ABSTRACT
Integrative and conjugative elements (ICEs) of the SXT/R391 family are the main contributors to acquired multidrug resistance in the seventh pandemic lineage of Vibrio cholerae, the etiological agent of the diarrheal disease cholera. Conjugative transfer of SXT/R391 ICEs is triggered by antibiotics and agents promoting DNA damage through RecA-dependent autoproteolysis of SetR, an ICE-encoded λ CI-like repressor. Here, we describe the role of CroS, a distant λ Cro homolog, as a key component contributing to the regulation of expression of the activator SetCD that orchestrates the expression of the conjugative transfer genes. We show that deletion of croS abolishes the SOS response-dependent induction of SXT despite the presence of a functional setR gene. Using quantitative reverse transcription-PCR and lacZ reporter assays, we also show that CroS represses setR and setCD expression by binding to operator sites shared with SetR. Furthermore, we provide evidence of an additional operator site bound by SetR and CroS. Finally, we show that SetCD expression generates a positive feedback loop due to SXT excision and replication in a fraction of the cell population. Together, these results refine our understanding of the genetic regulation governing the propagation of major vectors of multidrug resistance.
IMPORTANCE Healthcare systems worldwide are challenged by an alarming drug resistance crisis caused by the massive and rapid propagation of antibiotic resistance genes and the associated emergence of multidrug-resistant pathogenic bacteria. SXT/R391 ICEs contribute to this phenomenon not only in clinical and environmental vibrios but also in several members of the family Enterobacteriaceae. We have identified and characterized here the regulator CroS as a key factor in the stimulation of conjugative transfer of these ICEs in response to DNA-damaging agents. We have also untangled conflicting evidence regarding autoactivation of transfer by the master activator of SXT/R391 ICEs, SetCD. Discovery of CroS provides a clearer and more complete understanding of the regulatory network that governs the dissemination of SXT/R391 ICEs in bacterial populations.
INTRODUCTION
Cholera is a severe infectious disease caused by the ingestion of food or water contaminated by Vibrio cholerae. This disease remains widespread in regions with limited access to clean water and where poor sanitation allows easy dissemination of the bacterium in drinking water sources. Cholera is characterized by a profuse watery diarrhea that rapidly induces massive fluid loss, causing severe dehydration of the patient, which may lead to death within 24 h of symptom onset. Although epidemic cholera is usually caused by V. cholerae O1, the unusual serogroup O139 emerged in the early 1990s as the cause of a cholera outbreak in India (1). O139 clinical isolates were found to be resistant to sulfamethoxazole and trimethoprim, two antibiotics commonly used for the treatment of severe cases of cholera (2). This resistance was found to be transmissible and linked to an integrative and conjugative element (ICE) named SXT (3). ICEs are self-transmissible bacterial mobile elements that play a major role in gene exchange in bacterial populations, as they are horizontally transferred via conjugation by a process similar to that used by many conjugative plasmids (4, 5). Unlike plasmids, ICEs do not remain stably in an episomal form and are rather found integrated into the chromosome. SXT integrates itself into the chromosome of V. cholerae in a site-specific manner at the 5′ end of prfC, a gene coding for the peptide release factor RF3 (6). Since the discovery of SXT, SXT or related ICEs have been found to be prevalent in the seventh pandemic isolates of V. cholerae and sporadically present in other Vibrio species (7–9) and other Gammaproteobacteria of clinical origin or isolated from the aquatic environment, such as Photobacterium (10), Proteus (11), Alteromonas (12), Marinomonas (13), and Shewanella (9, 14) species. SXT is closely related to R391, an ICE conferring resistance to kanamycin and mercury, originally detected in a 1967 South African isolate of Providencia rettgeri (15, 16). All of the ICEs related to SXT and R391 are grouped into a single family, namely, the SXT/R391 family, because they all have the same chromosomal integration site and a set of conserved genes essential for site-specific integration, conjugative transfer, and regulation (6, 8). SXT/R391 ICEs also contain variable DNA insertions conferring adaptive traits, including resistance to antibiotics, heavy metals, and bacteriophage infection (8, 17, 18); synthesis of the second messenger c-di-GMP (19); and homologous recombination and mutagenic repair systems (20, 21). Beside their own transfer, SXT/R391 ICEs have been shown to mobilize plasmids, phylogenetically unrelated genomic islands (mobilizable genomic islands), and up to 1.5 Mb of chromosomal DNA by high-frequency recombination transfer (22, 23).
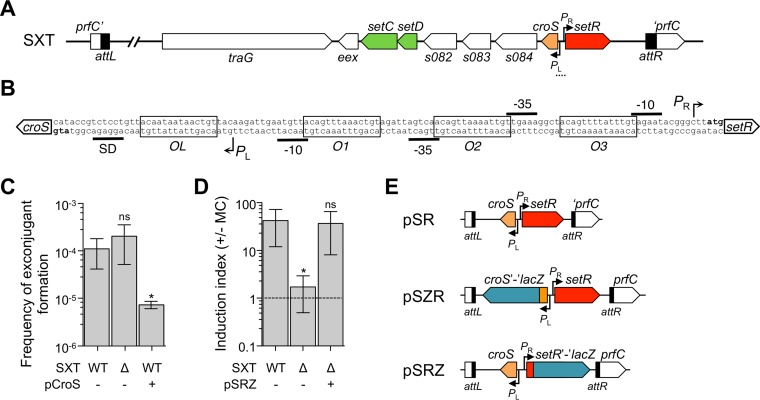
The most conserved genes (97% identity at the nucleotide level) shared by SXT/R391 ICEs are s086 and setR, which belong to the regulatory module located at the rightmost end of the integrated ICE, near the attR attachment site (Fig. 1A) (8). The regulatory module contains eight open reading frames (ORFs), seven of which are in the same orientation (s086, s084, s083, s082, setD, setC, and eex), while the last gene, setR, is divergently transcribed (Fig. 1A and B) (24). eex and the convergent gene traG code for the entry exclusion system of SXT/R391 ICEs (25). The overlapping genes setC and setD encode the SetCD activator complex. SetCD was shown to bind upstream of the −35 sequence of 11 promoters in SXT/R391 ICEs, activating the expression of >40 genes essential for site-specific and homologous recombination, ICE replication and partition, and conjugative transfer (26–28). The functions of s082, s083, s084, and s086 remain unknown. In silico analysis has revealed that s086 codes for a putative small basic protein of 9.4 kDa with a predicted helix-turn-helix (HTH) DNA-binding domain. setR codes for the main repressor of SXT/R391 ICEs (24, 29). SetR contains an HTH_XRE (PF01381) motif in its N-terminal moiety and a C-terminal LexA-like autoproteolysis domain (PF00717). SetR is related to phage 434 CI and other lambdoid phage repressors and has been shown to bind to four operator sites located between s086 and setR (Fig. 1B) (24, 30). These operators, O1, O2, O3, and OL, are part of PL and PR, two divergent overlapping promoters located in the intergenic region between s086 and setR. The relative affinities of SetR for these four operator sites and their positions suggest an autoregulation of setR expression from PR (30) and a repression of the operon containing s086 and setCD driven from PL (29). Repression at PL is alleviated when the cellular pool of SetR drops, most likely as a result of autoproteolysis stimulated by DNA damage-induced activation of RecA (29).
FIG 1.
Effects of croS on SXT conjugative transfer. (A) Schematic representation of the regulatory module of SXT. The dotted line indicates the region enlarged in panel B. (B) The intergenic region between croS, previously referred to as s086, and setR in SXT as represented by Beaber and Waldor (30). The PL and PR promoters, the OL, O1, O2, and O3 operators, the −10 and −35 elements, the Shine-Dalgarno (SD) sequence of croS, and the start codons of croS and setR are represented. (C) Effect of the deletion or overexpression of croS on SXT transfer. Conjugation assays were carried out with E. coli BW25113 SXT (VB17, WT) or its ΔcroS mutant (DPL353, Δ) as the donor. Overexpression assays were carried out by expressing croS from the arabinose-inducible PBAD promoter in pCroS (DPL525). E. coli CAG18439 (Tcr) was used as the recipient. Frequencies of exconjugant formation were expressed as the number of exconjugant CFU (Sur Tmr Tcr) per recipient CFU (Tcr). (D) Effect of croS deletion on the induction of SXT transfer by MC. The donor strains were VB17, DPL353, and DPL263 in which croS was expressed from its native PL promoter in pSRZ integrated in single copy in tandem with SXT ΔcroS. The recipient strain was CAG18439. The induction index corresponds to the ratio of the frequencies of exconjugant formation obtained with and without MC treatment prior to transfer. The dashed line indicates an induction index of 1, i.e., no difference between the induced and noninduced conditions. Each bar represents the mean and standard deviation of values obtained from at least three independent experiments. Statistical analyses of the logarithms of the mean values were performed with two-tailed unpaired t tests. Asterisks indicate that P values are <0.05 compared to the wild type (WT). (E) Schematic representation of the insert of the pSR, pSZR, and pSRZ plasmids site specifically integrated into the 5′ end of prfC alone or in tandem with SXT. The plasmid backbone is not represented. In pSZR and pSRZ, lacZ was translationally fused to the sixth codon of croS and setR, respectively.
The relative positions of setR and s086 are reminiscent of cI and cro carried by bacteriophage λ. CI and Cro form a pair that governs the transition between the lysogenic and lytic pathways of the λ life cycle (31, 32). To date, SXT/R391 ICEs have been known to be regulated by only two transcriptional regulators, the repressor SetR and the activator complex SetCD (24). In this study, we identify S086 as a third regulator, which will be referred to as CroS for Cro-like repressor of SXT. We demonstrate that CroS is a key factor in the alleviation of SetR repression and induction of conjugative transfer of SXT/R391 ICEs. By binding to five operator sites shared with SetR, CroS acts as a repressor of PL and PR, thereby repressing both setCD and setR by a mechanism that is evocative of λ Cro repression on CI and CII (33). We also show that SetCD increases its own expression not by activating PL but by indirectly generating a positive feedback loop triggered by activation of SXT replication, which increases the SXT copy number and ultimately the level of the setCD mRNA transcript. Finally, we propose a new model of SXT transfer regulation that includes CroS as a repressor of PL and PR, thereby promoting the SOS-dependent induction of SXT/R391 ICE transfer.
MATERIALS AND METHODS
Bacterial strains and media.
