Abstract
We report the first documented isolation of Wohlfahrtiimonas chitiniclastica from a human in the United States. Initially misidentified as Acinetobacter lwoffii by Vitek-2, the isolate was subsequently identified as W. chitiniclastica by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) and 16S rRNA gene sequencing. While the clinical significance of the isolate in this case is unclear, it highlights the superior performance of MALDI-TOF MS for bacterial identification.
CASE REPORT
A 26-year-old morbidly obese male courier worker was admitted with worsening right leg swelling and draining ulcers. He had developed lymphedema of the right leg during the preceding year after undergoing surgical correction of a right lower extremity varus deformity. A few weeks prior to the current admission, he had noticed right leg ulcers draining yellowish green serosanguineous fluid. On admission, he was afebrile and his white blood cell count was 11,100/μl. He was found to have right lower extremity cellulitis.
A swab was collected from the right leg and sent to the microbiology laboratory for wound cultures. The sample was inoculated onto blood, chocolate, and MacConkey agars and incubated under aerobic conditions at 37°C. Gram staining revealed many Gram-negative bacilli and moderate Gram-positive cocci. Four different organisms were isolated from the swab after growth. Microbial identification of each isolate was performed by using routine biochemical tests, including automated identification by the Vitek 2 system (bioMérieux, Marcy l'Etoile, France). The organisms were identified as Proteus vulgaris, Klebsiella pneumoniae, Acinetobacter lwoffii, and Staphylococcus aureus. The isolates were subsequently analyzed in triplicate by matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (RUO version 3.1, Bruker Biotyper; Bruker Daltonics, Bremen, Germany). Of note, the isolate identified as A. lwoffii by the Vitek 2 system was identified by MALDI-TOF MS as Wohlfahrtiimonas chitiniclastica, with identification log (score) values of 2.253, 2.296, and 2.229. Log (score) values of >2.0 are considered secure to the species level. The isolate was further confirmed to be W. chitiniclastica via 16S rRNA gene sequencing performed at an outside reference laboratory (ARUP Laboratories, Salt Lake City, UT).
By the Vitek 2 system, the W. chitiniclastica isolate was misidentified as A. lwoffii. The probability of identification was computed as 96%, which correlated with “excellent identification.” The organism was relatively biochemically inert on the Vitek 2 system, with positive reactions only for tyrosine arylamidase, Ellman's reagent, l-lactate assimilation, and oxidase. Both organisms are Gram-negative rods, although A. lwoffii often appears as a coccobacillus. The colonies were convex, nonpigmented, and small on blood agar and non-lactose fermenting on MacConkey agar. To compare the biochemical profile of the W. chitiniclastica isolate with that of A. lwoffii, we obtained a control strain of A. lwoffii (isolate group ATCC 17925-2) for analysis. The strain was correctly identified with 99% probability by the Vitek 2 system. The organism showed positive reactions for tyrosine arylamidase, urease, and l-lactate alkalization. However, unlike W. chitiniclastica, A. lwoffii was oxidase negative. Similar results were seen on API 20NE version 7.0 (bioMérieux, France), where W. chitinclastica was identified as A. lwoffii and Brevundimonas diminuta with 98.1% and 88.5% probability, respectively.
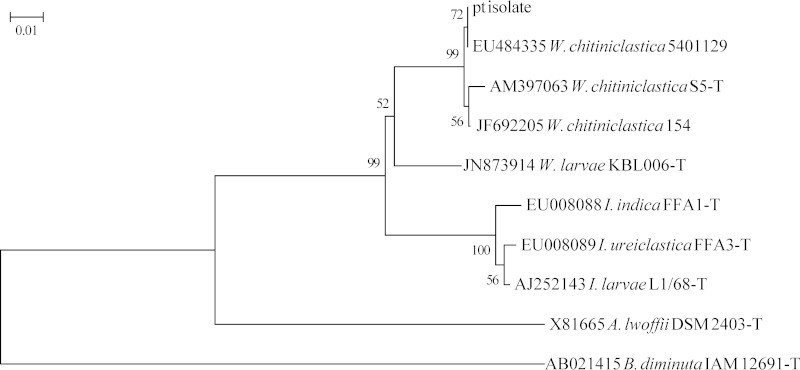
To verify the identification of our patient isolate, the 5′ approximately 500-bp region of the 16S rRNA gene was amplified with primers 5F-t and 534R-t (1), and M13 primers were used to sequence the amplicon bidirectionally. Sequences were assembled and analyzed with Ripseq (Isentio, Palo Alto, CA) and verified by using the GenBank nr database. Phylogenetic analysis of our patient's isolate based on 16S rRNA gene sequences (2, 3) clearly shows that it belongs to the W. chitiniclastica clade and exhibits a much closer relationship to other Wohlfahrtiimonas and Ignatzschineria sp. type strains than either A. lwoffii or B. diminuta (Fig. 1).
