Abstract
Based on bacterial genomic data, we developed a one-step multiplex PCR assay to identify Salmonella and simultaneously differentiate the two invasive avian-adapted S. enterica serovar Gallinarum biotypes Gallinarum and Pullorum, and the most frequent, specific, and asymptomatic colonizers of chickens, serovars Enteritidis, Heidelberg, and Kentucky.
TEXT
Strains of most Salmonella serovars are zoonotic. Approximately 90% of human salmonellosis is caused by ingestion of contaminated food products of animal or plant origin (1). With >19,000 reported cases in the United States in 2013, Salmonella remains the most frequently isolated bacterial food pathogen, as determined by the surveillance network FoodNet, which pools the data of 10 U.S. monitoring sites (2). In parallel with the rise in poultry consumption over the years in the United States, the commercial poultry industry has grown impressively, reaching >9 billion raised and processed broiler chickens and production of >77 billion table eggs per year, as indicated for 2009 (3). Salmonella is a frequent asymptomatic intestinal colonizer of poultry. Stress or underlying diseases in young birds create optimal conditions for productive horizontal transmission of Salmonella sp. Data from the USDA-FSIS suggest that every fourth raw chicken part is likely contaminated with Salmonella (2). Moreover, major Salmonella serovars can spread to reproductive organs, leading to vertical transfer of the bacteria and egg-related salmonellosis (4, 5). Accordingly, consumption of poultry and egg represents a significant source of Salmonella infections in the United States.
Four Salmonella serovars are of particular concern to the poultry industry, namely, Enteritidis, Heidelberg, Kentucky, and Gallinarum (6). S. Gallinarum is an invasive agent of chicken salmonellosis that results in high mortality and morbidity, with biotype Pullorum (S. Pullorum), which causes white diarrhea in young chickens (pullorum disease), and biotype Gallinarum, which is responsible for fowl typhoid (7). Although this serovar remains endemic in many countries, it has essentially been eradicated through culling programs in the domestic fowl industry of the United States and several other developed countries. S. Gallinarum can colonize and/or cause disease in various domestic and wild birds, which might explain its occasional detection in backyard birds of developed countries (8). In recent years, S. Enteritidis became the most frequently isolated serovar in poultry and from food-borne outbreaks linked to poultry products in developed countries (9). This serovar was suggested to have filled the ecological niche vacated by the eradicated S. Gallinarum biotypes Pullorum and Gallinarum (10). Lately, S. Heidelberg has become another major serovar responsible for food-borne infections from poultry products (11, 12) and one of the most common serovars obtained from nonclinical chicken isolates (9, 13, 14). S. Kentucky is the most common serotype isolated from chickens and the second most common serotype found among retail chicken products in the United States. It has rarely been reported in human cases in North America (15, 16), although this may change with the worldwide spread of the ciprofloxacin-resistant ST198 (17).
Here, we describe a simple one-step multiplex PCR method to identify major chicken S. enterica subsp. enterica serovars. The approach was based on designing primers that specifically amplify unique sets of Salmonella spp. and serovar-associated DNA sequences in one PCR tube (Table 1), taking advantage of 3,161 available Salmonella genomes, including strains from the serovars Enteritidis (369 genomes), Heidelberg (154), Kentucky (63), and Gallinarum (8 biotype Pullorum and 4 biotype Gallinarum) and of 2,563 genomes from 104 other serovars (http://www.ncbi.nlm.nih.gov/genome/genomes/152?). The desired specificities were checked by using BLAST (NCBI, nonredundant nucleotide collection). Strains of the Salmonella genus and Gallinarum biotypes were identified by primers for differently conserved DNA segments in their bcf and ste fimbrial usher genes, respectively (20). Specific primers for serovar Gallinarum biotype Gallinarum were made by taking advantage of a deletion of 4 nucleotides in steB of biotype Pullorum. Other specific DNA signatures served as primer targets to separate serovars Enteritidis, Heidelberg, and Kentucky. Briefly, for the multiplex PCR, pure template DNA (1 to 5 ng per reaction; MagNA Pure LC DNA isolation kit III; Roche Life Sci., Indianapolis, IN) or crude DNA (∼75 ng per reaction, from bacterial suspensions boiled for 5 min, 107 CFU/μl dH2O, using 1 μl of supernatant after centrifugation) was amplified with Taq DNA polymerase and a final concentration of 1.5 mM Mg2+ (Choice-Taq Blue; Denville Sci., Inc., South Plainfield, NJ) using standard protocols. The PCR (25 cycles with an annealing temperature of 56°C) was performed with a Hybaid thermal cycler (Thermo Fisher Sci., Waltham, MA). The specificity and compatibility of the primer sets in a multiplex PCR were assessed using genomic DNA from 128 Salmonella strains that included a total of 34 different serovars, as well as from 3 Escherichia coli and 2 Yersinia spp. as negative-control strains (see Table S1 in the supplemental material).
