Abstract
We compared carbapenemase detection among 266 Gram-negative bacilli (161 carbapenemase producers) using the Carba NP tests issued by the CLSI (CNPt-CLSI) and a novel protocol (CNPt-direct) designed for carbapenemase detection direct from bacterial cultures (instead of bacterial extracts required by the CLSI tests). The specificities were comparable (100%), but the CNPt-direct was more sensitive (98% versus 84%). The CNPt-direct was easier to perform due to the direct use of colonies and offered a more robust detection of carbapenemase producers.
TEXT
The Carba NP test (CNPt) is a biochemical test for rapid detection (≥2 h) of carbapenemase production on Gram-negative bacilli (1). It is based on in vitro hydrolysis of imipenem by a bacterial lysate, which is detected by changes in pH values using the indicator phenol red (red to orange/yellow). The CNPt was reported to be highly sensitive for detection of Klebsiella pneumoniae carbapenemase (KPC) and metallo-beta-lactamase (MBL) producers (1–4). However, consistent problems were reported for the detection of OXA-48-like carbapenemase producers, with sensitivities rates ranging from 11 to 100% (1–5). Another potential limitation of the CNPt, especially for low-income countries, is the reliance on a commercial extraction buffer (B-PER II), which is used to obtain bacterial extracts (1). A recent recommendation from the Clinical and Laboratory Standards Institute (CLSI) indicates that Enterobacteriaceae, Pseudomonas aeruginosa, and Acinetobacter spp. with elevated carbapenem MICs or reduced disk diffusion inhibition zones should be tested, for epidemiological or infection control purposes, for the production of carbapenemases by means of the CNPt (5). We evaluated a simplified protocol of the CNPt that allows direct use of colonies instead of bacterial extracts, which resulted in greater protocol simplicity and cost reduction per reaction. Additionally, the modification introduced into the protocol for direct sampling improved the detection of carbapenemase producers.
A total of 266 clinical isolates were included (161 carbapenemase producers and 105 carbapenemase nonproducers) (Table 1). The isolates, belonging to the collection of the Servicio Antimicrobianos, Instituto Nacional de Enfermedades Infecciosas, Argentina, and the Public Health Ontario Laboratories, Ontario, Canada, correspond to submissions of very diverse locations (22 countries) and thus should represent minimum clonal and enzyme bias. PCR analysis followed by DNA sequencing of the amplicons was considered the gold standard for β-lactamase characterization (2, 6). Carbapenemase nonproducers (carbapenem-resistant strains due to overexpression of chromosomal AmpC or expression of plasmid-mediated AmpC and/or extended-spectrum β-lactamase [ESBLs] coupled with impermeability or efflux hyperproducers) were characterized as described previously (2, 6). Panel isolates were previously identified using either conventional biochemical tests or matrix-assisted laser desorption ionization−time of flight (MALDI-TOF) mass spectrometry (Bruker, Germany).
TABLE 1.