The bacterial strains used in this study are described in Table 1. The strains were routinely grown in lysogeny broth (LB-Miller; EMD) at 37°C in an orbital shaker/incubator and maintained at −80°C in LB broth containing 15% (vol/vol) glycerol. Antibiotics were used at the following concentrations: ampicillin, 100 μg ml−1; chloramphenicol, 20 μg ml−1; erythromycin, 10 μg ml−1; kanamycin, 50 μg ml−1; spectinomycin, 50 μg ml−1; sulfamethoxazole (Su), 160 μg ml−1; tetracycline (Tc), 12 μg ml−1; trimethoprim (Tm), 32 μg ml−1. When required, bacterial cultures were supplemented with 0.1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), 0.02% l-arabinose, or 100 ng ml−1 mitomycin C (MC).
TABLE 1.
Strains and plasmids used in this study
| Strain or plasmid | Relevant genotype or description (phenotype)a | Reference(s) or source |
|---|---|---|
| E. coli strains | ||
| CAG18439 | MG1655 lacZU118 lacI42::Tn10 (Tcr) | 76 |
| HW220 | CAG18439 prfC::SXT (Sur Tmr) | 6 |
| DC147 | CAG18439 prfC::SXT ΔtraI::aph (Sur Tmr Knr) | This study |
| BW25113 | F− Δ(araD-araB)567 ΔlacZ4787(::rrnB-3) λ− rph-1 Δ(rhaD-rhaB)568 hsdR514 | 37, 77 |
| β2163 | (F−) RP4-2-Tc::Mu ΔdapA::(erm-pir) (Knr Emr) | 36 |
| BL21(DE3) | F− ompT ΔhsdS gal dcm λDE3 | Novagen |
| VB17 | BW25113 prfC::SXT (Sur Tmr) | 28 |
| DPL353 | BW25113 prfC::SXT ΔcroS (Sur Tmr) | This study |
| DPL263 | BW25113 prfC::[SXT ΔcroS]-pSRZ (Sur Tmr Cmr) | This study |
| DPL525 | BW25113 prfC::SXT pCroS (Sur Tmr Apr) | This study |
| DPL297 | BW25113 prfC::SXT setD′-′lacZ-aad7 ΔsetC (Sur Tmr Spr) | This study |
| DPL548 | BW25113 prfC::[SXT ΔtraI::aph setD′-′lacZ-aad7 ΔsetC] (Sur Tmr Spr Knr) | This study |
| DPL231 | BW25113 prfC::pSR (croS-setR) (Sur Tmr) | This study |
| DPL241 | BW25113 prfC::pSZR (croS′-′lacZ-setR) (Sur Tmr) | This study |
| DPL246 | BW25113 prfC::pSRZ (croS-setR′-′lacZ) (Sur Tmr) | This study |
| Plasmids | ||
| pVI36 | PCR template for one-step chromosomal gene inactivation (Spr) | 78 |
| pKD13 | PCR template for one-step chromosomal gene inactivation (Knr) | 37 |
| pVI42B | pVI36 BamHI::Plac-lacZ (Spr) | 20 |
| pKD46 | λRed recombination arabinose-inducible encoding plasmid (Apr) | 37 |
| pSW23T | oriTRP4 oriVR6Kγ (Cmr) | 36 |
| pSR | pSW23T croS-setR attPSXT (Cmr) | This study |
| pSZR | pSW23T croS′-′lacZ-aad7-setR attPSXT (Spr Cmr) | This study |
| pSRZ | pSW23T croS-setR′-′lacZ-aad7 attPSXT (Spr Cmr) | This study |
| pVI68 | pAH57 Δ(xisλ-intλ)::intSXT (Ts) | 20 |
| pBAD-TOPO | oripBR322 bla araC PBAD (Apr) | Invitrogen |
| pBAD30 | orip15A bla araC PBAD (Apr) | 79 |
| pGG2B | pBAD30::setDC (Apr) | 28 |
| pCroS | pBAD-TOPO::croS (5′-3′) (Apr) | This study |
| pCroS-inv | pBAD-TOPO::croS (3′-5′) (Apr) | This study |
| pSetR | pBAD-TOPO::setR (Apr) | This study |
| pTraI | pBAD-TOPO::traI (Apr) | 27 |
| pCR2.1-TOPO | oripUC (Apr Knr) | Invitrogen |
| pDPL128 | pCR2.1-TOPO::PL-PR (Apr Knr) | This study |
| pGEX-croS | pGEX-6P-1::croS (Apr) | This study |
| pGEX-setR | pGEX-6P-1::setR (Apr) | This study |
Apr, ampicillin resistant; Cmr, chloramphenicol resistant; Emr, erythromycin resistant; Knr, kanamycin resistant; Spr, spectinomycin resistant; Sur, sulfamethoxazole resistant; Tcr, tetracycline resistant; Tmr, trimethoprim resistant; Ts, thermosensitive.
Bacterial conjugations.
Conjugation assays were performed as described by Burrus and Waldor (34), with the following modifications. When induction with MC was performed, overnight cultures of donor and recipient cells were diluted 1:4 and the donor cultures were supplemented or not with MC and incubated for 1 h at 37°C with shaking. The cells were harvested by centrifugation for 3 min at 1,200 × g, washed in 1 volume of LB broth, and resuspended in 1/20 volume of LB broth. Mating mixtures were then deposited on LB agar plates and incubated at 37°C for 2 h.
Molecular biology methods.
Plasmid DNA was prepared with the EZ-10 Spin Column Plasmid DNA Minipreps kit (Bio Basic) according to the manufacturer's instructions. All of the enzymes used in this study were purchased from New England BioLabs. PCR assays were performed with the Taq, Q5, or Phusion polymerase according to the manufacturer's instructions. When necessary, PCR products were purified with an EZ-10 Spin Column PCR Products Purification kit (Bio Basic) according to the manufacturer's instructions. E. coli was transformed by electroporation as described by Dower et al. (35) in a Bio-Rad Gene Pulser Xcell apparatus set at 25 μF, 200 Ω, and 1.8 kV with 1-mm gap electroporation cuvettes. Sequencing reactions were performed by the Plateforme de Séquençage et de Génotypage du Centre de Recherche du CHUL (Québec, QC, Canada).
Plasmid and strain constructions.
The plasmids and oligonucleotides used in this study are listed in Table 1 (also see Table S1 in the supplemental material). Plasmids pCroS and pSetR were constructed by amplifying croS and setR with primer pair s086pBADf/s086pBADr.s or setRpBADf/setRpBADr.s and genomic DNA of E. coli HW220 as the template and cloning into the TA cloning expression vector pBAD-TOPO (Invitrogen) according to the manufacturer's instructions. pSR was constructed by PCR amplification of the croS-setR-attP region resulting from the excision of SXT in E. coli HW220 with the primer pair PEs086/VISLR3. The 2,022-bp PCR product was first cloned into pCR2.1-TOPO (Invitrogen) according to the manufacturer's instructions. The cloned fragment was then recovered by NotI/BamHI digestion and then ligated to NotI/BamHI-digested pSW23T, yielding pSR. pSW23T contains the RP4 mobilization locus, the R6K pir-dependent conditional origin of replication, and a chloramphenicol resistance gene (36). Plasmid pDPL128 was constructed by cloning into pCR2.1-TOPO (Invitrogen) the intergenic region between setR and croS, which was amplified by PCR with primer pair RRs086-4/RRs086setRr and genomic DNA of E. coli HW220 as the template.
ΔcroS and ΔtraI mutant derivatives of SXT were constructed in E. coli HW220 by the one-step chromosomal gene inactivation technique (37) with primer pairs delS086F/dels086R and TraIWF2/TraIWR3, respectively, and the pKD13 template. SXT ΔtraI::aph complemented with pTraI was transferred by conjugation into E. coli BW25113. To construct E. coli DPL548, the ΔtraI::aph mutation was then transduced into E. coli DPL297 (BW25113 prfC::SXT ΔsetCD::lacZ-aad7) by P1vir generalized transduction. Strains containing pSR integrated into the 5′ end of prfC were constructed by mating the RP4+ pir+ strain E. coli β2163/pSR with E. coli BW25113 containing pVI68 as the recipient strain. pVI68 expresses the integrase of SXT, thereby allowing site-specific integration of pSR, which was verified by amplifying the attL and attR sites by PCR with primer pairs EattBR/VISRF and EattBF/VISLR3, respectively. The absence of multiple integrations of pSR in tandem arrays was verified with primers VISLR3 and VISRF. The resulting strain, E. coli DPL231, contains a unique copy of the croS-setR region integrated site specifically into prfC. Translational lacZ fusions setR′-′lacZ, croS′-′lacZ, and setCD′-′lacZ were constructed by the one-step chromosomal gene inactivation technique (37) with primer pairs DATsetR-lacZf/DATsetR-lacZr, DATsetQ-lacZf/DATsetQ-lacZr, and DATsetCD-lacZf/DATsetCD-lacZr, respectively, and pVI42B as the template. In each construct, the sixth codon of the gene of interest was fused to the ninth codon of lacZ. The setR′-′lacZ and croS′-′lacZ mutations targeted BW25113 derivative strains containing pSR integrated into prfC (DPL231), whereas the setCD′-′lacZ mutation targeted SXT in E. coli VB17. SXT ΔcroS was transferred into DPL246 by conjugation to generate DPL263.
RNA isolation and quantitative reverse transcription (qRT)-PCR.
Total RNA was extracted and cDNA were synthesized as described previously (19, 38). RNA integrity was verified with a 2100 Bioanalyzer (Agilent). Quantitative amplification of setR, croS, setD, and setC was carried out with primer pairs RTsetR-F/RTsetR-R, RTcroS-F/RTcroS-R, RTsetD-F/RTsetD-R, and RTsetC-F/RTsetC-R, respectively. For normalization, rpoZ was amplified with primers RTrpoZcoli-F/RTrpoZcoli-R and the ΔΔCT calculation method. Experiments were repeated three times in triplicate and combined. Melting curves were determined with the final reaction products (156 to 165 bp) to confirm that amplification was specific to targets. All primer pairs exhibited efficiencies between 95 and 97%.
Real-time qPCR assays for relative quantification of attB and attP.