TABLE 1.
List of primers and concentrations used for PCR, with targeted DNA and amplicon sizes
| Primer | Sequence (5′ to 3′) | Final primer concn (pmol/ml) | Targeted gene or locusa | Targeted DNA (species, serovar) | Amplicon size (bp) | Accession no. and nucleotide segment | Reference |
|---|---|---|---|---|---|---|---|
| bcfC-F | GGGTGGGCGGAAAACTATTTC | 0.6 | bcfC | S. enterica | 993 | AM933172 | This study |
| bcfC-R | CGGCACGGCGGAATAGAGCAC | 25665–26657 | |||||
| heli-F | ACAGCCCGCTGTTTAATGGTG | 2 | ORF (predicted helicase) | Heidelberg | 782 | CP005995 | This study |
| heli-R | CGCGTAATCGAGTAGTTGCC | 3226024–3226805 | |||||
| steB-F | TGTCGACTGGGACCCGCCCGCCCGC | 2 | steB | Gallinarum biotype Gallinarumb | 636 | AM933173 | 18 |
| steB-R | CCATCTTGTAGCGCACCAT | 2976016–2976651 | |||||
| rhs-F | TCGTTTACGGCATTACACAAGTA | 2.6 | rhs locus | Gallinarum | 402 | AM933173 | This study |
| rhs-R | CAAACCCAGAGCCAATCTTATCT | 334109-334510 | |||||
| sdf-F | TGTGTTTTATCTGATGCAAGAG | 2.6 | sdf locus | Enteritidis | 293 | AF370716 | 19 |
| sdf-R | CGTTCTTCTGGTACTTCAGATGAC | 4950–5242 | |||||
| gly-F | TTCCAATTGAAACGAGTGCGG | 2.6 | ORF (putative membrane protein) | Kentucky | 170 | ABEI01000007 | This study |
| gly-R | ACTAACCGCTTGGGTTGTTGCTGT | 116981–117150 |
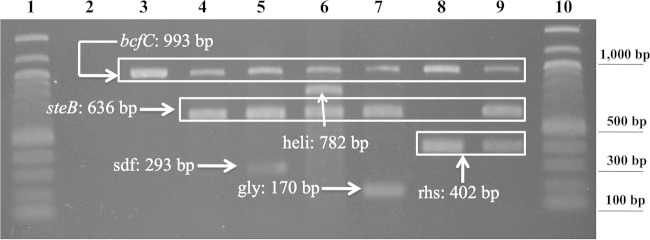
Agarose gel electrophoresis profiles for the different amplicon sets are shown with representative strains in Fig. 1, and the results for all the strains are listed in Table 2. All of the Salmonella strains were recognized as such, as were strains of the Gallinarum biotypes and the Enteritidis, Heidelberg, and Kentucky serovars. Thus, the obtained experimental results were in agreement with the genomic information used for the primer designs and validated the proposed identification of S. enterica and the serovar/biotype differentiation among major chicken isolates.
FIG 1.
Agarose gel (1.5%) of multiplex PCR amplicons from different bacterial strains. Representative gel from three comparable experiments. Lanes 1 and 10, 100-bp DNA ladder (NEB, Ipswich, MA); lane 2, Escherichia coli (DH5a, negative control); lane 3, S. enterica group 2, according to Table 2; lane 4, S. enterica group 1, according to Table 2; lane 5, S. enterica serovar Enteritidis; lane 6, S. enterica serovar Heidelberg; lane 7, S. enterica serovar Kentucky; lane 8, S. enterica serovar Gallinarum biotype Pullorum; lane 9, S. enterica serovar Gallinarum biotype Gallinarum.
TABLE 2.
Bacterial strains used to confirm the specificity of the multiplex PCR assay
| Salmonella enterica serovar and biotypea | Multiplex PCR positive for: |
|||||
|---|---|---|---|---|---|---|
| bcfC | heli | steB | rhs | sdf | gly | |
| Heidelberg (2) | + | + | + | − | − | − |
| Enteritidis (11) | + | − | + | − | + | − |
| Kentucky (4) | + | − | + | − | − | + |
| Gallinarum biotype Gallinarum (16) | + | − | + | + | − | − |
| Gallinarum biotype Pullorum (7) | + | − | − | + | − | − |
| Others | ||||||
| Group 1(68)b | + | − | + | − | − | − |
| Group 2 (20)c | + | − | − | − | − | − |
| Non-Salmonella strains (5)d | − | − | − | − | − | − |
Number of strains given in parentheses (see Table S1 in the supplemental material).
