Abstract
Purpose
Idiopathic asthenospermia is the most common type of male infertility. Although the mechanisms causing asthenospermia are complex, recent studies have indicated an important role of cation channel of sperm (CATSPER) gene downregulation or abnormality in the etiology of idiopathic asthenospermia.
Methods
In the present study, 192 patients with idiopathic asthenospermia and 288 healthy controls were enrolled, and a flight mass spectrometry using Sequenom’s MassArray biochip system was applied for genotyping 16 CATSPER gene SNPs reported in the human single nucleotide polymorphism (SNP) database.
Results
Our results indicated a correlation between CATSPER1 SNPs and idiopathic asthenospermia. In particular, the exonal SNP rs1893316 in CATSPER1 significantly correlated with idiopathic asthenospermia risk and is a potential important factor in determining an individual’s genetic susceptibility to idiopathic asthenospermia.
Conclusion
These finding will help to further elucidate the role of CATSPER1 in idiopathic asthenospermia pathogenesis.
Keywords: Cation channel of sperm (CATSPER), Single nucleotide polymorphisms (SNPs), Idiopathic asthenospermia
Introduction
Approximately 40 % of all infertility cases are attributed to male factors. Many male infertility cases only manifest as reduced sperm concentration or decreased sperm motility and are therefore diagnosed as idiopathic oligozoospermia or asthenospermia due to the lack of a clear clinical cause [1]. As the most common type of male infertility, idiopathic asthenospermia has recently become a research focus in the field.
The etiology of asthenospermia involves many factors, including infection, abnormal semen liquefaction, abnormal immunity, endocrine dysfunction, abnormal sperm structure, chromosomal abnormalities, and varicocele. The mechanisms causing asthenospermia are complex, as they can involve abnormal sperm structure and/or motility or defects in energy metabolism closely associated with sperm motility or abnormal signal transduction pathways [2]. Recent studies have identified specialized Ca2+ channel proteins expressed in the sperm flagellum that are involved in sperm hyperactivation [3] and affect sperm motility, egg-penetration ability, and fertilizability [4]. These proteins belong to the cation channel of sperm (CATSPER) protein family, which includes four members, CATSPER1–4. The CATSPERs are testicular germ cell-specific ionic channels localized to the main section of the sperm flagellum. CATSPER1 and 2 primarily localize to the flagellum’s plasma membrane and regulate its whip-like movement [5, 6]. Moreover, several techniques, including immunoprecipitation, in situ hybridization, and Southern blotting, have revealed that CATSPER3 and 4 also localize to the main section of the sperm flagellum and play important roles in male fertility [7]. Nikpoor et al. [8] used reverse transcription polymerase chain reaction (RT-PCR) to study the testis and found that asthenospermia patients displayed significantly reduced CATSPER1 expression compared to infertile patients with normal sperm motility. Animal model experiments showed that intraperitoneal selenium injection upregulated CATSPER gene expression in both aging and adult mice and improved their sperm motility and survival rate [9], further suggesting the causative role for decreased CATSPER1 expression or abnormality in idiopathic asthenospermia pathogenesis.
A single nucleotide polymorphism (SNP) refers to a DNA sequence polymorphism resulting from a single nucleotide mutation at the genome level [10]. SNPs are the most widely distributed and abundant type of genetic polymorphism. It became the third-generation genetic marker, after the first-generation restriction fragment length polymorphism (RFLP) and the second-generation microsatellite (i.e., simple tandem repeat markers). These genomic sequence variations may lead to individual differences in phenotypes, disease susceptibility (especially complex diseases), and responses to environmental factors or drugs. Therefore, SNPs serve as important genetics tools and are also important in functional genomics research. However, few studies to date have focused on the relationship between CATSPER SNPs and idiopathic asthenospermia, which was the aim of this study.
Materials and methods
Subjects, sample collection, and testing
All volunteers were informed of the objectives and gave informed written consent before beginning the study. All experimental manipulations and protocols were approved by the Ethics Committee of Nanfang Hospital, Southern Medical University (NFEC-200 909-K1). This study was performed in accordance with the principles of the Declaration of Helsinki.
All samples from the Department of Andrology, Nanfang Hospital, Southern Medical University, Guangzhou, China, were collected and analyzed between February 2012 and January 2014. A total of 192 idiopathic asthenoteratozoospermic patients with low sperm motility (rapid forward progressive motile sperm, grade A < 25 %, and forward progressive motile sperm, grade A + B < 50 %), who had failed to accomplish pregnancy in more than 1 year of unprotected sexual intercourse were enrolled in our study. Exclusion criteria were performed as described previously [11]. Partners of these infertile men enrolled in our study were healthy women who were not suffering from diseases, such as severe reproductive system infection and sexual hormone disturbance [11]. A total of 288 ethnically matched volunteers with normal sperm motility were enrolled and served as controls. All the subjects enrolled in our study were Han Chinese.
