Abstract
Purpose
Normally fertilized zygotes generally show two pronuclei (2PN) and the extrusion of the second polar body. Conventional in vitro fertilization (c-IVF) and intracytoplasmic sperm injection (ICSI) often result in abnormal monopronuclear (1PN), tripronuclear (3PN), or other polypronuclear zygotes. In this study, we performed combined analyses of the methylation status of pronuclei (PN) and the number of centrosomes, to reveal the abnormal fertilization status in human zygotes.
Method
We used differences in DNA methylation status (5-methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC)) to discriminate between male and female PN in human zygotes. These results were also used to analyze the centrosome number to indicate how many sperm entered into the oocyte.
Result
Immunofluorescent analysis shows that all of the normal 2PN zygotes had one 5mC/5hmC double-positive PN and one 5mC-positive PN, whereas a parthenogenetically activated oocyte had only 5mC staining of the PN. All of the zygotes derived from ICSI (1PN, 3PN) had two centrosomes as did all of the 2PN zygotes derived from c-IVF. Of the 1PN zygotes derived from c-IVF, more than 50 % had staining for both 5mC and 5hmC in a single PN, and one or two centrosomes, indicating fertilization by a single sperm. Meanwhile, most of 3PN zygotes derived from c-IVF had a 5mC-positive PN and two 5mC/5hmC double-positive PNs, and had four or five centrosomes, suggesting polyspermy.
Conclusions
We have established a reliable method to identify the PN origin based on the epigenetic status of the genome and have complemented these results by counting the centrosomes of zygotes.
Electronic supplementary material
The online version of this article (doi:10.1007/s10815-015-0568-1) contains supplementary material, which is available to authorized users.
Key words: Abnormal fertilization, Monopronuclear (1PN) zygote, Tripronuclear (3PN) zygote, 5-Hydroxymethylcytosine (5hmC), Centrosome
Introduction
Assisted reproductive technologies (ART) have enabled medical science to provide many couples with the opportunity of having a baby. In addition, ART enables scientists to investigate human embryonic development in vitro. These ART procedures include conventional in vitro fertilization (c-IVF) and intracytoplasmic sperm injection (ICSI). ART procedures also routinely assess if normal fertilization has occurred by determining whether the embryo has two pronuclei (PN) and if the second polar body (2nd PB) has been extruded. Abnormally fertilized human zygotes with one pronucleus (1PN) or three pronuclei (3PN) have been identified in previous studies [1–7]. Several hypotheses exist to explain such aneuploidies, but the underlying causes in human ART-derived zygotes remain unknown. This is partly due to the live births of babies developed from abnormal zygotes having been reported in clinical practice. As a result, it is difficult to verify the normality or abnormality of embryos [2]. Previously, we revealed by time-lapse cinematography (TLC) that the diameter of male PN (mPN) is approximately 2 μm larger than female PN (fPN) just before syngamy (unpublished data). This finding was established by highly accurate analysis of TLC and only in the limited circumstances where both mPN and fPN were in the same microscopic field. Therefore, more specific and distinct techniques are required to precisely discriminate between those PNs. This research has significant value because it could identify the developmental mechanism of human embryos as well as the processes of abnormal fertilization. However, the origin of PN in human zygotes with aneuploidies remains uncertain [8].
Recently, it was found that 5-methylcytosine (5mC) in the paternal genome is specifically converted to 5-hydroxymethylcytosine (5hmC) during the pronuclear stage [9–11]. Although these three studies were conducted in animals, we applied a similar approach in this study, using epigenetic divergence to determine the origin of mPN and fPN in abnormal human zygotes with 1PN and 3PN. We used immunofluorescent staining with anti-5mC and anti-5hmC antibodies to determine the origin of PN in human zygotes fertilized both normally and abnormally. This immunofluorescent technique stains mPN as 5mC/5hmC double-positive whereas fPN are stained as 5mC single-positive only.
Furthermore, we analyzed centrosome number using anti-pericentrin antibody to confirm the 5mC and 5hmC results. In humans, the centriole is transported by sperm into oocytes and the sperm centrosome forms a sperm aster after the centriole duplicates at the PN stage [12, 13]. Therefore, the number of centrosomes represents the number of sperm that have entered into oocyte because human oocytes lack centriolar structures [12].
This research project was undertaken with strict ethical review and patient’s agreement in view of the ethical restrictions on the studies with human fertilized embryos. We demonstrated the combined analysis of the methylation status of PN and the number of centrosomes. We also identified various states of abnormal human fertilization and the availability of 1PN zygotes containing a male genome.
Materials and methods
Ethical approval
This study was approved by the ethics committee of JISART (Japanese Institution for Standardizing Assisted Reproductive Technology).
