ABSTRACT
Mycobacterium tuberculosis, the etiological agent of tuberculosis, is a Gram-positive bacterium with a unique cell envelope composed of an essential outer membrane. Mycolic acids, which are very-long-chain (up to C100) fatty acids, are the major components of this mycomembrane. The enzymatic pathways involved in the biosynthesis and transport of mycolates are fairly well documented and are the targets of the major antituberculous drugs. In contrast, only fragmented information is available on the expression and regulation of the biosynthesis genes. In this study, we report that the hadA, hadB, and hadC genes, which code for the mycolate biosynthesis dehydratase enzymes, are coexpressed with three genes that encode proteins of the translational apparatus. Consistent with the well-established control of the translation potential by nutrient availability, starvation leads to downregulation of the hadABC genes along with most of the genes required for the synthesis, modification, and transport of mycolates. The downregulation of a subset of the biosynthesis genes is partially dependent on RelMtb, the key enzyme of the stringent response. We also report the phylogenetic evolution scenario that has shaped the current genetic organization, characterized by the coregulation of the hadABC operon with genes of the translational apparatus and with genes required for the modification of the mycolates.
IMPORTANCE Mycobacterium tuberculosis infects one-third of the human population worldwide, and despite the available therapeutic arsenal, it continues to kill millions of people each year. There is therefore an urgent need to identify new targets and develop a better understanding of how the bacterium is adapting itself to host defenses during infection. A prerequisite of this understanding is knowledge of how this adaptive skill has been implanted by evolution. Nutrient scarcity is an environmental condition the bacterium has to cope with during infection. In many bacteria, adaptation to starvation relies partly on the stringent response. M. tuberculosis's unique outer membrane layer, the mycomembrane, is crucial for its viability and virulence. Despite its being the target of the major antituberculosis drugs, only scattered information exists on how the genes required for biosynthesis of the mycomembrane are expressed and regulated during starvation. This work has addressed this issue as a step toward the identification of new targets in the fight against M. tuberculosis.
INTRODUCTION
Mycobacterium tuberculosis, the etiological agent of tuberculosis, infects one-third of humans worldwide, with 9 million new cases and 1.5 million people dying each year from this disease (77). A long-standing challenge to the fight against tuberculosis is the ability of the pathogen to develop a nonreplicating, persistent, drug-tolerant form (1–3). In addition, the emergence of multidrug-resistant (MDR), extremely drug-resistant (XDR) (4, 5), and total-drug-resistant (TDR) strains (6) underscores the urgent need to identify new therapeutic targets through a better understanding of the genetic programs developed by the bacterium to enable infection.
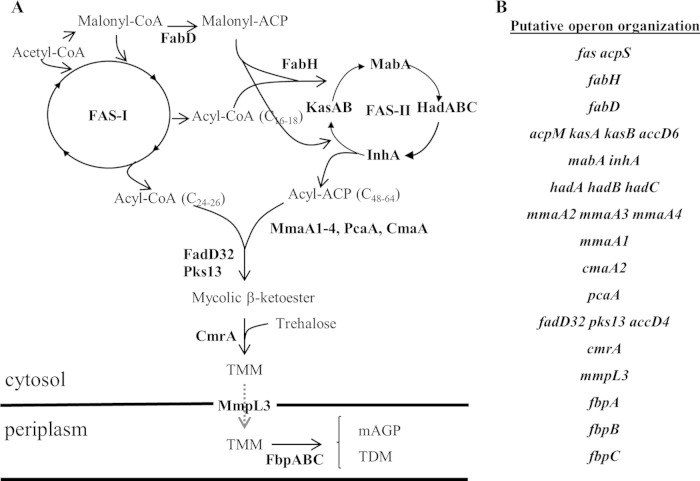
M. tuberculosis has an unusual cell envelope, characterized by an outer membrane (the mycomembrane) consisting largely of long-chain mycolic acids (MA) and a number of atypical lipids (7, 8). Most of these lipids are important to the viability and/or virulence of the bacteria (9), which is why much effort continues to be devoted to identifying, analyzing, and targeting enzymes involved in their metabolism (9–13). Consequently, much is known about the pathways responsible for biosynthesis of the mycolic acids and other components of the mycomembrane (Fig. 1A) (8, 14). This synthesis is centered on two fatty acid synthase (FAS) activities, the eukaryotic-type multifunctional single protein FAS-I and the bacterial-type multiprotein complex FAS-II. FAS-I initiates de novo synthesis from an acetyl coenzyme A (acetyl-CoA) precursor of two products, a C24-C26 acyl-CoA at the origin of the MA α-branch and a C16-C18 acyl-CoA that is subsequently elongated by FAS-II to produce meromycolates. Various methyltransferases add functional groups to the meromycolates (8). A final condensation step forms the mature α-alkyl β-ketoacyl-MA, which is then transported across the cytoplasmic membrane as a trehalose-monomycolate (TMM) molecule by the inner membrane Mmpl3 transporter. In the periplasmic compartment, the mycoloyltransferase Ag85 either transfers the mycoloyl moiety of TMM to the envelope arabinogalactan layer or generates a trehalose-dimycolate (TDM) molecule (Fig. 1A). Although some issues are not settled (notably, the step at which the methyltransferases introduce the functional groups into the meromycolate chains), the biochemistry of MA synthesis is fairly well documented. In contrast, less is known about the regulatory circuits that control the expression of the ∼30 genes involved in the biosynthesis of MA (Fig. 1B). A few transcription regulators that directly regulate the biosynthesis genes have been identified. FasR regulates the expression of the fas gene (15), while MabR (16) and FadR (17) regulate the expression of the fabD-accD6 cluster, the fasII operon (Fig. 1B). However, the environmental cues that trigger these regulatory pathways and their physiological roles are not yet known. Several genome-wide microarray studies of cells challenged with various stresses, including nutrient deprivation (18–20) and those experienced by bacteria during infection (21), have shown that genes involved in MA synthesis are likely to be regulated. However, with the exception of a predictive analysis (22), none of these studies specifically addressed the regulation of the whole set of MA genes. Therefore, only a partial picture of the regulation of MA biosynthesis is available. Two genome-wide microarray studies reported the expression profile of M. tuberculosis genes during starvation (23, 24). However, as far as MA biosynthesis is concerned, there were a number of discrepancies between these studies, confirming suggestions that conclusions based on genome-wide approaches should be experimentally verified (25).
FIG 1.
Overview of mycolate biosynthesis and transport. (A) Biosynthesis pathway. The names of enzymes involved in the biosynthesis or transport are in bold (see the text). The dashed arrow indicates transport across the inner membrane. TMM, trehalose-mono-mycolate; TDM, trehalose-dimycolate; mAGP, mycolyl-arabinogalactan-peptidoglycan. (B) Genetic organization of genes involved in the biosynthesis. The operon organization is from the Database of Prokaryotic Operons (76) (http://csbl.bmb.uga.edu/DOOR/).
Among the enzymes of the FAS-II multiprotein synthase, the dehydratases HadAB and HadBC were the last to be identified in M. tuberculosis (26) and in Mycobacterium smegmatis (27). The observation that the hadABC genes are localized downstream of genes encoding tRNAs and one ribosomal protein suggested a coordination of MA biosynthesis with translation potential. The stringent response (SR) is a universal adaptive response that regulates this potential under starvation conditions. The outcome of the SR is the upregulation of stress response genes and the downregulation of genes encoding the translational machinery and, more globally, of genes involved in energy-consuming processes, such as DNA replication and the synthesis of peptidoglycan and phospholipids (28–31). The SR is required for virulence of many pathogens (32). For M. tuberculosis, the SR, although not essential for survival inside macrophages, is critical for survival during long-term in vitro starvation (33, 34), virulence in the guinea pig model (35), and persistence in mice and artificial granulomas (24, 36, 37).
In this study, we report that the hadABC genes are part of an operon that includes genes required for translation and that as such they are coregulated by the SR. By phylogenetic reconstruction, the key evolutionary steps that have shaped the genetic organization at the hadABC locus in Actinobacteria have been identified. Finally, an integrated picture of the regulation of all MA synthesis genes by starvation and the SR is presented.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and the plasmids used in this study are listed in Table S1. For liquid cultures of mycobacteria strains, Middlebrook 7H9 medium supplemented with 0.05% Tween (Sigma-Aldrich), glycerol (0.2%) and ADC (bovine albumin, fraction V; dextrose; catalase) or OADC (oleic acid plus ADC) was used. The incubation temperature was 37°C. For solid medium, no Tween was added and Middlebrook 7H10 and 7H11 were used for M. smegmatis and M. tuberculosis, respectively. When required, antibiotics were added: zeocin at 15 μg/ml, kanamycin (Kan) at 37.5 μg/ml, and hygromycin (Hyg) at 150 μg/ml (M. smegmatis) and 50 μg/ml (M. tuberculosis). In liquid medium, zeocin was omitted, as we noticed that resistance to zeocin was efficient only in solid medium. To induce, respectively, the tetRO and PAMIE promoters, tetracycline (20 ng/ml) and acetamide (0.2%) were added. Luria-Bertani medium was used for the growth of Escherichia coli strains with antibiotics when required (Kan, 37.5 μg/ml; Hyg, 150 μg/ml).
Starvation assay.
For M. tuberculosis, standing cultures grown to mid-log phase in 7H9 plus Tween, glycerol, and ADC were harvested at mid-logarithmic growth phase, washed once, and resuspended in an equal volume of TBST buffer (50 mM Tris-HCl [pH 7], 150 mM NaCl, 0.05% Tween 80). The suspensions were then further incubated for 6 h before being processed for RNA preparation. Cultures for each strain were in triplicate (biological). For lipid preparation, the same growth conditions were used, except that Tween was replaced with 0.05% tyloxapol (Sigma-Aldrich) and the cell suspensions in TBST buffer were incubated for 96 h (a single biological and technical experiment).