Semen samples were collected after 2–7 days sexual abstinence duration through masturbation. Semen parameters were measured with SQA-V equipment (TECHNOPATH, Ballina, Ireland) according to the World Health Organization laboratory manual for the examination and processing of human semen (fifth edition). WHO criteria for sperm morphology were applied to all patients, and the percentage of sperm with normal morphology was assessed.
Genotype determination
Genomic DNA was extracted from sperm using a DNA extraction kit (BioTeke Corporation, Beijing, China) according to the manufacturer’s instructions. Assay Design 4.1 software (Sequenom, San Diego, CA, USA) was used to design the primers. SNP genotyping was performed using iPLEX genotyping assays on a MassARRAY platform (MassARRAY Workstation Version3.3, Sequenom). The DNA sample quality threshold was set at 90 %.
Fluorescence quantitative PCR
Total RNA of sperm was extracted using Trizol (Invitrogen, USA). Reverse transcription was performed with 1 μg of RNA after quantification. Quantitative PCR was conducted using the SYBR-Green dye (Toyobo) method with 100 ng of cDNA in a 20 μL system. The primer sequences are shown as follows: CATSPER1-F (5′-AAGAGGTGGCGAGTGAAG-3′), CATSPER1-R (5′-GCAGGTAATGGAACAGGAG-3′); GAPDH-F (5′-GGTATCGTGGAAGGACTC-3′), GAPDH-R (5′-GTAGAGGCAGGGATGATG-3′). The reaction conditions were as follows: 95 °C for 5 min; followed by 40 cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. All reactions were run in triplicate, and quantitative analysis was performed by comparing the 2-ΔΔCt values.
Western blot analysis
Sperm was homogenized in ice-cold CHAPS buffer (20 mM Hepes, pH 7.4, 140 mM NaCl, 10 mM CHAPS, 2 mM EDTA, 1 mM EGTA, and Complete protease inhibitor cocktail) and incubated on the thermoshaker for 30 min at 4 °C. The lysates were then subjected to 15 % SDS-PAGE, and proteins were transferred to a nitrocellulose membrane (Roche Biosciences, Germany). The membrane was blocked with 5 % nonfat milk powder in Tris-buffered saline (TBS; 10 mM Tris-HCl, pH 7.2, 150 mM NaCl) for 4 h. After being washed three times with TBS with 0.1 % Tween 20 (TBS-T), the membrane was then incubated with Anti-CATSPER1 (Abcam, China, 1:500) or GAPDH (Novus Biologicals, 1:1000) antibodies for 2 h. Antibodies binding was visualized by a colorimetric reaction catalyzed by peroxidase-conjugated goat anti-rabbit antibody (1:10,000 dilution in TBS; Promega).
Statistical analysis
The measurement data were presented as mean ± standard deviation, and the count data were expressed as percentages (%). The 16 SNPs were verified with Hardy-Weinberg equilibrium (HWE). Univariate analysis of each SNP and subsequent multivariate analysis of significant SNPs were performed using unconditional logistic regression. Linkage disequilibrium and haplotype analysis were performed with HaploView 4.2 (Daly Lab at the Broad Institute, Cambridge, MA, USA). Some data were adjusted for confounding factors, including age, semen volume, and sperm concentration. A value of p < 0.05 was considered statistically significant.
Results
Demographic and semen parameter comparisons between the two groups
We recruited a total of 251 patients in this study and enrolled 192 eligible subjects for subsequent experiments. Our analysis of demographic and clinical characteristics showed that there was no significant difference in age or sperm concentration between the two groups (p > 0.05). However, differences in semen volume, fast progressive motility, and normal morphology were statistically significant between the groups (p < 0.05, Table 1).
Table 1.