Isolates tested and test results
| Group and β-lactamase (no. of isolates) | Bacterial species included (no. of isolates) | No. of isolates with a positive test/no. of isolates tested (%)a |
|
|---|---|---|---|
| CNPt-CLSI | CNPt-direct | ||
| Class A carbapenemase producers | |||
| KPC-2 (27) | Klebsiella pneumoniae (11), Enterobacter cloacae (5), Serratia marcescens (3), Citrobacter freundii (2), Escherichia coli (2), Citrobacter braakii (1), Enterobacter aerogenes (1), Pseudomonas aeruginosa (1), Salmonella spp. (1) | 25/27 (93) | 27/27 (100) |
| KPC-3 (7) | K. pneumoniae (3), E. cloacae (2), C. freundii (1), E. coli (1) | 7/7 (100) | 7/7 (100) |
| IMI-1/NMC-A (4) | E. cloacae (3). Enterobacter asburiae (1) | 4/4 (100) | 4/4 (100) |
| Sme-1b (4) | S. marcescens (4) | 3/4 (75) | 4/4 (100) |
| GES-3 (2) | Enterobacter agglomerans (1), K. pneumoniae (1) | 1/2 (50) | 2/2 (100) |
| GES-5 (1) | P. aeruginosa (1) | 0/1 (0) | 1/1 (100) |
| All (45) | 40/45 (89) | 45/45 (100) | |
| Class B carbapenemase producers | |||
| NDM-1 (17) | Providencia rettgeri (4), Providencia stuartii (3), E. coli (3), K. pneumoniae (2), Acinetobacter baumannii (1), Acinetobacter pittii (1), E. cloacae (1), Morganella morganii (1), Proteus mirabilis (1) | 12/17 (71) | 17/17 (100) |
| IMP-1 (3) | Acinetobacter ursingii (1), Acinetobacter junii (1), Acinetobacter johnsonii (1) | 3/3 (100) | 3/3 (100) |
| IMP-7 (1) | P. aeruginosa (1) | 1/1 (100) | 1/1 (100) |
| IMP-8 (2) | E. cloacae (1), C. braakii (1) | 2/2 (100) | 2/2 (100) |
| IMP-13 (2) | P. aeruginosa (2) | 2/2 (100) | 2/2 (100) |
| IMP-16 (2) | P. aeruginosa (2) | 2/2 (100) | 2/2 (100) |
| IMP-18 (1) | P. aeruginosa (1) | 1/1 (100) | 1/1 (100) |
| IMP-27 (1) | P. mirabilis (1) | 0/1 (0) | 0/1 (0) |
| VIM-1 (3) | E. coli (2), E. cloacae (1) | 3/3 (100) | 3/3 (100) |
| VIM-2 (5) | P. aeruginosa (3), E. cloacae (1), P. rettgeri (1) | 4/5 (80) | 5/5 (100) |
| VIM-11 (1) | E. cloacae (1) | 0/1 (0) | 1/1 (100) |
| SPM-1 (2) | P. aeruginosa (2) | 2/2 (100) | 2/2 (100) |
| All (40) | 32/40 (80) | 39/40 (98) | |
| Class D carbapenemase producers | |||
| Enterobacteriaceae | |||
| OXA-48 (16) | K. pneumoniae (8), E. coli (7), Klebsiella oxytoca (1) | 15/16 (94) | 16/16 (100) |
| OXA-181 (10) | K. pneumoniae (6), E. coli (4) | 8/10 (80) | 10/10 (100) |
| OXA-232 (5) | K. pneumoniae (5) | 3/5 (60) | 4/5 (80) |
| OXA-244 (1) | E. coli (1) | 0/1 (0) | 0/1 (0) |
| OXA-247 (2) | E. coli (1), b K. pneumoniae (1) | 2/2 (100) | 2/2 (100) |
| OXA-438 (2) | E. coli (2) | 2/2 (100) | 2/2 (100) |
| All Enterobacteriaceae (36) | 30/36 (83) | 34/36 (94) | |
| Acinetobacter spp. | |||
| OXA-23 (20) | A. baumannii (20) | NAc | 19/20 (95) |
| OXA-58 (16) | A. baumannii (12), A. lwoffii (2), A. junii (1), A. guillouiae (1) | NA | 15/16 (94) |
| OXA-72 (1) | A. baumannii (1) | 1/1 (100) | |
| OXA-143 (1) | A. baumannii (1) | NA | 1/1 (100) |
| OXA-23 + OXA-58 (2) | A. baumannii (2) | NA | 2/2 (100) |
| All Acinetobacter (40) | NA | 38/40 (95) | |
| Carbapenemases nonproducers | |||
| ESBLs (35)d | K. pneumoniae (24), E. coli (4), P. aeruginosa (3), E. aerogenes (1), K. oxytoca (1), S. marcescens (1), S. flexneri (1) | 0/35 (0) | 0/35 (0) |
| AmpCs (15)e | E. cloacae (10), C. freundii (2), E. coli (2), E. aerogenes (1) | 0/15 (0) | 0/15 (0) |
| AmpCs + ESBLs (6)f | E. cloacae (6) | 0/6 (0) | 0/6 (0) |
| Narrow spectrum (9)g | A. baumannii (5), E. coli (2), P. aeruginosa (1), P. mirabilis (1) | 0/9 (0) | 0/9 (0) |
| None (nonenzymatic carbapenem-resistant isolates and QC strains) (40)h | P. aeruginosa (38), E. coli (2) | 0/40 (0) | 0/40 (0) |
| All nonproducers (105) | 0/105 (0) | 0/105 (0) | |
False-negative results are shown in italics.