Other S. enterica serovars (group 1) that have the same PCR profile: Paratyphi A (4 isolates), Paratyphi B var. Java (1), Agona (4), Abortusequi (2), Abortusovis (2), Saintpaul (3), Stanleyville (1), Typhisuis (2), Braenderup (5), Choleraesuis (24), Ohio (1), Thompson (1), Hadar (2), Muenchen (2), Newport (6), Berta (2), Dublin (2), Panama (1), Typhi (1), Agoueve (1), and Cerro (1).
Other S. enterica serovars (group 2) that have the same PCR profile: Schwarzengrund (3 isolates), Typhimurium (2), Bareilly (1), Hartford (1), Montevideo (2), Oranienburg (3), Javiana (6), Mississippi (1), and Pomona (1).
Three E. coli and two Yersinia strains.
Routine screening of flocks for the presence of Salmonella can be done by conventional serology, which is expensive, time consuming, and labor intensive. Based on the restricted number of major serovars found in chickens, extensive molecular techniques are not always cost-effective, and simpler, focused approaches may serve as rapid early diagnostic tests. Here, we took advantage of a small gap in the gene steB of biotype Pullorum that was predicted by genomic analysis (20) to design primers that hybridize to biotype Gallinarum but not Pullorum DNA, permitting a one-step PCR differentiation of the two biotypes (Table 2). This method shortened a previously described two-step technique (21). The addition of primers for additional chicken-associated serovars all in one multiplex PCR analysis is useful for the diagnosis of Salmonella in these birds. Although the designed probes are specific for the identification of serovars Heidelberg, Enteritidis, and Gallinarum, serovar Kentucky shares its PCR profile with serovar Albany, which is not a major chicken isolate in the United States (13, 14). If needed, the latter two serovars can be differentiated by a flagellin-specific PCR (see Fig. S1 in the supplemental material). Finally, rarer serovars for which genomic data are currently unavailable might share one of the described PCR profiles, but as such serovars are significantly less frequent in chicken isolates (13, 14), this would be of minor concern.
Taken together, this study used (i) genomic sequence data for Salmonella to design a chicken-specific multiplex PCR diagnostic test and (ii) an extensive library of Salmonella strains and serovars to validate the specificity of the method for the identification and differentiation of major avian-associated serovars. This simple and economical test should be useful for specific screening of poultry flocks, particularly in developing countries, and of backyard flocks and game birds in developed countries.
Supplementary Material
ACKNOWLEDGMENTS
We thank Leon De Masi for critical reading of the manuscript.
This work was supported by China Scholarship Council grant [2013]3018 to C.Z. and funds from NIH grant AI098041, USDA grant 2013-67015-21285, and the PennVet Center for Host-Microbial Interactions to D.M.S.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/JCM.01976-15.
REFERENCES
- 1.Cohen ML, Tauxe RV. 1986. Drug-resistant Salmonella in the United States: an epidemiologic perspective. Science 234:964–969. doi: 10.1126/science.3535069. [DOI] [PubMed] [Google Scholar]
- 2.Crim SM, Iwamoto M, Huang JY, Griffin PM, Gilliss D, Cronquist AB, Cartter M, Tobin-D'Angelo M, Blythe D, Smith K, Lathrop S, Zansky S, Cieslak PR, Dunn J, Holt KG, Lance S, Tauxe R, Henao OL, Centers for Disease Control and Prevention (CDC) . 2014. Incidence and trends of infection with pathogens transmitted commonly through food–Foodborne Diseases Active Surveillance Network, 10 U.S. sites, 2006–2013. MMWR Morb Mortal Wkly Rep 63:328–332. [PMC free article] [PubMed] [Google Scholar]
- 3.Foley SL, Nayak R, Hanning IB, Johnson TJ, Han J, Ricke SC. 2011. Population dynamics of Salmonella enterica serotypes in commercial egg and poultry production. Appl Environ Microbiol 77:4273–4279. doi: 10.1128/AEM.00598-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keller LH, Schifferli DM, Benson CE, Aslam S, Eckroade RJ. 1997. Invasion of chicken reproductive tissues and forming eggs is not unique to Salmonella enteritidis. Avian Dis 41:535–539. doi: 10.2307/1592142. [DOI] [PubMed] [Google Scholar]