Collection of zygotes and oocytes
Abnormal zygotes with 1PN (c-IVF: n = 14, ICSI: n = 2), 3PN (c-IVF: n = 31, ICSI: n = 6), and normal two-pronuclear (2PN) zygotes derived from c-IVF (n = 8; control) were donated from infertile couples and were cryopreserved by ultra-rapid vitrification (Kitazato Co., Japan). The number of zygotes and oocytes used for the analyses of 5mC/5hmC status and centrosome number was shown in Table 1. The zygotes were thawed immediately before immunofluorescent analysis. Unfertilized metaphase II (MII) oocytes (n = 2) were obtained from a couple with an infertile male after the oocyte pick-up procedure and testicular sperm extraction with the patients’ consent.
Table 1.
Number of zygotes and oocytes used in this study
| 5mC/5hmC analysis | Pericentrin analysis | |
|---|---|---|
| 1PN zygotes derived from c-IVF | 7 | 7 |
| 1PN zygotes derived from ICSI | – | 2 |
| 2PN zygotes derived from c-IVF | 5 | 3 |
| 3PN zygotes derived from c-IVF | 20 | 11 |
| 3PN zygotes derived from ICSI | 3 | 3 |
| Unfertilized MII oocytes | 1a | 1 |
| Total | 36 | 27 |
aParthenogenetic activated oocyte by electric stimulation
Electrical stimulation of oocyte
A non-inseminated oocyte was electrically stimulated to facilitate parthenogenesis to generate control 1PN zygotes containing the maternal genome only. The denuded oocyte was placed in an electrode wire chamber filled with 0.3 M mannitol solution containing 0.1 mM MgSO4 and 0.05 mM CaCl2. Electrical stimulation of each oocyte was applied using a single direct current (DC) pulse at 70 V/mm for a duration of 30 μs at an interval of 1 s using an electro-cell fusion generator (LF101; Nepa Gene, Chiba, Japan).
Immunofluorescent analysis
Zygotes and an electrically induced parthenogenetic oocyte were prepared for 5mC and 5hmC imaging by fixation in 4 % paraformaldehyde in phosphate-buffered saline (PBS) for 30 min, then washed with 0.1 % Tween 20 in PBS (PBST), and permeabilized with 0.25 % Triton X-100 for 30 min. The fixed specimens were then denatured with 4 N HCl for 10 min, neutralized with 100 mM Tris-HCl (pH 8.5) for 10 min, and blocked overnight at 4 °C in 3 % bovine serum albumin (BSA)/0.2 % Triton X-100 in PBS. The specimens were incubated overnight at 4 °C with the primary antibodies, anti-5mC (Calbiochem, USA) and anti-5hmC (Active Motif, Belgium). Zygotes and an unfertilized MII oocyte were prepared for centrosome imaging by fixation in 10 % trichloroacetic acid solution (WAKO) for 20 min, then washed with 0.1 % PBST, and permeabilized with 0.25 % Triton X-100 for 30 min. The specimens were blocked overnight at 4 °C in 3 % BSA in PBS, whereupon they were incubated for a further overnight period at 4 °C, with the primary anti-pericentrin antibody (Abcam, USA). Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 568 goat anti-rabbit IgG (Life Technologies, USA) were used as secondary antibodies. The specimens were then mounted on a glass-bottom dish in Vectashield anti-bleaching solution (Vector Laboratories, USA), and images were obtained using an FV1000 confocal microscope (Olympus, Japan). 3D reconstructions of confocal stacks were made using Volocity version 5.4.2 software (Perkin Elmer, USA).