Comparison of demographic characteristics and semen parameters between patient and control groups
| Variables | Patients | Control | t value | P value |
|---|---|---|---|---|
| Age (years) | 30.3 ± 5.9 | 29.4 ± 5.3 | −1.620 | 0.106 |
| Volume (mL) | 2.9 ± 1.3 | 3.2 ± 1.3 | 2.164 | 0.031 |
| Concentration (M/mL) | 90.1 ± 67.3 | 100.3 ± 59.2 | 1.756 | 0.080 |
| Fast progressive motility (%) | 17.6 ± 10.0 | 45.8 ± 9.9 | 30.341 | <0.001 |
| Normal morphology (%) | 4.7 ± 1.7 | 13.4 ± 5.1 | 26.864 | <0.001 |
Distribution of genotypes in the two groups
To investigate the correlation between idiopathic asthenospermia and CATSPER SNPs, we genotyped all of the samples for the following 16 known CATSPER SNPs: CATSPER1: rs1893316, rs1203998, rs2845570, rs35484336, rs3814747, rs3814748, and rs3829937; CATSPER2: rs8042868 and rs3853543; CATSPER3: rs3896260 and rs17167765; and CATSPER4: rs41284333, rs11247866, rs12138368, rs9970046, and rs17163674, using Sequenom’s MassARRAY time of flight mass spectrometry. The results are shown in Table 2. While rs3853543 (C > G) and rs35484336 (A > G) were inconsistent with the HWE in the patient group, rs3896260 (G > T), rs12138368 (G > T), and rs35484336 (A > G) were inconsistent with HWE in the control group. In other words, with the exception of these four sites (rs3853543 (C > G), rs3896260 (G > T), rs12138368 (G > T), and rs35484336 (A > G)), the remaining loci satisfied HWE (p > 0.05), indicating that the cohort of subjects in both groups of this study was representative. Therefore, our study was feasible.
Table 2.
Genotype distribution in patient and control groups
| SNP | Patients N (%) | Control N (%) | χ 2 patient | P patient | χ 2 control | P control |
|---|---|---|---|---|---|---|
| rs1893316 (C > T) | ||||||
| CC | 100 (52.1) | 183 (63.5) | 0.242 | 0.622 | 1.366 | 0.243 |
| CT | 79 (41.1) | 89 (30.9) | ||||
| TT | 13 (6.8) | 16 (5.7) | ||||
| rs1203998 (G > A) | ||||||
| GG | 50 (25.6) | 92 (31.9) | 3.390 | 0.065 | 0.006 | 0.941 |
| GA | 108 (55.4) | 141 (49.0) | ||||
| AA | 34 (19.0) | 55 (19.1) | ||||
| rs2845570 (G > T) | ||||||
| GG | 78 (40.6) | 131 (45.6) | 0.205 | 0.651 | 1.894 | 0.169 |
| GT | 91 (47.4) | 118 (41.1) | ||||
| TT | 23 (12.0) | 38 (13.3) | ||||
| rs3814747 (A > G) | ||||||
| AA | 159 (82.8) | 235 (81.6) | 1.243 | 0.265 | 3.644 | 0.056 |
| AG | 30 (15.6) | 47 (16.3) | ||||
| GG | 3 (1.6) | 6 (2.1) | ||||
| rs3814748 (A > G) | ||||||
| AA | 3 (1.6) | 6 (2.1) | 1.787 | 0.181 | 2.127 | 0.145 |
| AG | 28 (14.6) | 52 (18.1) | ||||
| GG | 161 (83.8) | 230 (79.8) | ||||
| rs3829937 (C > T) | ||||||
| CC | 156 (81.3) | 219 (76.0) | 0.650 | 0.420 | 1.678 | 0.195 |
| CT | 33 (17.2) | 67 (23.3) | ||||
| TT | 3 (1.5) | 2 (0.7) | ||||
| rs8042868 (C > T) | ||||||
| CC | 102 (53.1) | 169 (58.7) | 0.722 | 0.395 | 0.638 | 0.425 |
| CT | 79 (41.1) | 100 (34.7) | ||||
| TT | 11 (5.8) | 19 (6.6) | ||||
| rs17167765 (C > T) | ||||||
| CC | 156 (81.3) | 240 (83.3) | 2.055 | 0.152 | 0.674 | 0.412 |
| CT | 36 (18.7) | 47 (16.3) | ||||
| TT | 0 (0.0) | 1 (0.4) | ||||
| rs41284333 (A > G) | ||||||
| AA | 188 (97.9) | 278 (96.5) | 0.021 | 0.884 | 0.090 | 0.764 |
| AG | 4 (2.1) | 10 (3.5) | ||||
| GG | 0 (0.0) | 0 (0.0) | ||||
| rs11247866 (A > G) | ||||||
| AA | 186 (96.9) | 274 (95.1) | 0.048 | 0.826 | 3.495 | 0.062 |
| AG | 6 (3.1) | 13 (4.5) | ||||
| GG | 0 (0.0) | 1 (0.4) | ||||
| rs9970046 (A > G) | ||||||
| AA | 69 (35.9) | 95 (33.1) | 0.021 | 0.885 | 0.341 | 0.560 |
| AG | 93 (48.4) | 136 (47.4) | ||||
| GG | 30 (15.7) | 56 (19.5) | ||||
| rs17163674 (C > T) | ||||||
| CC | 164 (85.4) | 237 (82.3) | 0.689 | 0.407 | 0.934 | 0.334 |
| CT | 26 (13.5) | 50 (17.4) | ||||
| TT | 2 (1.1) | 1 (0.3) | ||||
Correlation of CATPSER SNPs with asthenospermia
We studied the correlation of the 16 SNPs with asthenospermia using univariate logistic regression analysis. Our results indicated that only rs1893316 (C > T) was significant and the risk of asthenospermia in individuals with the mutant genotype was 1.60 [95 % confidence interval (CI) 1.11–2.32] times of that of wild-type individuals. We observed similar results after adjustment for confounding variables, including age, semen volume, and sperm concentration. The other 15 SNPs showed no significant correlation with asthenospermia risk (Table 3).