Plasmid construction.
pGBT was constructed by cloning the C. glutamicum tetRO region from the pMIND vector into the pGB9.2 plasmid. The C. glutamicum tetRO region, including the upstream tandem rrn1 transcription terminators, was amplified by PCR, gel purified (Qiagen), cloned in pJET1.2 (Fermentas), digested with AseI and SpeI (Fermentas), gel purified, and cloned in pGB9.2 digested with the same restriction enzymes. Insertion was checked by sequencing (Eurofins). pLAM was constructed by replacing the Kanr gene with the Hygr gene from pJV53H. pLAM-rv0634A was constructed by PCR amplification of the rv0634A open reading frame (ORF) from strain H37Rv using the primers Rv0634Aup and Rv00634Adown. The up primer contains a 5′-end NdeI restriction site, and the down primer contains a 5′-end BamHI restriction site. The PCR product was digested with NdeI and BamHI and cloned into pLAM digested with the same enzymes. pGBT-relMtb was constructed by PCR amplification of the relMtb ORF from strain H37Rv using RelMtbup and RelMtbdown primers. The up primer contains a consensus Shine-Dalgarno sequence upstream of an ATG start codon and a 5′-end Bsu36I restriction site. The down primer contains a 5′-end BamH1 restriction site. Insertion was checked by sequencing (Eurofins). pGBG was constructed by cloning a Shine-Dalgarno egfp gene fragment between the Bsu36I and SpeI sites of pGBT. The tetRO promoter was then removed by an XhoI-BamHI deletion. Operon fusions were constructed by cloning PCR fragments between the XhoI and Bsu36I sites of pGBG.
Green fluorescent protein (GFP) activity measurements.
M. smegmatis strains were cultivated for 2 days in 7H9-based medium; then 4 ml was centrifuged, and the pellet was washed twice with 1× phosphate-buffered saline (PBS) plus 0.1% Tween and resuspended in 1 ml of the same buffer. Fluorescence at 520 nm (excitation at 485 nm) and the optical density at 590 nm (OD590) were measured in technical duplicates in a 96-well plate with the Clariostar reader (BMG). Relative fluorescence (in relative fluorescence units [RFU]) is the ratio of fluorescence to the OD value.
Construction of relMtb and rv0634A deletion mutants.
The mutants were constructed by the recombineering method, as described by van Kessel and Hatfull (38), with two modifications. The recombineering resulted in a hygromycin-resistant derivative (instead of the original kanamycin-resistant plasmid). The allelic exchange segment (AES) was generated with a two-step PCR procedure. In the first step, 3 PCRs were performed to amplify two fragments ∼500 to 600 bp long corresponding to the upstream and downstream regions of the ORF that was to be deleted and the ∼700-bp zeocin resistance cassette (the Streptoalloteichus hindustanus ble [sh ble] gene). The three products were gel purified (Qiagen) and used in the second step for a three-fragment fusion PCR to generate the AES. The product was gel purified and used to transform a competent recombineering strain. Selection was performed on 7H10 medium containing zeocin at 15 μg/ml. Incubation was done at 37°C. PCR with cell lysates was performed to check for the replacement of the target gene by the zeocin cassette and for the absence of the wild-type region. The absence of a specific transcript was also checked by reverse transcription-quantitative PCR (RT-qPCR). The recombineering plasmid (pJV53) was cured from the mutant by successive growth without selection, and its loss was checked by plating on hygromycin-containing solid medium.
Total-RNA preparation.
Cultures of the M. tuberculosis strains were grown to an A600 of ∼0.6. Total RNA was extracted using the RNeasy kit (Qiagen) following the manufacturer's instructions, with slight modifications. Briefly, 20 ml of culture was centrifuged for 5 min at 3,500 rpm, and the pellet was resuspended in 1.2 ml of 0.1% β-mercaptoethanol containing RLT lysis buffer (Qiagen) along with 0.1-mm-diameter glass beads. Cells were then lysed by two 120-s pulses at full speed in a bead beater device. The sample was centrifuged at 14,000 rpm for 30 s and filtered twice through 0.22-μm filter units (Millipore) to sterilize it before releasing it outside the biosafety level 3 (BSL-3) laboratory. One volume of absolute ethanol was then added to the filtrate, and total RNA was purified with an RNeasy column following the manufacturer's procedure. To remove all genomic DNA contamination, the RNA sample was treated twice for 45 min successively with 3 U and 2 U of Turbo DNase (Turbo DNA free kit; Ambion). Biological triplicates were performed for each condition.
RT-qPCR.
For each sample, 1 μg of RNA was reverse transcribed using random hexamers and Superscript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. qPCR of purified cDNA was performed using KAPA SYBRFAST qPCR master mix universal (CliniSciences) and primer sets (see Table S6 in the supplemental material). qPCR for each sample was performed in technical duplicate in a Bio-Rad CFX96 thermocycler with the following protocol: denaturation at 95°C for 3 min, followed by 40 cycles of denaturation at 95°C for 3 s and annealing and elongation with data collection at 60°C for 20 s. Standard curves and melting curves were obtained to check for the high amplification efficiency and the specificity of each primer pair, respectively. For qPCR, the mean threshold cycle (CT) values were normalized against sigA CT values to monitor the expression of the wild-type WT strain under test and control conditions. To calculate relMtb/WT expression ratios, CT values were normalized against dnaJ1 CT values, as dnaJ1 was previously reported to be a suitable control to compare expression between the WT and relMtb strains (24, 34). The difference in expression (n-fold) was calculated using the Pfaffl method (39). Statistical significance was determined by comparing CT values using a two-tailed unpaired Student t test.
5′ RNA linker-mediated random amplification of cDNA ends (5′ RLM-RACE).
For transcription start site (TSS) mapping, full-length mRNAs with 5′-triphosphate (5′-PPP) were enriched by incubating total RNA (3 μg) for 60 min with 3 U of Terminator 5′-phosphate-dependent exonuclease (Epicentre), which specifically digests degraded RNA with a 5′-monophosphate (5′-P). After a phenol extraction and ethanol precipitation, the remaining full-length 5′-PPP mRNAs were treated with 1 U of RNA 5′polyphosphatase (Epicentre) for 30 min in order to generate mRNA with a 5′-P end. The reaction was stopped by phenol extraction and ethanol precipitation. The GeneRacer kit (Ambion) RNA adapter was then ligated to the 5′-P end of the mRNA according to the manufacturer's instructions. The ligated mRNA was phenol extracted, ethanol precipitated, and reverse transcribed using a gene-specific primer (GSP) and Superscript III reverse transcriptase (Invitrogen). The amplification of 5′ cDNA ends was performed using the GeneRacer 5′ primer and a GSP, following by a second PCR amplification using the GeneRacer 5′ nested primer and a GSP to prevent nonspecific amplification. Each PCR product was gel purified (Qiagen), cloned in the pJET1.2 plasmid (Fermentas), and sequenced (Eurofins).
Phylogenomics analyses.
The complete Actinobacteria genome entries were retrieved from the EBI (European Bioinformatics Institute; http://www.ebi.ac.uk/genomes/bacteria.html) and processed by a set of Perl programs into a mySQL database. An RPS (reversed-position specific) BLAST database was computed by retrieving the profiles corresponding to the Conserved Domain Database from the NCBI server (http://www.ncbi.nlm.nih.gov/cdd). We used the RPS BLAST program and the RPS BLAST database to annotate the conserved domains identified in the protein sequences. We computed pairs of one-to-one ortholog proteins with BLASTP as follows: proteins a and b from genomes A and B are considered orthologs if a is the best hit for b in genome A and vice versa, and if a (or b) has a paralogous protein named c, then the score of a versus b should be greater than the score of a (or b) versus c.
Identification of orthologous loci.
By querying our Actinobacteria database, we retrieved, for each genome (one per species), the proteins orthologous to RPMG2 or HadB proteins from M. tuberculosis. The genetic organization around the rpmG2 (hadB) gene was analyzed in all genomes in which orthologs of RPMG2 or HadB were identified. Genes involved in microsynteny were identified, and the conserved domains (clusters of orthologous groups [COG]) characterizing their gene products were retrieved. The microsynteny was thus described by a cluster of COGs (see Table S5 in the supplemental material) that are present between two anchor genes: rmpG2 (upstream) and rpoC (downstream). Each Actinobacteria genome was screened for the presence of this cluster of COGs by delimitating the search in a region up to 40 kb between the two anchor genes. For each COG from our cluster, we verified that the corresponding genes identified at the locus in the different genomes form a set of orthologous genes.
Construction of a phylogenetic tree of Actinobacteria.
The Actinobacteria phylogeny was inferred from a concatenated sequence alignment obtained for a sample of 56 conserved protein families selected as follows. We used RPS BLAST (40) to functionally annotate the whole protein set of each complete genome with the COG profiles downloaded from the NCBI CDD (Conserved Domains Database) repository (41). We selected a sample of 131 complete Actinobacteria genomes which includes only one genome per species. A first set of 149 COG families conserved in this sample were selected according to the quality of the alignments (an E value of <1e−5 and an alignment coverage of at least 70% of the COG profile). The alignments for each COG family were created using the MUSCLE program (42) with the default parameters. We used the trimAl program (43) to remove spurious sequences and poorly aligned positions and to analyze the quality of the alignments according to gap numbers and residue conservation in the columns of the alignments. We used the automated parameters that were recommended by the authors to reconstruct maximum-likelihood trees. We retained the 56 top best sequence alignments. The maximum-likelihood trees were computed with PhyML (44), the ProtTest3 program, to select the optimal combinations of parameters (45). The most frequent combination was the LG model of sequence evolution with a γ correction (four categories of evolutionary rates) and shape parameter and proportion of invariant sites estimated from data. When a species did not have a record for a COG family, the missing sequence was replaced by gaps in the alignment. The aligned sequences of 56 COG families for the 131 species were concatenated to produce a single alignment of 16,329 aligned positions, including 2,685 sites without polymorphisms (16.44%). The concatenated tree was computed with PhyML with the parameters selected by ProtTest3, and the statistical branch support was inferred with the parametric bootstrap. The tree was rooted according to the work of Sen and collaborators (46). Our tree topology appears to be globally congruent with the one published by these authors. The tree was drawn with the online version of iTOL (47). Locus organizations were designed by using the protein domain symbolism provided by iTOL.
Phylogenetic tree of mycobacterium methyltransferases.
Protein sequences homologous to M. tuberculosis methyltransferases were identified through a BLASTP (40) search against the protein sequences encoded by each mycobacterium genome of our selection. The homologous methyltransferases identified in the genomes of Segniliparus rotundus, Tsukamurella paurometabola, and Nocardia brasiliensis were added to the analysis. The multiple alignments were performed using the MUSCLE program (42) with the default parameters. The maximum-likelihood tree was computed with PhyML (44) with the optimal combination of parameter selected by ProtTest3 (45), i.e., an LG model of sequence evolution with a γ correction (four categories of evolutionary rates) and the shape parameter and the proportion of invariant sites estimated from the data. The statistical branch supports were inferred with the parametric bootstrap. The tree was drawn with the online version of iTOL (47).
