Abstract
Gastrointestinal disease is a major cause of morbidity and mortality worldwide, especially among young children and immunocompromised patients. Diarrhea may result from infection with a variety of microbial pathogens, including bacteria, viruses, or parasites. Historically, the diagnosis of infectious diarrhea has been made using microscopy, antigen tests, culture, and real-time PCR. A combination of these traditional tests is often required due to the inability to distinguish between infectious etiologies based on the clinical presentation alone. Recently, several multiplex molecular assays have been developed for the detection of gastrointestinal pathogens directly from clinical stool samples. These panels allow for the detection and identification of up to 20 pathogens in as little as 1 h. This review will focus on the multiplex molecular panels that have received clearance from the FDA for the diagnosis of diarrheal disease and will highlight issues related to test performance, result interpretation, and cost-effectiveness of these new molecular diagnostic tools.
INTRODUCTION
Diarrheal disease remains a significant global health concern, with the World Health Organization estimating that there are ∼1.7 billion total cases each year, resulting in >750,000 deaths among children younger than 5 years old (1). Gastrointestinal (GI) infection with bacteria (e.g., Clostridium difficile, Escherichia coli, Shigella), viruses (e.g., norovirus, rotavirus), or parasites (e.g., Cryptosporidium, Giardia) is a major cause of diarrheal illness worldwide, often resulting from contaminated food/water and poor sanitation. Rapid and accurate detection of GI pathogens is important so that appropriate therapy can be initiated and proper infection control and epidemiologic measures can be taken to help reduce or prevent the spread of disease (2, 3). Traditionally, the laboratory diagnosis of GI infections has relied on a combination of conventional techniques, including microscopy, culture, antigen detection, and individual real-time PCR assays. These methods have demonstrated good performance (4–6), but they are labor intensive, are time consuming, and require health care providers to select the appropriate tests, as a result of clinically indistinguishable illness caused by many infectious agents (7). Recently, there has been substantial interest in the development of multiplex molecular assays for the detection and identification of infectious diseases, including pathogens responsible for causing diarrhea. These syndromic panels allow health care providers to cast a broad net and achieve a timely diagnosis, which may be important in certain patient populations, such as immunocompromised hosts (ICH) and the critically ill (8). This review highlights three commercial multiplex panels (FilmArray GI panel [BioFire Diagnostics, Salt Lake City, UT], Luminex xTag GI pathogen panel [GPP] [Luminex Corporation, Toronto, Canada], and Nanosphere Verigene enteric pathogen [EP] test [Nanosphere, Inc., Northbrook, IL]) that have been cleared by the Food and Drug Administration (FDA) for the detection of GI pathogens (bacteria, virus, and/or parasites) from clinical stool samples. A number of other commercial and laboratory developed multiplex GI assays are available outside the United States but are not cleared by the FDA, and these tests have been discussed previously (9, 10). In addition to reviewing the available FDA-cleared GI multiplex panels, this report will address issues related to test performance, result interpretation, and potential cost-effectiveness.
WHAT MULTIPLEX GI PANELS ARE COMMERCIALLY AVAILABLE AND FDA CLEARED?
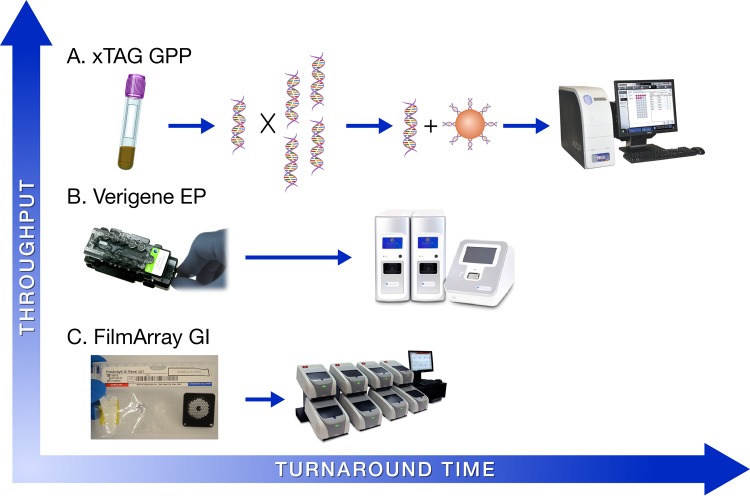
Multiplex (syndromic) panels have gained substantial interest in the field of infectious disease diagnostics due to several advantages over conventional methods, including (i) reduced sample volume requirements, (ii) broad coverage without the need to select specific tests, (iii) enhanced ability to detect coinfections, and in some cases, (iv) increased sensitivity and (v) higher throughput. In 2013, the xTag GPP (Luminex) became the first FDA-cleared multiplex test for the detection and identification of bacteria (n = 6), bacterial toxins (n = 2), viruses (n = 3), and parasites (n = 3) in clinical stool samples (Table 1 ). The GPP assay is performed on 100 μl of fresh or frozen stool, as well as on stool collected in transport medium (i.e., Cary-Blair). Testing of 24 samples can be completed in approximately 5 h, which involves (i) sample pretreatment, (ii) nucleic acid extraction, (iii) multiplex PCR, (iv) bead hybridization, and (v) data analysis on either the Magpix (Luminex) or the Luminex 100/200 analyzer (Fig. 1A). Importantly, the xTag GPP is an open system, which may increase the risk of amplicon contamination in the clinical laboratory.