Table 3.
Correlation of different CATSPER SNPs with asthenospermia
| CATSPER | Patient | Control | OR (95%CI) | OR§ (95%CI) | ||
|---|---|---|---|---|---|---|
| N | % | N | % | |||
| rs1893316 (C > T) | ||||||
| CC | 100 | 52.1 | 183 | 63.5 | 1.60 (1.11–2.32)* | 1.62 (1.02–2.60)* |
| CT/TT | 92 | 47.9 | 105 | 36.5 | ||
| rs1203998 (G > A) | ||||||
| GG | 50 | 26.0 | 92 | 31.9 | 1.33 (0.89–2.00) | 1.36 (0.77–2.40) |
| GA/AA | 142 | 74.0 | 196 | 68.1 | ||
| rs2845570 (G > T) | ||||||
| GG | 78 | 40.6 | 131 | 45.6 | 1.23 (0.85–1.78) | 1.50 (0.90–2.49) |
| GT/TT | 192 | 59.4 | 156 | 54.4 | ||
| rs3814747 (A > G) | ||||||
| AA | 159 | 82.8 | 235 | 81.6 | 0.92 (0.57–1.49) | 1.22 (0.69–2.19) |
| AG/GG | 33 | 17.2 | 53 | 18.4 | ||
| rs3814748 (A > G) | ||||||
| AA | 3 | 1.6 | 6 | 2.1 | 1.34 (0.33–5.43) | 0.96 (0.20–4.57) |
| AG/GG | 189 | 98.4 | 282 | 97.9 | ||
| rs3829937 (C > T) | ||||||
| CC | 156 | 81.2 | 219 | 76.0 | 0.73 (0.47–1.15) | 1.03 (0.58–1.81) |
| CT/TT | 36 | 18.8 | 69 | 24.0 | ||
| rs8042868 (C > T) | ||||||
| CC | 102 | 53.1 | 169 | 58.7 | 1.25 (0.87–1.81) | 1.27 (0.87–1.87) |
| CT/TT | 90 | 46.9 | 119 | 41.3 | ||
| rs17167765 (C > T) | ||||||
| CC | 156 | 81.2 | 240 | 83.3 | 1.15 (0.72–1.86) | 1.07 (0.64–1.79) |
| CT/TT | 36 | 18.8 | 48 | 16.7 | ||
| rs41284333 (A > G) | ||||||
| AA | 188 | 97.9 | 278 | 96.5 | 0.59 (0.18–1.91) | 0.47 (0.05–4.22) |
| AG/GG | 4 | 2.1 | 10 | 3.5 | ||
| rs11247866 (A > G) | ||||||
| AA | 186 | 96.9 | 274 | 95.5 | 0.68 (0.25–1.82) | 1.21 (0.19–7.62) |
| AG/GG | 6 | 3.1 | 13 | 4.5 | ||
| rs9970046 (A > G) | ||||||
| AA | 69 | 35.9 | 95 | 33.1 | 0.88 (0.60–1.30) | 0.95 (0.62–1.44) |
| AG/GG | 123 | 64.1 | 192 | 66.9 | ||
| rs17163674 (C > T) | ||||||
| CC | 164 | 85.4 | 237 | 82.3 | 0.79 (0.48–1.31) | 0.73 (0.43–1.26) |
| CT/TT | 28 | 14.6 | 51 | 17.7 | ||
§Logistic regression analysis, adjusted for confounders such as age, semen volume, and sperm concentration
*p < 0.05
Correlation between rs1893316 and CATSPER1 expression
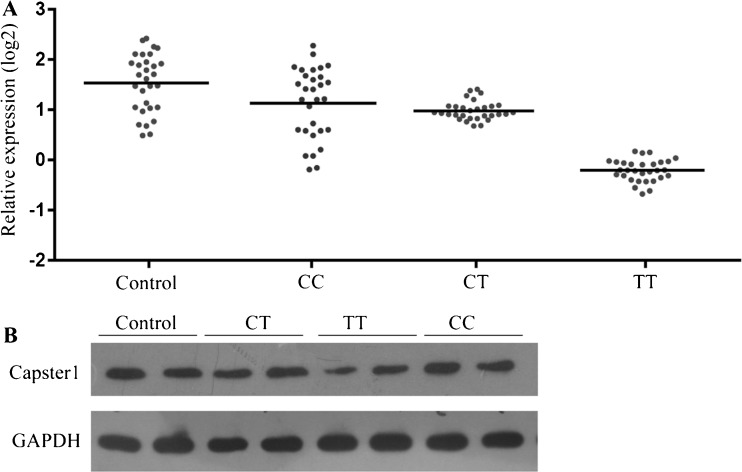