RESULTS
hadABC is coexpressed with genes involved in translation.
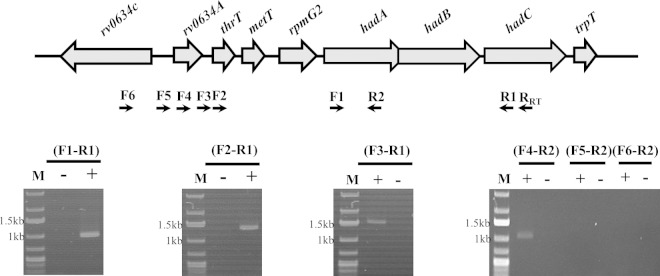
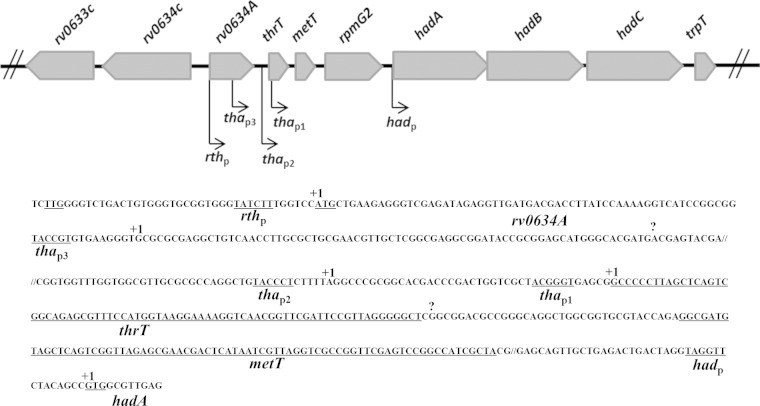
The location of hadABC downstream of genes encoding two tRNAs (thrT and metT) and the L33 ribosomal protein (rpmG2) suggested coordination between the translation potential and the biosynthesis of mycolic acids. As a first step toward revealing this relationship, the transcription organization of the cluster was analyzed. To identify the transcription units, specific cDNAs from total RNA preparations were synthesized by PCR using a downstream primer matching the 3′ end of hadC (RRT) and various upstream primers along the cluster (Fig. 2). A single transcription unit encompassing the three had genes was detected with the F1-R1 primer pair, confirming that the three genes form an operon, in agreement with both the operon browser of the TbDB database (http://tuberculosis.bu.edu/tbdb_sysbio/operon) and the database of prokaryotic operons (http://csbl.bmb.uga.edu/DOOR). As expected, a single transcription unit encompassing the hadABC genes plus the three translation-related genes (thrT, metT, and rpmG2) was also detected (primers F2-R1 and F3-R1), as was an additional transcription unit extending to rv0634A, located just upstream of thrT (F4-R2 primers). No amplification was obtained with primers upstream of rv0634A (F5-R2 and F6-R2). These results indicated that the hadA, hadB, and hadC genes were part of an operon composed of at least seven genes, in agreement with the MycoRRdb database (http://mycorrdb.uohbif.in). To better define the expression pattern of the whole cluster, transcription start sites (TSSs) were determined by 5′ RLM-RACE (Fig. 3). Five TSSs were mapped, one upstream of the hadA gene, two upstream of the metT gene, and two inside the rv0634A ORF. These results indicated that hadABC could be either independently expressed as a single operon (had operon), coexpressed with the translation genes (tha operon), or coexpressed with both the tha operon and the putative transcription regulator rv0634A gene (rth operon). The mapping of the most upstream TSS inside the rv0634A ORF suggested that the TTG codon start for this gene was misannotated. The ATG codon located 39 bp further downstream is likely the real start codon, with the TSS at the A of this codon (Fig. 3). The inside TSS mapped on the 74th nucleotide at a G. The metT-proximal +1 nucleotide is a G, the first nucleotide of the metT tRNA. The had operon +1 nucleotide is the G of the GTG start codon. The mapping of these TSSs was in agreement with the transcriptome shotgun sequencing (RNA-seq) global study reported in reference 48. Accordingly, a fairly conserved −10 box sequence could be found upstream these five TSSs (49). We identified two additional sites, 65 nucleotides before the end of rv0634A gene and 2 nucleotides downstream of the thrT sequence (Fig. 3). However, neither a −10 box nor promoter activity (see below) was associated with these sites, suggesting that they were false-positive TSSs and likely resulted from natural or experimental RNA cleavages.
FIG 2.
The hadABC genes are coexpressed with translation process and transcription regulator genes. Transcription units were identified by RT-PCR. The forward (F1 to -6) and reverse (R1 and -2) primers used to identify the transcription units as well as the primer used to synthesize the cDNA (RRT) are symbolized by arrows below the genetic map. Results of representative PCRs on reverse transcriptase-treated (+) or untreated (−) templates run on agarose gels are shown. M, DNA ladder.
FIG 3.
Identification of five transcription start sites in the rth cluster. Transcription start sites (TSSs) mapped by 5′ RLM-RACE are symbolized by broken arrows on the genetic map and are associated with promoter names. On the partial sequence extending from rv0634A to the 3′ end of hadA, the TSSs are shown (+1). The question marks indicate detected 5′-end RNAs that are unlikely to correspond to TSSs. Putative −10 boxes, proposed start codons, and tRNA-encoding gene sequences (thrT and metT) are underlined.
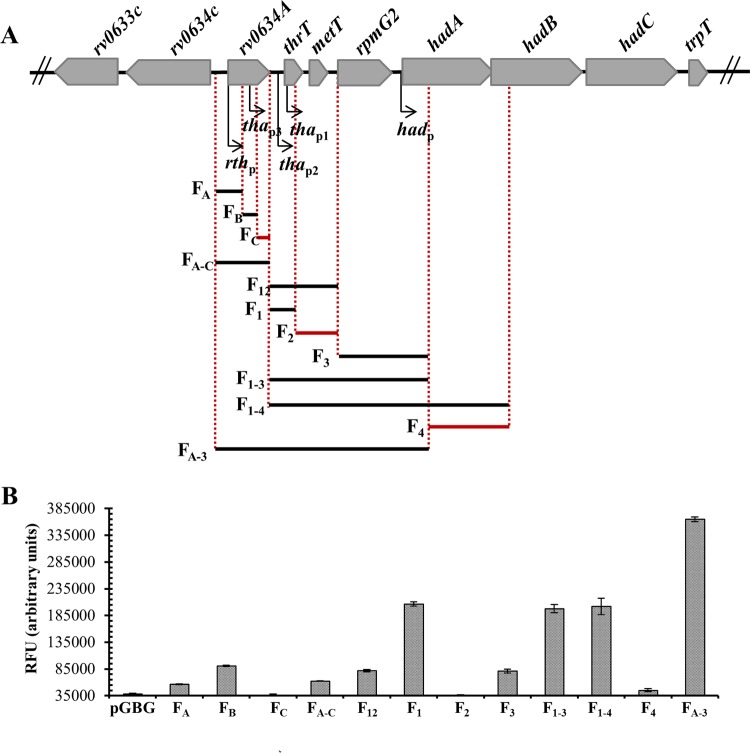
To correlate the transcription start sites with promoter activities, the whole region was subcloned downstream of two strong ribosomal T1T2 transcription terminators and upstream of the gfp reporter gene in a low-copy-number plasmid. The operon fusions were introduced into M. smegmatis and GFP fluorescence specific activities were measured in 2-day-old cultures (Fig. 4). The pGBG vector without any cloned fragment gave the background level. The FA and FB constructs indicated promoter activity specifically associated with the two TSSs located inside the predicted rv0634A ORF. In contrast, no promoter activity was detected in the FC fusion, suggesting that the 5′ end of the RNA detected at the 3′ end of rv0634A likely resulted from a cleavage. The F12 and F1 constructs confirmed the promoter activity associated with the 2 TSSs mapped upstream of thrT. The absence of any GFP activity in the F2 construct suggested that the 5′ end that mapped 2 nucleotides downstream of the thrT gene likely resulted from cleavage. The F3 construct pointed to promoter activity associated with the TSS that mapped at the beginning of the hadA sequence. In agreement with the absence of any other TSS in the hadABC sequence, no promoter activity was detected in fragments containing sequences beyond the beginning of hadA (F4) (data not shown). Taken together, these results indicated that the hadABC genes could be transcribed from five promoters (Fig. 4).
FIG 4.
Identification of five promoter activities in the rth cluster. (A) DNA fragments cloned in the GFP reporter vector are shown under the genetic map. Fragments which do not contain any promoter activity are in red. (B) The fluorescence specific activity is expressed in relative fluorescence units (RFU) and is the mean for biological triplicates. pGBG is the gfp promoterless control. Error bars show standard deviations.
The putative transcription regulator Rv0634A does not regulate the rth operon.