Results
Discrimination of PN origin by 5mC and 5hmC status in human zygotes
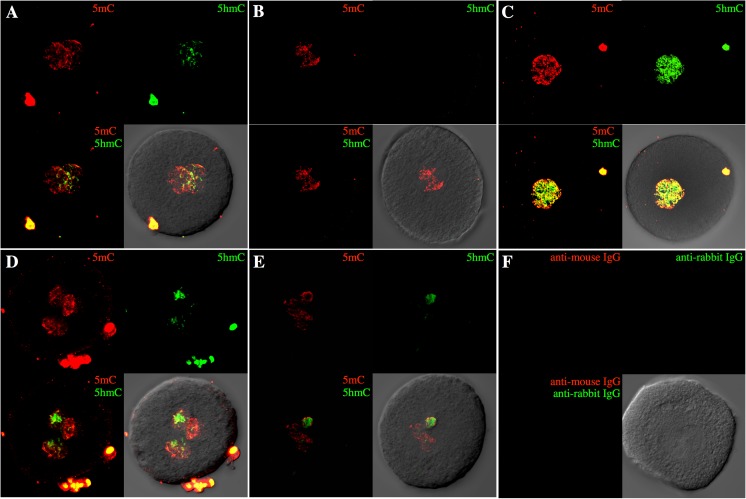
All of the normal 2PN zygotes (5/5) showed one 5mC/5hmC double-positive PN and one 5mC-positive PN. This result indicates that staining for 5mC and 5hmC is a reliable method to establish the origin of the PN (Fig. 1a and Supplementary Movie 1). In contrast, a parthenogenetically activated oocyte (an electrically induced parthenogenetic oocyte) showed only 5mC staining of the PN (Fig. 1b). More than 50 % (4/7) of 1PN zygotes derived from c-IVF had both 5mC and 5hmC staining in a single PN (Fig. 1c and Supplementary Movie 2). These zygotes were probably generated either by the fusion of the maternal and paternal genomes before the formation of 2PN or by androgenesis. The remaining 1PN zygotes (3/7) showed a 5mC-positive PN in less than 50 % (2/7) of cases or a 5mC-positive PN and 5hmC staining in the cytoplasm (1/7). These results indicate that the oocytes were produced by either parthenogenesis or the failure of mPN formation after fertilization. Most of the 3PN zygotes derived from c-IVF (19/20) showed a single 5mC-positive PN and two double-positive PNs, suggesting polyspermy (Fig. 1d and Supplementary Movie 3). On the other hand, 3PN zygotes after ICSI (3/3) showed two 5mC-positive PNs and a 5mC/5hmC double-positive PN, indicating failure of the extrusion of the 2nd PB (Fig. 1e and Supplementary Movie 4). These results were summarized in Fig. 3a. A 2PN zygote treated with secondary antibodies only as a negative control did not show any signals (Fig. 1f).
Fig. 1.
5mC and 5hmC status in normal and abnormal zygotes. Merged confocal Z-series of 5mC (red) and 5hmC (green) immunofluorescent staining is shown. a A normal zygote shows a single 5mC/5hmC double-positive PN and a 5mC-positive PN. b A 5hmC signal was not detected in a parthenogenetic oocyte activated by electric stimulation. c Both 5mC and 5hmC signals were detected in a single PN of a zygote derived from c-IVF. d Two 5mC/5hmC double-positive PNs were detected in a 3PN zygote derived from c-IVF. e A single 5mC/5hmC double-positive PN was detected in a 3PN zygote derived from ICSI. f A 2PN zygote was treated with the secondary antibody only as a negative control
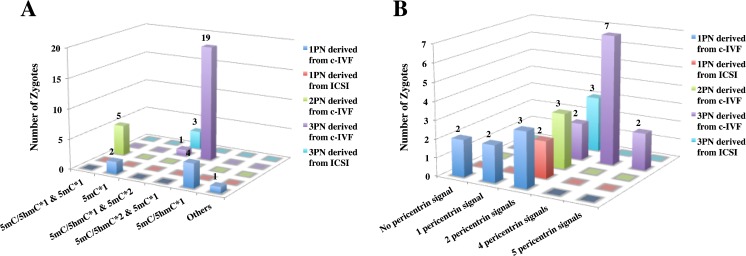
Fig. 3.
Summary of 5mC/5hmC and pericentrin analysis. 3D bar charts of the numbers of zygotes, which are classified according to PN numbers and ART procedure for each analysis, are shown. a The result disaggregated by 5mC and 5hmC status. b The result disaggregated by the number of pericentrin signals (equal to centrosome numbers)
Analyses of centrosome number in human zygotes with 2PN and aneuploidies
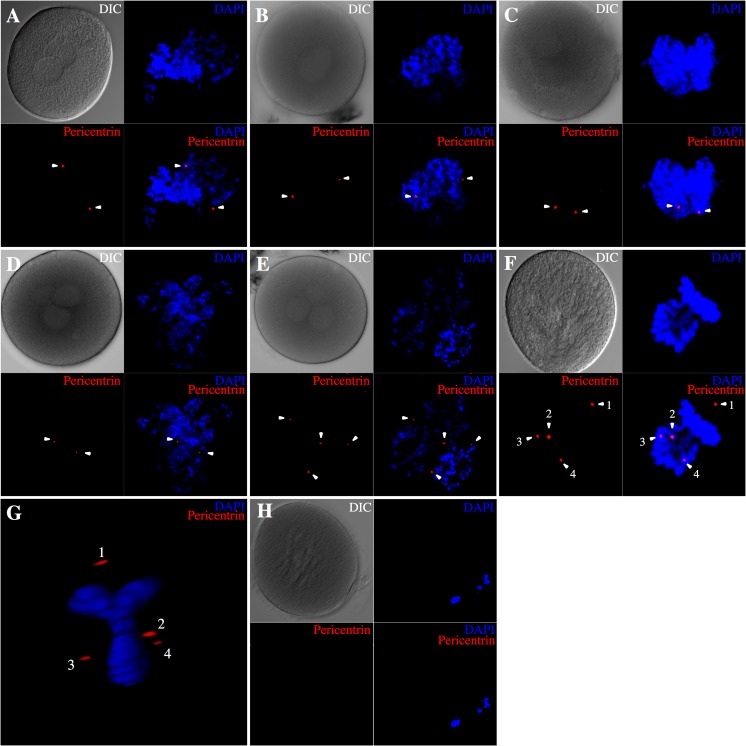
All of normal 2PN zygotes derived from c-IVF (3/3) had two centrosomes (Fig. 2a). All of 1PN and 3PN zygotes derived from ICSI (2/2 and 3/3, respectively) also had two centrosomes (Fig. 2b, d). Most of the 1PN zygotes (5/7) derived from c-IVF had one or two centrosomes (Fig. 2c). These results indicate that in these zygotes, a single sperm has fertilized the oocyte. In contrast, >50 % of 3PN zygotes derived from c-IVF (9/11) had four or five centrosomes, which was probably a result of polyspermy (Fig. 2e). Furthermore, the 3PN zygotes showed a Y-shaped metaphase and aberrant centrosome positions at syngamy (Fig. 2f, g and Supplementary Movie 5). We could not detect any centrosome in unfertilized MII oocytes (Fig. 2h). These results were summarized in Fig. 3b.