The mutant genotype of rs1893316 has been reported to increase the risk of asthenospermia. However, its exact role in asthenospermia development requires extensive investigation. SNP rs1893316 is a synonymous SNP and does not alter the CATSPER1 protein structure, which does not affect protein function. However, SNP rs1893316 is located in the first exon of the CATSPER1 gene, which usually contains CpG islands or regulator binding sites. The SNP rs1893316 mutation alters the nucleotide sequence, which may destroy the binding sites of some regulators, thereby affecting transcription. We collected and grouped clinical samples according to the SNP rs1893316 genotype and determined CATSPER1 mRNA and protein expression by RT-PCR assay and Western blotting analysis, respectively. We found that CATSPER1 mRNA and protein expression was significantly lower in patients with the TT genotype than in controls and patients with the CC and CT genotypes, while there were no significant differences in CATSPER1 mRNA and protein expression between controls and patients with the CC and CT genotypes (Fig. 1). The results demonstrate that SNP rs1893316 correlates with idiopathic asthenospermia, and it may be involved in the development of idiopathic asthenospermia by downregulating CATSPER1 expression at both the transcriptional and translation levels.
Fig. 1.
CATSPER1 expression. a Real-time qPCR analysis showed expression of CATSPER1 in normal tissue samples and idiopathic asthenospermia tissue samples, and they were significantly different (p <0.01). b Western blot analysis showed expression of CATSPER1 in normal tissue samples and idiopathic asthenospermia tissue samples
Haplotype analysis of CATSPER SNPs
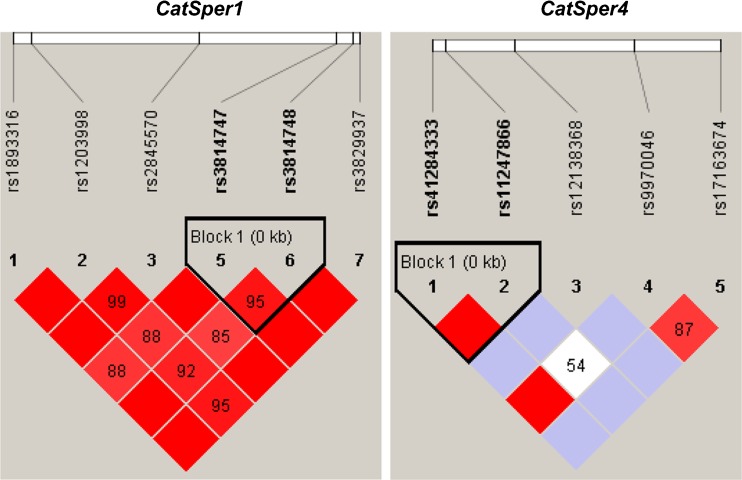
Finally, we analyzed linkage disequilibrium and haplotype using the HaploView software, and we found that among seven CATSPER1 SNPs (rs1893316, rs1203998, rs2845570, rs35484336, rs3814747, rs3814748, and rs3829937), rs3814747 and rs3814748 displayed linkage disequilibrium (D′ = 0.953, r2 = 0.877) (Fig. 2). However, the frequency of AG and GA haplotypes was 0.894 (p = 0.549) and 0.095 (p = 0.322), respectively, showing no statistically significant difference. The remaining CATSPER1 SNPs did not exhibit linkage disequilibrium. Among the five CATSPER4 SNPs (rs41284333, rs11247866, rs12138368, rs9970046, and rs17163674), rs41284333 and rs11247866 displayed linkage disequilibrium (D′ = 1, r2 = 0.733) (Fig. 2), and the frequency of AG and GG haplotypes was 0.980 (p = 0.448) and 0.015 (p = 0.379), respectively, which was not statistically significant. The remaining CATSPER4 SNPs did not show linkage disequilibrium.