TABLE 1.
Targets included on three commercial, FDA-cleared, multiplex assays for the detection of gastrointestinal pathogens
| Targeta | Multiplex panelb |
||
|---|---|---|---|
| Verigene EP | FilmArray GI | xTAG GPP | |
| Aeromonas | IUOc | ||
| Campylobacter | ✓ | ✓ | ✓ |
| Clostridium difficile (toxin A/B) | ✓ | ✓ | |
| Plesiomonas shigelloides | ✓ | ||
| Salmonella | ✓ | ✓ | ✓ |
| Yersinia enterocolitica | ✓ | ✓ | RUOd |
| Vibrio spp. | ✓ | ✓ | ✓ |
| EAEC | ✓ | ||
| EPEC | ✓ | ||
| ETEC | ✓ | ✓ | |
| STEC (stx1 and stx2) | ✓e | ✓ | ✓ |
| E. coli 0157 | ✓ | ✓ | |
| EIECf/Shigella | ✓ | ✓ | ✓ |
| Cryptosporidium | ✓ | ✓ | |
| Cyclospora cayetanensis | ✓ | ||
| Entamoeba histolytica | ✓ | ✓ | |
| Giardia lamblia | ✓ | ✓ | |
| Adenovirus 40/41 | ✓ | ✓ | |
| Norovirus GI/GII | ✓ | ✓ | ✓ |
| Rotavirus A | ✓ | ✓ | ✓ |
| Sapovirus | ✓ | ||
| Astrovirus | ✓ | ||
EAEC, enteroaggregative E. coli; EPEC, enteropathogenic E. coli; ETEC, enterotoxigenic E. coli (heat-labile toxin/heat-stable toxin); STEC, Shiga-like toxin-producing E. coli (stx1 and stx2); EIEC, enteroinvasive E. coli.
EP, enteric pathogen; GI, gastrointestinal; GPP, gastrointestinal pathogen panel.
IUO, investigational use only.
RUO, research use only.
The Nanosphere assay includes unique tests for Shiga toxin 1 (stx1) and Shiga toxin 2 (stx2).
The Luminex panel does not specifically target EIEC; however, the Luminex Shigella assay does cross-react with EIEC.
FIG 1.
Comparison of workflow, throughput, and turnaround time among three commercial, FDA-cleared multiplex GI platforms. (A) The Luminex xTag GPP assay requires an initial sample preparation step (45 to 60 min), followed by nucleic acid extraction (∼45 min) and multiplex PCR using a standard thermocycler (2.5 h). Subsequently, the amplified product is incubated with tagged beads to allow for hybridization (1 h), and, finally, data are acquired and analyzed on either the Magpix or Luminex 100/200 analyzers (∼10 min). (B) The Nanosphere Verigene enteric pathogens (EP) test involves pipetting 200 μl of sample into a supplied extraction tray. The extraction tray, reagents, and test cartridge are then loaded into the Verigene Processor SP (∼2 min). The Processor SP automates nucleic acid extraction, amplification, and hybridization with gold nanoparticles. Finally, the test cartridge is removed from the Processor and inserted into the Verigene Reader for data analysis, with a total turnaround time of ∼2 h per test. (C) The BioFire FilmArray GI panel uses a loading station to facilitate the addition of hydration solution and patient sample into the test pouch via a vacuum-controlled syringe (∼2 min). Once the sample has been added, the test pouch is inserted into the FilmArray instrument, which then automates nucleic acid extraction, nested PCR, and data acquisition using melting curve analysis (∼60 min).
