Abstract
Organ abscission is an important process in plant development and reproduction. During abscission, changes in cellular adhesion of specialized abscission zone cells ensure the detachment of infected organs or those no longer serving a function to the plant. In addition, abscission also plays an important role in the release of ripe fruits. Different plant species display distinct patterns and timing of organ shedding, most likely adapted during evolution to their diverse life styles. However, it appears that key regulators of cell separation may have conserved function in different plant species. Here, we investigate the functional conservation of the citrus ortholog of the Arabidopsis peptide ligand INFLORESCENCE DEFICIENT IN ABSCISSION (AtIDA), controlling floral organ abscission. We discuss the possible implications of modifying the citrus IDA ortholog for citrus fruit production.
Keywords: IDA, HAESA, receptor-like kinases, Citrus, floral abscission, fruit production, crop yield
Introduction
Small peptides are used as important signaling ligands to control plant growth and development and more than 1000 genes encoding putative peptides have been discovered in Arabidopsis thaliana (Arabidopsis; Lease and Walker, 2006; Murphy et al., 2012; Grienenberger and Fletcher, 2015). One such peptide ligand, INFLORESCENCE DEFICIENT IN ABSCISSION (AtIDA), was found to be important for the regulation of floral organ abscission in Arabidopsis as the ida mutant failed to abscise its floral organs (Butenko et al., 2003). In wild type (wt) plants the abscission process takes place at the boundary between the organ to be shed and the main plant body, in cell files constituting the abscission zone (AZ). After the formation of an AZ, which occurs early and simultaneously with the development of lateral organs from the apical meristem, the AZ cells can be distinguished from their neighbors by being small, densely cytoplasmic and lacking vacuoles (Addicott, 1982; Sexton and Roberts, 1982; Osborne, 1989; Roberts et al., 2000; Liljegren, 2012). Once the abscission process is initiated these cells go through three sequential developmental stages; (i) cell fate determination and acquisition of competence to respond to abscission signals; (ii) cell wall loosening and expansion by cell wall remodeling (CWR) enzymes followed by organ separation; and (iii) differentiation of a protective lignified layer (Patterson, 2001; Aalen et al., 2013; Gubert et al., 2014).
In the ida mutant, the cell separation event fails to take place (Butenko et al., 2003). In accordance with this, an IDA overexpression line (35S:AtIDA) exhibited ectopic abscission and displayed an enlarged floral AZ in Arabidopsis (Stenvik et al., 2006). Additionally, organ loss was observed at the bases of the pedicel, branches of the inflorescence, and cauline leaves; places where abscission normally does not occur in Arabidopsis. AtIDA signaling and the 35S:AtIDA phenotypes are dependent on the two leucine-rich repeat (LRR) receptor-like kinases (RLKs) HAESA (HAE) and HAESA-LIKE 2 (HSL2; Jinn et al., 2000; Cho et al., 2008; Stenvik et al., 2008). Upon receptor activation a MAP kinase signaling event is turned on ultimately leading to the induction of genes encoding CWR enzymes (Cai and Lashbrook, 2008; Cho et al., 2008; Kumpf et al., 2013). Recently, it was shown that a hydroxyprolinated AtIDA peptide of 12 amino acids was sufficient to mediate AtIDA signaling. Furthermore, this same peptide was shown to bind the HSL2 receptor with high affinity (Butenko et al., 2014).