Fig. 2.
CATSPER haploblocks in the Chinese population in relation to idiopathic asthenospermia-associated single nucleotide polymorphisms (SNPs). HaploView V4.2 was used to define linkage disequilibrium (LD) blocks represented by the triangular lines based on the current genotyping data from the Chinese population. Squares indicate pair-wise r 2 values on a red-scale with D′ = 1 (red) through to D′ = 0 (white)
Discussion
Recent studies have shown that the main sperm flagellar segment and acrosome contain a specific protein family known as cation channels of sperm (CATSPERs), which can mediate Ca2+ flow and thereby regulate sperm motility and hyperactivation. They play important roles in the acrosome reaction and capacitation [2, 7].
Studies have found that CATSPER1 is closely associated with sperm motility. CATSPER1 knockout mice exhibit poor sperm motility and lack the sperm’s fast progressive linear motility, powerful whip-like movement, and cAMP-induced Ca2+ flow [4]. In addition, CATSPER1 knockout sperm failed to show whip-like movement even after capacitation [12]; they also lost the ability to penetrate the zona pellucida as well as the Ca2+ flux in the sperm flagellum [13], suggesting that CATSPER channels not only regulate sperm movement but also function in sperm hyperactivation and the acrosome reaction [7, 14]. In addition, clinical studies have shown that decreased sperm motility in certain idiopathic infertility patients was associated with abnormal CATSPER1 expression due to mutations [8, 15]. Furthermore, Wang et al. [16] have found that CATSPER1 downregulation in epididymal spermatozoa contributes to asthenozoospermia pathogenesis, whereas Sheng-Jing-San treatment induces upregulation of the channel and improves sperm motility in a rat model of asthenozoospermia.
There have been recent studies on the correlation of gene SNPs with idiopathic asthenospermia. For example, Buldreghini et al. [17] reported that the T allele encoding for aspartic acid in eNOS (Glu298Asp) may contribute to poor sperm motility. Tronchon et al. [18] found that the frequency of the 2308 TNF A allele increased in patients with low sperm counts of testicular origin [p = 0.002; odds ratio (OR) = 2.93] or with normal production count but altered sperm motility (p = 0.003; OR = 2.32) compared to patients with normal sperm count and quality (morphology and motility). However, studies have rarely focused on the relationship between CATSPER SNPs and idiopathic asthenospermia. Although Visser et al. [19] reported relevant investigations, they only presented sequencing data and did not perform correlation analysis of a larger sample.
Therefore, we conducted an in-depth study on the basis of the previous study by Visser et al. [19] to analyze the 16 CATSPER SNPs selected from the human SNP database. We performed flight mass spectrometry using Sequenom’s MassArray biochip system for a case-control study of 192 patients with idiopathic asthenospermia and 288 healthy controls. Our results demonstrated a correlation between CATSPER1 SNPs and idiopathic asthenospermia; in particular, the exonal CATSPER1 rs1893316 SNP significantly correlated with the risk of idiopathic asthenospermia and may be an important factor in determining genetic susceptibility to idiopathic asthenospermia. As a synonymous SNP, rs1893316 does not alter CATSPER1 protein structure. Therefore, we speculated that rs1893316 does not affect the amino acid structure to correlate with idiopathic asthenospermia. However, rs1893316, which is located in the first exon of the CATSPER1 gene, may contribute to idiopathic asthenospermia by affecting transcription. We evaluated the association of rs1893316 with CATSPER1 mRNA and protein expression and found that both were significantly lower in patients with the TT genotype than controls and patients with the CC and CT genotypes. However, we did not detect any significant differences in CATSPER1 mRNA and protein expression between controls and patients with the CC and CT genotypes. These results demonstrate that rs1893316 is associated with idiopathic asthenospermia. By using denaturing high-performance liquid chromatography (DHPLC) and bidirectional sequence analysis in two consanguineous Iranian families segregating autosomal-recessive male infertility, Avenarius et al. [20] found two separate insertion mutations (c.539-540insT and c.948-949insATGGC) in CATSPER1 that are predicted to lead to frameshifts and premature stop codons (p.Lys180LysfsX8 and p.Asp317MetfsX18). Hildebrand et al. [21] thought that mutations in the CATSPER channel genes should be considered a potential cause in cases of recessively inherited male infertility and deafness-infertility syndrome (DIS), and CATSPER mutations may also be associated with sporadic cases of male infertility. Bhilawadikar et al. [22] found that the levels of Tektin 2 and CatSper 2 were significantly lower in spermatozoa of oligoasthenozoospermic men as compared to normozoospermic controls; the levels were also lower in immotile fraction as compared to motile fraction of spermatozoa obtained from normozoospermic individuals, and the levels of Tektin 2 and CatSper 2 were higher in individuals demonstrating sperm motility >60 % as compared to sperm motility <30 %. It is thought that CATSPER1 downregulation or abnormality may affect sperm motility and thereby induce the development of idiopathic oligozoospermia. Therefore, the clinical detection of CATSPER1 mutations, SNPs, or expression may facilitate of idiopathic oligozoospermia diagnosis and therapy.