In 2014, the FDA cleared the FilmArray GI panel (BioFire Diagnostics, LLC), which targets 22 analytes (bacteria [n = 11], bacterial toxins [n = 2], viruses [n = 5], and parasites [n = 4]) from stool in a Cary-Blair transport medium (Table 1). Testing and analysis are performed using a FilmArray analyzer (BioFire) inside a closed reaction pouch. After adding 200 μl of clinical sample and hydration solution to the FilmArray pouch, the system performs nucleic acid extraction, nested multiplex PCR, and, finally, melting curve analysis. Testing and result interpretation are completed in ∼60 min, but each FilmArray instrument processes 1 sample at a time. Notably, the FilmArray 2.0 system, allowing up to 8 instruments to be connected to a single computer and interfaced with the laboratory information system, received FDA clearance in 2015 (Fig. 1C).
The Verigene EP nucleic acid test (Nanosphere) was the third GI panel to receive FDA clearance, doing so in mid-2014. The Verigene EP assay targets 9 analytes (bacteria [n = 5], bacterial toxins [n = 2], and viruses [n = 2]) from stool in a Cary-Blair medium (Table 1). Testing requires 200 μl of sample and is performed on the Verigene Processor SP (Nanosphere). After the sample is pipetted into a supplied extraction tray, the Verigene Processor automates nucleic acid extraction, target amplification by PCR, and hybridization-based detection using microarray and gold nanoparticle, probe-based technology. Result interpretation is completed using the Verigene Reader (Nanosphere), and the total turnaround time per sample is ∼2 h (Fig. 1B). Up to 32 Verigene Processor SP units can be connected to a single Verigene Reader, allowing for scalability and random-access testing. It is important to note that each of these multiplex GI panels has been approved as an adjunct in the diagnosis of infectious diarrhea, and their use in screening asymptomatic patients has not been studied or approved.
HOW DO MULTIPLEX MOLECULAR TESTS PERFORM COMPARED TO CONVENTIONAL MICROBIOLOGY TESTS FOR THE DETECTION OF GI PATHOGENS?
Several studies have evaluated the performance of multiplex GI panels by comparing the results to those of conventional methods. A study published in 2013 (11) assessed the Luminex GPP assay using 440 stool samples collected from 329 patients, including immunosuppressed adults (n = 102) and children (n = 50). Of the 329 patients included in this study, 162 (49.2%) were positive for one or more pathogens by the xTag GPP panel, with rotavirus being the most commonly detected. Norovirus was also found to be a significant cause of disease in both immunosuppressed children (9.2%) and pediatric patients being evaluated in the emergency department (13.1%). The GPP assay was found to be more sensitive than the conventional tests (i.e., antigen testing, bacterial culture, microscopy) for several pathogens, including Campylobacter spp., toxigenic C. difficile, norovirus, rotavirus, and Salmonella. Interestingly, the xTag GPP assay detected 31 coinfections, which consisted of 2 pathogens in 23 (5.2%) samples and 3 pathogens in 8 (1.8%) specimens.
A study by Claas et al. (12) also evaluated the performance of the xTag GPP assay by testing 901 stool samples collected among four medical centers. Compared to routine individual real-time PCR, the xTag GPP showed a sensitivity of 100% for norovirus (9/9), rotavirus (18/18), Giardia (22/22), and Entamoeba histolytica (6/6); however, the sensitivity of the multiplex assay was lower for adenovirus (20%; 4/20), and Cryptosporidium (91.3%; 21/23). Compared to bacterial culture, the Luminex assays showed a sensitivity of 97.4% (111/114) for Campylobacter, 93.7% (15/16) for E. coli strain 0157, 82.7% (62/75) for Salmonella, and 100% (40/40) for Shigella, while demonstrating a specificity >96% for all targets. An important observation from this study was that, in 65% of samples positive by xTag GPP, the ordering health care provider had not specifically requested testing for the pathogen that was detected by the multiplex panel. In addition, the xTag GPP assay identified coinfections in 9.5% (n = 86) of positive samples, with C. difficile, norovirus, and Campylobacter being present in >25% of samples that had 2 or more pathogens detected (12). These data are similar to other studies that have evaluated the performance of the Luminex xTag GPP assay, which have also demonstrated that the Luminex GI panel increases both the positivity rate and the number of coinfections detected compared to conventional methods (13–15).