The frequency of excision and copy number of the excised circular form of SXT were assessed by real-time quantitative PCR (qPCR) as described elsewhere (39). Genomic DNA was obtained from cultures of E. coli VB17, DPL297/pGG2B, and DPL548/pGG2B cells grown under conditions described for the qRT-PCR assays. Bacterial cultures were grown in LB medium at 37°C to an optical density at 600 nm (OD600) of 0.2 and then for an additional 2 h at 37°C. prfC, attB, and attP were quantified with primer pairs prfC.qec.F1/prfC.qec.R1, attB.qec.F2/attB.qec.R2, and attP.qec.F2/attP.qec.R2, respectively (27) (see Table S1 in the supplemental material). Three biological replicates of qPCR experiments were performed on the RNomics platform of the Laboratoire de Génomique Fonctionnelle de l'Université de Sherbrooke, Sherbrooke, QC, Canada (http://lgfus.ca). Normalization was performed as described previously (39).
β-Galactosidase assays.
The substrate used to determine LacZ levels was o-nitrophenyl-β-d-galactopyranoside, and the assays were carried out as described previously (40) with LB medium and 100 μg ml−1 ampicillin for maintenance of pCroS and pGG2B. Induction of croS and setCD expression and of the SOS response was done by supplementing, respectively, with 0.2% arabinose and/or 100 ng ml−1 MC a refreshed culture grown to an OD600 of 0.2, followed by a 2-h incubation at 37°C with shaking prior to cell sampling.
Expression and purification of the SetR and CroS proteins.
Overnight-grown cultures of E. coli BL21(DE3) bearing pGEX-croS or pGEX-setR were diluted 1/1,000 in fresh 2× YT broth (80) supplemented with 100 µg ml−1 ampicillin and incubated at 37°C with agitation. Protein expression was induced at mid-exponential phase (OD600 of 0.6) with 0.1 mM IPTG. The cultures were grown for an additional 3 h at 37°C with agitation. Cells were collected by centrifugation, resuspended in phosphate-buffered saline containing 1% Triton X-100, 1 mM phenylmethylsulfonyl fluoride, and a cocktail of protease inhibitors (Protease Inhibitor Cocktail; Sigma). Cells were lysed by sonication and centrifuged to pellet the debris, and CroS and SetR were recovered by affinity chromatography with the glutathione S-transferase purification module (GE Healthcare) and the PreScission protease (GE Healthcare) according to the manufacturer's instructions. Protein concentration was estimated with the Bradford protein assay (Bio-Rad), and purity was determined by SDS-PAGE analysis.
Dimerization assay.
Prior to dimerization assay, the purity of CroS was estimated by SDS-PAGE analysis to be 95%. No contaminant proteins with a size similar to that of CroS was observed. A 4.7-μg sample of purified CroS was incubated with 0.625% glutaraldehyde for 30 min at room temperature. The reaction mixture was then denatured at 100°C for 3 min, loaded onto a 15% SDS-PAGE gel, and run for 75 min at 150 V (41). The gel was then stained with Coomassie brilliant blue R-250. The Precision Plus Protein Kaleidoscope Standards ladder (Bio-Rad) was used as a molecular weight marker.
EMSAs.
The probes used in electrophoretic mobility shift assays (EMSAs) were either annealed oligonucleotides or PCR products. Purified PCR products and a single oligonucleotide for each annealed pair were labeled with [γ-32P]dATP with the T4 polynucleotide kinase. An equimolar quantity of the complementary oligonucleotide was added, the volume was brought to 100 μl in 20 mM Tris-HCl (pH 8.0)–20 mM KCl, and the mixture was denatured at 95°C for 5 min and then cooled to room temperature over 1 h. The γ-32P-labeled double-stranded DNA fragments were separated from unincorporated [γ-32P]dATP with Illustra MicroSpin G-25 columns (GE Healthcare) according to the manufacturer's instructions. Purified CroS and SetR proteins were preincubated in 24 μl of buffer I (20 mM Tris-HCl [pH 8.0], 20 mM KCl, 1 mM MgCl2, 5% glycerol) for 20 min at 4°C. For PCR probes 1 to 6 (Fig. 4D), nonspecific competitor DNA was added by supplementing buffer I with sonicated salmon sperm DNA at 50 ng ml−1. Radiolabeled probes (2,000 cpm) were added, and samples were incubated for 30 min at 37°C and then for 10 min at 4°C. Samples were immediately loaded onto a prerun 4% polyacrylamide gel containing 0.5× Tris-borate-EDTA buffer and subjected to electrophoresis at a constant voltage of 120 V at 4°C for 1 h 15 min. Gels were dried and exposed to a Phosphor Screen (Kodak), and results were visualized with a Typhoon FLA 9500 imager (GE Healthcare). The densitometry values (intensity units per millimeter squared) of each shifted operator were quantified with the Quantity One software (Bio-Rad). The background was subtracted, and then percentages of maximal intensity were calculated by dividing the density of each band by the highest density, both obtained with 1 μM SetR or CroS.
FIG 4.
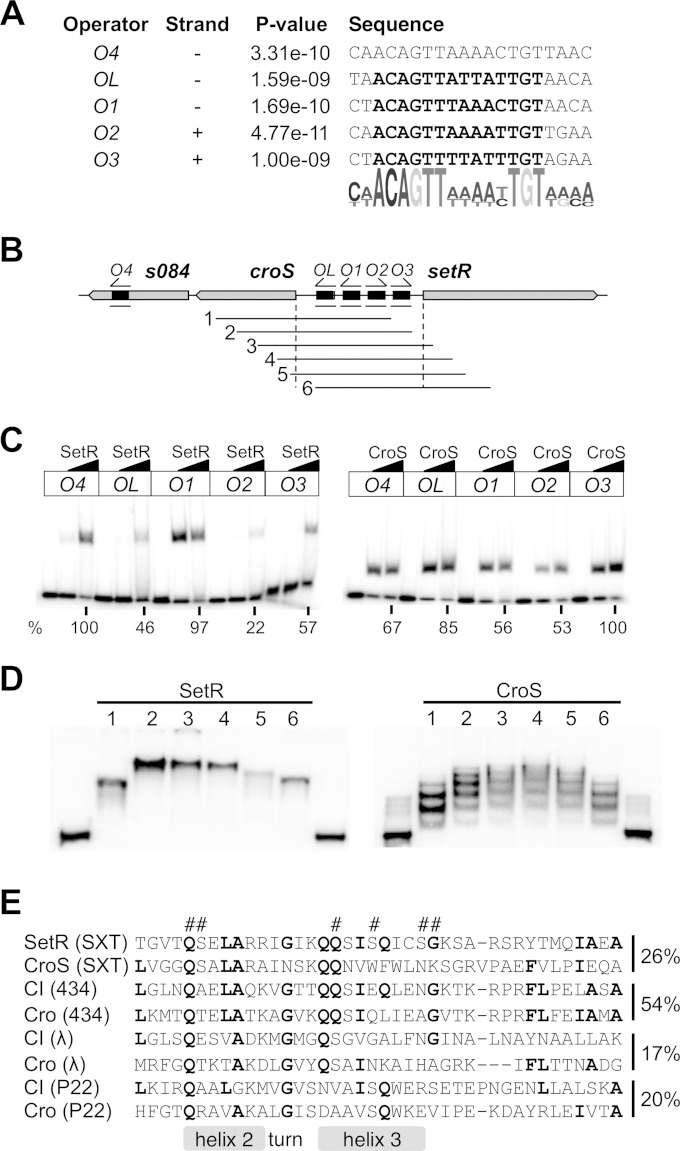
CroS and SetR binding to the switch locus of SXT. (A) Sequences of the five SetR and CroS operators and the associated logo computed by the MEME motif-based sequence analysis tool. The sites previously described by Beaber and Waldor (30) are in bold letters. (B) Representation of the switch locus and the 32P-labeled probes used for the gel shift assays in panels C and D. The orientations of the motifs are indicated above the operators. (C) Binding of 0.6 and 1 μM purified SetR or CroS to annealed oligonucleotides containing a single operator (length of 31 to 36 bp). The leftmost lane for each operator contains the probe alone. The percent intensity of each shifted band in relation to the band with maximal intensity (O4 for SetR and O3 for CroS with 1 μM SetR or CroS) is indicated below the autoradiograph. (D) Bending assays with 1 μM SetR or CroS and permuted PCR probes 1 to 6. The border lanes contain probe 1 with no added protein. (E) Alignment of the HTH_XRE domains (74) of corepressor pairs computed by Clustal Omega (72). Amino acids conserved in at least four of the aligned repressors are in bold. Hash marks (#) above the alignment indicate the specific residues of SetR involved in DNA binding that have been annotated in an NCBI-curated HTH_XRE domain (74). The HTH components are indicated as described elsewhere (51, 75). Percentages of identity between the represented amino acids of the corepressors are indicated on the right of the alignment.
DNase I protection assays.
To analyze the top strand, the probe was prepared first by digesting pDPL128 with HindIII. The linearized plasmid was then dephosphorylated with Antarctic phosphatase and subsequently digested with NsiI. To analyze the bottom strand, the probe was prepared by digestion of pDPL128 with XhoI, Antarctic phosphatase dephosphorylation, and digestion with KpnI. The probes were then end labeled with the T4 polynucleotide kinase and [γ-32P]ATP at 6,000 Ci mmol−1. Fifty-microliter binding reaction mixtures containing CroS and the radiolabeled probe (20,000 cpm) were prepared as described for EMSAs. Fifty microliters of cofactor solution (10 mM MgCl2, 5 mM CaCl2) and 0.1 U of DNase I were added, and the mixture was incubated for 2 min at room temperature. The reaction was terminated by the addition of 100 μl of stop solution (1% SDS, 200 mM NaCl, 20 mM EDTA [pH 8.0], tRNA at 40 μg ml−1) and extracted with 200 μl of phenol-chloroform-isoamyl alcohol (25:24:1). DNA was precipitated with 3 volumes of ethanol, washed with 70% ethanol, dried, resuspended in sequencing gel loading buffer, and denatured for 5 min at 95°C. Sequencing reaction mixtures that serve as a ladder were prepared with the Sequenase 2.0 DNA sequencing kit (Affymetrix-usb) and primers FP-frag2-HindIII and FP-frag2-XhoI for the top and bottom strands, respectively. After electrophoresis through a denaturing 10% polyacrylamide sequencing gel in 0.8× glycerol-tolerant gel buffer, the gel was dried and detection was carried out with a Phosphor Screen (Kodak) and a Typhoon FLA 9500 imager (GE Healthcare).