The gene encoding the AtIDA peptide belongs to a family of five AtIDA-LIKE (AtIDL) genes in Arabidopsis that are expressed in a variety of tissues in the Arabidopsis plant body, including the base of the pedicel, in the floral tissue AZ, in the funicle AZ and in the main root tip (Stenvik et al., 2008). Interestingly, some of the regions of expression represent cell files where abscission takes place in other plants such as in Citrus (citrus) species (Lewis et al., 2006; Estornell et al., 2013), where, unlike for Arabidopsis, leaf abscission does take place. Flower and fruit abscission in citrus occurs at the pedicel AZ (called AZ-A), located close to the boundary between the pedicel and the twig, and at the ovary/fruit AZ (called AZ-C), located in the calyx between the pericarp and the nectary or floral disk, respectively (Tadeo et al., 2008). Cultivated citrus trees usually display heavy flowering. However, a high number of flowers and young fruits are shed during the fruit set period (physiological drop) thus maintaining only those fruits that can be nurtured until maturity. The attachment force by which a fruit is held to the calyx shows a large reduction in value at the end of the maturation phase of fruit development in almost all citrus species. However, early and mid-season varieties of sweet orange are especially prone to a premature decline in the attachment force of the fruit leading to pre-harvest fruit abscission. This problem has a serious economic impact especially in those citrus producing areas dedicated to the fresh fruit market as the Mediterranean Basin. The premature decline in the attachment force of the fruit prevents on-tree storage of fruit which shortens the harvesting season and hinders the orderly fruit marketing. Therefore, understanding the mechanisms controlling fruit abscission in citrus, with the prospect of genetic engineering (transgenic or CRISPR gene editing technologies), is of importance for two reasons; one for controlling fruit loss during the physiological drop and pre-harvest abscission, and two to facilitate shedding of unmarketable fruits remaining on the tree once the harvesting season is over.
INFLORESCENCE DEFICIENT IN ABSCISSION and the IDL peptides are evolutionary conserved across the plant kingdom and they exhibit sequence similarity in their conserved C-terminal domain (PIP domain) containing the highly active AtIDA peptide (Butenko et al., 2003; Stenvik et al., 2008; Butenko et al., 2014). Furthermore, the HSL receptors are found across the plant kingdom and in dicots the HSL receptor sequences are conserved (Stø et al., 2015). It has previously been proposed that the presence of IDL and HSL transcripts at sites where cell separation occur in Arabidopsis may indicate that these genes play a more general role in cell separation (Butenko et al., 2009). Here, we explore on the possibility that this may not only be the case in Arabidopsis but also in citrus.
We show that the C- terminal domain of AtIDA is highly conserved in the IDA and IDL citrus orthologs. We also provide evidence that the citrus IDA (CitIDA) most similar to AtIDA, CitIDA3, has a function in abscission. We discuss the potential agriculture implications of modifying CitIDA3 and additional downstream signaling components of the IDA pathway identified in Arabidopsis.
Materials and Methods
Plant Material and Generation of Transgenic Lines
The CitIDA3 CDS was amplified from clementine cDNA, obtained from total RNA isolated from post-anthesis floral buds, by amplification of a 270 bp fragment with primers: 5′ CACCATGGCTTCTTCTTCTTCTTC 3′, 5′ TCAATTTTGAGTAGAATCCACAACAGA 3′. In parallel, screening of a clementine BAC collection (Terol et al., 2008) was carried out, and as a result BAC clones CCL005I18 and CCH3006E10 were identified to contain the genomic region of CitIDA3. A 3 kb CitIDA3 promoter fragment was amplified from BAC clone CCL005I18 using 5′ CACCGAATTTGTAATTAACTTGTCTTCTT 3′ and 5′ ATAAATTGTTTGTTTTGGGTTGGC 3′ and cloned into pKGWFS7.0 (Plant Systems Biology, Ghent, Belgium). 35S:CitIDA3 lines were made by cloning the CDS fragments into pK2GW7.0 (Plant Systems Biology, Ghent, Belgium). Sixteen independent lines were obtained and two independent lines were investigated in detail. The Agrobacterium tumefaciens C58 strain was used for Arabidopsis transformation by flower dipping (Clough and Bent, 1998).
Plant Material for Complementation of the ida Mutation
Two independent 35S:CitIDA3 lines were crossed into the ida-2 mutant background (Cho et al., 2008). The F2 progeny of both lines were phenotypically scored based on their ability to abscise and genotyped by the ida-2 genotyping primers 5′ CGGTGTTGGTGGATCCAAGTC 3′ and 5′ CCCTCATTTCCGCCACACTTA 3′ and the T-DNA LBb1 primer 5′ ATTTTGCCGATTTCGGAAC 3′. The presence of the 35S:CitIDA3 transgene was verified with primers: 5′ CGCACAATCCCACTATCCTT 3′, 5′ TCAATTTTGAGTAGAATCCACAACAGA 3′.