Fig. 2.
Centrosome number in normal and abnormal zygotes. Confocal Z-series projection images of immunofluorescent staining for pericentrin (red) are shown. The pericentrin signal is indicated with an arrowhead, and DNA was counterstained with DAPI (blue). The panels labeled DIC show the differential interference contrast images. a Two pericentrin signals were detected around two PNs in normal zygotes. b, d Both 1PN (b) and 3PN (d) zygotes derived from ICSI show two pericentrin signals. c Some 1PN zygotes derived from c-IVF showed two pericentrin signals. e Many 3PN zygotes derived from c-IVF had four centrosomes. f, g Y-shaped metaphase plate and four centrosomes are apparent in 3PN zygotes derived from c-IVF at syngamy. h No pericentrin signal was detected in an unfertilized MII oocyte
Discussion
The most important practical assay for oocytes that undergo normal fertilization in ART procedures is to observe the 2PN and 2PB on day 1 after oocyte pick-up (OPU). Abnormal zygotes with either 1PN or three or more PNs are often visualized at that stage. A review of chromosome abnormalities in human embryos after c-IVF and ICSI identified numerous reports that used fluorescence in situ hybridization to study chromosomal status [14–18]. More recently, we developed an in vitro culture system for TLC to analyze human embryonic development from fertilization to the hatched blastocyst stage [7, 8, 19–21]. Dynamic analyses of TLC images demonstrated several aberrant zygotes with 1PN and uneven 2PN [7].
A previous study has reported that a third PB-like material was extruded into the perivitelline space in some zygotes with 1PN and in uneven 2PN zygotes [7]. Surprisingly, an fPN-like body formed within the third PB-like material in uneven 2PN zygotes and was reabsorbed into the cytoplasm. This event was followed by the migration of a small fPN-like body towards the mPN and subsequent abutment with the mPN. Zygotes with 3PN that resulted from ICSI often occurred because of the failure of the 2nd PB extrusion. These findings indicate that there is an increased likelihood of aberrant fertilization occurring during these processes in patients.
Several possible fertilization patterns can occur in abnormal zygotes, and four plausible mechanisms for the formation of 1PN zygotes are as follows: (1) parthenogenesis, (2) incorporation of mPN and fPN, (3) asynchrony in PN formation, and (4) the third PB-like material extrusion [2, 17, 22, 23]. Pronuclear fusion was not observed in an investigation using time-lapse cinematography from the appearance of 2PN until syngamy [24]. The following three patterns could be considered for the formation of 3PN zygotes: (1) polyspermy normally occurs only in IVF cases; however, it might be possible in rare cases that a diploid spermatozoon was chosen in ICSI; (2) failure of the 2nd PB extrusion in both IVF and ICSI cycles; and (3) fertilization of a diploid oocyte in IVF or ICSI [25–28]. There are other complicated patterns in addition to the above, such as uneven 2PN zygote formation that we observed using TLC analysis.
