrichments of specific populations at targeted sites are observed, we find a large ‘background’ of complexes of various sizes bound nearly uniformly on DNA strands. As noted above, this distribution of binding sites could suggest that these complexes are undergoing random diffusion on the DNA, which is consistent with the fluorescence microscopy experiments in vitro demonstrating that MutS at low concentrations diffuses along the DNA after binding mismatches (39). As it was previously reported that DNA molecules extended using magnetic tweezers (in such a manner that prevented looping) still experienced mismatch-dependent nicking by MutH (54,55), diffusion of MutS(LH) complexes could also be occurring in conjunction with the specific looping we observe such that the two search methods improve the efficiency of strand discrimination signal discovery. As an example, protein ‘roadblocks’ between d(GATC) sites and a mismatch were found to hinder MutH nicking (56), however it was recently reported that E. coli Dam methyltransferase can loop DNA via intersegmental transfer if there are bound proteins hindering its diffusion pathway (57)—a similar mechanism where both looping and diffusive search mechanisms occur simultaneously could explain the results here.
Additionally, the presence of large multimeric structures on mismatched DNA which lacked a true stand discrimination signal might suggest that after extensive search, if no strand discrimination signal is quickly identified, additional MMR proteins are recruited to sites of mismatches. Consistent with this, Elez et al. identified large foci of MutS and MutL at unrepaired mismatch sites (30), while we find that these foci of MutSLH dissociate or rarely form when a strand discrimination signal is present and can be identified. The time-dependent effects (related to whether or not a strand discrimination signal can be found) may be related to the slow hydrolysis of ATP by MutS and MutL, the function of which is complex and currently subject of debate (5,6). Ultimately, the diversity of multicomponent complexes observed, and their dynamic nature as mismatch repair progresses, highlights some of the challenges faced by single molecule fluorescence studies of MMR (13) and suggests a versatility of MutSLH proteins in their search for errors or strand discrimination signals while initiating mismatch repair.
There is an additional possibility to explain the heterogeneity of complexes observed in these studies, which is that these in vitro experiments containing only MutS, MutL, and MutH with DNA lack the natural spatial and temporal constraints expected to be found during the operation of normal mismatch repair. MutS (58) and MutL (59) have been shown to directly interact with the β clamp of DNA polymerase II and MutL foci were observed to localize frequently with the replisome (60), suggesting that MutSLH operate close to the replication fork. During replication, there is a limited window of time during which to correct these errors as d(GATC) sites become occupied by SeqA foci or are fully-methylated by Dam methyltransferase (61) and as nicks at the boundaries of Okazaki fragments (suggested to also be a strand discrimination signal in vivo(62)) are sealed by ligases. Furthermore, in vivo, endogenous errors in replication which can result in a mutation without repair occur extremely infrequently (2.75 × 10−8 per nucleotide per generation (63)), which is a significant difference from most in vitro studies in which all DNA molecules possess a mismatch. Both the paucity of mismatches in vivo and an interaction with the replisome may limit or regulate the extensive diversity of complexes observed here. Continuing efforts to move single-molecule studies of MMR (13) into living cells (64) will be essential to unravel the dynamic heterogeneity of the MMR pathway.
4.3 A dual-search / proofreading model for the initiation of mismatch repair
An additional notable finding of this study is that loops in DNA formed by MutSLH complexes preferentially bridge mismatched sites and a strand discrimination signal. Taken together with the observation of MutSLH anchored at d(GATC) sites on homoduplex DNA preferentially formed loops originating at the d(GATC) sites, this finding raises an interesting possibility for ‘dual’ roles of DNA looping / diffusive search by MutSLH in MMR. The first role may be that loops are formed by MutSL(H) complexes at discovered mismatched sites to rapidly find a strand discrimination signal where additional proteins may block diffusive search, as discussed above. Additionally, MutSL(H), trailing the replisome, may be arrested at strand discrimination signals (hemi-methylated d(GATC) sites or nicked DNA formed at the boundaries of Okazaki fragments)then form loops in the DNA in order to scan and ‘proofread’ newly replicated DNA and identify any mismatches in their immediate vicinity (Figure 12). Anchoring of MutL(H) by MutS at hemi-methylated d(GATC) and nicks sites also provides a means to briefly forestall displacement by SeqA or ligase, respectively. A recent report found that very close proximity (<50 bp) to a d(GATC) site suppressed the local mutation rate (62). This report also provided evidence of methylation-independent mismatch repair by MutS and MutL in E. coli, raising the possibility that nicks in the lagging strand may serve quite an important role in directing MMR. Further study of the dynamics of these complexes at strand discrimination signals, as well as investigations into the dynamics of mismatch generation (63)and the relationship with thein vivo repair of those errors (62), will be necessary to elucidate the role of the observed binding and looping from d(GATC) and nicked sites by MMR complexes.