Petal Breakstrength Measurements
The measurements were performed using a petal breakstrength meter, as previously described (Stenvik et al., 2008). The petal breakstrength was quantified as the force, in gram equivalents, required to remove a petal from a flower (Butenko et al., 2003).
Identification of IDL and HSL Citrus Orthologs
We employed two different strategies to identify IDL citrus orthologs. We first performed TBLASTN searches against the citrus (sweet orange and clementine) genome at the Join Genome Institute (JGI1) using the amino acid sequences corresponding to the variable region and the EPIP motif of the Arabidopsis IDL proteins. Second, we used only the amino acid sequences of the EPIP motifs of the Arabidopsis IDL proteins as query to perform TBLASTN searches in the NCBI GenBank dbEST database for the expressed sequence tag (EST) set of citrus and Poncirus trifoliata. Selected EST sequences (query coverage > 85%; max identity > 55%) were downloaded from the GenBank database and assembled using the CAP3 program (Huang and Madan, 1999). We used the sequences of the unigenes to query against the citrus (sweet orange and clementine) genome at the JGI.1 Signal peptide predictions of all putative citrus IDL proteins were carried out using the SignalP 4.0 server2.
To identify the citrus HSL orthologs TBLASTN (six-frame translation) searches against the citrus (sweet orange and clementine) genome were performed at the JGI1 using the amino acid sequences of the Arabidopsis HAE (AT4G28490), HSL1 (AT1G28440), and HSL2 (AT5G65710) proteins.
Protein Alignments and Phylogenetic Analysis
Multiple sequence alignments of the IDL and HSL proteins were performed using ClustalW tools offered by GenomeNet3 with default parameters and displayed with GENEDOC. Based on the aligned sequences of the citrus IDL and HSL proteins, Neighbor Joining trees were constructed using MEGA version 6.0 (Tamura et al., 2013) with a bootstrap of 1000 replicates.
Results
Identification of the Citrus Members of the IDA-HAE/HSL2 Signaling Module
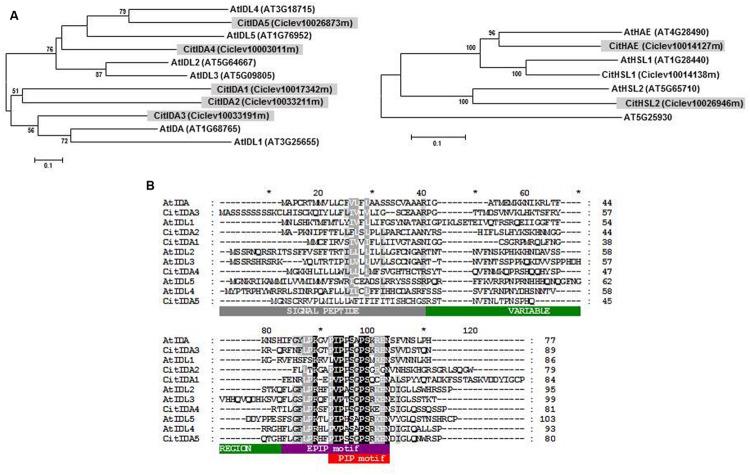
In the citrus (sweet orange and clementine) genome five IDL genes were identified. They were named CitIDA1 to CitIDA5 depending on the location of their sequences in the citrus chromosomes or scaffolds (Supplementary Table S1). The phylogenetic relationship between the six Arabidopsis and five citrus IDL proteins were investigated, showing CitIDA3 to be the closest related to AtIDA (Figure 1A). This gave rise to the possibility that CitIDA3 could have a function in regulating cell separation during organ abscission. Supporting this idea the complete nucleotide sequence of CitIDA3 can be reconstructed from two ESTs derived from AZ libraries (Supplementary Table S3).