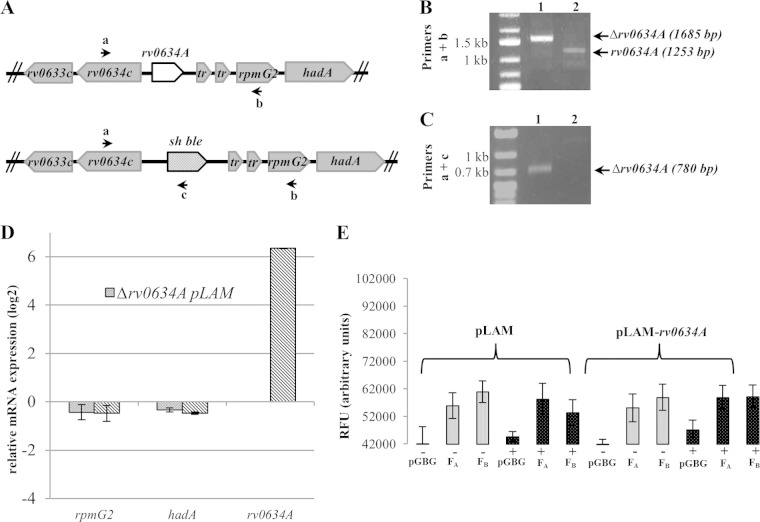
Rv0634A belongs to the mycobacterial core genome (50), and its Mycobacterium marinum ortholog MMAR_0966 is annotated as a putative Arc/MetJ class transcription regulator (http://mycobrowser.epfl.ch/marinolist.html) (51). Many transcription regulators regulate neighboring genes as well as their own (52). The genetic organization at the hadABC locus suggested that Rv0643A might regulate the expression of the whole cluster. To test this possibility, the gene along with its promoters was deleted from the chromosome (Fig. 5A; also, see Materials and Methods). The deletion was verified by PCR (Fig. 5B and C), confirming the previous prediction that the gene is not essential (53), and the absence of rv0634A mRNA was verified by RT-qPCR (Fig. 5D). The amount of rpmG2 and hadA mRNAs of the Δrv0634A strain measured by RT-qPCR was not significantly different from that of the WT strain, indicating that neither the putative Rv0634A regulator nor the rv0634A promoter (rthp and thap3) contributes to the expression of the thrT-hadC cluster in log phase (Fig. 5D). When rv0634A was overexpressed 64-fold from a plasmid-borne acetamide-inducible promoter, no effect on the expression of either rpmG2 or hadA was observed (Fig. 5D), indicating that Rv0634A does not regulate the rpmG2-hadABC operon. In addition, in M. smegmatis, the overexpression of rv0634A did not affect the expression of either the FA gfp fusion, which indicates the absence of autoregulation of rv0634A, or the FB fusion, showing that Rv0634A did not regulate the promoter inside the gene (Fig. 5E). Together, these results indicated that Rv0634A does not regulate the transcription of the rv0634A-hadC cluster.
FIG 5.
The putative transcription regulator Rv0634A does not regulate the tha operon. (A) Genetic map showing the rv0634A and sh ble allelic exchange. Primers for the deletion analysis by PCR are symbolized by arrows. (B and C) PCR products with the a+b and a+c primer pairs to confirm the deletion of the rv0634A gene. Lysates of a Δrv0634A strain (1) and of a WT clone (2) were used as PCR templates. (D) Monitoring by RT-qPCR of the expression (log2) of the rpmG2, hadA, and rv0634A genes relative to the WT strain values (WT/pLAM) in the rv0634A mutant (Δrv0634A/pLAM) and in the rv0634A-overexpressing background (Δrv0634A/pLAM-rv0634A). Each strain was cultivated in the presence of 0.2% acetamide. Error bars show standard errors of the means (SEM) from three biological triplicates. (E) Expression, in relative fluorescent units (RFU), of rthp::gfp (FA) and thap3::gfp (FB) fusions in M. smegmatis carrying a plasmid expressing rv0634A from an acetamide-inducible promoter (pLAM12-rv0634A) or not (pLAM12). Each strain was cultivated in either the presence (+) or absence (−) of 0.2% acetamide. pGBG was the gfp promoterless control.
The mycolic acid biosynthesis genes hadABC are negatively coregulated with the ribosomal protein gene rpmG2 by the stringent response.
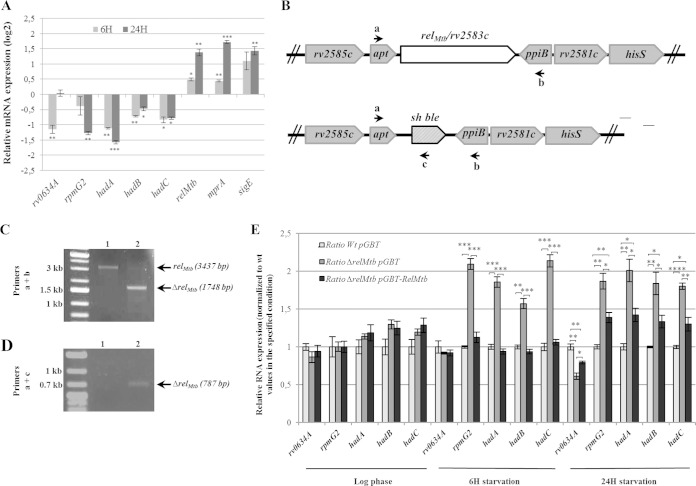
The stringent response (SR) is a conserved adaptive response that downregulates translation potential in starvation conditions. The molecular hallmark of the SR is the synthesis of the alarmone (p)ppGpp by the RelA/SpoT homolog (RSH), which leads to dramatic reprogramming of cell transcription, including a rapid inhibition of stable RNA synthesis (54). The coexpression of the hadABC genes with genes coding for tRNAs and a ribosomal protein suggested that the hadABC genes might also be regulated by the SR. To test this proposition, the expression of genes in the rv0634A-hadC cluster was then measured by RT-qPCR after starvation for 6 and 24 h (approximately one-fourth and one generation time, respectively). As shown in Fig. 6A, in the WT background, the expression of relMtb was upregulated, as well as that of sigE and mprA, in agreement with previous reports (23, 24). In contrast, the five genes of the operon were downregulated, either after 6 h starvation only (rv0634A), 24 h starvation only (rpmG2) or both starvation periods (hadA, hadB, and hadC) (Fig. 6A). To determine the contribution of the SR to this regulation, we deleted relMtb, the M. tuberculosis RSH-encoding gene. The construction was checked by PCR (Fig. 6B to D) and by RT-qPCR for the absence of relMtb mRNA (see Tables S2 to S4 in the supplemental material). The expression level was then measured in the ΔrelMtb background, and the mutant-to-WT expression ratio was determined (Fig. 6E). In log phase, the relative expression of rv0634A, rpmG2, and the three had genes in the ΔrelMtb background was similar to that in the WT strain and in the complemented strain expressing relMtb from a low-copy-number plasmid (pGBT-relMtb). After starvation, a similar result was observed for rv0634A, whereas for rpmG2, hadA, hadB, and hadC, the expression ratio was higher for the ΔrelMtb strain but not for the complemented strain (Fig. 6E). These results indicated that rpmG2 and hadABC were negatively coregulated by the SR. The magnitude of the repression due to the SR for the four genes was similar (∼2-fold) (Fig. 6E), suggesting that the SR exerted its control through the promoter(s) thap2 and/or thap1, located just upstream of thrT (Fig. 4). Thus, in agreement with the genetic organization, these results suggested coordination of the MA biosynthesis with translation potential.
FIG 6.
The tha operon is regulated by the stringent response. (A) Monitoring by RT-qPCR of the relative expression (log2) of the rv0634A, rpmG2, hadABC, relMtb, mprA, and sigE genes in the WT strain (WT/pGBT) upon 6 and 24 h of starvation. The values are expressed as a ratio to WT values in unstarved log-phase cultures. (B) Genetic map showing the relMtb and sh ble allelic exchange. Primers for the deletion analysis by PCR are symbolized by arrows. (C and D) PCR products obtained with the a+b and a+c primer pairs to confirm the deletion of the relMtb gene. Lysates of the WT strain (lanes 1) and of a ΔrelMtb clone (lanes 2) were used as PCR templates. (E) Gene expression ratio relative to values for the WT/pGBT strain from log phase, upon 6 and 24 h of starvation, respectively. In panels A and E, error bars represent SEM from three biological triplicates. P values were <0.05 (*), <0.01 (**), <0.001 (***), and < 0.0001 (****).
Starvation leads to a global downregulation of genes required for mycolic acid synthesis.
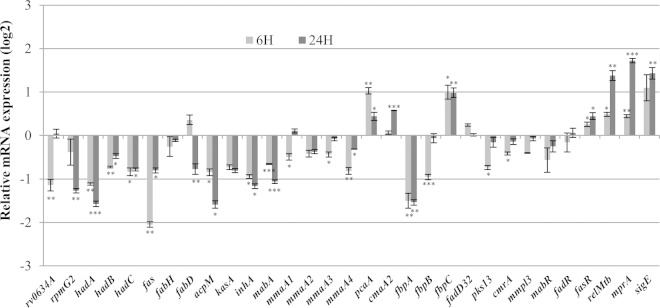
One might have expected the regulation of the hadABC genes by starvation to be coordinated with that of other genes involved in the biosynthesis of MAs. Thus, in order to get an integrated picture of the regulation of MA biosynthesis, the expression of a subset of genes representative of each step of the biosynthesis and transport of MAs was measured after 6 and 24 h starvation. The results are presented in Fig. 7. The expression of fas, which is required for the synthesis of the MA α chain as well as the acyl-CoA substrates for the FAS-II complex (Fig. 1), was downregulated. The gene fabH acting at the intermediate step between FAS-I and FAS-II activities was not affected (Fig. 1) (55). Both fabD and acpM, which are required for the synthesis of FAS-II malonyl-acyl carrier protein (malonyl-ACP) elongation units, were downregulated (at 24 h starvation for fabD) (Fig. 1). Along with hadABC, the genes involved in the four reaction steps driven by the FAS-II complex, kasA, mabA, and inhA, were also downregulated (Fig. 1). Of the two genes involved in the condensation of the FAS-I and FAS-II products, fabD32 appeared not to be regulated, whereas pks13 was downregulated after 6 h of starvation (Fig. 1). The genes required for the subsequent reduction step (cmrA) and the transport of TMM across the inner membrane (mmpL3) were also downregulated at 6 h starvation (Fig. 1). Among the three genes involved, the synthesis of the mycoloyltransferase antigen Ag85 complex gene, fbpA, was downregulated at 6 and 24 h, that of fbpB was downregulated at 6 h, and the expression of the third and most active Ag85 subunit gene, fbpC (Fig. 1) (56, 57), was upregulated at both 6 and 24 h. Among the six methyltransferase genes known to be involved in decorating the meromycolic chain in M. tuberculosis (Fig. 1), pcaA and cmaA2 were upregulated (at 24 h for cmaA2), whereas mmaA1 to -4 were downregulated (at 6 h for mmaA1 and mmaA3). Therefore, upon starvation, the expression of most of the MA biosynthesis genes was affected, and with few exceptions, the expression was lower. To determine the contribution of the SR to this regulation, gene expression was measured in the ΔrelMtb mutant as well, and the ratio to the WT values was determined. The complete data are presented in Tables S2 to S4 in the supplemental material. With an arbitrary cutoff ratio of 1.4, Table 1 lists the genes whose expression was affected by the ΔrelMtb mutation after 24 h starvation. This set included acpM, the FAS-II complex genes hadABC and kasA, the Ag85 fpbA gene, and the methyltransferase genes mmaA2, mmaA3, cmaA2, and pcaA. It is worth noting that acpM and kasA were the only genes to be positively regulated by the SR, despite the fact that the interplay between the different regulatory pathways that have been induced by our experimental starvation conditions ultimately led to the downregulation of both genes. Therefore, during severe starvation, most of the mycolate biosynthesis genes were regulated, whereas environmental signals specifically triggering the SR would regulate only a small gene subset. This later observation was in agreement with microarray studies of Dahl and coworkers showing that only 5 genes involved in MA biosynthesis were regulated by RelMtb following 6 h of starvation (24). These genes also include hadB, hadC, and fbpA, along with the FAS-II gene mabA and the Ag85 gene fbpC.