5. Conclusions
AFM imaging for the first time of all three essential proteins of the initiation of MMR provides new clues to its mechanism of initiation and allows us to further examine the model of MMR. Furthermore, temporally-resolved studies will be essential to see how the population of MutSLH complexes move over time to elucidate the relationship between the looping (between the mismatch and d(GATC) site) and dwelling at the unlooped d(GATC) or nicked sites. Understanding a precise kinetic relationship between MutH nicking rate and the mismatchd (GATC) site distance (9,65,66) should also help to elucidate this this behavior, and work testing the implications of the described model in Figure 12 using biochemical, single-molecule, and in particular, time-resolved methods is presently underway.
The work presented here paves the way for future studies reconstituting the entire MMR pathway at the single molecule level. In particular, while it is known in human MMR how excision terminates after the mismatch, (67)it is not known in MutH-dependent MMR; rigorous characterization of MMR protein complexes via AFM appears suitable to identify further mechanisms of methyl-dependent MMR.
Supplementary Material
Highlights.
DNA molecules have been engineered to mimic the initial stages of mismatch repair.
Complexes of MutS, MutL, and MutH are imaged on individual DNA molecules via AFM.
Protein complexes are identified that otherwise would be difficult to discern.
AFM reveals marked structural heterogeneity for proteins and DNA during repair.
DNA is preferentially looped to bridge mismatches and a strand discrimination site.
Acknowledgements
We wish to thank Dr. Paul Modrich (Duke University Medical Center, Durham, NC, USA, and Howard Hughes Medical Institute) for helpful discussions and gift of monovalent streptavidin as well as Prof. Malcolm Winkler and Tiffany Ho-Ching Tsui (Indiana University Bloomington) for plasmids and bacterial strains. This work was supported by the National Science Foundation [MCB-1244297 to PEM] and the National Institute of General Medical Sciences of the National Institutes of Health [F32GM112502 to EAJ].
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Modrich P, Lahue R. Mismatch repair in replication fidelity, genetic recombination, and cancer biology. Annual Review of Biochemistry. 1996;65:101–133. doi: 10.1146/annurev.bi.65.070196.000533. [DOI] [PubMed] [Google Scholar]
- 2.Jiricny J. Post-replicative Mismatch Repair. Cold Spring Harbor perspectives in biology. 2013;5 doi: 10.1101/cshperspect.a012633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kunkel TA, Erie DA. DNA Mismatch Repair*. Annu. Rev. Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- 4.Au KG, Welsh K, Modrich P. Initiation of methyl-directed mismatch repair. Journal of Biological Chemistry. 1992;267:12142–12148. [PubMed] [Google Scholar]
- 5.Lee J-B, Cho W-K, Park J, Jeon Y, Kim D, Lee SH, Fishel R. Single-molecule views of MutS on mismatched DNA. DNA repair. 2014;20:82–93. doi: 10.1016/j.dnarep.2014.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Molecular cell. 2003;12:233–246. doi: 10.1016/s1097-2765(03)00219-3. [DOI] [PubMed] [Google Scholar]
- 7.Grilley M, Griffith J, Modrich P. Bidirectional excision in methyl-directed mismatch repair. The Journal of biological chemistry. 1993;268:11830–11837. [PubMed] [Google Scholar]
- 8.Cooper DL, Lahue RS, Modrich P. Methyl-directed mismatch repair is bidirectional. The Journal of biological chemistry. 1993;268:11823–11829. [PubMed] [Google Scholar]