The FilmArray GI panel targets 22 pathogens responsible for diarrheal disease, and this assay has been evaluated in several recent studies. A large, multicenter study in Europe tested 709 stool samples by the FilmArray GI panel and identified that 384 (54.2%) of these were positive for at least 1 pathogen (16). In contrast, conventional laboratory methods were positive in only 128 (18.1%) samples. Among all 10 participating countries, enteropathogenic E. coli (EPEC), enteroaggregative E. coli (EAEC), enterotoxigenic E. coli (ETEC), Campylobacter, C. difficile, and norovirus were the most commonly detected pathogens. Interestingly, the authors observed that the pathogen distribution varied by patient age, with children <5 years old being most frequently infected with norovirus and EPEC, whereas individuals >60 years old were most commonly infected with C. difficile and EPEC. The rate of coinfections was also high in this study, with 16.4% (116/709) of samples being positive for multiple (≥2) targets. The patient groups most likely to be positive for at least 2 pathogens were children <5 years of age and hospital outpatients, and the most common coinfections were with Campylobacter and EPEC (16).
Stockmann et al. (17) also compared the FilmArray GI panel to routine testing using 378 diarrheal stool specimens. This study identified that a median of 3 (range, 1 to 10) routine tests were ordered on each specimen. Despite the relatively high number of conventional tests performed, the FilmArray GI panel was positive in ∼20% more samples than was routine testing (65% versus 46%). When a subanalysis was performed on those patients for whom only C. difficile testing was ordered, an alternate pathogen was detected in 29% by the FilmArray GI panel, with norovirus being identified in a substantial percentage (12%) of children in whom only C. difficile testing was requested.
Finally, a recent report by Khare et al. (18) directly compared the FilmArray GI and xTag GPP panels using Cary-Blair stool samples (n = 500) submitted for routine diagnostic testing. Among the 500 total samples, 270 (54%) were stored specimens that were previously shown to be positive for at least 1 pathogen, while the remaining 230 (46%) were prospectively tested. Of the 270 characterized samples, both multiplex panels showed a sensitivity >90% for the majority of targets; however, the FilmArray was positive in only 5 of 21 (23.8%) samples that grew Aeromonas spp. in bacterial culture. All of these samples had been stored at −20°C for >1 month prior to testing by FilmArray, and only 2 of 21 specimens regrew Aeromonas by culture. Therefore, it is possible that nucleic acid degradation accounted for the negative FilmArray results. Interestingly, Aeromonas was not included in the final FilmArray panel that was cleared by the FDA. The xTag GPP assay showed lower sensitivity for Yersinia enterocolitica, detecting the organism in only 13 of 27 (48.1%) culture-positive samples. Among prospective stool samples (n = 230), the FilmArray and Luminex GI panels were positive in 76 (33%) and 69 (30.3%) specimens, respectively, compared to only 19 (8.3%) specimens by routine testing. C. difficile and norovirus were two of the most common pathogens detected by both multiplex assays. Importantly, sapovirus was detected by the FilmArray GI panel in 5.7% of the prospective samples, suggesting that this viral pathogen may be significantly underreported by many clinical microbiology labs. The FilmArray and Luminex panels detected mixed infections in 21.1% and 13% of positive prospective samples, respectively, compared to 8.3% by routine methods (18). These data corroborate other published findings and underscore that the implementation of multiplex panels increases the positivity rate and the number of coinfections detected compared to conventional techniques (19, 20). Future studies evaluating the use of these panels in developing nations are needed, as the performance characteristics of certain targets (i.e., Vibrio spp., Plesiomonas shigelloides, Entamoeba histolytica) have not been adequately defined due to the low prevalence of certain GI pathogens in developed countries.
WHAT ARE THE CHALLENGES ASSOCIATED WITH RESULT INTERPRETATION OF MULTIPLEX GI PANELS?
More is always better, right? Unfortunately, this may not be the case in regard to multiplex GI panels, as the routine use of these assays is likely to introduce several significant challenges. First, each of the FDA-cleared multiplex GI panels targets microbial nucleic acid (DNA or RNA), and, therefore, cannot distinguish between viable, replicating organisms that are responsible for disease and nonviable pathogens or remnant nucleic acid. This may be especially important in the diagnosis of GI infections, as a number of pathogens (e.g., norovirus, rotavirus, adenovirus, astrovirus, Salmonella) on these panels have been shown to be present in the stool of asymptomatic individuals or shed for long periods following the resolution of disease (21–26). For example, a recent study used real-time PCR to demonstrate nearly constant shedding of a number of enteric viruses, including adenovirus, rotavirus, astrovirus, and enterovirus, in 2 healthy infant siblings during their first 12 months of life (22).