Many studies of aberrant fertilization patterns have used cellular morphology, cytogenetics, and TLC investigations of morphokinetics. Nevertheless, it is still difficult to identify the underlying mechanisms of those fertilization patterns in detail. Therefore, we established an evidence-based method to reveal the status of abnormal fertilization for the management of ART. This will enable us to decide, for example, if it is necessary to reduce the amount of insemination in c-IVF for patients with an increased risk polyspermy in the next treatment cycle.
Previous studies have demonstrated that only one PN is demethylated in normally fertilized human zygotes [29–31]. Furthermore, a study of the DNA methylation pattern in human 3PN zygotes established that active demethylation of the paternal PN occurs post-fertilization in both c-IVF and ICSI [32]. In addition, recent studies have shown that the demethylation of paternal PN is induced by conversion from 5mC to 5hmC by ten-eleven translocation methylcytosine dioxygenases (TETs) [9–11]. Among TET proteins, TET3 is intensely expressed in oocytes and zygotes. On the other hand, maternal PN is protected from this demethylation by PGC7 [33]. Recently, it has been reported that the DNA methylation patterns were different between male- and female-derived nuclei in human pronuclear stage embryos [34]. We utilized this phenomenon to identify the origin of the PN in order to reveal the fertilization pattern of abnormal zygotes.
Here, we should be fully considered about the effect of vitrification to epigenetic modification in the zygotes. It has been reported that the vitrification procedure reduces the methylation levels of the Oct4, Nanog, and Cdx2 promoters at mouse blastocyst stage [35]. On the other hand, a different study reported that global DNA (hydroxy)-methylation patterns were not significantly different between day 3 embryos obtained from fresh or from vitrified oocytes [36]. Since we detected global DNA methylation status in this study, it seems that vitrified/warmed procedures did not affect critically our results.
In addition, we complemented the results based on this epigenetic status by measuring the centrosome numbers retained in abnormal zygotes. Transmission electron microscopy (TEM) analysis revealed that centrosomes are duplicated at the PN stage, when male and female PNs are in close association [12, 13]. Human oocytes lack centriolar structures [12] whereas human sperm have centrioles, and as a result, the zygotic centrosome must be derived from the sperm in human zygotes. Therefore, the centrosome number can be used to determine the number of sperm incorporated into the zygote. If a single sperm has entered into the oocyte, then two pericentrin signals should be detected in zygote stages. Indeed, both 2PN zygotes derived from c-IVF and abnormal zygotes derived from ICSI showed two pericentrin signals, whereas unfertilized MII oocyte did not show any signal.
In this study, we describe the development of methods utilizing the 5mC and 5hmC content of PN and the analysis of centrosome number to identify the fertilization pattern of abnormal human zygotes. In particular, the results for the 1PN zygotes showed that about half of 1PN zygotes had both 5mC and 5hmC in a single pronucleus or contained a centrosome. In the case of 1PN zygotes derived from ICSI, it is suggested that these are from PN fusion or androgenesis because a single sperm should have contributed to those 1PN zygotes. It is impossible to distinguish between PN fusion and androgenesis with our current technique. However, only 13.1 % of 1PN zygote from c-IVF shows haploidy [18]. Therefore, these zygotes are likely to contain both paternal and maternal genomes and be diploid. Karyotype analyses have shown that between 46 and 80 % of 1PN zygote are diploid [2, 15]. Indeed, some zygotes with a single pronucleus are able to develop to the blastocyst stage in humans [37]. However, the embryos that develop from abnormal zygotes frequently have chromosomal abnormalities [18]. Therefore, the clinical use of these 1PN zygotes for transfer must be carefully considered. 3PN zygotes derived from ICSI cannot have more than two centromeres in the cytoplasm because they are derived from a single injected sperm. Their formation may be a consequence of the failure of the 2nd PB extrusion. However, this explanation is not completely accurate because we have observed 2nd PB or fragmented PB in 3PN zygotes from ICSI (3/3). Additional experiments are needed to determine the possible causes of this fertilization pattern.
In summary, we have established a novel method to identify the PN origin based on the epigenetic status of the genome. In addition, we complemented these results by counting the centrosomes of zygotes. In this study, we have demonstrated the existence of 1PN containing a diploid genome, which would be discarded using only morphological assessment. This finding is expected to lead to the efficient use of 1PN containing a diploid genome in clinical practice.