E. coli J53 transconjugant strain.
NA, not applicable (an alternative procedure is required for optimal detection of OXA-type carbapenemases among Acinetobacter spp., as described in reference 8.
CTXM-2, n = 15; CTXM-15, n = 9; CTXM-13, n = 1; CTXM-14, n = 1; GES-1, n = 3; PER-2, n = 1; SHV-1, n = 1; SHV-5, n = 2; SHV-18, n = 1; SHV-27, n = 1.
Hyperproduction of chromosomally mediated cephalosporinase, n = 15; CMY-2, n = 2; DHA-1, n = 1.
Hyperproduction of chromosomally mediated cephalosporinase plus CTXM-2, n= 2; TEM-3, n = 1; TEM-5, n = 1; TEM-26, n = 1; SHV-3, n = 1.
OXA-51 (without upstream ISAba1), n = 5; TEM-1, n = 1; TEM-2, n = 1; OXA-1, n = 2.
Nonenzymatically carbapenem-resistant isolates and quality control (QC) strains, i.e., Pseudomonas aeruginosa carbapenem resistant because of efflux overproduction (n = 5), porin loss (n = 5), or dual mechanisms (n = 18), and quality control strains P. aeruginosa ATCC 27853 and Escherichia coli ATCC 25922.
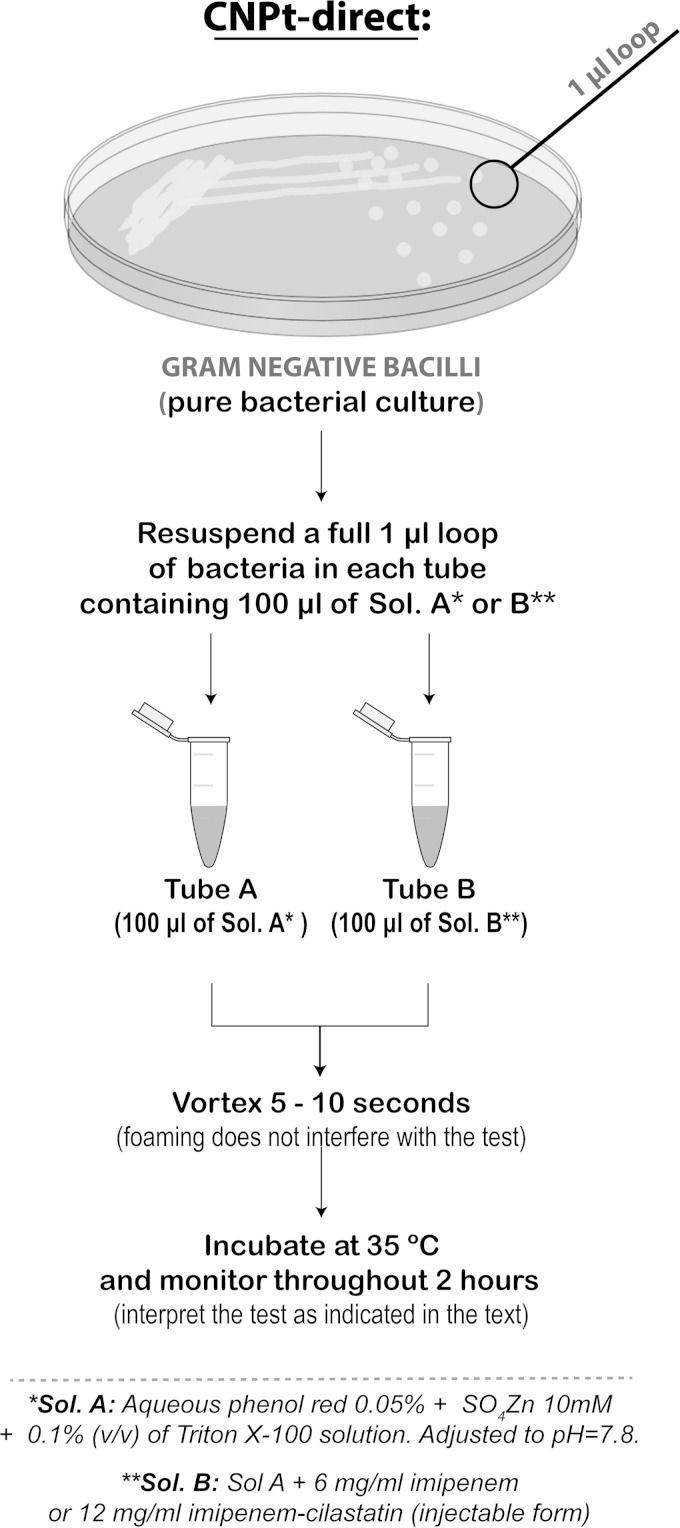
The CNPt was performed following the protocol recommended by CLSI (CNPt-CLSI) (5). Briefly, bacteria were grown overnight on Mueller-Hinton agar (MHA) (BD BBL). The bacterial mass was scraped off with a 1-μl loop and suspended in a 1.5-ml Eppendorf tube containing 100 μl of 20 mM Tris-HCl lysis buffer and mixed using a vortex device during 5 s. This lysate was mixed with 100 μl of an aqueous indicator solution consisting of 0.05% phenol red with 0.1 mmol/liter ZnSO4, previously adjusted to pH 7.8 and 6 mg/ml imipenem (Sigma) or 12 mg/ml imipenem-cilastatin injectable form (equivalent to 6 mg of imipenem standard powder) (reaction tube) and, as a control tube, the phenol red solution without antibiotic. A modified protocol was attempted for the direct use of colonies (instead of bacterial extracts) (CNPt-direct), as follows (Fig. 1): 0.1% (vol/vol) of Triton X-100 (Mallinckrodt, St. Louis, MO) was added to the aqueous indicator mix referred to above (0.05% phenol red-0.1 mmol/liter ZnSO4) before the pH adjustment. This Triton X-100 concentration was selected after preliminary assays using 2-fold dilutions from 0.2% to 0.0125% (vol/vol) (not shown). A full 1-μl loop of a pure bacterial culture recovered from Mueller-Hinton agar was directly suspended in 1.5-ml Eppendorf tubes containing 100 μl of