A second challenge associated with the use of multiplex GI panels may be the increased detection of C. difficile and the potential impact this might have on isolation and treatment decisions. The xTag GPP and FilmArray GI panels are both FDA cleared for testing stool specimens collected in Cary-Blair transport media. Although the manufacturer's instructions specify that these tests should only be performed on patients with signs and/or symptoms of GI infection, it will be difficult for clinical laboratories to confirm that stool specimens were unformed prior to being placed in a liquid transport media. The potential for laboratories to inappropriately test nondiarrheal stool samples may render the interpretation of results difficult, as numerous evaluations of these multiplex panels have demonstrated that C. difficile is one of the most common organisms detected (12, 16–18). Whether detection of C. difficile represents true infection that is associated with disease or represents colonization will be an important area for future research.
The third, and final, challenge presented here relates to how health care providers will use and interpret the large amount of data that will be made available with the broad implementation of multiplex GI panels. Studies have demonstrated that the use of these multiplex panels will increase the positivity rates of GI pathogens by 2- to 4-fold compared to conventional methods (17, 18). Not only will the number of positive results increase, but also health care providers will be faced with how to interpret the presence of organisms (e.g., EAEC, EPEC, sapovirus) that have not been routinely tested for in the past. For example, the study by Khare et al. (18) showed that 14.1% (38/270) of characterized samples were unexpectedly positive for EAEC (n = 11) or EPEC (n = 27) by the FilmArray GI panel. Furthermore, the rate of coinfections reported will almost certainly increase, and, to date, insufficient data are available to guide laboratorians and clinicians on how to interpret these findings. Studies will be needed to address several important issues related to coinfections, including the impact of multiple pathogens on disease severity, and how, and if, mixed infections should be managed differently than infections involving a single organism.
ARE MULTIPLEX GI PANELS COST-EFFECTIVE?
To date, limited data are available on the cost-effectiveness of multiplex GI panels, both in terms of their impact on laboratory costs and the downstream expense of managing patients with diarrhea. Several studies have shown that, on average, a patient with diarrhea has ∼3 routine microbiology tests ordered by their health care provider (17, 18). Therefore, it is possible that performing a single, multiplex assay might reduce the total cost (i.e., reagents, instrumentation, technologist time) associated with completing a number of routine tests. In addition, it seems reasonable that performing a broad, molecular panel may allow health care providers to make more informed decisions regarding isolation of patients and the discontinuation of antibiotic therapy if a viral pathogen (i.e., norovirus or sapovirus) is detected. To address these issues, a study in the United Kingdom assessed the potential economic benefit of testing hospitalized patients with the xTag GPP assay (27). The investigators conducted an 8-month parallel diagnostic study, in which the financial impact of testing with either the Luminex panel or conventional testing was measured (or estimated) for 800 patients. Testing by the xTag GPP assay was estimated to cost an additional $34,800 in laboratory expense; however, use of the multiplex assay reduced the total number of patient isolation days by 755, yielding a total cost savings of ∼$69,500 during the study period. While these findings are promising, additional studies are needed to confirm the results as well as to assess the financial impact of multiplex GI panels on antimicrobial stewardship and, ultimately, patient outcomes.
CONCLUSIONS
The development and implementation of highly multiplexed molecular panels will allow clinical laboratories to more rapidly and sensitively diagnose GI infections. The decision of which commercial platform to implement will ultimately depend on a number of factors, including (i) test performance, (ii) cost, (iii) workflow, (iv) throughput, and (v) the patient population to be tested. Each of the FDA-cleared multiplex GI panels has unique advantages and limitations that a clinical microbiology lab must consider. The xTag GPP assay offers an extensive test menu (14 FDA-cleared targets) with the ability to perform high-throughput testing, which may be beneficial to large reference laboratories. However, this assay has the longest hands-on (∼45 min) and turnaround (∼5 h) times, and it is an open system, which poses a risk for amplicon contamination in the laboratory. The Verigene EP assay is a closed, sample-to-answer platform that includes 9 FDA-cleared targets. Results are available in as little as 2 h, but the system is designed to test 1 sample per Verigene Processor SP. This limitation can be overcome by connecting additional Processors (up to 32) to the Verigene Reader. At the time of preparing this review, no peer-reviewed publications were available describing the performance of the Verigene EP assay; therefore, this is an area for future research. Finally, the FilmArray GI panel represents the most comprehensive test menu (22 FDA-cleared targets), and the results are available in ∼60 min. Hands-on time is minimal (∼2 min), and testing is completed in a closed reaction vessel (Table 2). The availability of the FilmArray 2.0 system now allows up to 8 instruments to be connected to a single computer, thereby increasing throughput capabilities (Fig. 1).