RESULTS
CroS is important for the DNA damage-induced activation of SXT transfer.
To investigate the role of CroS in the biology of SXT/R391 ICEs, we first monitored the effect of its deletion or overexpression on the transfer of SXT under noninduced conditions and in the presence of the DNA-damaging agent MC. First, we observed that the basal level of SXT transfer remained unaffected by croS deletion under noninduced conditions (Fig. 1C). However, while transfer of wild-type SXT was induced more than 30-fold by MC, the ΔcroS mutant remained virtually unresponsive to MC (Fig. 1D). This phenotype could result from a polar effect of croS deletion on the expression of the downstream genes setCD, which are part of the same operon. To rule out this possibility, we carried out a complementation assay by expressing croS from its native promoter PL from pSRZ inserted as a single copy into the chromosomal gene prfC in a tandem fashion with SXT ΔcroS (Fig. 1E). pSRZ restored the 30-fold MC-dependent induction to SXT ΔcroS, thereby confirming that the lack of response to MC exhibited by the ΔcroS mutant was not due to a polar effect on the expression of setCD (Fig. 1D). Moreover, overexpression of croS from a PBAD promoter provided by pCroS led to a 10-fold reduction of SXT transfer (Fig. 1C). Altogether, these results suggest that CroS is a crucial regulator for stimulation of SXT transfer in response to DNA damage and suggest the existence of a λ-like SetR-CroS switch in the regulatory network of SXT/R391 ICEs.
Predicted structure and DNA-binding domain of CroS.
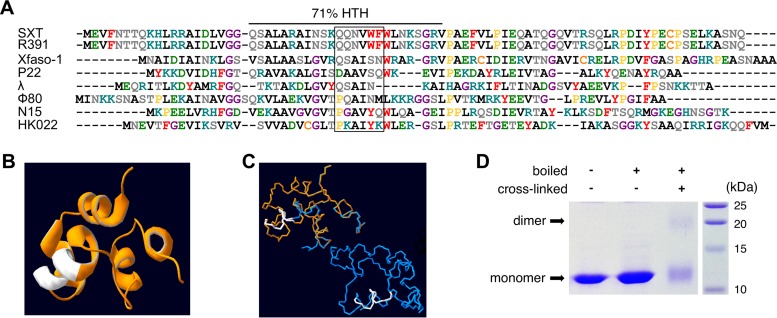
Extensive studies of many members of the Cro-like family have provided valuable insights for predicting distinctive features of newly discovered Cro-like repressors, which allowed this family to be used as a model in evolutionary studies of protein structure and DNA recognition (42, 43). CroS is predicted to contain a DNA-binding HTH domain (HTH_XRE, COG4197) (Fig. 2A). HHpred analyses revealed that CroS is 25% identical to λ Cro and 35% identical to Xfaso 1, a putative regulator encoded by a prophage of Xylella fastidiosa. Although these two homologous regulators have similar regulatory functions and are encoded by genes that are found in similar contexts, λ Cro and Xfaso 1 have been shown to be structurally different. λ Cro is a dimeric protein with a mixed β-sheet/α-helix fold, while Xfaso 1 is a monomeric all-α-helical fold protein in solution (42). A high-accuracy model of the predicted structure of CroS based on the crystal structure of Xfaso 1 was obtained with Phyre2 (44) (Fig. 2B and C). This model suggests that CroS exhibits an all-α structure and exists in solution either as a monomer or as a dimer. Despite extensive similarities, CroS does not contain the cysteine residues C42 and C55 forming the intrasubunit cystine bond observed in Xfaso 1 (42).
FIG 2.
Predicted structure and DNA-binding domain of CroS. (A) Amino acid sequence alignment of Cro-like repressors computed by Clustal Omega (72). The source ICE or phage is indicated on the left. Sequences are sorted according to phylogenetic proximity. Amino acids are color coded as follows according to their chemical properties: pink, positively charged; green, negatively charged; gray, no charge, polar, and hydrophilic; red, aromatic; black, hydrophobic; orange, cysteine; yellow, proline; purple, glycine. DNA recognition helices as described by Hall et al. (43) are indicated by a black box. The Dodd and Egan method predicted a 71% chance of an HTH motif (73). (B) Three-dimensional ribbon structure of CroS as modeled by Phyre2 with template Protein Data Bank (PDB) entry 3BD1 (Cro protein from Xfaso 1 in X. fastidiosa strain ann-1) with 99.9% confidence, 76% coverage, and 38% identity (44). The putative DNA recognition helix is shown in white. (C) Superposition of the modeled monomer of CroS (orange) and a dimer of Xfaso 1 (blue) as crystallized by Roessler et al. (42) with the predicted DNA recognition helices shown in white. (D) SDS-PAGE analysis of purified CroS protein cross-linked or not with 0.625% glutaraldehyde.
We carried out an oligomerization assay with glutaraldehyde cross-linking and observed two bands with sizes compatible to either monomeric or dimeric forms of CroS in solution (Fig. 2D), indicating that CroS can form higher-order complexes even in the absence of target DNA.
CroS acts as a repressor of setR.
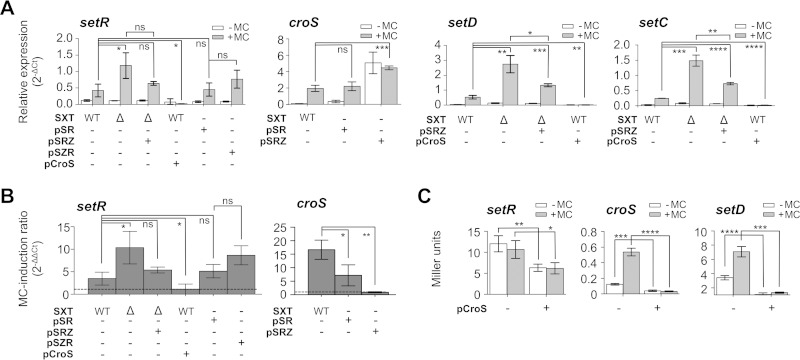
CroS similarity to other Cro-like repressors and the relative positions of croS and setR led us to hypothesize that the inability of a ΔcroS mutant to respond to MC induction was at least in part due to the lack of repression of the PR promoter that drives setR expression. To test this hypothesis, we used qRT-PCR to measure and compare the relative mRNA levels of setR in wild-type SXT and its ΔcroS mutant, with or without overexpression of croS and in the presence or absence of MC (Fig. 3A and B, setR). We observed that while setR expression increased only 3-fold upon MC induction in wild-type SXT, it increased 10-fold in the ΔcroS mutant (Fig. 3A, setR). Complementation with pSRZ expressing croS from its native promoter restored the induction of setR expression to nearly the wild-type level. In contrast, overexpression of croS abolished the MC-mediated induction of setR expression (Fig. 3B, setR). This phenotype results from croS overexpression, as control plasmid pCroS-inv, in which the ORF of croS is in the reverse orientation, did not lead to abolition of MC-dependent induction (see Fig. S1 in the supplemental material). To isolate the CroS-SetR locus from the rest of the SXT-encoded functions, we also quantified setR from pSR to mimic wild-type SXT and from pSZR to mimic SXT ΔcroS. Although the differences observed were not statistically significant in these simplified genetic contexts, MC-mediated induction of setR expression followed trends similar to those observed with complete SXT, suggesting that, besides CroS and SetR, no other ICE-encoded factor regulates setR expression (Fig. 3A and B, setR). To confirm the repression of setR expression by CroS, we also measured the β-galactosidase activity of a setR′-′lacZ fusion (pSRZ) in the presence or absence of arabinose-induced pCroS (Fig. 3C, setR). Expression of the setR′-′lacZ fusion was decreased 2-fold when croS was overexpressed. Similar results were obtained regardless of the absence or presence of MC, indicating that setR expression is not altered by MC in the absence of the SetR protein. Together, these results confirm that CroS represses setR, likely by acting on the PR promoter.
FIG 3.
CroS represses expression from PL and PR. (A) Effects of croS deletion and overexpression on setR, croS, setD, and setC mRNA levels. setR, croS, setD, and setC mRNA transcript levels in the presence or absence of MC were measured by qRT-PCR. The strains carried SXT (VB17, WT [wild type]) or its ΔcroS mutant (DPL353, Δ) and/or the simplified pSR (DPL231) construct or its pSZR (DPL241) or pSRZ (DPL263, DPL246) derivative. (B) Effects of croS deletion and overexpression on the SOS-dependent induction of setR and croS expression. The strains used are the same as those in panel A for setR and croS, but the results are shown as induction ratios obtained by ΔΔCT calculations. The level of expression without MC was subtracted from the level with MC. The dashed line indicates an MC induction ratio of 1, i.e., no difference between the induced and noninduced conditions. (C) Effect of croS overexpression on the translation of setR, croS, and setD. Shown are β-galactosidase assays with strains harboring translational lacZ fusions to setR (DPL246) and croS (DPL241) derived from pSR or to setD derived from SXT (DPL297). Each bar represents the mean and standard deviation of values obtained from at least three independent experiments. Statistical analyses of the mean values were performed with two-tailed unpaired t tests. Statistical significance is indicated as follows: ns (not significant), P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
CroS represses the promoter PL.
In λ, Cro not only represses the expression of the associated cI repressor gene but also represses the expression of lytic genes (33). In SXT, integration/excision and conjugative-transfer genes are the counterpart of λ lytic genes. Their expression is activated by the class II transcriptional activator complex SetCD (24, 26). Previously, expression of SetCD was shown to be driven from PL in the same mRNA transcript that harbors croS (30). To assess whether CroS can repress PL, we measured the relative mRNA level of transcripts containing setC and setD in cells bearing wild-type SXT or its ΔcroS mutant derivative in the presence or absence of pSRZ, pCroS, and MC (Fig. 3A, setD and setC). In an MC-induced ΔcroS mutant context, we observed that the level of mRNA containing setD and setC rose 5-fold compared to that of wild-type SXT. This enhanced expression of setCD was partially complemented by croS provided from its native promoter in pSRZ. Moreover, overexpression of croS from pCroS virtually abolished setD and setC expression (Fig. 3A, setD and setC). The repressive effect of overexpression of CroS was also observed at the translational level since the croS′-′lacZ and setD′-′lacZ fusions yielded very low β-galactosidase activities even when the cells were exposed to MC (Fig. 3C, croS and setD). Expression of croS was similar in the SXT and pSR contexts, while removal of the setR gene in pSRZ boosted the expression of croS, confirming the repressive function of SetR on PL (Fig. 3A, croS). Abolition of the MC responsiveness in pSRZ confirms the pivotal role of SetR for integration of the SOS signal (RecA*) to the regulatory network. croS expressed from its native promoter is not sufficient to completely repress PL upon MC induction. These expression assays confirm the importance of SetR for the MC-dependent induction of SXT/R391 ICE transfer and that CroS acts as a mild repressor of the PL promoter, which drives the expression of the master activator complex SetCD.