FIGURE 1.
Proteins encoded by IDL and HSL genes from Arabidopsis and citrus. (A) Phylogenetic relationships between IDL and HSL proteins. At5g25930 is a protein kinase not related to HSL proteins. (B) Alignment of the IDL full-length proteins showing the N-terminal secretion signal, the variable region, and the short proline-rich EPIP and PIP motives.
All of the citrus IDL genes showed uninterrupted open reading frames encoding translation products with a predicted N-terminal signal peptide sequence followed by a variable region and a highly conserved C-terminal PIP domain (Stenvik et al., 2008; Butenko et al., 2014; Figure 1B). Within the twelve amino acid residues that make up the PIP domain, there were invariant amino acid residues (positions 3, 5, 7, 8, and 12) and others in which the substitutions were conservative (positions 1, 2, and 6). CitIDA3 was the IDA-like protein with highest similarity to the PIP motif of AtIDA differing only in one amino acid (Figure 1B). Given these similarities we hypothesized that CitIDA3 has a conserved function similar to that of AtIDA.
Our genome-wide analysis identified three HSL proteins in the citrus (sweet orange and clementine) genome highly similar to each of the Arabidopsis proteins (Figure 1A and Supplementary Table S2). Therefore, the IDA-HAE/HSL2 signaling module could also work in citrus.
Function and Expression of CitIDA3 Phenocopies AtIDA
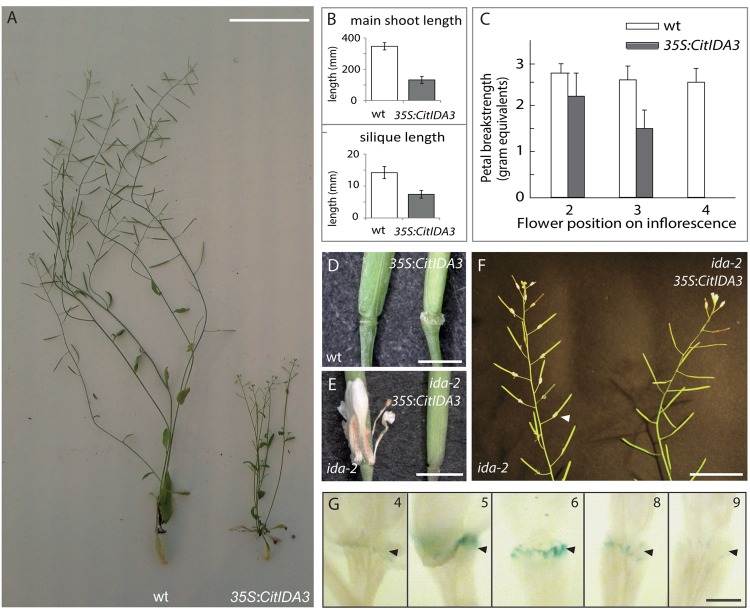
To investigate whether CitIDA3 has a conserved function to AtIDA in regulating cell separation during organ abscission, Arabidopsis plants were transformed with a construct driving CitIDA3 expression with the strong constitutive cauliflower mosaic virus 35S promoter. Fully developed 35S:CitIDA3 plants showed reduced stature and shorter siliques in comparison to wild-type plants (Figures 2A,B). The 35S:CitIDA3 transgenic plants abscised their floral organs at an earlier stage than wild-type plants. Petal breakstrength (pBS), the force needed to remove petals from a flower, decreases along the length of the inflorescence. Changes in progression of abscission are determined by pBS (Bleecker and Patterson, 1997). In wt pBS reaches zero at floral position 8, while for 35S:CitIDA3 this occurs at position 4, comparable to that observed for 35S:AtIDA (Figure 2C; Stenvik et al., 2006). Furthermore, the premature floral organ abscission observed for 35S:CitIDA was associated with an increase in the size of the AZ similar to that observed for 35S:AtIDA (Figure 2D; Stenvik et al., 2006; Shi et al., 2011).