- 9.Langle-Rouault F, Maenhaut-Michel G, Radman M. GATC sequences, DNA nicks and the MutH function in Escherichia coli mismatch repair. The EMBO journal. 1987;6:1121–1127. doi: 10.1002/j.1460-2075.1987.tb04867.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lahue RS, Au KG, Modrich P. DNA mismatch correction in a defined system. Science (Washington) 1989;245:160–164. doi: 10.1126/science.2665076. [DOI] [PubMed] [Google Scholar]
- 11.Lahue RS, Su SS, Modrich P. Requirement for d(GATC) sequences in Escherichia coli mutHLS mismatch correction. Proceedings of the National Academy of Sciences of the United States of America. 1987;84:1482–1486. doi: 10.1073/pnas.84.6.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matson SW, Robertson AB. The UvrD helicase and its modulation by the mismatch repair protein MutL. Nucleic acids research. 2006;34:4089–4097. doi: 10.1093/nar/gkl450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Erie DA, Weninger KR. Single molecule studies of DNA mismatch repair. DNA repair. 2014 doi: 10.1016/j.dnarep.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kolodner RD, Mendillo ML, Putnam CD. Coupling distant sites in DNA during DNA mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12953–12954. doi: 10.1073/pnas.0705698104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Feng G, Tsui HC, Winkler ME. Depletion of the cellular amounts of the MutS and MutH methyl-directed mismatch repair proteins in stationary-phase Escherichia coli K-12 cells. Journal of Bacteriology. 1996;178:2388–2396. doi: 10.1128/jb.178.8.2388-2396.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Sass LE, Du C, Hsieh P, Erie DA. Determination of protein-DNA binding constants and specificities from statistical analyses of single molecules: MutS-DNA interactions. Nucleic acids research. 2005;33:4322–4334. doi: 10.1093/nar/gki708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiang Y, Marszalek PE. Atomic force microscopy captures MutS tetramers initiating DNA mismatch repair. The EMBO journal. 2011;30:2881–2893. doi: 10.1038/emboj.2011.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang H, Yang Y, Schofield MJ, Du C, Fridman Y, Lee SD, Larson ED, Drummond JT, Alani E, Hsieh P, et al. DNA bending and unbending by MutS govern mismatch recognition and specificity. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:14822–14827. doi: 10.1073/pnas.2433654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng G, Winkler ME. Single-step purifications of His6-MutH, His6-MutL and His6-MutS repair proteins of escherichia coli K-12. BioTechniques. 1995;19:956–965. [PubMed] [Google Scholar]
- 20.Howarth M, Chinnapen DJF, Gerrow K, Dorrestein PC, Grandy MR, Kelleher NL, El-Husseini A, Ting AY. A monovalent streptavidin with a single femtomolar biotin binding site. Nature methods. 2006;3:267–273. doi: 10.1038/NMETHXXX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Robertson AB, Matson SW. Reconstitution of the very short patch repair pathway from Escherichia coli. The Journal of biological chemistry. 2012;287:32953–32966. doi: 10.1074/jbc.M112.384321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao Y, Jung C, Marx AD, Winkler I, Wyman C, Lebbink JHG, Friedhoff P, Cristovao M. Generation of DNA nanocircles containing mismatched bases. BioTechniques. 2011;51:259. doi: 10.2144/000113749. [DOI] [PubMed] [Google Scholar]
- 23.Volkmer B, Heinemann M. Condition-dependent cell volume and concentration of Escherichia coli to facilitate data conversion for systems biology modeling. PLoS One. 2011;6:e23126. doi: 10.1371/journal.pone.0023126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shlyakhtenko LS, Gall AA, Filonov A, Cerovac Z, Lushnikov A, Lyubchenko YL. Silatrane-based surface chemistry for immobilization of DNA, protein-DNA complexes and other biological materials. Ultramicroscopy. 2003;97:279–287. doi: 10.1016/S0304-3991(03)00053-6. [DOI] [PubMed] [Google Scholar]