Electronic supplementary material
3D image reconstruction of a 2PN zygote derived from c-IVF. Red signal means 5mC-positive and green signal means 5hmC-positive. It provides images relating to Fig. 1a. This normal 2PN zygote showed a 5mC/5hmC double-positive PN and a 5mC-positive PN. (MOV 847 kb)
3D image reconstruction of a 1PN zygote derived from c-IVF. It provides images relating to Fig. 1c. This 1PN zygote showed both 5mC and 5hmC staining in a single PN. (MOV 1074 kb)
3D image reconstruction of a 3PN zygote derived from c-IVF. It provides images relating to Fig. 1d. This 3PN zygote showed a single 5mC-positive PN and two 5mC/5hmC double-positive PNs. (MOV 856 kb)
3D image reconstruction of a 3PN zygote derived from ICSI. It provides images relating to Fig. 1e. This 3PN zygote showed two 5mC-positive PNs and a 5mC/5hmC double-positive PN. (MOV 849 kb)
3D image reconstruction of a 3PN zygote derived from c-IVF at syngamy. It provides images relating to Fig. 2g. This movie shows a Y-shaped metaphase plate and aberrant centrosome positions of this 3PN zygote. (MOV 288 kb)
Acknowledgments
We are grateful to Keitaro Yumoto, Minako Sugishima, Chizuru Mizoguchi, Sayako Furuyama, and Yuka Matoba for the collection of the samples. This work was supported by all the staff in the Reproductive Centre, Mio Fertility Clinic, Japan. We also thank S.N. for the technical advice and are particularly grateful for the assistance given by Motokazu Tsuneto.
Abbreviations
- 5mC
5-Methylcytosine
- 5hmC
5-Hydroxymethylcytosine
- ART
Assisted reproductive technologies
- BSA
Bovine serum albumin
- PBS
Phosphate-buffered saline
- PN
Pronucleus (plural: pronuclei)/pronuclear
- mPN
Male pronuclei
- fPN
Female pronuclei
- 2nd PB
Second polar body
- c-IVF
Conventional in vitro fertilization
- ICSI
Intracytoplasmic sperm injection
- MII
Metaphase II
Footnotes
Heading
Identifying the fertilization status in abnormal human zygotes
Capsule
A novel method was established to identify PN origin based on the epigenetic status of the genome utilizing a molecular biological technique. The results were also used to analyze the centrosome numbers in human zygotes.
References
- 1.Kola I, Trounson A, Dawson G, Rogers P. Tripronuclear human oocytes: altered cleavage patterns and subsequent karyotypic analysis of embryos. Biol Reprod. 1987;37:395–401. doi: 10.1095/biolreprod37.2.395. [DOI] [PubMed] [Google Scholar]
- 2.Staessen C, Janssenwillen C, Devroey P, Van Steirteghem AC. Cytogenetic and morphological observations of single pronucleated human oocytes after in-vitro fertilization. Hum Reprod. 1993;8:221–23. doi: 10.1093/oxfordjournals.humrep.a138026. [DOI] [PubMed] [Google Scholar]
- 3.Taylor AS, Braude PR. The early development and DNA content of activated human oocytes and parthenogenetic human embryos. Hum Reprod. 1994;9:2389–97. doi: 10.1093/oxfordjournals.humrep.a138457. [DOI] [PubMed] [Google Scholar]
- 4.Palermo GD, Munné S, Colombero LT, Cohen J, Rosenwaks Z. Genetics of abnormal human fertilization. Hum Reprod. 1995;1:120–7. doi: 10.1093/humrep/10.suppl_1.120. [DOI] [PubMed] [Google Scholar]
- 5.Balakier H, Cadesky K. The frequency and developmental capability of human embryos containing multinucleated blastomeres. Hum Reprod. 1997;12:800–4. doi: 10.1093/humrep/12.4.800. [DOI] [PubMed] [Google Scholar]
- 6.Rosenbusch B, Schneider M, Kreienberg R, Brucker C. Cytogenetic analysis of human zygotes displaying three pronuclei and one polar body after intracytoplasmic sperm injection. Hum Reprod. 2001;16:2362–7. doi: 10.1093/humrep/16.11.2362. [DOI] [PubMed] [Google Scholar]
- 7.Mio Y, Iwata K, Yumoto K, Maeda K. Human embryonic behavior observed with time-lapse cinematography. J Health Med Inf. 2014;5:143. [Google Scholar]
- 8.Mio Y. Morphological analysis of human embryonic development using time-lapse cinematography. J Mamm Ova Res. 2006;23:27–35. doi: 10.1274/jmor.23.27. [DOI] [Google Scholar]
- 9.Iqbal K, Jin SG, Pfeifer GP, Szabó PE. Reprogramming of the paternal genome upon fertilization involves genome-wide oxidation of 5-methylcytosine. Proc Natl Acad Sci U S A. 2011;108:3642–7. doi: 10.1073/pnas.1014033108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wossidlo M, Nakamura T, Lepikhov K, Marques CJ, Zakhartchenko V, Boiani M, et al. 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming. Nat Commun. 2011;2:241. doi: 10.1038/ncomms1240. [DOI] [PubMed] [Google Scholar]
- 11.Gu TP, Guo F, Yang H, Wu HP, Xu GF, Liu W, et al. The role of Tet3 DNA dioxygenase in epigenetic reprogramming by oocytes. Nature. 2011;477:606–610. doi: 10.1038/nature10443. [DOI] [PubMed] [Google Scholar]
- 12.Sathananthan AH, Kola I, Osborne J, Trouson A, Ng SC, Bongso A, et al. Centrioles in the beginning of human development. Proc Natl Acad Sci USA. 1991;88:4806–10. doi: 10.1073/pnas.88.11.4806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sathananthan AH, Ratnam SS, Ng SC, Tarín JJ, Gianaroli L, Trouson A. The sperm centriole: its inheritance, replication and perpetuation in early human embryos. Hum Reprod. 1996;11:345–56. doi: 10.1093/HUMREP/11.2.345. [DOI] [PubMed] [Google Scholar]
- 14.Munné S, Cohen J. Chromosome abnormalities in human embryos. Hum Reprod Update. 1998;4:842–55. doi: 10.1093/humupd/4.6.842. [DOI] [PubMed] [Google Scholar]
- 15.Plachot M. Chromosome analysis of oocytes and embryos. In Verlinsky Y, Kuliev A. (eds), Preimplantation Genetics. Plenum Press; New York, 1991: 103–12.