FIGURE 2.
CitIDA3 phenocopies the function of AtIDA. (A) Phenotype of fully developed Arabidopsis wt (Col-0) plants and Arabidopsis plants ectopically overexpressing the citrus IDA ortholog, 35S:CitIDA3. Scalebar = 5 cm. (B) Comparison between main shoot (upper panel) and silique (lower panel) length in wt and 35S:CitIDA3 plants. Values represent mean ± SD, n = 10 (main shoot length) n = 25 (silique length). (C) Petal breakstrength measurements of wt and 35S:CitIDA3 petals. Values represent mean ± SD, n = 15. (D) Comparison of wt and 35S:CitIDA3 AZ. Scalebar = 2 mm. (E) Comparison of ida-2 and ida-2 35S:CitIDA3 floral abscission. Scalebar = 3 mm. (F) Comparison of the inflorescence of ida-2 and ida-2 35S:CitIDA3. Arrow head indicates attached petal. Scalebar = 3 cm. (G) ProCitIDA3:GUS expression in Arabidopsis floral AZ. The numbers indicate flower position along the inflorescence, where position 1 refers to the flower at anthesis. Black arrow heads indicate the AZ. Scalebar = 1 mm.
In an additional experiment performed to investigate the spatial and temporal activity of the CitIDA3 promoter, we monitored the expression of a ProCitIDA3:GUS construct in Arabidopsis plants. In the flowers, CitIDA3 was expressed in the style and in floral organ AZs (Supplementary Figure S1A). GUS expression was absent in flowers from positions 1–4 but detected in the AZ of flowers from positions 5–8 (Figure 2G). CitIDA3 was also expressed in cotyledons, developing leaves and roots of 1-week-old transgenic Arabidopsis seedlings (Supplementary Figure S1B). The expression of CitIDA3 in Arabidopsis flowers is similar to that of AtIDA (Butenko et al., 2003).
35SCitIDA3 Complements the Abscission Deficiency of the ida Mutant
In order to investigate to what extent CitIDA3 could replace the function of AtIDA, 35S:CitIDA3 plants were crossed into the ida-2 mutant background. The F2 progeny of two independent lines was genotyped in order to identify ida-2 plants. 6 out of 29 and 5 out of 27 plants were homozygous for the ida-2 allele. Furthermore, the plants were scored for their abscission phenotype. Two and three of the ida-2 plants from each cross, respectively, showed the ida-2 phenotype, with attached petals after anthesis (Figures 2E,F), The remaining ida-2 homozygotes showed wt abscission (Figures 2E,F). All of these plants contained the 35S:CitIDA3 transgene indicating that the presence of 35S:CitIDA3 is sufficient to induce abscission in ida-2 mutant plants.
Discussion
Pre-harvest abscission is a problem in citrus producing areas dedicated to the fresh fruit market, preventing on-tree storage of fruit and thereby shortening the harvesting and marketing season. On the other hand, the force required to remove mature fruits from the trees in citrus varieties dedicated to the processing market (juices and concentrates) is too large for effective mechanical harvesting. Therefore, new citrus genotypes with different abscission behavior or plants derived from genetic engineering (transgenic or CRISPR gene editing technologies) aiding in timing or execution of citrus fruit specific abscission would be favorable.
In Arabidopsis, the small peptide IDA forms a ligand-receptor module with the two RLKs HAE and HSL2 to initiate a signaling pathway in floral organ abscission (Stenvik et al., 2008). Even though IDA was first found to regulate floral organ abscission, it has later been shown to play a role in cell separation during lateral root emergence, providing this peptide with a more general function in the control of cell separation (Kumpf et al., 2013). The discovery that IDA, HSL and additional downstream components of the IDA signaling system are found across the plant kingdom (Butenko and Simon, 2015; Stø et al., 2015) makes it plausible that orthologs in citrus have a conserved function. Indeed, several auxin response factor (ARF) genes conserved between citrus and Arabidopsis have been shown to have a role in fruit abscission in citrus and floral abscission in Arabidopsis (Ellis et al., 2005; Tadeo et al., 2015). Thus, different cell separation processes appear to share common signaling elements that are conserved across plant species.