CNPt-direct mix, supplemented with 6 mg/ml imipenem or 12 mg/ml imipenem-cilastatin injectable form (reaction tube) or without antibiotic (control tube). Tubes were vigorously mixed during 5 to 10 s using a vortex device before incubation (foaming does not affect the test). Finally, regardless of the protocol used, tubes were incubated at 35°C and monitored throughout 2 h for color change from red to orange/yellow in the antibiotic-containing tube, which was interpreted as a positive result. Panel strains were tested in duplicate with both protocols. Unsupplemented CNPt-CLSI and CNPt-direct mix solutions were stored at 4°C (pH adjustments were needed after 8 weeks of storage). Test solutions were supplemented with imipenem (standard powder or the injectable form) immediately before being used.
FIG 1.
Schematic representation of the CNPt-direct protocol.
Using the CNPt-CLSI, most class A carbapenemases were detected (Table 1), with the exception of one Pseudomonas aeruginosa KPC-2 producer, one Klebsiella pneumoniae KPC-2 producer with an unusual imipenem MIC of 0.03 μg/ml (M13403 in reference 7) and producers of enzymes with slow imipenem hydrolysis, such as SME or GES. The results were less than optimal with class B and class D carbapenemase producers. Most MBL producers that remained undetected with the CNPt-CLSI were isolates of the tribe Proteeae (Table 1): two Proteus mirabilis isolates, expressing IMP-27 or NDM-1, all three NDM-1-producing Providencia stuartii isolates, and one NDM-1-producing P. rettgeri isolate. Additionally, two Enterobacter cloacae isolates (VIM-2 or VIM-11 producers) with unusual imipenem MICs of 0.25 μg/ml resulted undetected with the CNPt-CLSI. Among the OXA producers, false-negative results with the CNPt-CLSI were found for two OXA-181-producing Escherichia coli isolates, one OXA-48-producing E. coli isolate, one OXA-244-producing E. coli isolate, and two OXA-232-producing K. pneumoniae isolates (Table 1). Most of these false-negative results were consistent with what was demonstrated in previous works (2, 4, 5). These results reduced the sensitivity and negative predictive value (NPV) of the test to 84%. All noncarbapenemase producers were negative by the CNPt-CLSI (Table 1), which confirmed its specificity and positive predictive value (PPV) (100%) (1–5).
