Abstract
Vancomycin-variable enterococcus (VVE) is an emerging pathogen. VVE isolates initially appear phenotypically susceptible to vancomycin but possesses the vanA gene and can develop in vitro and in vivo resistance to vancomycin. We report a case of VVE bacteremia and describe how VVE poses diagnostic and therapeutic dilemmas.
CASE REPORT
In February 2014, a 77-year-old female with a history of coronary artery disease and dyslipidemia underwent an elective hepatic resection and Whipple's procedure for cholangiocarcinoma. Two weeks after the operation, she complained of increased abdominal pain, and a computed tomography (CT) scan demonstrated necrosis of her liver with associated gas, consistent with a leak from her duodenal stump. She was treated with empirical piperacillin-tazobactam for 8 days, and her condition stabilized.
Two months later, she developed tachycardia and leukocytosis. A repeat CT scan of her abdomen demonstrated evolving hepatic abscesses. Blood cultures were positive for Escherichia coli and Bacteroides fragilis, and she was treated again with piperacillin-tazobactam on the basis of susceptibility results. She went to the operating room 1 week later for closure of the duodenal stump leak and drainage of intraabdominal abscesses. The following day, she developed abdominal compartment syndrome and required two additional surgical procedures on consecutive days to evacuate an intraabdominal hematoma. Two sets of blood cultures were obtained because of ongoing fever and hemodynamic instability. Both sets grew an Enterococcus faecium strain reported as resistant to ampicillin (MIC, >32 mg/liter) and susceptible to vancomycin (MIC, 1 mg/liter) by Vitek 2 (bioMérieux, Marcy l'Etoile, France). Intravenous vancomycin was added to the antimicrobial regimen. Persisting hemodynamic instability and fever led to another blood culture being obtained on day 8 of vancomycin therapy (day 21 of therapy with piperacillin-tazobactam). Once again, E. faecium was isolated. On this occasion, Vitek initially failed to produce a susceptibility result; when the test was repeated, the result was a vancomycin MIC of >32 mg/liter. This isolate was sent to the provincial reference laboratory for confirmation of the susceptibility test result; the isolate was confirmed by agar dilution to be a vancomycin-variable enterococcus (VVE) susceptible to vancomycin (MIC, 1 mg/liter). The patient continued to deteriorate despite multiple surgical interventions and the use of broad-spectrum antibiotics. After discussions with the family, care was transitioned to comfort measures and the patient died.
Rectal swabs are routinely obtained from all of the adults admitted to our institution to screen for vancomycin-resistant enterococcus (VRE) by chromogenic agar VRESelect (Bio-Rad, Montreal, Canada). Testing of the index patient's admission rectal swab did not detect VRE. When VVE was clinically identified in the index patient, a total of three point prevalence screens were performed in the two units where the patient had resided; two point prevalence screenings were performed in the intensive care unit (ICU; 12 patients were screened during the first point prevalence screening, and 15 were screened during a follow-up point prevalence screening), and one point prevalence screening was performed on a surgery ward (20 patients were screened). Rectal swabs obtained during the point prevalence screening were tested directly with GeneXpert vanA/vanB PCR (Cepheid). Standard methodology was used to isolate E. faecium in culture and confirm the VVE phenotype and genotype (1). VVE was isolated from the rectal swabs of two patients during the first ICU point prevalence screening, and these were included with three blood culture isolates from the case patient for analysis. Isolates were characterized by traditional biochemical testing, as well as by molecular assays to identify the presence of the vanA, vanB, vanC, vanD, vanE, and vanG genes (2). In addition, the transposon Tn1546-like element was genetically characterized with previously described overlapping primer sets (2). Susceptibility testing was performed by agar dilution, and MIC results were interpreted in accordance with Clinical and Laboratory Standards Institute (CLSI) guidelines (3). Isolates were also typed by pulsed-field gel electrophoresis (PFGE) and multilocus sequence typing (MLST) (2). Informed consent to publish the case report was obtained from the family.
The three blood culture isolates collected from the index patient before and during vancomycin therapy were identical by PFGE, suggesting persistent bacteremia despite effective antimicrobial therapy (Fig. 1). The isolates were resistant to ampicillin but susceptible to vancomycin, daptomycin, and linezolid (Table 1). Molecular assays confirmed that these isolates contained the vanA gene but were negative for the vanB, vanC, vanD, vanȨ and vanG genes. Amplification and sequencing of the genes in transposon Tn1546 identified the presence of vanH, vanA, vanX, vanY, and vanZ. However, none of the isolates contained the orf1, orf2, vanR, or vanS gene. On the basis of these results, it was confirmed that the patient was infected with the recently discovered VVE isolate (1, 2). With respect to the two colonized patients, one of the ICU patients was colonized with a strain identical to the index patient's VVE when PFGE analysis was performed and the other patient was colonized with a strain genetically closely related to the index patient's VVE, thus demonstrating transmission among patients (Fig. 1). These additional isolates had susceptibility profiles identical to that of the index patient's isolates, and all of the isolates were of sequence type 18 (ST18) by MLST (Table 1). Isolates recovered from the colonized patients were also positive for vanHAXYZ gene clusters but negative for the orf1, orf2, and vanRS genes. Patients colonized with VVE were placed on contact precautions (4, 5), and enhanced cleaning of high-touch surfaces with bleach was performed. The subsequent point prevalence screenings (one in the ICU and one on the surgical ward) did not identify additional cases. Neither of the two patients colonized with VVE developed a clinical infection.