The results shown here indicate that CitIDA3 plays a role in floral abscission when overexpressed in Arabidopsis. The Arabidopsis 35S:CitIDA3 transgenic plants were characterized by reduced plant and silique growth and precocious floral organ abscission together with an increased size of the AZ (Figures 2B–D). Overexpression of CitIDA3 resulted in a highly similar phenotype to that observed in Arabidopsis 35S:AtIDA plants (Stenvik et al., 2006). Furthermore expression of the CitIDA3 transgene in an ida-2 mutant background was sufficient to rescue the abscission defect of the mutant and the ProCitIDA3:GUS expression pattern in Arabidopsis plants is also consistent with a role of this gene in organ abscission (Figures 2D–G). Additionally, we identified two LRR-RLKs highly homologous in sequence to the Arabidopsis HAE and HSL2 in the citrus (sweet orange and clementine) genome (Figure 1A and Supplementary Table S2). All these results strongly suggest that the IDA-HAE/HSL2 abscission-signaling pathway characterized in Arabidopsis is conserved in citrus and therefore it could be feasible to manipulate CitIDA3 in order to prevent or stimulate organ abscission in this fruit crop.
Downstream components of the IDA-HAE/HSL2 abscission-signaling pathway could also be potential candidates for manipulation in order to control abscission in citrus. There is genetic evidence for several KNOTTED LIKE HOMEOBOX (KNOX) transcription factors working downstream of the activated IDA-HAE/HSL2 pathway. These include BREVIPEDICELLUS (BP)/KNOTTED-LIKE FROM ARABIDOPSIS THALIANA1 (KNAT1), KNAT2, and KNAT6 (Shi et al., 2011). There are two genes in the citrus (sweet orange and clementine) genome (LOC102615394/Ciclev10001508m and LOC102628261/Ciclev10001779m, respectively) highly homologous to KNAT1 and KNAT2/KNAT6 suggesting that the known downstream components of the IDA-HAE/HSL2 abscission-signaling pathway also exist in citrus.
The MADS-domain transcription factor AGAMOUS-like 15 (AGL15) that previously was reported to play a role in regulating floral abscission in Arabidopsis has recently been shown to act downstream of the IDA-HAE/HSL2 and MAP kinase signaling module to regulate HAE expression (Fernandez et al., 2000; Patharkar and Walker, 2015). Phosphorylation of AGL15 relieves repression of HAE expression leading to production of HAE transcript in a positive feedback loop, thereby increasing the expression of the receptor (Patharkar and Walker, 2015). Manipulation of a citrus version of AGL15 (LOC102612882/Ciclev10032519m) could be a target to control abscission as could the CWR enzymes induced by IDA. Several members of different classes of CWR enzymes have been found in Arabidopsis, tomato and citrus (Estornell et al., 2013). By investigating the enzymes that are expressed in the citrus AZs, fruit abscission prevention or stimulation could be further modified.
Considering all the above, we can glimpse the possibilities available to manipulate the expression of CitIDA3 and/or the citrus orthologs of the downstream components of the IDA-HAE/HSL2 abscission-signaling pathway as an agronomic tool that would affect positively on the economic benefit of growers and the citrus industry in general.
Author Contributions
LE, MW, MP-A, MT, FT, and MB designed the research. LE, MW, MP-A, MT, FT, and MB performed the research. MW, FT, and MB analyzed the data. MW, FT, and MB wrote the article.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work at the Centre de Genómica was supported by Ministerio de Economia e Innovación grants PSE-060000-2009-8, IPT-010000-2010-43, and AGL2011-30240 and by Grants PSE-060000-2009-8, IPT-010000-2010-43 and AGL2011-30240 to MT and BIO2011-26302 to MP-A from the Ministerio de Economia e Innovación of Spain. Work at UiO was supported by Grants 13785/F20 and 230849/F20 to MB, 348256/F20 to MW from the Research Council of Norway. The help and expertise of Clara Fuster, Isabel Sanchís, and Ángel Boix are gratefully acknowledged.