TABLE 2.
Comparison of features between three commercial, FDA-cleared, multiplex platforms for the detection of gastrointestinal pathogens
| Feature | Platforma |
||
|---|---|---|---|
| Verigene EP | FilmArray GI | xTAG GPP | |
| No. of FDA-cleared targets | 9 | 22 | 14 |
| Processing (hands-on) time per run, min | <5 | 2 | 45 |
| Separate extraction required? | No | No | Yes (∼45 min) |
| Time/run, h | ∼2 | ∼1 | ∼5 |
| Throughput, specimens/run | 1b | 1b | 96 |
| Technology | PCR + gold nanoparticle hybridization | Nested PCR + melting curve | PCR + xTag (fluorescent bead-based detection) |
| Open or closed system | Closed | Closed | Open |
| Footprint | Small to moderate | Small | Moderate |
| List price per instrument, $ | 40,000c | 39,500 | 37,000 |
| List price reagent cost per specimen, $ | 80d | 155d | 80–90d |
EP, enteric pathogen; GI, gastrointestinal; GPP, gastrointestinal pathogen panel.
Scalable to increase throughput with additional instruments/processing modules. The FilmArray 2.0 system can connect up to 8 instruments to a single computer. The Verigene System allows for up to 32 Processor SP units to be connected to a single Verigene Reader.
Includes the list price for both the Verigene Processor SP ($20,000) and the Verigene Reader ($20,000).
Actual reagent price may be discounted based on the volume of samples tested.
It is almost certain that multiplex panels will be increasingly employed in diagnostic laboratories in the future; however, how these assays will be used and the impact that they will have on patient care remains unclear. Although multiplex GI panels will increase both the positivity rate and the number of coinfections that are detected, routine bacterial culture will continue to be needed for antimicrobial susceptibility testing and epidemiologic investigations. It will be critical for clinical laboratories to work closely with their colleagues in public health to ensure that a system is in place to provide these necessary services. It is essential that future studies address several important clinically related issues, including the impact of multiplex GI panels on antibiotic use and the influence the results have on management decisions and patient outcomes. A final consideration is whether the use of these panels should be restricted to certain patient populations (e.g., ICH) or made available to all patients. In the opinion of the author, reserving highly multiplexed GI panels to ICH, the critically ill, or patients with prolonged diarrhea may improve test utilization and reduce the inappropriate use of antibiotics.
ACKNOWLEDGMENTS
M.J.B. has no conflicts of interest to disclose related to the products mentioned.
Approval of the product information and images included herein was granted by BioFire, Inc., Luminex, and Nanosphere.
Biography

Matthew J. Binnicker is the Director of Clinical Virology and an Associate Professor of Laboratory Medicine and Pathology at Mayo Clinic in Rochester, Minnesota. He completed his postdoctoral training in Clinical Microbiology at Mayo Clinic and now serves as the program director for the Ph.D. fellowship training program at Mayo. Dr. Binnicker's research interests focus on the molecular detection of viral infections, including the use of next generation sequencing.
REFERENCES
- 1.World Health Organization. Diarrheal disease. http://www.who.int/mediacentre/factsheets/fs330/en/ Accessed 9 July 2015.