In this study, we examined 16 CATSPER1 SNPs, which did not cover all SNP loci. It is currently thought that four CATSPER family members may directly or indirectly form a functional tetramer. Further studies to investigate other idiopathic oligozoospermia-associated SNPs are necessary. Subsequent linkage disequilibrium and haplotype analysis using HaploView software detected linkage disequilibrium in rs3814747 of CATSPER1 and in rs3814748, rs41284333, and rs11247866 of CATSPER4, but the difference in haplotypes between groups was not statistically significant.
In summary, there were several SNPs in the CATSPER genes among the subjects in this study, but only the CATSPER1 SNP rs1893316 significantly correlated with the pathogenesis of idiopathic asthenospermia. This finding will help us to further decipher the role of CATSPER1 in idiopathic asthenospermia pathogenesis.
Footnotes
Capsule The exonal SNP rs1893316 in CATSPER1 significantly correlated with idiopathic asthenospermia risk and is a potential important factor in determining an individual’s genetic susceptibility to idiopathic asthenospermia.
Contributor Information
Xuexi Yang, Phone: +86-20-61641765, Email: shirongphd@126.com, Email: 13802503635@126.com.
Rong Shi, Phone: +86-20-61641765, Email: shirongphd@126.com, Email: 13802503635@126.com.
Xiangming Mao, Phone: +86-20-61641765, Email: shirongphd@126.com, Email: 18620050609@163.com.
References
- 1.Brugh VM, Lipshultz LI. Male factor infertility: evaluation and management. Med Clin North Am. 2004;88(2):367–85. doi: 10.1016/S0025-7125(03)00150-0. [DOI] [PubMed] [Google Scholar]
- 2.Tamburrino L, Marchiani S, Minetti F, Forti G, Muratori M, Baldi E. The CatSper calcium channel in human sperm: relation with motility and involvement in progesterone-induced acrosome reaction. Hum Reprod. 2014;29(3):418–28. doi: 10.1093/humrep/det454. [DOI] [PubMed] [Google Scholar]
- 3.Quill TA, Ren D, Clapham DE, Garbers DL. A voltage-gated ion channel expressed specifically in spermatozoa. Proc Natl Acad Sci U S A. 2001;98(22):12527–31. doi: 10.1073/pnas.221454998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ren D, Navarro B, Perez G, Jackson AC, Hsu S, Shi Q, et al. A sperm ion channel required for sperm motility and male fertility. Nature. 2001;413(6856):603–9. doi: 10.1038/35098027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Quill TA, Sugden SA, Rossi KL, Doolittle LK, Hammer RE, Garbers DL. Hyperactivated sperm motility driven by CatSper2 is required for fertilization. Proc Natl Acad Sci U S A. 2003;100(25):14869–74. doi: 10.1073/pnas.2136654100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jin JL, O'Doherty AM, Wang S, Zheng H, Sanders KM, Yan W. Catsper3 and catsper4 encode two cation channel-like proteins exclusively expressed in the testis. Biol Reprod. 2005;73(6):1235–42. doi: 10.1095/biolreprod.105.045468. [DOI] [PubMed] [Google Scholar]
- 7.Qi H, Moran MM, Navarro B, Chong JA, Krapivinsky G, Krapivinsky L, et al. All four CatSper ion channel proteins are required for male fertility and sperm cell hyperactivated motility. Proc Natl Acad Sci U S A. 2007;104(4):1219–23. doi: 10.1073/pnas.0610286104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nikpoor P, Mowla SJ, Movahedin M, Ziaee SA, Tiraihi T. CatSper gene expression in postnatal development of mouse testis and in subfertile men with deficient sperm motility. Hum Reprod. 2004;19(1):124–8. doi: 10.1093/humrep/deh043. [DOI] [PubMed] [Google Scholar]