DNA-binding motif recognized by SetR.
To confirm the motif bound by SetR (30), the ∼4.8-kb nucleotide sequence overlapping s084 and attR was submitted to the motif-based sequence analysis tool Multiple EM for Motif Elicitation (MEME) (45). This analysis led to the identification of the same operators previously described for SetR located between croS and setR (OL, O1, O2, and O3), as well as the remote fifth operator O4 (Fig. 4A) (30). A motif alignment and search tool (46) analysis revealed that this motif is not found anywhere else in the SXT sequence.
SetR binding to each independent operator, including O4, was confirmed by EMSAs with purified SetR and a DNA probe corresponding to the single operator (Fig. 4B and C). We observed differential affinities of SetR for its operator sites in the following order: O4 ≈ O1 > O3 > OL > O2. To gain a better understanding of the SetR DNA-binding pattern, we mixed purified SetR with permutated radiolabeled probes, i.e., probes identical in size and partially overlapping each other, containing OL, O1, O2, and O3 (Fig. 4D). SetR binding led to the formation of one predominant complex for all of the probes tested, suggesting that it binds cooperatively to the operators. The variation in the migration distances of the protein-DNA complexes suggests that SetR binding to its operator leads to DNA bending, as shown for other CI-like repressors (47, 48). Indeed, the position of the bending angle along the length of the DNA probe modifies the overall structure of the migrating protein-DNA complex and hence its migration distance (49).
DNA-binding motif recognized by CroS.
It has been reported for numerous lambdoid bacteriophages that CI-like and cognate Cro-like repressors have operator sites in common because of similarity in their HTH DNA-binding domains (50–52). The HTH domains of CroS and SetR show 26% identity, which is higher than that of other well-studied λ-like repressor pairs such as λ and P22 (Fig. 4E), suggesting that both proteins have the same operator sites. We tested this hypothesis by carrying out EMSAs with purified CroS and DNA probes corresponding to individual SetR operator sites and confirmed that CroS independently binds to the exact same five operators (Fig. 4C). We observed a differential affinity of CroS for its operator sites in the following order: O3 > OL > O4 > O1 ≈ O2. Like SetR, purified CroS specifically binds PL and PR, as neither protein was able to shift probes containing the promoter Ps089 or Ps003 in the presence of a specific competitor DNA (annealed oligonucleotides containing either O1 or O2) (see Fig. S2 in the supplemental material). Incubation of purified CroS with permutated radiolabeled probes containing OL, O1, O2, and O3 (Fig. 4D) led to the formation of multiple shifted CroS-DNA complexes, suggesting that, unlike SetR, CroS binds multiple sites in a noncooperative manner. The large footprints observed following CroS binding to the OL, O1, and O2 operators confirm the concomitant binding of CroS to these sites (see Fig. S3 in the supplemental material). Altogether, the results of our SetR and CroS binding assays suggest that both repressors bind the same operators located in the SXT locus that contains the promoters PL and PR.
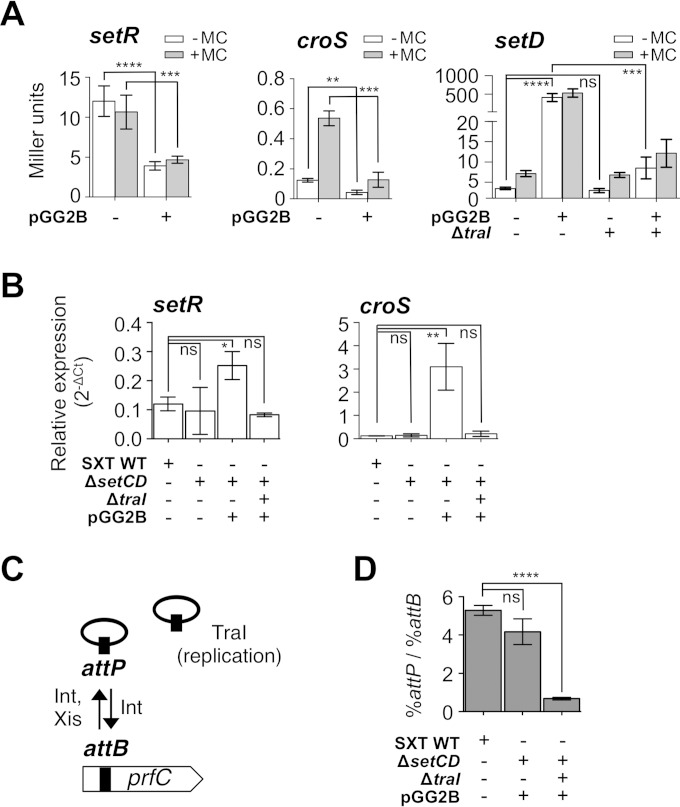
SetCD indirectly generates a positive feedback loop by activating SXT replication.
Previously published work suggests that SetCD regulates its own expression (24). To test this hypothesis, we quantified the β-galactosidase activity of a setD′-′lacZ fusion in SXT (DPL297) with or without overexpression of setCD from a PBAD promoter (pGG2B) (Fig. 5A, setD). Upon overexpression of setCD, the β-galactosidase activity of the setD′-′lacZ fusion rose by 75-fold. Although croS and setD are part of the same mRNA transcript driven by PL (Fig. 5A, croS), this drastic increase was not observed for the setR′-′lacZ and croS′-′lacZ fusions measured from pSRZ (DPL246) and pSZR (DPL241), which are integrated and locked into the chromosome. We also measured the setR and croS mRNA transcript levels in cells bearing SXT or its ΔsetCD mutant form in the absence of MC with or without pGG2B (Fig. 5B). Deletion of setCD did not significantly alter the expression levels of setR and croS. In contrast, overexpression of setCD increased both setR and croS mRNA levels, which is seemingly inconsistent with the results obtained by measuring β-galactosidase activities from pSRZ or pSZR (Fig. 5A and B). Altogether, these results suggested that setCD expression could be driven from an unidentified alternative SetCD-dependent promoter located between PL and setD. However, despite several attempts, we failed to detect any additional promoter between these two loci with 5′ rapid amplification of cDNA ends (RACE) and primer extension assays (data not shown). Furthermore, genome-wide 5′ RACE and chromatin immunoprecipitation-exonuclease experiments recently confirmed that no promoter is detectable downstream of PL and that PL itself is not a SetCD-dependent promoter (26). Another SXT-encoded factor absent from pSZR and pSRZ could explain this discrepancy.
FIG 5.
Overexpression of setCD leads to an indirect positive feedback loop boosting the expression from PL and PR. (A) Effect of SetCD on the expression of setR, croS, and setD translationally fused to lacZ measured by β-galactosidase assay in the presence or absence of MC. The strains contain pSRZ (DPL246), pSZR (DPL241), SXT setD′-′lacZ ΔsetC (DPL297), or SXT ΔtraI setD′-′lacZ ΔsetC (DPL548). Overexpression assays were carried out by expressing setCD from the arabinose-inducible PBAD promoter in pGG2B. (B) Effects of setCD deletion and overexpression on setR and croS mRNA levels. qRT-PCR assays were carried out with E. coli BW25113 carrying SXT (VB17, WT [wild type]) or its ΔsetCD or ΔtraI ΔsetCD (DPL297 and DPL548) derivative without MC induction. Overexpression assays were carried out by expressing setCD from pGG2B. (C) Representation of integration/excision and replication of SXT. (D) Measurement of SXT copy number by qRT-PCR (percent attP/percent attB with prfC as a reference) in SXT derivatives (identical to those in panels A and B) with or without overexpression of setCD. Each bar represents the mean and standard deviation of values from three independent experiments. Statistical analyses of the mean values were performed with two-tailed unpaired t tests, with the exception of the setD results in panel A, for which the logarithms of the mean values were used. Statistical significance is indicated as follows: ns (not significant), P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001.
Recent work has established that under inducing conditions, SXT/R391 ICEs replicates by a rolling-circle mechanism initiated by the relaxase TraI. This replication step is important for conjugative transfer, as well as the stability of the ICE in the cell and in the bacterial population (27). To test whether activation of traI expression by SetCD (26) is responsible for the increased setCD expression in an SXT background, we measured the β-galactosidase activity of the setD′-′lacZ fusion in a strain carrying SXT ΔtraI (DPL548) and pGG2B. We observed that deletion of traI abolished the pGG2B-induced increase in setR, croS, and setD expression (Fig. 5A and B). To confirm that traI deletion has an effect on the copy number of SXT, we measured the mean copy number of extrachromosomal circular forms of SXT per cell by establishing the ratio of the amount of attP recombination sites resulting from SXT excision to the amount of unoccupied chromosomal attB sites by real-time qPCR. In theory, each SXT excision event yields one unoccupied attB site on the chromosome and one attP site on the circular excised SXT if it does not replicate (attP/attB ratio of 1). Consistent with previous reports (27, 39), the attP/attB ratio was 5 for wild-type SXT. While this ratio was similar at 4 for SXT ΔsetD′-′lacZ/pGG2B, it decreased to 0.7 for SXT ΔtraI setD′-′lacZ/pGG2B, confirming the importance of TraI in SXT replication (Fig. 5D). Altogether, these results demonstrate that the apparent involvement of SetCD in the expression of genes in the regulatory region is not due to direct activation of PL and PR by SetCD. Instead, SetCD triggers int, xis, and traI expression, thereby driving SXT excision and replication, which increases the copy number of SXT molecules in a larger fraction of the cell population, leading to increased setR and croS-setCD mRNA levels.