- 2.Schlenker C, Surawicz CM. 2009. Emerging infections of the gastrointestinal tract. Best Pract Res Clin Gastroenterol 23:89–99. doi: 10.1016/j.bpg.2008.11.014. [DOI] [PubMed] [Google Scholar]
- 3.Hatchette TF, Farina D. 2011. Infectious diarrhea: when to test and when to treat. CMAJ 183:339–344. doi: 10.1503/cmaj.091495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cunningham SA, Sloan LM, Nyre LM, Vetter EA, Mandrekar J, Patel R. 2010. Three-hour molecular detection of Campylobacter, Salmonella, Yersinia, and Shigella species in feces with accuracy as high as that of culture. J Clin Microbiol 48:2929–2933. doi: 10.1128/JCM.00339-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Karre T, Sloan L, Patel R, Mandrekar J, Rosenblatt J. 2011. Comparison of two commercial molecular assays to a laboratory-developed molecular assay for diagnosis of Clostridium difficile infection. J Clin Microbiol 49:725–727. doi: 10.1128/JCM.01028-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buckwalter SP, Teo R, Espy MJ, Sloan LM, Smith TF, Pritt BS. 2012. Real-time qualitative PCR for 57 human adenovirus types from multiple specimen sources. J Clin Microbiol 50:766–771. doi: 10.1128/JCM.05629-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hennessy TW, Marcus R, Deneen V, Reddy S, Vugia D, Townes J, Bardsley M, Swerdlow D, Angulo FJ. 2004. Survey of physician diagnostic practices for patients with acute diarrhea: clinical and public health implications. Clin Infect Dis 38 (suppl 3):S203–S211. doi: 10.1086/381588. [DOI] [PubMed] [Google Scholar]
- 8.Fasel D, Mellmann A, Cernela N, Hachler H, Fruth A, Khanna N, Egil A, Beckmann C, Hirsch HH, Goldenberger D, Stephan R. 2014. Hemolytic uremic syndrome in a 65-year-old male linked to a very unusual type of stx2e- and eae-harboring 051:H49 Shiga toxin-producing Escherichia coli. J Clin Microbiol 52:1301–1303. doi: 10.1128/JCM.03459-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Verweij JJ, Blange RA, Templeton K, Schinkel J, Brienen EA, van Rooyen MA, van Lieshout L, Polderman AM. 2004. Simultaneous detection of Entamoeba histolytica, Giardia lamblia, and Cryptosporidium parvum in fecal samples by using multiplex real-time PCR. J Clin Microbiol 42:1220–1223. doi: 10.1128/JCM.42.3.1220-1223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang H, Morrison S, Tang YW. 2015. Multiplex polymerase chain reaction tests for detection of pathogens associated with gastroenteritis. Clin Lab Med 35:461–486. doi: 10.1016/j.cll.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mengelle C, Mansuy JM, Prere MF, Grouteau Claudet EI, Kamar N, Huynh A, Plat G, Benard M, Marty N, Valentin A, Berry A, Izopet J. 2013. Simultaneous detection of gastrointestinal pathogens with multiplex Luminex-based molecular assay in stool samples from diarrheic patients. Clin Microbiol Infect 19:E458–E465. doi: 10.1111/1469-0691.12255. [DOI] [PubMed] [Google Scholar]
- 12.Claas EC, Burnham CD, Mazzulli T, Templeton K, Topin F. 2013. Performance of the xTag gastrointestinal pathogen panel, a multiplex molecular assay for simultaneous detection of bacterial, viral, and parasitic causes of infectious gastroenteritis. J Microbiol Biotechnol 23:1041–1045. doi: 10.4014/jmb.1212.12042. [DOI] [PubMed] [Google Scholar]
- 13.Wessels E, Rusman LG, van Bussel M, Claas EC. 2014. Added value of multiplex Luminex gastrointestinal pathogen panel (xTag GPP) testing in the diagnosis of infectious gastroenteritis. Clin Microbiol Infect 20:O182–O287. doi: 10.1111/1469-0691.12364. [DOI] [PubMed] [Google Scholar]
- 14.Beckmann C, Heininger U, Marti H, Hirsch HH. 2014. Gastrointestinal pathogens detected by multiplex nucleic acid amplification testing in stools of pediatric patients and patients returning from the tropics. Infection 42:961–970. doi: 10.1007/s15010-014-0656-7. [DOI] [PubMed] [Google Scholar]