- 25.Bjornson KP, Blackwell LJ, Sage H, Baitinger C, Allen D, Modrich P. Assembly and molecular activities of the MutS tetramer. The Journal of biological chemistry. 2003;278:34667–34673. doi: 10.1074/jbc.M305513200. [DOI] [PubMed] [Google Scholar]
- 26.Calmann MA, Nowosielska A, Marinus MG. Separation of mutation avoidance and an tire combination functions in an Escherichia coli mutS mutant. Nucleic acids research. 2005;33:1193–1200. doi: 10.1093/nar/gki263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tham K-C, Hermans N, Winterwerp HHK, Cox MM, Wyman C, Kanaar R, Lebbink JHG. Mismatch repair inhibits homeologous recombination via coordinated directional unwinding of trapped DNA structures. Molecular cell. 2013;51:326–337. doi: 10.1016/j.molcel.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mendillo ML, Putnam CD, Kolodner RD. Escherichia coli MutS tetramerization domain structure reveals that stable dimers but not tetramers are essential for DNA mismatch repair in vivo. The Journal of biological chemistry. 2007;282:16345–16354. doi: 10.1074/jbc.M700858200. [DOI] [PubMed] [Google Scholar]
- 29.Grilley M, Welsh KM, Su SS, Modrich P. Isolation and characterization of the Escherichia coli mutL gene product. The Journal of biological chemistry. 1989;264:1000–1004. [PubMed] [Google Scholar]
- 30.Elez M, Radman M, Matic I. Stoichiometry of MutS and MutL at unrepaired mismatches in vivo suggests a mechanism of repair. Nucleic acids research. 2012;40:3929–3938. doi: 10.1093/nar/gkr1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ahrends R, Kosinski J, Kirsch D, Manelyte L, Giron-Monzon L, Hummerich L, Schulz O, Spengler B, Friedhoff P. Identifying an interaction site between MutH and the C-terminal domain of MutL by crosslinking, affinity purification, chemical coding and mass spectrometry. Nucleic acids research. 2006;34:3169–3180. doi: 10.1093/nar/gkl407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ratcliff GC, Erie DA. A novel single-molecule study to determine protein-protein association constants. Journal of the American Chemical Society. 2001;123:5632–5635. doi: 10.1021/ja005750n. [DOI] [PubMed] [Google Scholar]
- 33.Yang Y, Wang H, Erie DA. Quantitative characterization of biomolecular assemblies and interactions using atomic force microscopy. Methods. 2003;29:175–187. doi: 10.1016/s1046-2023(02)00308-0. [DOI] [PubMed] [Google Scholar]
- 34.Li Y-L, Meng Y-F, Zhang Z-M, Jiang Y. Detecting the Oligomeric State of Escherichia coli MutS from Its Geometric Architecture Observed by an Atomic Force Microscope at a Single Molecular Level. The Journal of Physical Chemistry B. 2014;118:9218–9224. doi: 10.1021/jp504644r. [DOI] [PubMed] [Google Scholar]
- 35.Su SS, Modrich P. Escherichia coli mutS-encoded protein binds to mismatched DNA base pairs. Proceedings of the National Academy of Sciences of the United States of America. 1986;83:5057–5061. doi: 10.1073/pnas.83.14.5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA Mismatch Repair: Functions and Mechanisms. Chem. Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- 37.Cristovao M, Sisamakis E, Hingorani MM, Marx AD, Jung CP, Rothwell PJ, Seidel CA, Friedhoff P. Single-molecule multiparameter fluorescence spectroscopy reveals directional MutS binding to mismatched bases in DNA. Nucleic acids research. 