The amendments introduced to the CNPt-direct protocol to allow direct use of bacterial colonies helped reverse almost all of the false negatives observed with the CNPt-CLSI technique (Table 1). Only one MBL producer (an IMP-27-producing P. mirabilis isolate) and two OXA-producers (one OXA-244-producing E. coli isolate and one OXA-232-producing K. pneumoniae isolates) remained undetected with the CNPt-direct. These results increased the sensitivity and NPV of the CNPt-direct up to 98% and 97%, respectively. One possible cause of this improved sensitivity is the absence of residual buffer in the CNPt-direct mix compared with the buffer remnant in the CNPt-CLSI reaction mix (about 10 mM Tris-HCl, which results from the 2-fold dilution of the 20 mM Tris-HCl lysate solution after addition of the phenol red solution). A previous report had suggested that this buffer might hinder the color change, even at lower residual buffer concentrations (4.3 mM) obtained with an outdated CNPt protocol (4). To further validate this hypothesis, we conducted a simple titration assay (volumetric analysis) and measured the volume of the titrator (5 mM acetic acid [Sigma]) needed to produce a decrease of 0.2 pH unit (Adwa AD100 pH meter; Hungary) of both a 0.05% phenol red-10 mM Tris-HCl solution (equivalent to the CNPt-CLSI mix) or the aqueous phenol red solution without buffer (as the CNPt-direct mix). The pH endpoint was selected because it was associated with a color change undoubtedly perceived by the human eye. We observed in triplicate experiments that 5.4 times more volume of titrator was needed in the CNPt-CLSI mix than in the CNPt-direct mix (The titration volume was 25 μl versus 135 μl, respectively). Therefore, the absence of buffer in the CNPt-direct mix might significantly contribute to the improved sensitivity, especially for those enzymes with slow imipenem hydrolysis such as GES and OXA-48-like, which are more likely to be hidden by the buffer. The absence of remnants of the buffer in the CNPt-direct mix did not affect the stability of the mix solution during incubation, as we did not observe invalid results (defined as an orange/yellow control tube). Another possible reason for the superior performance of the CNPt-direct is improved (quantitative and/or qualitative) release of beta-lactamases with Triton X-100 than with the commercial buffer, although we cannot probe this hypothesis because the buffer composition is proprietary. The inclusion of small amounts of Triton X-100 in the reaction mix did not affect the specificity and PPV of the CNPt-direct, which was maintained at 100%.
The CNPt-direct was further challenged with the most frequently acquired OXA-type carbapenemases identified in Acinetobacter spp. clinical isolates (Table 1). The CNPt-CLSI results were excluded at this point (only 16/40 [40%] isolates tested positive) since it has already been determined that the test requires modified lysis conditions and an increased bacterial inoculum compared to those stated by the CLSI for optimal detection of this activity (8). Results obtained with the CNPt-direct were very promising as most of the OXA-type producers tested positive (exceptions were two A. baumannii isolates expressing OXA-23 or OXA-58) (Table 1).
Besides the marked improvement in sensitivity (Table 1), the CNPt-direct was easier to perform than the CNPt-CLSI due to the direct use of colonies (instead of bacterial extracts), inexpensive (Triton X-100 is ca. 1,500 times cheaper than the extraction buffer on the market), and, for some producers (35% of the NDM producers), the color indicator turned yellow much faster than with the CNPt-CLSI (usually 30 to 45 min previously). Furthermore, we obtained identical results using either imipenem monohydrate powder or the pharmaceutical form.