FIG 7.
Regulation of mycolic acid biosynthesis, transport, and regulator genes upon starvation. The WT strain (WT/pGBT), was grown to log phase and was starved for 6 or 24 h before total-RNA sampling. (A) The relative expression of the genes representative of the major steps of the mycolic acid biosynthesis and transport was measured by RT-qPCR. The values are expressed in log2 as a ratio to values for unstarved log-phase WT cultures. Error bars show SEM from three biological triplicates. P values were <0.05 (*), <0.01 (**), and <0.001 (***).
TABLE 1.
Genes regulated by the stringent response
| Gene | Expression ratio (mean ± SEM) in strain with genotypea |
||
|---|---|---|---|
| WT (pGBT) | ΔrelMtb (pGBT) | ΔrelMtb (pGBT)-relMtb | |
| rv0634A | 1 ± 0.04 | 0.61 ± 0.04 | 0.79 ± 0.02 |
| rpmG2 | 1 ± 0.03 | 1.87 ± 0.10 | 1.39 ± 0.07 |
| hadA | 1 ± 0.04 | 2.01 ± 0.15 | 1.42 ± 0.09 |
| hadB | 1 ± 0.01 | 1.84 ± 0.15 | 1.33 ± 0.08 |
| hadC | 1 ± 0.02 | 1.80 ± 0.04 | 1.30 ± 0.09 |
| acpM | 1 ± 0.06 | 0.55 ± 0.01 | 1.05 ± 0.10 |
| kasA | 1 ± 0.02 | 0.46 ± 0.02 | 1.17 ± 0.14 |
| mmaA2 | 1 ± 0.02 | 2.08 ± 0.29 | 1.26 ± 0.08 |
| mmaA3 | 1 ± 0.01 | 1.42 ± 0.12 | 1.18 ± 0.08 |
| cmaA2 | 1 ± 0.04 | 1.46 ± 0.12 | 1.13 ± 0.09 |
| pcaA | 1 ± 0.06 | 1.54 ± 0.14 | 1.23 ± 0.06 |
| fbpA | 1 ± 0.07 | 1.73 ± 0.02 | 1.19 ± 0.07 |
| mprA | 1 ± 0.07 | 1.55 ± 0.02 | 1.39 ± 0.09 |
| sigE | 1 ± 0.07 | 1.42 ± 0.02 | 1.10 ± 0.10 |
Expression relative to the wild-type value after 24 h of starvation.
Few transcription regulators of MA genes have been characterized at the molecular level, namely, FasR, which activates the fas gene (15), MabR, which represses the fasII operon (fabD-acpM-kasA-kasB-accD6) and the fas gene (16), and FadR, a repressor of the fasII operon (17). As shown in Fig. 7, neither mabR nor fadR was regulated by starvation, whereas fasR was slightly upregulated during 6 and 24 h starvation independently of RelMtb. The expression of relMtb was reported to be indirectly activated by MprA, which regulates the response of the MprAB two-component system through the sigma factor SigE (58–60). Both the corresponding genes, mprA and sigE, were also upregulated by starvation, suggesting that these genes might also contribute to the upregulation of the relMtb gene during starvation. Interestingly, the expression of both mprA and sigE was lower in the WT than in the relMtb mutant (Table 1), suggesting negative feedback control of both genes by RelMtb.
Phylogenetic reconstruction of the rth operon–mmaA1-mmaA4 cluster.
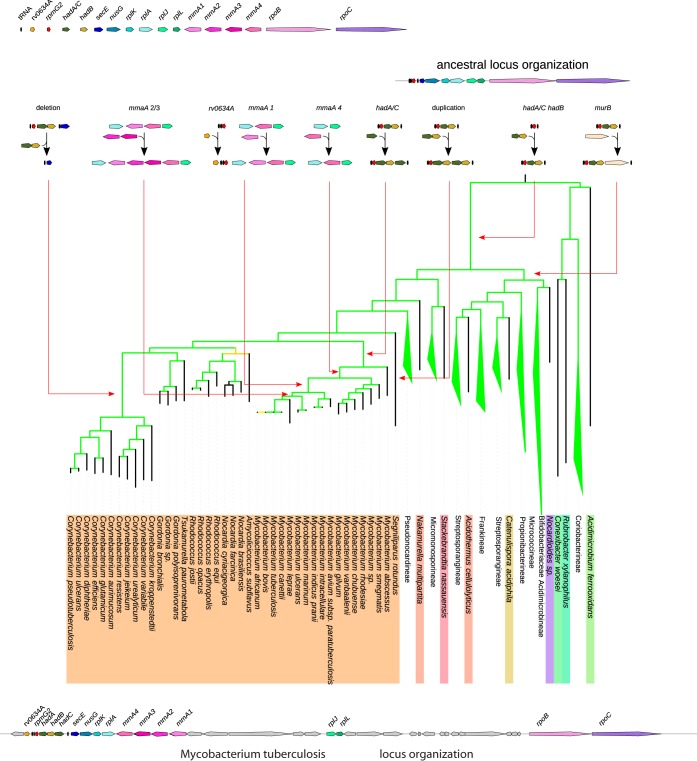
We undertook a detailed analysis of the rth locus in 131 genomes of Actinobacteria (one strain per species) to understand the key evolutionary steps that have shaped the current genetic organization of this locus in mycobacterial genomes and, by extension, in Actinobacteria genomes. First, we retrieved for every genome the protein orthologous either to M. tuberculosis RpmG2 or to M. tuberculosis HadB when the rpmG2 gene was absent from the studied genome. The genetic context of both genes in all genomes was further analyzed, and microsynteny was detected only downstream of them, with, in about 20 genomes, the following gene order conservation: rpmG2-hadA-hadB-secE-nusG-rplK-rplA-rplJ-rplL-rpoB-rpoC. The products of these genes are characterized by specific conserved domains (see the corresponding COG number in Table S5 in the supplemental material). This microsynteny could be disrupted by the insertion of a few other genes, among which murB was found at the locus in 54 genomes. In addition, three tRNA genes upstream of rpmG2 were found to be conserved in order and nature (tyrT-thrT-metT) and one upstream gene, secE (trpT). Thus, we carried out a systematic search of this cluster of COGs on a chromosomal region whose maximal size was set to 40 kb using rpmG2 or hadA as an upstream landmark and rpoB and rpoC as a downstream limit. The result of this search was plotted over our Actinobacteria species tree in order to identify the key evolutionary steps involved in shaping the locus genetic organization (Fig. 8; also, see Fig. S1 in the supplemental material).
FIG 8.
Key evolutionary steps that have shaped the current genetic organization of the rpmG2-hadA/CB locus in Actinobacteria. Except for Corynebacterineae, the different species subtrees have been collapsed for clarity. The ancestral locus organization is shown at the top right. The dating of the various evolutionary events (gene acquisition, gene deletion, and gene duplication) is indicated by an arrow pointing to the branch of the tree where they probably occurred.
From the analysis of the phylogenetic distribution of the locus genes, the organization rpmG2-secE-nusG-rplK-rplA-rplJ-rplL-rpoB-rpoC should have been the ancestral locus organization present in the last common ancestor (LCA) of all current Actinobacteria species, as (i) those genes are found in almost all genomes, whatever their taxonomic classification (see Fig. S2 in the supplemental material), and (ii) this organization is observed in the genomes of the most ancient groups on the Actinobacteria species tree (Conexibacteraceae and the Rubrobacterineae) (see Fig. S1 in the supplemental material) as well as in genomes outside the Actinobacteria, notably in 123 of 219 Firmicutes genomes (see Fig. S3 in the supplemental material). From this ancestral organization, the evolutionary scenario that would best explain the data in relation to the Actinobacteria species tree is summarized in Fig. 8. The genes hadA/C (the ancestor gene for the hadA and hadC genes) and hadB were acquired once and inserted just downstream of rpmG2 along the branch leading to the LCA of the Actinobacteria at the origin of all taxonomic groups except Conexibacteraceae and Rubrobacterineae. In the branch leading to the LCA of Mycobacterium, a duplication of the hadA/C ancestor gene should have occurred, giving rise to the actual hadA-hadB-hadC locus organization found in those species. The absence of the genes rpmG2, hadA, and hadB from the region upstream of secE in the Corynebacterium genomes corresponds clearly to a loss of these genes in the genome of their LCA, as the three tRNA genes, tyrT, thrT, and metT, always located upstream of rpmG2, are found conserved and located just upstream of the tRNA gene trpT, invariably located upstream of secE. The absence of both hadA/C and hadB genes in the Bifidobacteriaceae genomes and of rpmG2 in a group of Kineosporiineae genomes forming a subtree on the species tree (see Fig. S1 in the supplemental material) is probably due to the loss of those genes in the LCA genome of each group.
The gene rv0634A would have been acquired once on the branch leading to the LCA of the Mycobacterium avium and M. tuberculosis complexes and of M. ulcerans, M. marinum, and M. leprae (Fig. 8).
Finally, the four methyltransferase genes found at the M. tuberculosis locus were acquired through three independent events (Fig. 8). The gene mmaA4 was acquired first after the speciation of M. abscessus on the branch leading to the LCA of the other mycobacterium species and was probably subsequently replaced in M. smegmatis. Then, mmaA1 was gained on the branch leading to the LCA of the M. avium and M. tuberculosis complexes and M. ulcerans, M. marinum, and M. leprae. Finally, mmaA2 and mmaA3 were acquired on the branch leading to the LCA of M. ulcerans, M. marinum, and M. leprae and the M. tuberculosis complex. The different acquisition events do not result from successive duplications of the inserted genes, as the subtrees corresponding to MmaA4, MmaA3, MmaA2, and MmaA1 gene products are not sister groups on the tree built on the homologous methyltransferase from mycobacterium (see Fig. S4A in the supplemental material). However, once acquired, those genes were inherited vertically, since both species and protein trees are congruent (see Fig. S4B in the supplemental material). In some species, some methyltransferase genes became pseudogenes (see Fig. S4B in the supplemental material).