DISCUSSION
SXT/R391 ICEs are major vectors of multidrug resistance dissemination across a plethora of bacterial genera encountered in clinical and environmental settings (7–9). Their highly conserved regulatory region governs the transition between the ICE quiescent and conjugative states (24, 29, 30). Here, we report several new levels of genetic regulation controlling SXT/R391 ICE behavior. Using SXT as a prototypical member of the family, we have identified and characterized the function of CroS, an ICE-encoded Cro-like repressor that serves as a key component of this genetic switch together with SetR, the main repressor of SXT/R391 ICEs (24). We also show that SetR and CroS are able to bind the O4 operator, likely consolidating the repressed state of these ICEs. Finally, expression of SetCD results in a positive feedback mechanism that is dependent on traI, which encodes the presumed relaxase of SXT/R391 ICEs. Given the high conservation of the core genes, especially those of the regulatory region of SXT/R391 ICEs (8), our findings are most likely applicable to all of the members of the family.
The SetR-CroS switch.
In λ genetic regulation, the nature of the patterns of CI and Cro binding to the OR1, OR2, and OR3 operators leads either to the activation or repression of cI expression from PRM or to the repression or derepression of cro from PR (52). The relative affinities of both repressors for each operator and their respective cell abundance, combined with intrinsic cellular stochasticity, determine the fate of the switch. The closest structural relative of SetR is the CI repressor encoded by lambdoid bacteriophage 434. The N-terminal dimer interface of phage 434 CI was shown to drive the central base preference, which defines the specific affinity of the repressor for each operator independently (53). Even though extensive thermodynamic studies are mandatory to infer quantitative relative affinities, the conditions used in binding reactions allowed us to observe variations in complex formation between the independent operators. With these as an indicators of affinity, the inferred relative affinities of SetR for the five operator sites are O4 ≈ O1 > O3 > OL > O2, which are consistent with what was observed by DNase I footprinting (30). The relative affinities of CroS for the same operators were O3 > OL > O4 > O1 ≈ O2. This binding pattern is consistent with λ Cro, which binds more tightly to OR3 that to OR1 and OR2.
In SXT, the center-to-center O1-O2 and O2-O3 spacings are 24 and 23 bp (Fig. 1B), the ideal distances for cooperative binding, given that two adjacent bound repressor dimers are located on the same side of the DNA helix (54). The OL-O1 spacing of 30 bp suggests that binding of SetR to O1 cannot yield cooperative binding to OL, the orientation of the adjacent repressors being inadequate. Given the observed affinity of SetR, and in agreement with λ regulation, the likely preferred pattern of SetR binding is to O1-O2. The organization of operator sites and promoters of the regulatory locus of SXT (Fig. 1B) suggests that SetR activates its own expression from PR when cooperatively bound to O1-O2, likely by contacting the σ70 subunit of the RNA polymerase (RNAP) (30, 55–57). Binding of SetR to O1-O2 not only triggers its own expression from PR but also blocks access of the RNAP to PL, which drives the expression of setCD and the subsequent activation of conjugative genes of SXT (26, 29, 30, 58). By analogy with λ cI regulation, SetR would repress its own expression by further binding to O3 in addition to O1-O2 when its intracellular level increases (30, 55, 56).
In our assays, the level of SetR translation appeared to be higher than that of CroS translation, suggesting that SetR protein levels are higher than CroS levels (Fig. 3C). In fact, under noninduced conditions, only 1 cell out of 11,500, on average, acts as a donor cell (frequency of exconjugant formation = 8.65 × 10−5 ± 9.88 × 10−6 exconjugant CFU/donor CFU, n = 3), suggesting that the quantity of croS mRNA transcripts per cell is lower than the quantity of SetR mRNA transcripts in most cells of the population. The primer efficiency bias in qRT-PCR precludes us from directly comparing the amounts of mRNA containing setR and croS (Fig. 3A). CroS is presumably dominant in only 1 out of 11,500 cells transitioning from the quiescent to the transfer-proficient state of SXT. We have established that CroS is mandatory for SOS induction of SXT transfer, which is consistent with λ regulation (33). CroS represses PL and PR when SetR repression is alleviated. This double repression likely delays the recovery of the SetR pool and displaces the balance toward PL expression long enough for activation of conjugative transfer to occur properly. In our assays, PL repression by CroS was observed only upon the overexpression of croS, not when croS was expressed from its native promoter (Fig. 3A, croS). This observation suggests that CroS is a weak repressor of PL. In wild-type induced SXT, conditional replication mediated by TraI (27) increases the croS mRNA level (Fig. 5B), which could allow partial repression of PL. CroS would then act at PL to mitigate the expression of setCD, which has been shown to be toxic when overexpressed (24, 59).
The closest structural relative of CroS is Xfaso 1, a Cro-like repressor whose structure has been determined (42). To our knowledge, experimental data supporting the functionality of Xfaso 1 remain limited (60). Secondary-structure predictions suggest that Xfaso 1 and CroS have an all-α-helix structure, as does the well-studied Cro protein encoded by lambdoid bacteriophage P22 isolated from Salmonella (Fig. 2B) (61). The genetic regulation network of P22 is more complex than that of λ, as P22 codes for additional repressors to maintain lysogeny (62). Despite their structural differences (all-α versus α-β folds), Cro proteins likely have highly similar interactions with the PR and PRM promoters in P22 and λ (63, 64). Indeed, opposite affinities of Cro-like (OR3 > OR1 > OR2) and CI-like (OR1 > OR2 > OR3) repressors for each operator were observed in both phages. Consistent with our results, the affinity of the Cro-like repressors for each operator exhibits less variation than the affinity of CI-like repressors (63). Study of many natural and synthetic versions of the λ genetic network led to the conclusion that while most components are not essential, altogether they contribute to the overall efficiency and stability of the system (65). We showed that the role of CroS is particularly important in SXT regulation, as SXT lacks two components that are important for noise reduction and stabilization of the λ system, the activator CII and two additional operators in the OL region.
Pleiotropic effect of SetCD overexpression.
In λ and in SXT, the cro genes are cotranscribed with genes coding for transcriptional activators, cII in λ and setCD in SXT. These activators are not related. Instead, SetCD is related to FlhCD and AcaCD, the master activators of flagellum synthesis in Gram-negative bacteria (66) and of conjugative transfer functions of IncA/C plasmids (67), respectively. One role of CII is the activation of cI by acting at the promoter PRE (52). The intracellular level of CII ultimately makes the decision between lysis and lysogeny. Similarly, our results show that setR mRNA levels rise upon setCD overexpression (Fig. 5B, setR). However, our results revealed that this is not due to direct activation of PR by SetCD. Instead, expression of setCD markedly enhances int, xis, and traI mRNA levels, presumably leading to ICE excision and conjugation-associated replication in a greater portion of the cell population (26, 27). The elevated setR and croS mRNA levels observed in SetCD-induced bacterial cultures (Fig. 5B) also support this scenario. Paradoxically, overexpression of setCD lowered the β-galactosidase activity of the setR-lacZ and croS-lacZ fusions constructed in the minimal pSR construct (without the SXT backbone) (Fig. 5A). This collateral repressive effect of SetCD overexpression could be caused by illegitimate SetCD DNA binding. In addition to being overexpressed, none of the SetCD target promoters was present in such cells to titrate SetCD excess. Alternatively, overexpression of SetCD might reduce lacZ expression levels through nonspecific effects on cellular growth rates.
The level of translation was higher for setD than for croS (Fig. 3C). This difference could be attributable to the spacing between their respective Shine-Dalgarno sequences and start codons, which is closer to the optimum for setD than for croS (68).
A revised model of SXT early regulation.
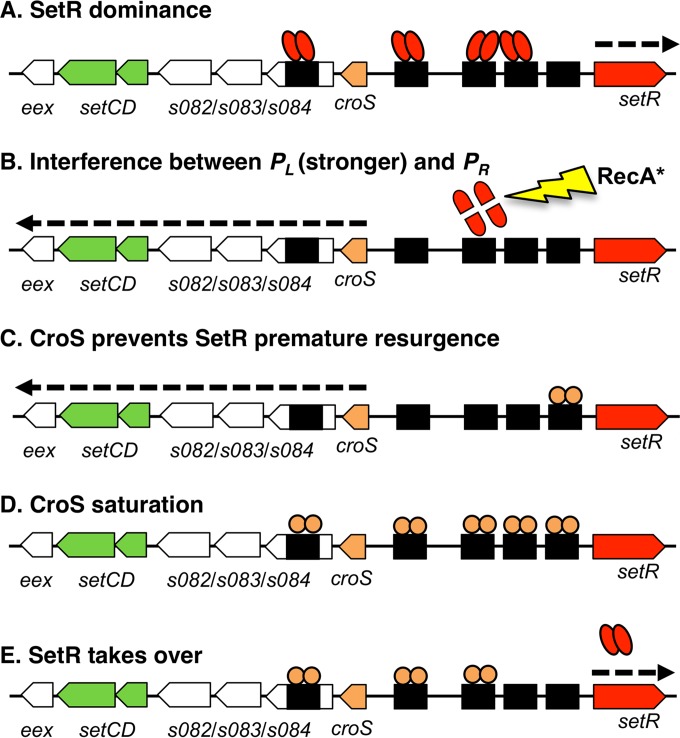
Since the preferred substrate for RecA*-activated self-cleavage of phage 434 CI is the DNA-bound dimer (69), one could argue that, upon DNA damage, the cooperatively bound SetR-DNA complex becomes the substrate of RecA*, unleashing SetR's latent autoproteolysis activity (29). Release of the operators alleviates the repression exerted by SetR on PL, allowing croS and setCD expression (Fig. 6B). CroS, the first translated protein, would bind to O3 to prevent premature resurgence of the SetR pool (Fig. 6C). CroS would then bind in a noncooperative fashion to OL, O1, O2, and O4, thereby preventing adverse overexpression of setCD (Fig. 6D). This repression by CroS would allow exclusion of SetR's strong binding to the intergenic region between croS and setR long enough to observe the SOS-mediated induction of transfer. At some point, CroS would vacate OL, O1, O2, and O3, allowing SetR to take over the intergenic region and reset the quiescent state (Fig. 6E). Our results show that both CroS and SetR are able to bind to O4. Their binding could generate a roadblock that compromises the transcription of the whole croS-eex operon driven from PL beyond O4. Alternatively, because SetR is able to bind cooperatively, its binding could lead to the formation of a long-range loop between a SetR-bound O4 and the intergenic region between croS and setR. Such a role is unlikely for CroS, which lacks the C-terminal domain mediating self-assembly, cooperative DNA binding, and concomitant loop formation ability (70, 71). This long-range loop or roadblock at O4 mediated by SetR could stabilize the SetR-dominant state. Additional experiments are required to decipher the regulatory mechanism(s) involving the O4 operator site.