- 9.Mohammadi S, Movahedin M, Mowla SJ. Up-regulation of CatSper genes family by selenium. Reprod Biol Endocrinol. 2009;7:126. doi: 10.1186/1477-7827-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sachidanandam R, Weissman D, Schmidt SC, Kakol JM, Stein LD, Marth G, et al. A map of human genome sequence variation containing 1.42 million single nucleotide polymorphisms. Nature. 2001;409(6822):928–33. doi: 10.1038/35057149. [DOI] [PubMed] [Google Scholar]
- 11.Yu Q, Zhang Y, Xia Y, Yang X, Li N, Ye L, et al. Analysis of endothelial nitric oxide synthase (eNOS) G894T polymorphism and semen parameters in a Chinese Han population. Andrologia. 2014;46(5):541–6. doi: 10.1111/and.12113. [DOI] [PubMed] [Google Scholar]
- 12.Carlson AE, Westenbroek RE, Quill T, Ren D, Clapham DE, Hille B, et al. CatSper1 required for evoked Ca2+ entry and control of flagellar function in sperm. Proc Natl Acad Sci U S A. 2003;100(25):14864–8. doi: 10.1073/pnas.2536658100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kirichok Y, Navarro B, Clapham DE. Whole-cell patch-clamp measurements of spermatozoa reveal an alkaline-activated Ca2+ channel. Nature. 2006;439(7077):737–40. doi: 10.1038/nature04417. [DOI] [PubMed] [Google Scholar]
- 14.Xia J, Ren D. Egg coat proteins activate calcium entry into mouse sperm via CATSPER channels. Biol Reprod. 2009;80(6):1092–8. doi: 10.1095/biolreprod.108.074039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li HG, Liao AH, Ding XF, Zhou H, Xiong CL. The expression and significance of CATSPER1 in human testis and ejaculated spermatozoa. Asian J Androl. 2006;8(3):301–6. doi: 10.1111/j.1745-7262.2006.00132.x. [DOI] [PubMed] [Google Scholar]
- 16.Wang YN, Wang B, Liang M, Han CY, Zhang B, Cai J, et al. Down-regulation of CatSper1 channel in epididymal spermatozoa contributes to the pathogenesis of asthenozoospermia, whereas up-regulation of the channel by Sheng-Jing-San treatment improves the sperm motility of asthenozoospermia in rats. Fertil Steril. 2013;99(2):579–87. doi: 10.1016/j.fertnstert.2012.10.030. [DOI] [PubMed] [Google Scholar]
- 17.Buldreghini E, Mahfouz RZ, Vignini A, Mazzanti L, Ricciardo-Lamonica G, Lenzi A, et al. Single nucleotide polymorphism (SNP) of the endothelial nitric oxide synthase (eNOS) gene (Glu298Asp variant) in infertile men with asthenozoospermia. J Androl. 2010;31(5):482–8. doi: 10.2164/jandrol.109.008979. [DOI] [PubMed] [Google Scholar]
- 18.Tronchon V, Vialard F, El Sirkasi M, Dechaud H, Rollet J, Albert M, et al. Tumor necrosis factor-alpha-308 polymorphism in infertile men with altered sperm production or motility. Hum Reprod. 2008;23(12):2858–66. doi: 10.1093/humrep/den277. [DOI] [PubMed] [Google Scholar]
- 19.Visser L, Westerveld GH, Xie F, van Daalen SK, van der Veen F, Lombardi MP, et al. A comprehensive gene mutation screen in men with asthenozoospermia. Fertil Steril. 2011;95(3):1020–4. doi: 10.1016/j.fertnstert.2010.11.067. [DOI] [PubMed] [Google Scholar]
- 20.Avenarius MR, Hildebrand MS, Zhang Y, Meyer NC, Smith LL, Kahrizi K, et al. Human male infertility caused by mutations in the CATSPER1 channel protein. Am J Hum Genet. 2009;84(4):505–10. doi: 10.1016/j.ajhg.2009.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hildebrand MS, Avenarius MR, Fellous M, Zhang Y, Meyer NC, Auer J, et al. Genetic male infertility and mutation of CATSPER ion channels. Eur J Hum Genet. 2010;18(11):1178–84. doi: 10.1038/ejhg.2010.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bhilawadikar R, Zaveri K, Mukadam L, Naik S, Kamble K, Modi D, et al. Levels of Tektin 2 and CatSper 2 in normozoospermic and oligoasthenozoospermic men and its association with motility, fertilization rate, embryo quality and pregnancy rate. J Assist Reprod Genet. 2013;30(4):513–23. doi: 10.1007/s10815-013-9972-6. [DOI] [PMC free article] [PubMed] [Google Scholar]