Footnotes
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.01003
References
- Aalen R. B., Wildhagen M., Sto I. M., Butenko M. A. (2013). Ida: a peptide ligand regulating cell separation processes in Arabidopsis. J. Exp. Bot. 64 5253–5261. 10.1093/jxb/ert338 [DOI] [PubMed] [Google Scholar]
- Addicott F. T. (1982). Abscission. Berkeley, CA: University of California Press. [Google Scholar]
- Bleecker A. B., Patterson S. E. (1997). Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9 1169–1179. 10.1105/tpc.9.7.1169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M. A., Patterson S. E., Grini P. E., Stenvik G.-E., Amundsen S. S., Mandal A., et al. (2003). INFLORESCENCE DEFICIENT IN ABSCISSION controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15 2296–2307. 10.1105/tpc.014365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko M. A., Simon R. (2015). Beyond the meristems: similarities in the CLV3 and IDA peptide mediated signalling pathways. J. Exp. Bot. 66 5195–5203. 10.1093/jxb/erv310 [DOI] [PubMed] [Google Scholar]
- Butenko M. A., Vie A. K., Brembu T., Aalen R. B., Bones A. M. (2009). Plant peptides in signalling: looking for new partners. Trends Plant Sci. 14 255–263. 10.1016/j.tplants.2009.02.002 [DOI] [PubMed] [Google Scholar]
- Butenko M. A., Wildhagen M., Albert M., Jehle A., Kalbacher H., Aalen R. B., et al. (2014). Tools and strategies to match peptide-ligand receptor pairs. Plant Cell 26 1838–1847. 10.1105/tpc.113.120071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai S., Lashbrook C. C. (2008). Stamen abscission zone transcriptome profiling reveals new candidates for abscission control: enhanced retention of floral organs in transgenic plants overexpressing Arabidopsis ZINC FINGER PROTEIN2. Plant Physiol. 146 1305–1321. 10.1104/pp.107.110908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S. K., Larue C. T., Chevalier D., Wang H., Jinn T. L., Zhang S., et al. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 105 15629–15634. 10.1073/pnas.0805539105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Ellis C. M., Nagpal P., Young J. C., Hagen G., Guilfoyle T. J., Reed J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132 4563–4574. 10.1242/dev.02012 [DOI] [PubMed] [Google Scholar]
- Estornell L. H., Agusti J., Merelo P., Talon M., Tadeo F. R. (2013). Elucidating mechanisms underlying organ abscission. Plant Sci. 19 48–60. 10.1016/j.plantsci.2012.10.008 [DOI] [PubMed] [Google Scholar]
- Fernandez D. E., Heck G. R., Perry S. E., Patterson S. E., Bleecker A. B., Fang S. C. (2000). The embryo MADS domain factor AGL15 acts postembryonically. Inhibition of perianth senescence and abscission via constitutive expression. Plant Cell 12 183–198. 10.1105/tpc.12.2.183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grienenberger E., Fletcher J. C. (2015). Polypeptide signaling molecules in plant development. Curr. Opin. Plant Biol. 23 8–14. 10.1016/j.pbi.2014.09.013 [DOI] [PubMed] [Google Scholar]
- Gubert C. M., Christy M. E., Ward D. L., Groner W. D., Liljegren S. J. (2014). ASYMMETRIC LEAVES 1 regulates abscission zone placement in Arabidopsis flowers. BMC Plant Biol. 14:195 10.1186/s12870-014-0195-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Madan A. (1999). Cap3: a DNA sequence assembly program. Genome Res. 9 868–877. 10.1101/gr.9.9.868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jinn T. L., Stone J. M., Walker J. C. (2000). Haesa, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14 108–117. [PMC free article] [PubMed] [Google Scholar]