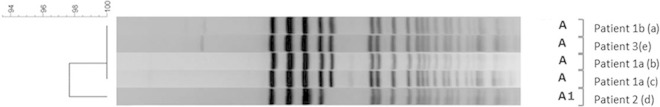
FIG 1.
PFGE of isolates from the index patient bacteremic with VVE (patient 1; isolate 1a was obtained prior to vancomycin use, and isolate 1b was obtained while she was receiving vancomycin) and two patients colonized with VVE (patients 2 and 3). Isolates a, b, c, and e are labeled A since they are genetically identical to each other, whereas isolate d is labeled A1 because it is genetically closely related to the isolates above.
TABLE 1.
Summary of susceptibility profiles, typing results, and the presence of vanA operon genes in VVE isolates recovered from three patients
| Parameter | Resulta for patient (isolate): |
||||
|---|---|---|---|---|---|
| 1a (a)b | 1a (b)b | 1b (c)c | 2 (d) | 3 (e) | |
| Specimen type | Blood | Blood | Blood | Rectal swab | Rectal swab |
| MIC (mg/liter) of: | |||||
| Ampicillin | >8 (R) | >8 (R) | >8 (R) | >8 (R) | >8 (R) |
| Penicillin | >8 (R) | >8 (R) | >8 (R) | >8 (R) | >8 (R) |
| Daptomycin | 4 (S) | 4 (S) | 4 (S) | 4 (S) | 4 (S) |
| Linezolid | 2 (S) | 2 (S) | 2 (S) | 2 (S) | 2 (S) |
| Vancomycin | 1 (S) | 1 (S) | 1 (S) | 1 (S) | 1 (S) |
| PFGE type | A | A | A | A1 | A |
| ST | 18 | 18 | 18 | 18 | 18 |
| Presence of: | |||||
| vanA gene | + | + | + | + | + |
| vanHAXYZ genes | + | + | + | + | + |
| vanRS genes | − | − | − | − | − |
S, susceptibility; R, resistance.
Isolate obtained before vancomycin use.
Isolate obtained during vancomycin use.
VRE colonization and infection rates have increased over the last several decades (6, 7). Among VRE isolates, E. faecium is the most common species and causes both colonization and infection (8). Resistance to vancomycin in E. faecium is conferred primarily by the vanA gene, which encodes a ligase that changes the structure of terminal peptides in its cell wall from d-alanine–d-alanine to d-alanine–d-lactate, thereby reducing the binding affinity of glycopeptides (8). vanA is located on transposon Tn1546, along with several other regulatory and structural genes necessary to confer resistance, i.e., vanR, vanS, vanH, and vanX (8).
A glycopeptide-susceptible, vanA-bearing E. faecium strain was first described in 2011 in Quebec (9). Six isolates were found to lack vanR and vanS, two components of the regulatory system involved in vanHAX gene expression. More recently, an outbreak of VVE was described in Ontario involving 44 patients with isolates once again lacking the vanR and vanS genes (2). More concerning was a case report published at the same time describing the emergence of vancomycin resistance in a patient colonized and infected with VVE after treatment with vancomycin (1). Currently, there is little guidance in terms of the management of patients colonized or infected with VVE.
To our knowledge, this is the first case report of bacteremia associated with VVE. This case raises several important questions. First, should vancomycin be avoided in patients with serious VVE infections? In our case, vancomycin failed to clear VVE from the blood, as the patient continued to be bacteremic after 8 days of vancomycin therapy. However, this may have been due to poor source control, given that the isolate was still susceptible to vancomycin on a repeat blood culture. A patient colonized with this organism was recently described, and it was shown in that case that the isolate became resistant to vancomycin in vivo over time during vancomycin therapy (1). A recent study reported that exposure of VVE strains to vancomycin in vitro leads to the development of vancomycin resistance by the constitutive expression of vanHAXYZ gene clusters (10). The use of vancomycin to treat VVE infections could provide selective pressure that may lead to the development of resistance on therapy and resultant treatment failure. Therefore, it seems prudent to avoid the administration of vancomycin to patients infected with VVE strains.
Second, if vancomycin should be avoided when treating VVE infections, should molecular testing be the standard of care for identifying VVE in all clinical specimens positive for phenotypically vancomycin-susceptible E. faecium? The isolate in this case would not have been identified as having the vanA gene if not for the Vitek system's failure to report a MIC on the repeat blood culture, triggering further testing. Our hospital now performs vanA/vanB PCR testing of any E. faecium isolates obtained from a sterile site. Further surveillance data are needed to better understand the prevalence of VVE in clinical specimens. If VVE becomes an established nosocomial pathogen, it may be appropriate to recommend routine genotypic testing of E. faecium clinical isolates, particularly those from sterile sites, to determine the presence of vancomycin resistance genes.