2012;40:5448–5464. doi: 10.1093/nar/gks138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorman J, Wang F, Redding S, Plys AJ, Fazio T, Wind S, Alani EE, Greene EC. Single-molecule imaging reveals target-search mechanisms during DNA mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E3074–E3083. doi: 10.1073/pnas.1211364109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Qiu R, DeRocco VC, Harris C, Sharma A, Hingorani MM, Erie DA, Weninger KR. Large conformational changes in MutS during DNA scanning, mismatch recognition and repair signalling. The EMBO journal. 2012;31:2528–2540. doi: 10.1038/emboj.2012.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sass LE, Lanyi C, Weninger K, Erie DA. Single-Molecule FRET TACKLE Reveals Highly Dynamic Mismatched DNA− MutS Complexes. Biochemistry. 2010;49:3174–3190. doi: 10.1021/bi901871u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jeong C, Cho W-K, Song K-M, Cook C, Yoon T-Y, Ban C, Fishel R, Lee J-B. MutS switches between two fundamentally distinct clamps during mismatch repair. Nature structural & molecular biology. 2011;18:379–385. doi: 10.1038/nsmb.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cho W-K, Jeong C, Kim D, Chang M, Song K-M, Hanne J, Ban C, Fishel R, Lee J-B. ATP alters the diffusion mechanics of MutS on mismatched DNA. Structure. 2012;20:1264–1274. doi: 10.1016/j.str.2012.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Allen DJ, Makhov A, Grilley M, Taylor J, Thresher R, Modrich P, Griffith JD. MutS mediates heteroduplex loop formation by a translocation mechanism. The EMBO journal. 1997;16:4467–4476. doi: 10.1093/emboj/16.14.4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Giron-Monzon L, Manelyte L, Ahrends R, Kirsch D, Spengler B, Friedhoff P. Mapping protein-protein interactions between MutL and MutH by cross-linking. The Journal of biological chemistry. 2004;279:49338–49345. doi: 10.1074/jbc.M409307200. [DOI] [PubMed] [Google Scholar]
- 45.Selmane T, Schofield MJ, Nayak S, Du C, Hsieh P. Formation of a DNA mismatch repair complex mediated by ATP. Journal of Molecular Biology. 2003;334:949–965. doi: 10.1016/j.jmb.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 46.Lee JY, Chang J, Joseph N, Ghirlando R, Rao DN, Yang W. MutH complexed with hemi-and unmethylated DNAs: coupling base recognition and DNA cleavage. Molecular cell. 2005;20:155–166. doi: 10.1016/j.molcel.2005.08.019. [DOI] [PubMed] [Google Scholar]
- 47.Junop MS, Yang W, Funchain P, Clendenin W, Miller JH. In vitro and in vivo studies of MutS, MutL and MutH mutants: correlation of mismatch repair and DNA recombination. DNA repair. 2003;2:387–405. doi: 10.1016/s1568-7864(02)00245-8. [DOI] [PubMed] [Google Scholar]
- 48.Waldminghaus T, Skarstad K. The <i>Escherichia coli</i> SeqA protein. Plasmid. 2009;61:141–150. doi: 10.1016/j.plasmid.2009.02.004. [DOI] [PubMed] [Google Scholar]
- 49.Rotman E, Khan SR, Kouzminova E, Kuzminov A. Replication fork inhibition in seqA mutants of Escherichia coli triggers replication fork breakage. Molecular microbiology. 2014 doi: 10.1111/mmi.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winkler I, Marx AD, Lariviere D, Heinze RJ, Cristovao M, Reumer A, Curth U, Sixma TK, Friedhoff P. Chemical trapping of the dynamic MutS-MutL complex formed in DNA mismatch repair in Escherichia coli. The Journal of biological chemistry. 2011;286:17326–17337. doi: 10.1074/jbc.M110.187641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Robertson A, Pattishall SR, Matson SW. The DNA binding activity of MutL is required for methyl-directed mismatch repair in Escherichia coli. The Journal of biological chemistry. 2006;281:8399–8408. doi: 10.1074/jbc.M509184200. [DOI] [PubMed] [Google Scholar]