- 5.Keller LH, Benson CE, Krotec K, Eckroade RJ. 1995. Salmonella enteritidis colonization of the reproductive tract and forming and freshly laid eggs of chickens. Infect Immun 63:2443–2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Foley SL, Johnson TJ, Ricke SC, Nayak R, Danzeisen J. 2013. Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol Mol Biol Rev 77:582–607. doi: 10.1128/MMBR.00015-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shivaprasad HL, Methner U, Barrow PA. 2013. Salmonella infections in the domestic fowl, p 162–192. In Barrow PA, Methner U (ed), Salmonella in domestic animals, 2nd ed CABI, Wallingford, United Kingdom. [Google Scholar]
- 8.Barrow PA, Freitas Neto OC. 2011. Pullorum disease and fowl typhoid–new thoughts on old diseases: a review. Avian Pathol 40:1–13. doi: 10.1080/03079457.2010.542575. [DOI] [PubMed] [Google Scholar]
- 9.Andino A, Hanning I. 2015. Salmonella enterica: survival, colonization, and virulence differences among serovars. ScientificWorldJournal 2015:520179. doi: 10.1155/2015/520179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baumler AJ, Hargis BM, Tsolis RM. 2000. Tracing the origins of Salmonella outbreaks. Science 287:50–52. doi: 10.1126/science.287.5450.50. [DOI] [PubMed] [Google Scholar]
- 11.Routh JA, Pringle J, Mohr M, Bidol S, Arends K, Adams-Cameron M, Hancock WT, Kissler B, Rickert R, Folster J, Tolar B, Bosch S, Barton Behravesh C, Williams IT, Gieraltowski L. 2015. Nationwide outbreak of multidrug-resistant Salmonella Heidelberg infections associated with ground turkey: United States, 2011. Epidemiol Infect 13 April 2015:1–8. doi: 10.1017/S0950268815000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Centers for Disease Control and Prevention (CDC). 2013. Outbreak of Salmonella Heidelberg infections linked to a single poultry producer–13 states, 2012–2013. MMWR Morb Mortal Wkly Rep 62:553–556. [PMC free article] [PubMed] [Google Scholar]
- 13.Waltman D, Sellers R. 2014. Report of the Committee on Salmonella, p 351–368. In Richey B, Janicek K (ed), Proceedings 118th Annual Meeting of the United States Animal Health Association. U.S. Animal Health Association, Saint Joseph, MO http://www.usaha.org/Portals/6/Proceedings/2014%20USAHA%20Proceedings%20web.pdf. [Google Scholar]
- 14.Waltman D, Sellers R. 2013. Report of the Committee on Salmonella, p 328–338. In Richey B, Janicek K (ed), Proceedings 117th Annual Meeting of the United States Animal Health Association. U.S. Animal Health Association, Saint Joseph, MO http://www.usaha.org/Portals/6/Proceedings/USAHAProceedings-2013-117th.pdf. [Google Scholar]
- 15.Mulvey MR, Boyd DA, Finley R, Fakharuddin K, Langner S, Allen V, Ang L, Bekal S, El Bailey S, Haldane D, Hoang L, Horsman G, Louis M, Robberts L, Wylie J. 2013. Ciprofloxacin-resistant Salmonella enterica serovar Kentucky in Canada. Emerg Infect Dis 19:999–1001. doi: 10.3201/eid1906.121351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rickert-Hartman R, Folster JP. 2014. Ciprofloxacin-resistant Salmonella enterica serotype Kentucky sequence type 198. Emerg Infect Dis 20:910–911. doi: 10.3201/eid2005.131575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Hello S, Hendriksen RS, Doublet B, Fisher I, Nielsen EM, Whichard JM, Bouchrif B, Fashae K, Granier SA, Jourdan-Da Silva N, Cloeckaert A, Threlfall EJ, Angulo FJ, Aarestrup FM, Wain J, Weill FX. 2011. International spread of an epidemic population of Salmonella enterica serotype Kentucky ST198 resistant to ciprofloxacin. J Infect Dis 204:675–684. doi: 10.1093/infdis/jir409. [DOI] [PubMed] [Google Scholar]
- 18.Pugliese N, Circella E, Pazzani C, Pupillo A, Camarda A. 2011. Validation of a seminested PCR approach for rapid detection of Salmonella enterica subsp. enterica serovar Gallinarum. J Microbiol Methods 85:22–27. [DOI] [PubMed] [Google Scholar]
- 19.Agron PG, Walker RL, Kinde H, Sawyer SJ, Hayes DC, Wollard J, Andersen GL. 2001. Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. Appl Environ Microbiol 67:4984–4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yue M, Rankin SC, Blanchet RT, Nulton JD, Edwards RA, Schifferli DM. 2012. Diversification of the Salmonella Fimbriae: a model of macro- and microevolution. PLoS One 7:e38596. doi: 10.1371/journal.pone.0038596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheraghchi N, Khaki P, Moradi Bidhendi S, Sabokbar A. 2014. Identification of isolated Salmonella enterica serotype Gallinarum biotype Pullorum and Gallinarum by PCR-RFLP. Jundishapur J Microbiol 7:e19135. doi: 10.5812/jjm.19135. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.