FIG 6.
Representation of proposed key intermediate states (A to E) of O4, OL, O1, O2, and O3 binding by SetR or CroS. Panel A is a presumed intermediate of the dominant quiescent behavior, while panels B to E are most likely transient. The dotted arrow indicates active transcription.
The genetic network of SXT/R391 ICEs elegantly sets at play regulatory components reminiscent of lambdoid phages (SetR-CroS epigenetic switch) and bacterial motility (SetCD activation) to rule the propagation of DNA material. The diversity of bacterial hosts in which SXT/R391 ICEs dwell suggests that the regulation system they encode was selected for minimal interactions with host genetic networks apart from their induction stimulated by RecA*. SXT/R391 ICEs take advantage of a host regulatory stress response that can be caused by DNA-targeting antibiotics and UV light to trigger the dissemination of multidrug resistance and other adaptive functions through Gammaproteobacteria populations.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by a Discovery Grant and Discovery Acceleration Supplement from the Natural Sciences and Engineering Council of Canada (326810 and 412288 to V.B.). V.B. holds a Canada Research Chair in molecular bacterial genetics. D.P.-L. was supported by scholarships from the Fonds de Recherche du Québec.
We are grateful to C. Jouogo Noumsi, V. Côté-Breton, M. Paquette-D'Avignon, D. Ceccarelli, and the Centre de Calcul Scientifique of the Université de Sherbrooke for their technical assistance and to N. Carraro, A. Lavigueur, and J. P. Brousseau for their insightful comments on the manuscript.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JB.00638-15.
REFERENCES
- 1.Ramamurthy T, Garg S, Sharma R, Bhattacharya SK, Nair GB, Shimada T, Takeda T, Karasawa T, Kurazano H, Pal A, et al. . 1993. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet 341:703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
- 2.Kaper JB, Morris JG Jr, Levine MM. 1995. Cholera. Clin Microbiol Rev 8:48–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Waldor MK, Tschape H, Mekalanos JJ. 1996. A new type of conjugative transposon encodes resistance to sulfamethoxazole, trimethoprim, and streptomycin in Vibrio cholerae O139. J Bacteriol 178:4157–4165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wozniak RA, Waldor MK. 2010. Integrative and conjugative elements: mosaic mobile genetic elements enabling dynamic lateral gene flow. Nat Rev Microbiol 8:552–563. doi: 10.1038/nrmicro2382. [DOI] [PubMed] [Google Scholar]
- 5.Burrus V, Pavlovic G, Decaris B, Guedon G. 2002. Conjugative transposons: the tip of the iceberg. Mol Microbiol 46:601–610. doi: 10.1046/j.1365-2958.2002.03191.x. [DOI] [PubMed] [Google Scholar]
- 6.Hochhut B, Waldor MK. 1999. Site-specific integration of the conjugal Vibrio cholerae SXT element into prfC. Mol Microbiol 32:99–110. doi: 10.1046/j.1365-2958.1999.01330.x. [DOI] [PubMed] [Google Scholar]
- 7.Spagnoletti M, Ceccarelli D, Rieux A, Fondi M, Taviani E, Fani R, Colombo MM, Colwell RR, Balloux F. 2014. Acquisition and evolution of SXT-R391 integrative conjugative elements in the seventh-pandemic Vibrio cholerae lineage. mBio 5:e01356-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wozniak RA, Fouts DE, Spagnoletti M, Colombo MM, Ceccarelli D, Garriss G, Dery C, Burrus V, Waldor MK. 2009. Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet 5:e1000786. doi: 10.1371/journal.pgen.1000786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rodríguez-Blanco A, Lemos ML, Osorio CR. 2012. Integrating conjugative elements as vectors of antibiotic, mercury, and quaternary ammonium compound resistance in marine aquaculture environments. Antimicrob Agents Chemother 56:2619–2626. doi: 10.1128/AAC.05997-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Osorio CR, Marrero J, Wozniak RA, Lemos ML, Burrus V, Waldor MK. 2008. Genomic and functional analysis of ICEPdaSpaI, a fish-pathogen-derived SXT-related integrating conjugative element that can mobilize a virulence plasmid. J Bacteriol 190:3353–3361. doi: 10.1128/JB.00109-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harada S, Ishii Y, Saga T, Tateda K, Yamaguchi K. 2010. Chromosomally encoded blaCMY-2 located on a novel SXT/R391-related integrating conjugative element in a Proteus mirabilis clinical isolate. Antimicrob Agents Chemother 54:3545–3550. doi: 10.1128/AAC.00111-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.López-Pérez M, Gonzaga A, Rodríguez-Valera F. 2013. Genomic diversity of “deep ecotype” Alteromonas macleodii isolates: evidence for pan-Mediterranean clonal frames. Genome Biol Evol 5:1220–1232. doi: 10.1093/gbe/evt089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badhai J, Kumari P, Krishnan P, Ramamurthy T, Das SK. 2013. Presence of SXT integrating conjugative element in marine bacteria isolated from the mucus of the coral Fungia echinata from Andaman Sea. FEMS Microbiol Lett 338:118–123. doi: 10.1111/1574-6968.12033. [DOI] [PubMed] [Google Scholar]
- 14.Pembroke JT, Piterina AV. 2006. A novel ICE in the genome of Shewanella putrefaciens W3-18-1: comparison with the SXT/R391 ICE-like elements. FEMS Microbiol Lett 264:80–88. doi: 10.1111/j.1574-6968.2006.00452.x. [DOI] [PubMed] [Google Scholar]
- 15.Coetzee JN, Datta N, Hedges RW. 1972. R factors from Proteus rettgeri. J Gen Microbiol 72:543–552. doi: 10.1099/00221287-72-3-543. [DOI] [PubMed] [Google Scholar]
- 16.Beaber JW, Burrus V, Hochhut B, Waldor MK. 2002. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell Mol Life Sci 59:2065–2070. doi: 10.1007/s000180200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Peters SE, Hobman JL, Strike P, Ritchie DA. 1991. Novel mercury resistance determinants carried by IncJ plasmids pMERPH and R391. Mol Gen Genet 228:294–299. [DOI] [PubMed] [Google Scholar]
- 18.Balado M, Lemos ML, Osorio CR. 2013. Integrating conjugative elements of the SXT/R391 family from fish-isolated vibrios encode restriction-modification systems that confer resistance to bacteriophages. FEMS Microbiol Ecol 83:457–467. doi: 10.1111/1574-6941.12007. [DOI] [PubMed] [Google Scholar]
- 19.Bordeleau E, Brouillette E, Robichaud N, Burrus V. 2010. Beyond antibiotic resistance: integrating conjugative elements of the SXT/R391 family that encode novel diguanylate cyclases participate to c-di-GMP signalling in Vibrio cholerae. Environ Microbiol 12:510–523. doi: 10.1111/j.1462-2920.2009.02094.x. [DOI] [PubMed] [Google Scholar]
- 20.Garriss G, Waldor MK, Burrus V. 2009. Mobile antibiotic resistance encoding elements promote their own diversity. PLoS Genet 5:e1000775. doi: 10.1371/journal.pgen.1000775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kulaeva OI, Wootton JC, Levine AS, Woodgate R. 1995. Characterization of the umu-complementing operon from R391. J Bacteriol 177:2737–2743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hochhut B, Marrero J, Waldor MK. 2000. Mobilization of plasmids and chromosomal DNA mediated by the SXT element, a constin found in Vibrio cholerae O139. J Bacteriol 182:2043–2047. doi: 10.1128/JB.182.7.2043-2047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Daccord A, Ceccarelli D, Burrus V. 2010. Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol Microbiol 78:576–588. doi: 10.1111/j.1365-2958.2010.07364.x. [DOI] [PubMed] [Google Scholar]
- 24.Beaber JW, Hochhut B, Waldor MK. 2002. Genomic and functional analyses of SXT, an integrating antibiotic resistance gene transfer element derived from Vibrio cholerae. J Bacteriol 184:4259–4269. doi: 10.1128/JB.184.15.4259-4269.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marrero J, Waldor MK. 2005. Interactions between inner membrane proteins in donor and recipient cells limit conjugal DNA transfer. Dev Cell 8:963–970. doi: 10.1016/j.devcel.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 26.Poulin-Laprade D, Matteau D, Jacques PE, Rodrigue S, Burrus V. 2015. Transfer activation of SXT/R391 integrative and conjugative elements: unraveling the SetCD regulon. Nucleic Acids Res 43:2045–2056. doi: 10.1093/nar/gkv071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Carraro N, Poulin D, Burrus V. 2015. Replication and active partition of integrative and conjugative elements (ICEs) of the SXT/R391 family: the line between ICEs and conjugative plasmids is getting thinner. PLoS Genet 11:e1005298. doi: 10.1371/journal.pgen.1005298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Garriss G, Poulin-Laprade D, Burrus V. 2013. DNA-damaging agents induce the RecA-independent homologous recombination functions of integrating conjugative elements of the SXT/R391 family. J Bacteriol 195:1991–2003. doi: 10.1128/JB.02090-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beaber JW, Hochhut B, Waldor MK. 2004. SOS response promotes horizontal dissemination of antibiotic resistance genes. Nature 427:72–74. doi: 10.1038/nature02241. [DOI] [PubMed] [Google Scholar]