The region extending from rpmG2 to rpoC seems to be a hot spot of recombination, as it has suffered many gene gains and losses. Thus, in a few genomes, the rpmG2 locus has been disrupted with the organization secE-nusG-rplK-rplA-rplJ-rplL-rpoB-rpoC found at a distant chromosomal location (see Fig. S1 in the supplemental material). However, those genomes are found scattered along the species tree, except for the Bifidobacteriaceae species, where the disruption events may have occurred in their last common ancestor.
DISCUSSION
The M. tuberculosis mycomembrane is a highly efficient permeability barrier containing many antigenic determinants and is essential to cell viability and virulence. During the process of infection, the bacteria must adapt to various environmental conditions, and little is known of how the biosynthesis of mycolic acids, the major components of the mycomembrane, responds to these conditions, notably to the nutrient scarcity that is believed to prevail inside the host (53, 61). About 30 genes, including hadA, hadB, and hadC, are required for the synthesis and transport of MAs. This study shows that upon starvation, the had genes are coexpressed and downregulated with three neighboring genes, thrT, metT, and rpmG2, involved in translation, in accordance with their genetic organization. This result suggests that MA biosynthesis is coordinated with the translation potential of the cell. The regulation of synthesis of stable RNAs and ribosomal proteins is a hallmark of the stringent response (31), and accordingly, the downregulation was seen to be dependent on a functional RelMtb protein, the key enzyme of the stringent response. The hadABC genes could also be coexpressed with rv0634A, the first gene of a seven-gene operon that includes thrT, metT, and rpmG2. Although the M. marinum Rv0634A homolog is recorded as a transcription regulator of the Arc/MetJ family (http://mycobrowser.epfl.ch/marinolist.html), we have no evidence that Rv0634A was involved in the regulation of this operon. The gene rv0634A was reported to belong to the mycobacterial core genes (50) and is conserved in only some slow-growing pathogenic and opportunistic mycobacterial species (Fig. 8; also, see Fig. S1 and S2 in the supplemental material) (62). Despite these observations, the deletion of the gene did not affect the fitness of the bacteria, including the ability to grow in human THP-1 macrophages (our unpublished results). The function of this gene remains to be discovered.
In coordination with the expression profile of the hadABC genes, starvation downregulated expression of most genes representative of each step of the synthesis and transport of the MAs. Because MA biosynthesis is a highly energy-consuming process, it makes physiological sense for the cell to slow down its synthesis during nutrient scarcity. Rohde et al. (21) reported that in a comprehensive transcription profiling study over the course of a 14-day infection in resting macrophages, one-third of the genes involved in MA biosynthesis were downregulated. These results agree with the pioneering observations of Lacave and coworkers, i.e., the arrest of MA biosynthesis in stationary phase (63), and are consistent with the observation that during infection, M. tuberculosis switches from a carbon to a lipid regimen during which the fatty acid catabolism and the storage compound triacylglycerol (TAG) synthesis pathways are activated instead (64, 65). The expression profile of genes involved in the first (fas, fabH, and fabD) and last (fadD32, pks13, cmrA, mmpL3, and fbpBC) steps of the biosynthesis pathway and of two of the six genes coding for methyltransferases (mmaA1 and mmaA4) was independent of RelMtb (Table 1; also, see Tables S2 to S4 in the supplemental material). In contrast, among the genes encoding FAS-II, the hadABC operon and kasA are regulated by the SR, with the latter being upregulated. Such an uncoupling between the expression of FAS-II genes and that of genes involved in upstream steps such as fas has been previously reported during growth arrest. This uncoupling would allow the rerouting of the carbon flux toward the synthesis of storage and stress-protective compounds, such as TAG and glutamate (65). A similar explanation might also apply to our results. A reverse enzymatic pathway during the SR primed at the KasA step would return the meromycolate intermediate products resulting from the activity of FAS-II to FAS-I for the synthesis of alternative fatty acids.
There are several types of MA species, as defined by the functional groups found at two positions on the meromycolate chain. In M. tuberculosis, six methyltransferase genes are involved in the introduction of these groups. Two, pcaA and cmaA2, were upregulated upon starvation, whereas mmaA1, mmaA2, mmaA3, and mmaA4 were downregulated. Hence, upon starvation, all mycolic acid species should be enriched in decoration stemming from PcaA and CmaA2 activities, i.e., with a cyclopropanation group at the proximal position (8, 66). Cyclopropanation by PcaA and CmaA2 has been shown to be crucial for the virulence of M. tuberculosis, implying that specifically enriching MAs in cyclopropanated species might contribute to the adaptive response during infection (67, 68). With the exception of mmaA1 and mmaA4, the expression of the methyltransferase genes was negatively controlled by RelMtb. Therefore, environmental signals specifically triggering the SR should lead to enrichment of MA species with a methyl group at the distal position of oxygenated species due to MmaA4 activity and at the proximal position of trans-methoxy and trans-keto species due to MmaA1 activity (8). The activity of MmaA4 is crucial for envelope impermeability and for full virulence in mice (69). Loss of MmaA1 has been shown to enhance the virulence-associated cording phenotype of M. tuberculosis (66). One unresolved issue in the biosynthesis of MA is the step at which decoration is added to the MA mero chain; addition could accompany elongation by either FAS-II or, following synthesis, the full-length meromycolate chain. The former possibility is favored by previous reports that methyltransferases interact with subunits of the FAS-II complex (70, 71). The observation that mmaA1 to -4 are clustered with the hadABC genes would also agree with the hypothesis of a functional coordination between methyltransferases and FAS-II. Moreover, it was recently shown that during macrophage infection by M. bovis BCG, the hadABC genes were the only ones contributing to the FAS-II complex to be regulated. Interestingly, this regulation was coordinated with that of the methyltransferase genes, arguing again for a close relationship between the HadABC enzymes and activity of the methyltransferases (72). Our observation that the regulation of both sets of genes—mmaA2/mmaA3 and hadABC—is RelMtb dependent further strengthens this relationship. Recently we have shown that mutations in the gene hadC influence the methyltransferase-dependent decoration profiles of meromycolate chains, reinforcing this hypothesis (our unpublished results).
There is a regulatory signaling cascade linking the two-component signal transduction system MprAB, the sigma factor SigE, and RelMtb. Upon activation, the response regulator MprA directly upregulates sigE, which then directly stimulates expression of relMtb (58, 73). Both mprA and sigE activate their own expression. Data from microarray analysis suggest additional positive feedback loops, with SigE upregulating mprAB (74) and RelMtb activating its own expression (Table 1) (23, 24). The outcome of this complex regulatory circuit design is phenotypic heterogeneity, with two subpopulations expressing relMtb at different levels (59, 60). This heterogeneity is part of a “bet-hedging strategy” contributing to the survival of the population (1). Our results, showing that RelMtb might also negatively regulate the expression of both mprB and sigE, would therefore add two negative feedback loops to the positive feedback design of the mprB-sigE-relMtb circuit. These negative loops may contribute to turning off the MprBA-SigE-RelMtb signalization pathway once the stress is over.
For the MA biosynthesis genes whose regulation was partly or fully independent of RelMtb, other transcription regulators must be involved. With the exception of the transcription activator FasR, which might contribute to the downregulation of its single target fas gene, neither MabR nor FadR, the two other well-characterized regulators, is likely to contribute to the regulation of the MA biosynthesis genes during starvation. Hence, further actors in the regulatory circuitry leading to global repression of MA synthesis genes during starvation remain to be identified.
Despite the downregulation of the MA biosynthesis genes, the qualitative and quantitative profiles of the MAs were not affected after 96 h of starvation in either WT (see Fig. S5 in the supplemental material) or relMtb (data not shown) strains, indicating that MAs are stable molecules. This result suggested that during starvation, the integrity of the mycomembrane would not be dramatically compromised by sparing the energy cost of synthesizing de novo MAs. The differential expression observed among the methyltransferase-encoding genes suggests that adaptation of the cell to starvation is mainly through subtle modifications of the meromycolic acid chains.
Following a comprehensive analysis of currently available actinobacterial genomes, we were able to draw an evolutionary scenario (Fig. 8) leading to the current organization of the locus encompassing hadABC and mmaA1 to -4 in Mycobacterium species. The hadA and hadB genes have been found to be associated in all the genomes analyzed so far. Conservation of the gene order in distantly related species has been shown to result from a functional constraint and could be a fingerprint of physical interaction between the encoded proteins (75). This is certainly the case for hadA or hadC and hadB, which are cotranscribed and for which physical interactions between their products have been described—the active complexes are either HadA-HadB or HadB-HadC heterodimers (26). The analysis of the methyltransferase tree (see Fig. S4 in the supplemental material) leads to the conclusion that the presence at the locus of the four neighboring methyltransferase genes (mmaA1 to -4) is due to their acquisition through three independent events rather than the result of successive duplication events from an ancestral inserted gene. This intriguing observation suggests that further investigation will reveal the identity of recombination insertion sites. Considering the numerous rearrangements that have occurred, it is likely that this locus is a recombination hot spot. However, the microsynteny observed in the pathogenic mycobacterium genomes suggests that a functional constraint acts at this locus to maintain the methyltransferase genes in the vicinity of the hadABC genes. This is in agreement with the fact that their gene products are involved in the same biosynthesis pathway. The efficiency of interaction between the Had and methyltransferase proteins previously reported (70, 71) would certainly benefit from such a genetic organization, since clustering of genes at the same locus would increase the local protein concentrations. Furthermore, this is consistent with our proposed evolutionary scenario, as the duplication of the hadA/C gene predated the acquisition of the first methyltransferase gene at the locus.