Last, should hospitals screen for VVE colonization? Routine VRE screening methods do not reliably identify VVE and did not detect the two colonized cases epidemiologically linked to the index case that we identified by molecular testing. A large hospital outbreak due to VVE has also been previously described in Ontario; thus, it is clear that VVE is able to spread within health care facilities and is circulating in the Toronto area (2). We suspect that our patient acquired this organism locally while in our hospital since the patient (and the two colonized patients subsequently identified by point prevalence screening) did not have recent hospitalizations in other health care facilities. In addition, we have shown that VVE can cause a clinically significant infection similar to that caused by VRE, suggesting that VVE should be managed from an infection control perspective as VRE; in our jurisdiction, screening and isolation for VRE are recommended (5). A prior study has shown that some VVE isolates can grow on Brilliance agar selective medium (Oxoid, Canada) and outlines an approach using Brilliance agar selective medium in combination with molecular detection methods (2). However, further research is required to determine the sensitivity of various selective agars in detecting VVE and whether this strategy in combination with molecular testing is cost-effective.
In conclusion, we report the first case of VVE bacteremia. This organism has been shown to develop resistance to vancomycin and raises new questions about screening for VVE and the diagnosis and treatment of VVE infections.
ACKNOWLEDGMENTS
We thank Jenny Seah for her valuable input during the management of the case as the St. Joseph's Health Center intensive care pharmacist and a member of the antimicrobial stewardship team. We also thank Helen Kwan and the St. Joseph's Health Center Microbiology Laboratory for their dedicated work on the identification of this VVE, as well as Bianche Shum at Mount Sinai Hospital Microbiology Laboratory for her valuable input.
REFERENCES
- 1.Coburn B, Low DE, Patel SN, Poutanen SM, Shahinas D, Eshaghi A, Willey BM, McGeer A. 2014. Vancomycin-variable Enterococcus faecium: in vivo emergence of vancomycin resistance in a vancomycin-susceptible isolate. J Clin Microbiol 52:1766–1767. doi: 10.1128/JCM.03579-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Szakacs TA, Kalan L, McConnell MJ, Eshaghi A, Shahinas D, McGeer A, Wright GD, Low DE, Patel SN. 2014. Outbreak of vancomycin-susceptible Enterococcus faecium containing the wild-type vanA gene. J Clin Microbiol 52:1682–1686. doi: 10.1128/JCM.03563-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clinical and Laboratory Standards Institute. 2014. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. CLSI document M100-S24. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 4.Ontario Agency for Health Protection and Promotion Provincial Infectious Diseases Advisory Committee. 2012. Routine practices and additional precautions in all health care settings, 3rd edition Queen's Printer for Ontario, Toronto, ON, Canada. [Google Scholar]
- 5.Ontario Agency for Health Protection and Promotion Provincial Infectious Diseases Advisory Committee. 2013. Annex A—screening, testing and surveillance for antimicrobial resistant organisms (AROs). Annexed to: routine practices and additional precautions in all health care settings, 3rd edition Queen's Printer for Ontario, Toronto, ON, Canada. [Google Scholar]
- 6.Gravel D, Archibald CP, Pelude L, Mulvey M, Golding G, Canadian Nosocomial Infection Surveillance Program . 2014. Antimicrobial resistance surveillance in Canadian hospitals, 2007–2012. Canada Commun Dis Rep 40:S-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ramsey AM, Zilberberg MD. 2009. Secular trends of hospitalization with vancomycin-resistant enterococcus infection in the United States, 2000–2006. Infect Control Hosp Epidemiol 30:184–186. doi: 10.1086/593956. [DOI] [PubMed] [Google Scholar]
- 8.Cetinkaya Y, Falk P, Mayhall CG. 2000. Vancomycin-resistant enterococci. Clin Microbiol Rev 13:686–707. doi: 10.1128/CMR.13.4.686-707.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gagnon S, Lévesque S, Lefebvre B, Bourgault AM, Labbé AC, Roger M. 2011. vanA-containing Enterococcus faecium susceptible to vancomycin and teicoplanin because of major nucleotide deletions in Tn1546. J Antimicrob Chemother 66:2758–2762. doi: 10.1093/jac/dkr379. [DOI] [PubMed] [Google Scholar]
- 10.Thaker M, Kalan L, Waglechner N, Eshaghi A, Patel SN, Poutanen S, Willey B, Coburn B, McGeer A, Low DE, Wright GD. 2015. Vancomycin-variable enterococci can give rise to constitutive resistance during antibiotic therapy. Antimicrob Agents Chemother 59:1405–1410. doi: 10.1128/AAC.04490-14. [DOI] [PMC free article] [PubMed] [Google Scholar]