- 16.Palermo GD, Munné S, Cohen J. The human zygote inherits its mitotic potential from the male gamete. Hum Reprod. 1994;9:1220–5. doi: 10.1093/oxfordjournals.humrep.a138682. [DOI] [PubMed] [Google Scholar]
- 17.Levron J, Munné S, Willadsen S, Rosenwaks Z, Cohen J. Male and female genomes associated in a single pronucleus in human zygotes. Biol Reprod. 1995;52:653–7. doi: 10.1095/biolreprod52.3.653. [DOI] [PubMed] [Google Scholar]
- 18.Staessen C, Van Steirteghem AC. The chromosomal constitution of embryos developing from abnormally fertilized oocytes after intracytoplasmic sperm injection and conventional in-vitro fertilization. Hum Reprod. 1997;12:321–7. doi: 10.1093/humrep/12.2.321. [DOI] [PubMed] [Google Scholar]
- 19.Mio Y, Maeda K. Time-lapse cinematography of dynamic changes occurring during in vitro development of human embryos. Am J Obstet Gynecol. 2008;199:1–5. doi: 10.1016/j.ajog.2008.03.040. [DOI] [PubMed] [Google Scholar]
- 20.Mio Y, Iwata K, Yumoto K, Kai Y, Sargant HC, Mizoguchi C, et al. Possible mechanism of polyspermy block in human oocytes observed by time-lapse cinematography. J Assist Reprod Genet. 2012;29:951–6. doi: 10.1007/s10815-012-9815-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Iwata K, Yumoto K, Sugishima M, Mizoguchi C, Kai Y, Iba Y, et al. Analysis of compaction initiation in human embryos by using time-lapse cinematography. J Assist Reprod Genet. 2014;31:421–6. doi: 10.1007/s10815-014-0195-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Balakier H, Squire J, Casper RF. Characterization of abnormal one pronuclear human oocytes by morphology, cytogenetics and in-situ hybridization. Hum Reprod. 1993;8:402–8. doi: 10.1093/oxfordjournals.humrep.a138060. [DOI] [PubMed] [Google Scholar]
- 23.Nagy ZP, Liu J, Joris H, Devroey P, Van Steirteghem A. Time-course of oocyte activation, pronucleus formation and cleavage in human oocytes fertilized by intracytoplasmic sperm injection. Hum Reprod. 1994;9:1743–8. doi: 10.1093/oxfordjournals.humrep.a138786. [DOI] [PubMed] [Google Scholar]
- 24.Payne D, Flaherty SP, Barry MF, Matthews CD. Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum Reprod. 1997;12:532–41. doi: 10.1093/humrep/12.3.532. [DOI] [PubMed] [Google Scholar]
- 25.Plachot M, Mandelbaum J, Junca AM, De Grouchy J, Salta-Baroux J, Cohen J. Cytogenetic analysis and development capacity of normal and abnormal embryos after IVF. Hum Reprod. 1989;4:99–103. doi: 10.1093/humrep/4.suppl_1.99. [DOI] [PubMed] [Google Scholar]
- 26.Parelmo G, Joris H, Derde MP, Camus M, Devroey P, Van Steirteghem A. Sperm characteristics and outcome of human assisted fertilization by subzonal insemination and intracytoplasmic sperm injection. Fertil Steril. 1993;59:826–35. doi: 10.1016/s0015-0282(16)55867-1. [DOI] [PubMed] [Google Scholar]
- 27.Grossman M, Calafell JM, Brandy N, Vanrell JA, Rubio C, Pellicer A, et al. Origin of tripronucleate zygotes after intracytoplasmic sperm injection. Hum Reprod. 1997;12:2762–5. doi: 10.1093/humrep/12.12.2762. [DOI] [PubMed] [Google Scholar]
- 28.Rosenbusch B, Schneider M, Kreienberg R, Brucker C. Cytogenetic analysis of giant oocytes and zygotes to assess their relevance for the development of digynic triploidy. Hum Reprod. 2002;17:2388–93. doi: 10.1093/humrep/17.9.2388. [DOI] [PubMed] [Google Scholar]