- 15.Vocale C, Rimoldi SG, Pagani C, Grande R, Pedna F, Arghittu M, Lunghi G, Maraschini A, Gismondo MR, Landini MP, Torresani E, Topin F, Sambri V. 2015. Comparative evaluation of the new xTag GPP multiplex assay in the laboratory diagnosis of acute gastroenteritis: clinical assessment and potential application from a multicentre Italian study. Int J Infect Dis 34:33–37. doi: 10.1016/j.ijid.2015.02.011. [DOI] [PubMed] [Google Scholar]
- 16.Spina A, Kerr KG, Cormican M, Barbut F, Eigentler A, Zerva L, Tassios P, Popescu GA, Rafila A, Eerola E, Batista J, Maass M, Aschbacher R, Olsen KE, Allerberger F. 2015. Spectrum of enteropathogens detected by the FilmArray GI panel in a multicentre study of community-acquired gastroenteritis. Clin Microbiol Infect 21:719–728. doi: 10.1016/j.cmi.2015.04.007. [DOI] [PubMed] [Google Scholar]
- 17.Stockmann C, Rogatcheva M, Harrel B, Vaughn M, Crisp R, Poritz M, Thatcher S, Korgenski EK, Barney T, Daly J, Pavia AT. 2015. How well does physician selection of microbiologic tests identify Clostridium difficile and other pathogens in pediatric diarrhea? Insights using multiplex PCR-based detection. Clin Microbiol Infect 21:179.e9–179.e15. doi: 10.1016/j.cmi.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Khare R, Espy MJ, Cebelinski E, Boxrud D, Sloan LM, Cunningham SA, Pritt BS, Patel R, Binnicker MJ. 2014. Comparative evaluation of two commercial multiplex panels for detection of gastrointestinal pathogens by use of clinical stool specimens. J Clin Microbiol 52:3667–3673. doi: 10.1128/JCM.01637-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buss SN, Leber A, Chapin K, Fey PD, Bankowski MJ, Jones MK, Rogatcheva M, Kanack KJ, Bourzac KM. 2015. Multicenter evaluation of the BioFire FilmArray gastrointestinal panel for etiologic diagnosis of infectious gastroenteritis. J Clin Microbiol 53:915–925. doi: 10.1128/JCM.02674-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rand KH, Tremblay EE, Hoidal M, Fisher LB, Grau KR, Karst SM. 2015. Multiplex gastrointestinal pathogen panels: implications for infection control. Diagn Microbiol Infect Dis 82:154–157. doi: 10.1016/j.diagmicrobio.2015.01.007. [DOI] [PubMed] [Google Scholar]
- 21.Medus C, Smith KE, Bender JB, Besser JM, Hedberg CW. 2006. Salmonella outbreaks in restaurants in Minnesota, 1995 through 2003: evaluation of the role of infected foodworkers. J Food Prot 69:1870–1878. [DOI] [PubMed] [Google Scholar]
- 22.Kapusinszky B, Minor P, Delwart E. 2012. Nearly constant shedding of diverse enteric viruses by two healthy infants. J Clin Microbiol 50:3427–3434. doi: 10.1128/JCM.01589-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukhopadhya I, Sarkar R, Menon VK, Babji S, Paul A, Rajendran P, Sowmyanarayanan TV, Moses PD, Iturriza-Gomara M, Gray JJ, Kang G. 2013. Rotavirus shedding in symptomatic and asymptomatic children using reverse transcription-quantitative PCR. J Med Virol 85:1661–1668. doi: 10.1002/jmv.23641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lemes LG, Correa TS, Fiaccadori FS, Cardoso D, Arantes AM, Souza M. 2014. Prospective study on norovirus infection among allogeneic stem cell transplant recipients: prolonged viral excretion and viral RNA in the blood. J Clin Virol 61:329–333. doi: 10.1016/j.jcv.2014.08.004. [DOI] [PubMed] [Google Scholar]
- 25.Corcoran MS, van Well GT, van Loo IH. 2014. Diagnosis of viral gastroenteritis in children: interpretation of real-time PCR results and relation to clinical symptoms. Eur J Clin Microbiol Infect Dis 33:1663–1673. doi: 10.1007/s10096-014-2135-6. [DOI] [PubMed] [Google Scholar]
- 26.Teunis PF, Sukhrie FH, Vennema H, Bogerman J, Beersma MF, Koopmans MP. 2015. Shedding of norovirus in symptomatic and asymptomatic infections. Epidemiol Infect 143:1710–1717. doi: 10.1017/S095026881400274X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldenberg SD, Bacelar M, Brazier P, Bisnauthsing K, Edgeworth JD. 2015. A cost-benefit analysis of the Luminext xTag gastrointestinal pathogen panel for detection of infectious gastroenteritis in hospitalized patients. J Infect 70:504–511. doi: 10.1016/j.jinf.2014.11.009. [DOI] [PubMed] [Google Scholar]