- 52.Hall MC, Jordan JR, Matson SW. Evidence for a physical interaction between the Escherichia coli methyl-directed mismatch repair proteins MutL and UvrD. The EMBO journal. 1998;17:1535–1541. doi: 10.1093/emboj/17.5.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Dao V, Modrich P. Mismatch-, MutS-, MutL-, and helicase II-dependent unwinding from the single-strand break of an incised heteroduplex. The Journal of biological chemistry. 1998;273:9202–9207. doi: 10.1074/jbc.273.15.9202. [DOI] [PubMed] [Google Scholar]
- 54.Monnet J, Quessada-Vial A, Hermans N, Graves E, Winterwerp HHK, Friedhoff P, Sixma TK, Lebbink JHG, Strick TR. Nicking Single DNA Molecules to Study Initiation of Mismatch Repair. Biophysical journal. 2014;106:692a. [Google Scholar]
- 55.Graves ET, Térence S. Combining Magnetic Tweezers and Fluorescence to Study DNA Mismatch Repair by MutS, MutL and MutH. Biophysical journal. 2012;102:284a. [Google Scholar]
- 56.Pluciennik A, Modrich P. Protein roadblocks and helix discontinuities are barriers to the initiation of mismatch repair. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:12709–12713. doi: 10.1073/pnas.0705129104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pollak AJ, Reich NO. DNA Adenine Methyltransferase Facilitated Diffusion Is Enhanced by Protein–DNA"Roadblock” Complexes That Induce DNA Looping. Biochemistry. 2015;54:2181–2192. doi: 10.1021/bi501344r. [DOI] [PubMed] [Google Scholar]
- 58.Pluciennik A, Burdett V, Lukianova O, O'Donnell M, Modrich P. Involvement of the beta clamp in methyl-directed mismatch repair in vitro. The Journal of biological chemistry. 2009;284:32782–32791. doi: 10.1074/jbc.M109.054528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pillon MC, Miller JH, Guarné A. The endonuclease domain of MutL interacts with the β sliding clamp. DNA repair. 2011;10:87–93. doi: 10.1016/j.dnarep.2010.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith BT, Grossman AD, Walker GC. Visualization of mismatch repair in bacterial cells. Molecular cell. 2001;8:1197–1206. doi: 10.1016/s1097-2765(01)00402-6. [DOI] [PubMed] [Google Scholar]
- 61.Løbner-Olesen A, Skovgaard O, Marinus MG. Dam methylation: coordinating cellular processes. Current opinion in microbiology. 2005;8:154–160. doi: 10.1016/j.mib.2005.02.009. [DOI] [PubMed] [Google Scholar]
- 62.Sneppen K, Semsey S. Mismatch repair at stop codons is directed independent of GATC methylation on the Escherichia coli chromosome. Scientific reports. 2014;4 doi: 10.1038/srep07346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H, Popodi E, Tang H, Foster PL. Rate and molecular spectrum of spontaneous mutations in the bacterium Escherichia coli as determined by whole-genome sequencing. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:E2774–E2783. doi: 10.1073/pnas.1210309109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Stracy M, Uphoff S, de Leon FG, Kapanidis AN. In vivo single-molecule imaging of bacterial DNA replication, transcription, and repair. FEBS letters. 2014;588:3585–3594. doi: 10.1016/j.febslet.2014.05.026. [DOI] [PubMed] [Google Scholar]
- 65.Bruni R, Martin D, Jiricny J. d(GATC) sequences influence Escherichia coli mismatch repair in a distance-dependent manner from positions both upstream and downstream of the mismatch. Nucleic acids research. 1988;16:4875–4890. doi: 10.1093/nar/16.11.4875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu AL. Influence of GATC sequences on Escherichia coli DNA mismatch repair in vitro. Journal of Bacteriology. 1987;169:1254–1259. doi: 10.1128/jb.169.3.1254-1259.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhang Y, Yuan F, Presnell SR, Tian K, Gao Y, Tomkinson AE, Gu L, Li G-M. Reconstitution of 5′-directed human mismatch repair in a purified system. Cell. 2005;122:693–705. doi: 10.1016/j.cell.2005.06.027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.