- 29.Mayer W, Niveleau A, Walter J, Fundele R, Haaf T. Demethylation of the zygotic paternal genome. Nature. 2000;403:501–2. doi: 10.1038/35000656. [DOI] [PubMed] [Google Scholar]
- 30.Santos F, Hendrich B, Reik W, Dean W. Dynamic reprogramming of DNA methylation in the early mouse embryo. Dev Biol. 2002;241:172–82. doi: 10.1006/dbio.2001.0501. [DOI] [PubMed] [Google Scholar]
- 31.Beaujean N, Harttshorne G, Cavilla J, Taylor J, Gardner J, Wilmut I, et al. Non-conservation of mammalian preimplantation methylation dynamics. Curr Biol. 2004;14:266–7. doi: 10.1016/j.cub.2004.03.019. [DOI] [PubMed] [Google Scholar]
- 32.Xu Y, Zhang JJ, Grifo JA, Krey LC. DNA methylation patterns in human tripronucleate zygotes. Mol Hum Reprod. 2005;11:167–71. doi: 10.1093/molehr/gah145. [DOI] [PubMed] [Google Scholar]
- 33.Nakamura T, Arai Y, Umehara H, Masuhara M, Kimura T, Taniguchi H, et al. PGC7/Stella protects against DNA demethylation in early embryogenesis. Nat Cell Biol. 2007;9:64–71. doi: 10.1038/ncb1519. [DOI] [PubMed] [Google Scholar]
- 34.Guo H, Zhu P, Yan L, Li R, Hu B, Lian Y, et al. The DNA methylation landscape of human early embryos. Nature. 2014;511:606–10. doi: 10.1038/nature13544. [DOI] [PubMed] [Google Scholar]
- 35.Zhao XM, Du WH, Hao HS, Wang D, Qin T, Liu Y, et al. Effect of vitrification on promoter methylation and the expression of pluripotency and differentiation genes in mouse blastocysts. Mol Reprod Dev. 2012;79:445–50. doi: 10.1002/mrd.22052. [DOI] [PubMed] [Google Scholar]
- 36.De Munck N, Petrussa L, Verheyen G, Staessen C, Vandeskelde Y, Sterckx J, et al. Chromosomal meiotic segregation, embryonic developmental kinetics and DNA (hydroxy)methylation analysis consolidate the safety of human oocyte vitrification. Mol Hum Reprod. 2015;21:535–44. doi: 10.1093/molehr/gav013. [DOI] [PubMed] [Google Scholar]
- 37.Otsu E, Sato A, Nagaki M, Araki Y, Utsunomiya T. Developmental potential and chromosomal constitution of embryos derived from larger single pronuclei of human zygotes used in in vitro fertilization. Fertil Steril. 2004;81:723–4. doi: 10.1016/j.fertnstert.2003.07.042. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
3D image reconstruction of a 2PN zygote derived from c-IVF. Red signal means 5mC-positive and green signal means 5hmC-positive. It provides images relating to Fig. 1a. This normal 2PN zygote showed a 5mC/5hmC double-positive PN and a 5mC-positive PN. (MOV 847 kb)
3D image reconstruction of a 1PN zygote derived from c-IVF. It provides images relating to Fig. 1c. This 1PN zygote showed both 5mC and 5hmC staining in a single PN. (MOV 1074 kb)
3D image reconstruction of a 3PN zygote derived from c-IVF. It provides images relating to Fig. 1d. This 3PN zygote showed a single 5mC-positive PN and two 5mC/5hmC double-positive PNs. (MOV 856 kb)
3D image reconstruction of a 3PN zygote derived from ICSI. It provides images relating to Fig. 1e. This 3PN zygote showed two 5mC-positive PNs and a 5mC/5hmC double-positive PN. (MOV 849 kb)
3D image reconstruction of a 3PN zygote derived from c-IVF at syngamy. It provides images relating to Fig. 2g. This movie shows a Y-shaped metaphase plate and aberrant centrosome positions of this 3PN zygote. (MOV 288 kb)