- Kumpf R. P., Shi C. L., Larrieu A., Sto I. M., Butenko M. A., Peret B., et al. (2013). Floral organ abscission peptide IDA and its Hae/Hsl2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. U.S.A. 110 5235–5240. 10.1073/pnas.1210835110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lease K. A., Walker J. C. (2006). The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142 831–838. 10.1104/pp.106.086041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis M. W., Leslie M. E., Liljegren S. J. (2006). Plant separation: 50 ways to leave your mother. Curr. Opin. Plant Biol. 9 59–65. 10.1016/j.pbi.2005.11.009 [DOI] [PubMed] [Google Scholar]
- Liljegren S. J. (2012). Organ abscission: exit strategies require signals and moving traffic. Curr. Opin. Plant Biol. 15 670–676. 10.1016/j.pbi.2012.09.012 [DOI] [PubMed] [Google Scholar]
- Murphy E., Smith S., De Smet I. (2012). Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24 3198–3217. 10.1105/tpc.112.099010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osborne D. J. (1989). Abscission. Crit. Rev. Plant Sci. 8 103–129. 10.1080/07352688909382272 [DOI] [Google Scholar]
- Patharkar O. R., Walker J. C. (2015). Floral organ abscission is regulated by a positive feedback loop. Proc. Natl. Acad. Sci. U.S.A. 112 2906–2911. 10.1073/pnas.1423595112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson S. E. (2001). Cutting loose. Abscission and dehisence in Arabidopsis. Plant Physiol. 126 494–500. 10.1104/pp.126.2.494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts J. A., Whitelaw C. A., Gonzalez-Carranza Z. H., Mcmanus M. T. (2000). Cell separation processes in plants– models, mechanisms and manipulation. Ann. Bot. (Lond.) 86 223–235. 10.1006/anbo.2000.1203 [DOI] [Google Scholar]
- Sexton R., Roberts J. A. (1982). Cell biology of abscission. Ann. Rev. Plant Physiol. 33 133–162. 10.1146/annurev.pp.33.060182.001025 [DOI] [Google Scholar]
- Shi C. L., Stenvik G. E., Vie A. K., Bones A. M., Pautot V., Proveniers M., et al. (2011). Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the Ida-Hae/Hsl2 floral abscission signaling pathway. Plant Cell 23 2553–2567. 10.1105/tpc.111.084608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G. E., Butenko M. A., Urbanowicz B. R., Rose J. K., Aalen R. B. (2006). Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18 1467–1476. 10.1105/tpc.106.042036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G. E., Tandstad N. M., Guo Y., Shi C. L., Kristiansen W., Holmgren A., et al. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in Arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20 1805–1817. 10.1105/tpc.108.059139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stø I. M., Orr R. J. S., Fooyontphanich K., Jin X., Knutsen J. M. B., Fischer U., et al. (2015). Conservation of the abscission signaling peptide IDA during Angiosperm evolution: withstanding genome duplications and gain and loss of the receptors HAE/HSL2. Front. Plant Sci. 6:931 10.3389/fpls.2015.00931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadeo F. R., Agustí J., Merelo P., Estornell L. H., Cercós M., Terol J., et al. (2015). “To fall or not to fall, that’s the question!” Molecular mechanisms underlying organ abscission in Citrus. Acta Hortic. 1065 1189–1195. [Google Scholar]
- Tadeo F. R., Cercós M., Colmenero-Flores J. M., Iglesias D. J., Naranjo M. A., Ríos G., et al. (2008). Molecular physiology of development and quality of citrus. Adv. Bot. Res. 47 148–202. [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. (2013). Mega6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terol J., Naranjo M. A., Ollitrault P., Talon M. (2008). Development of genomic resources for Citrus clementina: characterization of three deep-coverage BAC libraries and analysis of 46,000 BAC end sequences. BMC Genomics 9:423 10.1186/1471-2164-9-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.