In conclusion, we propose here a simplified technique of the CNPt, called CNPt-direct, which facilitates its implementation in routine laboratories due to the dispensability of the extraction buffer with a consequent reduced cost per reaction. For laboratories concerned with the widely disseminated OXA-48-like carbapenemase or the escalation of NDM-1-producing Proteeae species (9–11), this updated protocol enables a more robust detection of these (and other types) of carbapenemase producers, minimizing the number of negative isolates that require confirmation by other methods.
ACKNOWLEDGMENTS
We declare no conflicts of interest.
This work was supported by the regular federal budget of the Ministry of Health of Argentina and by Public Health Ontario Laboratory internal funding.
REFERENCES
- 1.Nordmann P, Poirel L, Dortet L. 2012. Rapid detection of carbapenemase-producing Enterobacteriaceae. Emerg Infect Dis 18:1503–1507. doi: 10.3201/eid1809.120355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tijet N, Boyd D, Patel S, Mulvey M, Melano R. 2013. Evaluation of the Carba NP test for rapid detection of carbapenemase-producing Enterobacteriaceae and Pseudomonas aeruginosa. Antimicrob Agents Chemother 57:4578–4580. doi: 10.1128/AAC.00878-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vasoo S, Cunningham SA, Kohner PC, Simner PJ, Mandrekar JN, Lolans K, Hayden MK, Patel R. 2013. Comparison of a novel, rapid chromogenic biochemical assay, the Carba NP test, with the modified Hodge test for detection of carbapenemase producing gram-negative bacilli. J Clin Microbiol 51:3097–3101. doi: 10.1128/JCM.00965-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Osterblad M, Hakanen AJ. 2014. Evaluation of the Carba NP test for carbapenemase detection. Antimicrob Agents Chemother 58:7553–7556. doi: 10.1128/AAC.02761-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing: 25th informational supplement. CLSI M100-S25. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 6.Pasteran F, Veliz O, Ceriana P, Lucero C, Rapoport M, Albornoz E, Gomez E, Corso A. 2015. Evaluation of the Blue-Carba test for rapid detection of carbapenemases in gram-negative bacilli. J Clin Microbiol 53:1996–1998. doi: 10.1128/JCM.03026-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Veliz O, Pasteran F, Guerriero L, Gomez S, Faccone D, Corso A. 2012. Emergence of ertapenem susceptible (ETP S) KPC-producing Enterobacteriaceae (KPE-PE) with conflicting susceptibility results by reference and routine methods. Abstr 53rd Intersci Conf Antimicrob Agents Chemother, abstr D-598. [Google Scholar]
- 8.Dortet L, Poirel L, Errera C, Nordmann P. 2014. CarbAcineto NP test for rapid detection of carbapenemase-producing Acinetobacter spp. J Clin Microbiol 52:2359–2364. doi: 10.1128/JCM.00594-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pasteran F, Meo A, Gomez S, Derdoy L, Albornoz E, Faccone D, Guerriero L, Archuby D, Tarzia A, Lopez M, Corso A. 2014. Emergence of genetically related NDM-1-producing Providencia rettgeri strains in Argentina. J Glob Antimicrob Resist 2:344–345. doi: 10.1016/j.jgar.2014.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Carvalho-Assef AP, Pereira PS, Albano RM, Berião GC, Chagas TP, Timm LN, Da Silva RC, Falci DR, Asensi MD. 2013. Isolation of NDM-producing Providencia rettgeri in Brazil. J Antimicrob Chemother 68:2956–2957. doi: 10.1093/jac/dkt298. [DOI] [PubMed] [Google Scholar]
- 11.Gefen-Halevi S, Hindiyeh MY, Ben-David D, Smollan G, Gal-Mor O, Azar R, Castanheira M, Belausov N, Rahav G, Tal I, Mendelson E, Keller N. 2013. Isolation of genetically unrelated blaNDM-1-positive Providencia rettgeri strains in Israel. J Clin Microbiol 51:1642–1643. doi: 10.1128/JCM.00381-13. [DOI] [PMC free article] [PubMed] [Google Scholar]