- 30.Beaber JW, Waldor MK. 2004. Identification of operators and promoters that control SXT conjugative transfer. J Bacteriol 186:5945–5949. doi: 10.1128/JB.186.17.5945-5949.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Svenningsen SL, Costantino N, Court DL, Adhya S. 2005. On the role of Cro in lambda prophage induction. Proc Natl Acad Sci U S A 102:4465–4469. doi: 10.1073/pnas.0409839102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnson A, Meyer BJ, Ptashne M. 1978. Mechanism of action of the cro protein of bacteriophage lambda. Proc Natl Acad Sci U S A 75:1783–1787. doi: 10.1073/pnas.75.4.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schubert RA, Dodd IB, Egan JB, Shearwin KE. 2007. Cro's role in the CI Cro bistable switch is critical for λ's transition from lysogeny to lytic development. Genes Dev 21:2461–2472. doi: 10.1101/gad.1584907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burrus V, Waldor MK. 2004. Formation of SXT tandem arrays and SXT-R391 hybrids. J Bacteriol 186:2636–2645. doi: 10.1128/JB.186.9.2636-2645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dower WJ, Miller JF, Ragsdale CW. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res 16:6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demarre G, Guerout AM, Matsumoto-Mashimo C, Rowe-Magnus DA, Marliere P, Mazel D. 2005. A new family of mobilizable suicide plasmids based on broad host range R388 plasmid (IncW) and RP4 plasmid (IncPalpha) conjugative machineries and their cognate Escherichia coli host strains. Res Microbiol 156:245–255. doi: 10.1016/j.resmic.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 37.Datsenko KA, Wanner BL. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Daccord A, Mursell M, Poulin-Laprade D, Burrus V. 2012. Dynamics of the SetCD-regulated integration and excision of genomic islands mobilized by integrating conjugative elements of the SXT/R391 family. J Bacteriol 194:5794–5802. doi: 10.1128/JB.01093-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrus V, Waldor MK. 2003. Control of SXT integration and excision. J Bacteriol 185:5045–5054. doi: 10.1128/JB.185.17.5045-5054.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miller JF. 1992. A short course in bacterial genetics. Cold Spring Harbor Laboratory Press, Plainview, NY. [Google Scholar]
- 41.Pradervand N, Delavat F, Sulser S, Miyazaki R, van der Meer JR. 2014. The TetR-type MfsR protein of the integrative and conjugative element (ICE) ICEclc controls both a putative efflux system and initiation of ICE transfer. J Bacteriol 196:3971–3979. doi: 10.1128/JB.02129-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Roessler CG, Hall BM, Anderson WJ, Ingram WM, Roberts SA, Montfort WR, Cordes MH. 2008. Transitive homology-guided structural studies lead to discovery of Cro proteins with 40% sequence identity but different folds. Proc Natl Acad Sci U S A 105:2343–2348. doi: 10.1073/pnas.0711589105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hall BM, Lefevre KR, Cordes MH. 2005. Sequence correlations between Cro recognition helices and cognate O(R) consensus half-sites suggest conserved rules of protein-DNA recognition. J Mol Biol 350:667–681. doi: 10.1016/j.jmb.2005.05.025. [DOI] [PubMed] [Google Scholar]
- 44.Kelley LA, Sternberg MJ. 2009. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc 4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 45.Bailey TL, Elkan C. 1994. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc Int Conf Intell Syst Mol Biol 2:28–36. [PubMed] [Google Scholar]
- 46.Bailey TL, Gribskov M. 1998. Combining evidence using p-values: application to sequence homology searches. Bioinformatics 14:48–54. doi: 10.1093/bioinformatics/14.1.48. [DOI] [PubMed] [Google Scholar]
- 47.Griffith J, Hochschild A, Ptashne M. 1986. DNA loops induced by cooperative binding of lambda repressor. Nature 322:750–752. doi: 10.1038/322750a0. [DOI] [PubMed] [Google Scholar]
- 48.Koudelka GB. 1991. Bending of synthetic bacteriophage 434 operators by bacteriophage 434 proteins. Nucleic Acids Res 19:4115–4119. doi: 10.1093/nar/19.15.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim J, Zwieb C, Wu C, Adhya S. 1989. Bending of DNA by gene-regulatory proteins: construction and use of a DNA bending vector. Gene 85:15–23. doi: 10.1016/0378-1119(89)90459-9. [DOI] [PubMed] [Google Scholar]
- 50.Koudelka GB, Lam CY. 1993. Differential recognition of OR1 and OR3 by bacteriophage 434 repressor and Cro. J Biol Chem 268:23812–23817. [PubMed] [Google Scholar]
- 51.Sauer RT, Yocum RR, Doolittle RF, Lewis M, Pabo CO. 1982. Homology among DNA-binding proteins suggests use of a conserved super-secondary structure. Nature 298:447–451. doi: 10.1038/298447a0. [DOI] [PubMed] [Google Scholar]
- 52.Ptashne M. 2004. A genetic switch: phage lambda revisited, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 53.Koudelka GB. 1998. Recognition of DNA structure by 434 repressor. Nucleic Acids Res 26:669–675. doi: 10.1093/nar/26.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mao C, Carlson NG, Little JW. 1994. Cooperative DNA-protein interactions. Effects of changing the spacing between adjacent binding sites. J Mol Biol 235:532–544. [DOI] [PubMed] [Google Scholar]
- 55.Li M, Moyle H, Susskind MM. 1994. Target of the transcriptional activation function of phage lambda cI protein. Science 263:75–77. doi: 10.1126/science.8272867. [DOI] [PubMed] [Google Scholar]
- 56.Xu J, Koudelka GB. 2000. DNA sequence requirements for the activation of 434 P(RM) transcription by 434 repressor. DNA Cell Biol 19:621–630. doi: 10.1089/104454900750019380. [DOI] [PubMed] [Google Scholar]
- 57.Kuldell N, Hochschild A. 1994. Amino acid substitutions in the −35 recognition motif of sigma 70 that result in defects in phage lambda repressor-stimulated transcription. J Bacteriol 176:2991–2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Meyer BJ, Maurer R, Ptashne M. 1980. Gene regulation at the right operator (OR) of bacteriophage lambda. II. OR1, OR2, and OR3: their roles in mediating the effects of repressor and cro. J Mol Biol 139:163–194. [DOI] [PubMed] [Google Scholar]
- 59.Armshaw P, Pembroke JT. 2013. Generation and analysis of an ICE R391 deletion library identifies genes involved in the element encoded UV-inducible cell-sensitising function. FEMS Microbiol Lett 342:45–53. doi: 10.1111/1574-6968.12107. [DOI] [PubMed] [Google Scholar]
- 60.de Mello Varani A, Souza RC, Nakaya HI, de Lima WC, Paula de Almeida LG, Kitajima EW, Chen J, Civerolo E, Vasconcelos AT, Van Sluys MA. 2008. Origins of the Xylella fastidiosa prophage-like regions and their impact in genome differentiation. PLoS One 3:e4059. doi: 10.1371/journal.pone.0004059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Newlove T, Konieczka JH, Cordes MH. 2004. Secondary structure switching in Cro protein evolution. Structure 12:569–581. doi: 10.1016/j.str.2004.02.024. [DOI] [PubMed] [Google Scholar]
- 62.Susskind MM, Botstein D. 1978. Molecular genetics of bacteriophage P22. Microbiol Rev 42:385–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Poteete AR, Hehir K, Sauer RT. 1986. Bacteriophage P22 Cro protein: sequence, purification, and properties. Biochemistry 25:251–256. doi: 10.1021/bi00349a035. [DOI] [PubMed] [Google Scholar]
- 64.Poteete AR, Ptashne M. 1982. Control of transcription by the bacteriophage P22 repressor. J Mol Biol 157:21–48. doi: 10.1016/0022-2836(82)90511-3. [DOI] [PubMed] [Google Scholar]
- 65.Little JW. 2010. Evolution of complex gene regulatory circuits by addition of refinements. Curr Biol 20:R724-734. doi: 10.1016/j.cub.2010.06.028. [DOI] [PubMed] [Google Scholar]
- 66.Soutourina OA, Bertin PN. 2003. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev 27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 67.Carraro N, Matteau D, Luo P, Rodrigue S, Burrus V. 2014. The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet 10:e1004714. doi: 10.1371/journal.pgen.1004714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen H, Bjerknes M, Kumar R, Jay E. 1994. Determination of the optimal aligned spacing between the Shine-Dalgarno sequence and the translation initiation codon of Escherichia coli mRNAs. Nucleic Acids Res 22:4953–4957. doi: 10.1093/nar/22.23.4953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Pawlowski DR, Koudelka GB. 2004. The preferred substrate for RecA-mediated cleavage of bacteriophage 434 repressor is the DNA-bound dimer. J Bacteriol 186:1–7. doi: 10.1128/JB.186.1.1-7.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Bell CE, Frescura P, Hochschild A, Lewis M. 2000. Crystal structure of the lambda repressor C-terminal domain provides a model for cooperative operator binding. Cell 101:801–811. doi: 10.1016/S0092-8674(00)80891-0. [DOI] [PubMed] [Google Scholar]
- 71.Bell CE, Lewis M. 2001. Crystal structure of the lambda repressor C-terminal domain octamer. J Mol Biol 314:1127–1136. doi: 10.1006/jmbi.2000.5196. [DOI] [PubMed] [Google Scholar]
- 72.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, López R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. 2011. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Dodd IB, Egan JB. 1990. Improved detection of helix-turn-helix DNA-binding motifs in protein sequences. Nucleic Acids Res 18:5019–5026. doi: 10.1093/nar/18.17.5019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Marchler-Bauer A, Zheng C, Chitsaz F, Derbyshire MK, Geer LY, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Lanczycki CJ, Lu F, Lu S, Marchler GH, Song JS, Thanki N, Yamashita RA, Zhang D, Bryant SH. 2013. CDD: conserved domains and protein three-dimensional structure. Nucleic Acids Res 41:D348-352. doi: 10.1093/nar/gks1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Harrison SC, Aggarwal AK. 1990. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem 59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- 76.Singer M, Baker TA, Schnitzler G, Deischel SM, Goel M, Dove W, Jaacks KJ, Grossman AD, Erickson JW, Gross CA. 1989. A collection of strains containing genetically linked alternating antibiotic resistance elements for genetic mapping of Escherichia coli. Microbiol Rev 53:1–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grenier F, Matteau D, Baby V, Rodrigue S. 2014. Complete genome sequence of Escherichia coli BW25113. Genome Announc 2:e01038-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ceccarelli D, Daccord A, Rene M, Burrus V. 2008. Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J Bacteriol 190:5328–5338. doi: 10.1128/JB.00150-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guzman LM, Belin D, Carson MJ, Beckwith J. 1995. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol 177:4121–4130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Miller H. 1987. Practical aspects of preparing phage and plasmid DNA: growth, maintenance, and storage of bacteria and bacteriophage. Methods Enzymol 152:145–170. doi: 10.1016/0076-6879(87)52016-X. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.