The issue that triggered the present study was determining the significance, if any, of the localization of the had genes downstream of genes involved in the translation process. We have demonstrated that this organization has allowed both types of genes to be coregulated by the SR. Although the driving force for the gene rearrangement in this region was probably the recombinogenic nature of the locus, the coregulation together with the subsequent acquisition of the methyltransferase genes might have imposed a selection pressure strong enough to shape and maintain the current organization at this locus.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Graham F. Hatfull for providing the mycobacterial recombineering system, Marcus A. Horwitz for the pGB9.2 plasmid, Brian D. Robertson for the pMIND plasmid, and Wladimir Malaga for the pJV53::hyg plasmid. We thank Ingrid Mercier, Alexandre Gouzy, Jeanne Pilaire, Clément Carel, and Aïcha Bah for technical advice. We are grateful to Dave Lane for critical reading of the manuscript and to the members of the Daffé team for their constructive criticisms.
This work was supported by institutional grants from the CNRS (Centre National de la Recherche Scientifique). S.J. was supported by an MESR (Ministère de l'Enseignement Supérieur et de la Recherche) fellowship.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00433-15.
REFERENCES
- 1.Balaban NQ, Gerdes K, Lewis K, McKinney JD. 2013. A problem of persistence: still more questions than answers? Nat Rev Microbiol 11:587–591. doi: 10.1038/nrmicro3076. [DOI] [PubMed] [Google Scholar]
- 2.Gengenbacher M, Kaufmann SH. 2012. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol Rev 36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Young DB, Gideon HP, Wilkinson RJ. 2009. Eliminating latent tuberculosis. Trends Microbiol 17:183–188. doi: 10.1016/j.tim.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Falzon D, Jaramillo E, Wares F, Zignol M, Floyd K, Raviglione MC. 2013. Universal access to care for multidrug-resistant tuberculosis: an analysis of surveillance data. Lancet Infect Dis 13:690–697. doi: 10.1016/S1473-3099(13)70130-0. [DOI] [PubMed] [Google Scholar]
- 5.Migliori GB, Sotgiu G, Gandhi NR, Falzon D, DeRiemer K, Centis R, Hollm-Delgado MG, Palmero D, Perez-Guzman C, Vargas MH, D'Ambrosio L, Spanevello A, Bauer M, Chan ED, Schaaf HS, Keshavjee S, Holtz TH, Menzies D. 2013. Drug resistance beyond extensively drug-resistant tuberculosis: individual patient data meta-analysis. Eur Respir J 42:169–179. doi: 10.1183/09031936.00136312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Velayati AA, Masjedi MR, Farnia P, Tabarsi P, Ghanavi J, Ziazarifi AH, Hoffner SE. 2009. Emergence of new forms of totally drug-resistant tuberculosis bacilli: super extensively drug-resistant tuberculosis or totally drug-resistant strains in iran. Chest 136:420–425. doi: 10.1378/chest.08-2427. [DOI] [PubMed] [Google Scholar]
- 7.Daffe M. 2015. The cell envelope of tubercle bacilli. Tuberculosis 95(Suppl 1):S155–S158. doi: 10.1016/j.tube.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 8.Marrakchi H, Laneelle MA, Daffe M. 2014. Mycolic acids: structures, biosynthesis, and beyond. Chem Biol 21:67–85. doi: 10.1016/j.chembiol.2013.11.011. [DOI] [PubMed] [Google Scholar]
- 9.Daffe M, Crick DC, Jackson M. 2014. Genetics of capsular polysaccharides and cell envelope (glyco)lipids. Microbiol Spectr 2:MGM2-0021-2013. [DOI] [PubMed] [Google Scholar]
- 10.Guenin-Mace L, Simeone R, Demangel C. 2009. Lipids of pathogenic mycobacteria: contributions to virulence and host immune suppression. Transbound Emerg Dis 56:255–268. doi: 10.1111/j.1865-1682.2009.01072.x. [DOI] [PubMed] [Google Scholar]
- 11.Jackson M, McNeil MR, Brennan PJ. 2013. Progress in targeting cell envelope biogenesis in Mycobacterium tuberculosis. Future Microbiol 8:855–875. doi: 10.2217/fmb.13.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Favrot L, Ronning DR. 2012. Targeting the mycobacterial envelope for tuberculosis drug development. Expert Rev Anti Infect Ther 10:1023–1036. doi: 10.1586/eri.12.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Neyrolles O, Guilhot C. 2011. Recent advances in deciphering the contribution of Mycobacterium tuberculosis lipids to pathogenesis. Tuberculosis (Edinb) 91:187–195. doi: 10.1016/j.tube.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Goude R, Parish T. 2008. The genetics of cell wall biosynthesis in Mycobacterium tuberculosis. Future Microbiol 3:299–313. doi: 10.2217/17460913.3.3.299. [DOI] [PubMed] [Google Scholar]
- 15.Mondino S, Gago G, Gramajo H. 2013. Transcriptional regulation of fatty acid biosynthesis in mycobacteria. Mol Microbiol 89:372–387. doi: 10.1111/mmi.12282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzman V, Mondino S, Sala C, Cole ST, Gago G, Gramajo H. 2010. Transcriptional regulation of lipid homeostasis in mycobacteria. Mol Microbiol 78:64–77. [DOI] [PubMed] [Google Scholar]
- 17.Biswas RK, Dutta D, Tripathi A, Feng Y, Banerjee M, Singh BN. 2013. Identification and characterization of Rv0494: a fatty acid-responsive protein of the GntR/FadR family from Mycobacterium tuberculosis. Microbiology 159:913–923. doi: 10.1099/mic.0.066654-0. [DOI] [PubMed] [Google Scholar]
- 18.Provvedi R, Boldrin F, Falciani F, Palu G, Manganelli R. 2009. Global transcriptional response to vancomycin in Mycobacterium tuberculosis. Microbiology 155:1093–1102. doi: 10.1099/mic.0.024802-0. [DOI] [PubMed] [Google Scholar]
- 19.Boshoff HI, Myers TG, Copp BR, McNeil MR, Wilson MA, Barry CE III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J Biol Chem 279:40174–40184. doi: 10.1074/jbc.M406796200. [DOI] [PubMed] [Google Scholar]
- 20.Hampshire T, Soneji S, Bacon J, James BW, Hinds J, Laing K, Stabler RA, Marsh PD, Butcher PD. 2004. Stationary phase gene expression of Mycobacterium tuberculosis following a progressive nutrient depletion: a model for persistent organisms? Tuberculosis 84:228–238. doi: 10.1016/j.tube.2003.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rohde KH, Veiga DF, Caldwell S, Balazsi G, Russell DG. 2012. Linking the transcriptional profiles and the physiological states of Mycobacterium tuberculosis during an extended intracellular infection. PLoS Pathog 8:e1002769. doi: 10.1371/journal.ppat.1002769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Colijn C, Brandes A, Zucker J, Lun DS, Weiner B, Farhat MR, Cheng TY, Moody DB, Murray M, Galagan JE. 2009. Interpreting expression data with metabolic flux models: predicting Mycobacterium tuberculosis mycolic acid production. PLoS Comput Biol 5:e1000489. doi: 10.1371/journal.pcbi.1000489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol Microbiol 43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 24.Dahl JL, Kraus CN, Boshoff HI, Doan B, Foley K, Avarbock D, Kaplan G, Mizrahi V, Rubin H, Barry CE III. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc Natl Acad Sci U S A 100:10026–10031. doi: 10.1073/pnas.1631248100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Barbacioru C, Hyland F, Xiao W, Hunkapiller KL, Blake J, Chan F, Gonzalez C, Zhang L, Samaha RR. 2006. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics 7:59. doi: 10.1186/1471-2164-7-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sacco E, Covarrubias AS, O'Hare HM, Carroll P, Eynard N, Jones TA, Parish T, Daffe M, Backbro K, Quemard A. 2007. The missing piece of the type II fatty acid synthase system from Mycobacterium tuberculosis. Proc Natl Acad Sci U S A 104:14628–14633. doi: 10.1073/pnas.0704132104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brown AK, Bhatt A, Singh A, Saparia E, Evans AF, Besra GS. 2007. Identification of the dehydratase component of the mycobacterial mycolic acid-synthesizing fatty acid synthase-II complex. Microbiology 153:4166–4173. doi: 10.1099/mic.0.2007/012419-0. [DOI] [PubMed] [Google Scholar]
- 28.Boutte CC, Crosson S. 2013. Bacterial lifestyle shapes stringent response activation. Trends Microbiol 21:174–180. doi: 10.1016/j.tim.2013.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolz C, Geiger T, Goerke C. 2010. The synthesis and function of the alarmone (p)ppGpp in firmicutes. Int J Med Microbiol 300:142–147. doi: 10.1016/j.ijmm.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Potrykus K, Cashel M. 2008. (p)ppGpp: still magical? Annu Rev Microbiol 62:35–51. doi: 10.1146/annurev.micro.62.081307.162903. [DOI] [PubMed] [Google Scholar]
- 31.Cashel M, Gentry DR, Hernandez VJ, Vinella D. 1996. The stringent response, p 1458–1496. In Neidhardt FC, Curtiss R III, Ingraham JL, Lin ECC, Low KB, Magasanik B, Reznikoff WS, Riley M, Schaechter M, Umbarger HE (ed), Escherichia coli and Salmonella: cellular and molecular biology. ASM Press, Washington, DC. [Google Scholar]
- 32.Dalebroux ZD, Swanson MS. 2012. ppGpp: magic beyond RNA polymerase. Nat Rev Microbiol 10:203–212. doi: 10.1038/nrmicro2720. [DOI] [PubMed] [Google Scholar]
- 33.Primm TP, Andersen SJ, Mizrahi V, Avarbock D, Rubin H, Barry CE 3rd. 2000. The stringent response of Mycobacterium tuberculosis is required for long-term survival. J Bacteriol 182:4889–4898. doi: 10.1128/JB.182.17.4889-4898.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dahl JL, Arora K, Boshoff HI, Whiteford DC, Pacheco SA, Walsh OJ, Lau-Bonilla D, Davis WB, Garza AG. 2005. The relA homolog of Mycobacterium smegmatis affects cell appearance, viability, and gene expression. J Bacteriol 187:2439–2447. doi: 10.1128/JB.187.7.2439-2447.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klinkenberg LG, Lee JH, Bishai WR, Karakousis PC. 2010. The stringent response is required for full virulence of Mycobacterium tuberculosis in guinea pigs. J Infect Dis 202:1397–1404. doi: 10.1086/656524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Karakousis PC, Yoshimatsu T, Lamichhane G, Woolwine SC, Nuermberger EL, Grosset J, Bishai WR. 2004. Dormancy phenotype displayed by extracellular Mycobacterium tuberculosis within artificial granulomas in mice. J Exp Med 200:647–657. doi: 10.1084/jem.20040646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weiss LA, Stallings CL. 2013. Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J Bacteriol 195:5629–5638. doi: 10.1128/JB.00759-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Kessel JC, Hatfull GF. 2007. Recombineering in Mycobacterium tuberculosis. Nat Methods 4:147–152. doi: 10.1038/nmeth996. [DOI] [PubMed] [Google Scholar]
- 39.Pfaffl MW. 2001. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res 29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Jackson JD, Ke Z, Lanczycki CJ, Lu F, Marchler GH, Mullokandov M, Omelchenko MV, Robertson CL, Song JS, Thanki N, Yamashita RA, Zhang D, Zhang N, Zheng C, Bryant SH. 2011. CDD: a Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res 39:D225-229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Capella-Gutierrez S, Silla-Martinez JM, Gabaldon T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 45.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sen A, Daubin V, Abrouk D, Gifford I, Berry AM, Normand P. 2014. Phylogeny of the class Actinobacteria revisited in the light of complete genomes; the orders ‘Frankiales’ and Micrococcales should be split into coherent entities: proposal of Frankiales ord nov, Geodermatophilales ord nov, Acidothermales ord nov and Nakamurellales ord nov Int J Syst Evol Microbiol 64:3821–3832. [DOI] [PubMed] [Google Scholar]
- 47.Letunic I, Bork P. 2011. Interactive Tree Of Life v2: online annotation and display of phylogenetic trees made easy. Nucleic Acids Res 39:W475–478. doi: 10.1093/nar/gkr201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cortes T, Schubert OT, Rose G, Arnvig KB, Comas I, Aebersold R, Young DB. 2013. Genome-wide mapping of transcriptional start sites defines an extensive leaderless transcriptome in Mycobacterium tuberculosis. Cell Rep 5:1121–1131. doi: 10.1016/j.celrep.2013.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Newton-Foot M, Gey van Pittius NC. 2013. The complex architecture of mycobacterial promoters. Tuberculosis (Edinb) 93:60–74. doi: 10.1016/j.tube.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Marmiesse M, Brodin P, Buchrieser C, Gutierrez C, Simoes N, Vincent V, Glaser P, Cole ST, Brosch R. 2004. Macro-array and bioinformatic analyses reveal mycobacterial ‘core’ genes, variation in the ESAT-6 gene family and new phylogenetic markers for the Mycobacterium tuberculosis complex. Microbiology 150:483–496. doi: 10.1099/mic.0.26662-0. [DOI] [PubMed] [Google Scholar]
- 51.Aravind L, Anantharaman V, Balaji S, Babu MM, Iyer LM. 2005. The many faces of the helix-turn-helix domain: transcription regulation and beyond. FEMS Microbiol Rev 29:231–262. [DOI] [PubMed] [Google Scholar]
- 52.Kepes F. 2004. Periodic transcriptional organization of the E. coli genome. J Mol Biol 340:957–964. doi: 10.1016/j.jmb.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 53.Zhang YJ, Ioerger TR, Huttenhower C, Long JE, Sassetti CM, Sacchettini JC, Rubin EJ. 2012. Global assessment of genomic regions required for growth in Mycobacterium tuberculosis. PLoS Pathog 8:e1002946. doi: 10.1371/journal.ppat.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hauryliuk V, Atkinson GC, Murakami KS, Tenson T, Gerdes K. 2015. Recent functional insights into the role of (p)ppGpp in bacterial physiology. Nat Rev Microbiol 13:298–309. doi: 10.1038/nrmicro3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choi KH, Kremer L, Besra GS, Rock CO. 2000. Identification and substrate specificity of beta-ketoacyl (acyl carrier protein) synthase III (mtFabH) from Mycobacterium tuberculosis. J Biol Chem 275:28201–28207. [DOI] [PubMed] [Google Scholar]
- 56.Jackson M, Raynaud C, Laneelle MA, Guilhot C, Laurent-Winter C, Ensergueix D, Gicquel B, Daffe M. 1999. Inactivation of the antigen 85C gene profoundly affects the mycolate content and alters the permeability of the Mycobacterium tuberculosis cell envelope. Mol Microbiol 31:1573–1587. doi: 10.1046/j.1365-2958.1999.01310.x. [DOI] [PubMed] [Google Scholar]
- 57.Belisle JT, Vissa VD, Sievert T, Takayama K, Brennan PJ, Besra GS. 1997. Role of the major antigen of Mycobacterium tuberculosis in cell wall biogenesis. Science 276:1420–1422. doi: 10.1126/science.276.5317.1420. [DOI] [PubMed] [Google Scholar]
- 58.Sureka K, Dey S, Datta P, Singh AK, Dasgupta A, Rodrigue S, Basu J, Kundu M. 2007. Polyphosphate kinase is involved in stress-induced mprAB-sigE-rel signalling in mycobacteria. Mol Microbiol 65:261–276. doi: 10.1111/j.1365-2958.2007.05814.x. [DOI] [PubMed] [Google Scholar]
- 59.Sureka K, Ghosh B, Dasgupta A, Basu J, Kundu M, Bose I. 2008. Positive feedback and noise activate the stringent response regulator rel in mycobacteria. PLoS One 3:e1771. doi: 10.1371/journal.pone.0001771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Ghosh S, Sureka K, Ghosh B, Bose I, Basu J, Kundu M. 2011. Phenotypic heterogeneity in mycobacterial stringent response. BMC Syst Biol 5:18. doi: 10.1186/1752-0509-5-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hingley-Wilson SM, Sambandamurthy VK, Jacobs WR Jr. 2003. Survival perspectives from the world's most successful pathogen, Mycobacterium tuberculosis. Nat Immunol 4:949–955. doi: 10.1038/ni981. [DOI] [PubMed] [Google Scholar]
- 62.Gutierrez MC, Supply P, Brosch R. 2009. Pathogenomics of mycobacteria. Genome Dyn 6:198–210. doi: 10.1159/000235772. [DOI] [PubMed] [Google Scholar]
- 63.Lacave C, Laneelle MA, Daffe M, Montrozier H, Laneelle G. 1989. Mycolic acid metabolic filiation and location in Mycobacterium aurum and Mycobacterium phlei. Eur J Biochem 181:459–466. doi: 10.1111/j.1432-1033.1989.tb14747.x. [DOI] [PubMed] [Google Scholar]
- 64.Russell DG, VanderVen BC, Lee W, Abramovitch RB, Kim MJ, Homolka S, Niemann S, Rohde KH. 2010. Mycobacterium tuberculosis wears what it eats. Cell Host Microbe 8:68–76. doi: 10.1016/j.chom.2010.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Shi L, Sohaskey CD, Pfeiffer C, Datta P, Parks M, McFadden J, North RJ, Gennaro ML. 2010. Carbon flux rerouting during Mycobacterium tuberculosis growth arrest. Mol Microbiol 78:1199–1215. doi: 10.1111/j.1365-2958.2010.07399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Barkan D, Rao V, Sukenick GD, Glickman MS. 2010. Redundant function of cmaA2 and mmaA2 in Mycobacterium tuberculosis cis cyclopropanation of oxygenated mycolates. J Bacteriol 192:3661–3668. doi: 10.1128/JB.00312-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rao V, Fujiwara N, Porcelli SA, Glickman MS. 2005. Mycobacterium tuberculosis controls host innate immune activation through cyclopropane modification of a glycolipid effector molecule. J Exp Med 201:535–543. doi: 10.1084/jem.20041668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Corrales RM, Molle V, Leiba J, Mourey L, de Chastellier C, Kremer L. 2012. Phosphorylation of mycobacterial PcaA inhibits mycolic acid cyclopropanation: consequences for intracellular survival and for phagosome maturation block. J Biol Chem 287:26187–26199. doi: 10.1074/jbc.M112.373209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dubnau E, Chan J, Raynaud C, Mohan VP, Laneelle MA, Yu K, Quemard A, Smith I, Daffe M. 2000. Oxygenated mycolic acids are necessary for virulence of Mycobacterium tuberculosis in mice. Mol Microbiol 36:630–637. [DOI] [PubMed] [Google Scholar]
- 70.Cantaloube S, Veyron-Churlet R, Haddache N, Daffe M, Zerbib D. 2011. The Mycobacterium tuberculosis FAS-II dehydratases and methyltransferases define the specificity of the mycolic acid elongation complexes. PLoS One 6:e29564. doi: 10.1371/journal.pone.0029564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Veyron-Churlet R, Guerrini O, Mourey L, Daffe M, Zerbib D. 2004. Protein-protein interactions within the fatty acid synthase-II system of Mycobacterium tuberculosis are essential for mycobacterial viability. Mol Microbiol 54:1161–1172. doi: 10.1111/j.1365-2958.2004.04334.x. [DOI] [PubMed] [Google Scholar]
- 72.Rienksma RA, Suarez-Diez M, Mollenkopf HJ, Dolganov GM, Dorhoi A, Schoolnik GK, Martins Dos Santos V, Kaufmann S, Schaap PJ, Gengenbacher M. 2015. Comprehensive insights into transcriptional adaptation of intracellular mycobacteria by microbe-enriched dual RNA sequencing. BMC Genomics 16:34. doi: 10.1186/s12864-014-1197-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manganelli R. 2007. Polyphosphate and stress response in mycobacteria. Mol Microbiol 65:258–260. doi: 10.1111/j.1365-2958.2007.05819.x. [DOI] [PubMed] [Google Scholar]
- 74.Manganelli R, Voskuil MI, Schoolnik GK, Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol Microbiol 41:423–437. doi: 10.1046/j.1365-2958.2001.02525.x. [DOI] [PubMed] [Google Scholar]
- 75.Dandekar T, Snel B, Huynen M, Bork P. 1998. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem Sci 23:324–328. doi: 10.1016/S0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- 76.Mao F, Dam P, Chou J, Olman V, Xu Y. 2009. DOOR: a database for prokaryotic operons. Nucleic Acids Res 37:D459–D463. doi: 10.1093/nar/gkn757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.World Health Organization. 2014. Global tuberculosis report 2014. World Health Organization, Geneva, Switzerland. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.