FIG 1.
Neighbor-joining phylogenetic tree of partial 16S rRNA gene sequences. Sequences from GenBank are shown with accession numbers, species names, and strain numbers (T indicates a type strain) and were trimmed to the length of the sequence (492 nucleotides) of the patient's (pt) isolate. Bootstrap values from 1,000 replicates are shown at internal nodes. Evolutionary distance is shown as the number of base substitutions per site and was computed in MEGA6 by the maximum composite likelihood method. W. chitiniclastica = Wohlfartiimonas chitiniclastica; W. larvae = Wohlfartiimonas larvae; I. indica = Ignatzschineria indica; I. ureiclastica = Ignatzschineria ureiclastica; I. larvae = Ignatzschineria larvae; A. lwoffii = Acinetobacter lwoffii; B. diminuta = Brevundimonas diminuta.
Testing for susceptibility to seven antimicrobial agents was performed on the Vitek 2 system in accordance with the manufacturer's instructions (AST-GN67). The drugs selected for testing were based on the original identification of the isolate as A. lwoffii. The results are summarized in Table 1, with all of the drugs showing MICs lower than the lowest concentration on the Vitek 2 panel, indicating broad in vitro susceptibility.
TABLE 1.
Antimicrobial susceptibility profile of W. chitiniclastica
| Antimicrobial agent | MIC (μg/ml) |
|---|---|
| Ampicillin-sulbactam | ≤2 |
| Cefepime | ≤1 |
| Ciprofloxacin | ≤0.25 |
| Gentamicin | ≤1 |
| Imipenem | ≤0.25 |
| Levofloxacin | ≤0.12 |
| Trimethoprim-sulfamethoxazole | ≤1/19 |
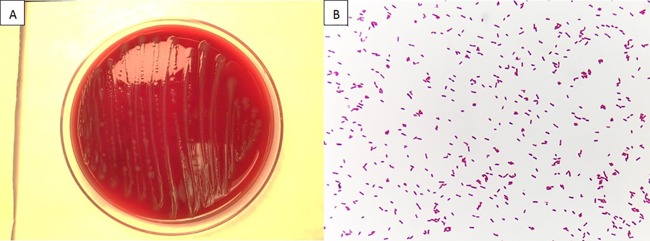
W. chitiniclastica is a Gram-negative, short, straight rod in the class Gammaproteobacteria (Fig. 2B). A. lwoffii belongs to the same class but is a Gram-negative coccobacillus. On blood agar, colonies appear nonpigmented, small, convex, smooth, and glistening (Fig. 2A), which is similar to the appearance of A. lwoffii. The organism is strictly aerobic and catalase and oxidase positive. It was first isolated from the fly Wohlfahrtia magnifica in 2008 (4). W. magnifica is known to carry W. chitiniclastica in its normal gut flora. The fly transmits bacteria to a host animal by laying eggs that subsequently hatch into larvae inside an open wound. Bacteria associated with W. magnifica gut flora are being increasingly isolated and identified by clinical laboratories in recent years, namely, the phylogenetically related Ignatzschineria spp. in human blood and urinary tract infections of myiasis patients (5, 6, 7).
FIG 2.
Sheep blood agar colony (A) and Gram stain (B) morphologies of W. chitiniclastica.
The W. magnifica fly has never been identified in the United States. However, W. chitiniclastica may colonize other species of Wohlfahrtia flies that are geographically distributed in North America, such as Wohlfahrtia vigil and Wohlfahrtia opaca (8). The only previous isolate of W. chitiniclastica described in the United States is from wound and blood cultures in a white-tailed deer that died of septicemia (8).
There are few reported cases of W. chitiniclastica infection worldwide. Most cases occur in countries with warm climates like Morocco and Egypt (9). Known risk factors include poor personal hygiene, alcoholism, peripheral vascular disease, and the presence of a chronic open wound. Many of the patients affected are homeless. Maggots have been identified in some, but not all, of the cases described (8–11). In one previously described case, a patient from Estonia presented similarly to the patient described here (10). Other reported cases that have been described include septicemia and skin and soft tissue infections (8–11).
In our case, no clear history of the route of transmission was found. The isolate was recovered by swabbing a chronic open leg wound of a 26-year-old male. The patient works as a courier, placing him at higher risk of coming into contact with insects. He is also morbidly obese and likely had difficulty cleaning his wound. He did not follow the wound care recommendations of his health care provider, which exacerbated his symptoms. The wound infection was secondary to poor lymphatic drainage of the legs due to previous surgical correction of the right lower extremity complicated by lymphedema. Due to the polymicrobial infection of the ulcers, we cannot assign clear clinical significance to the isolation of W. chitiniclastica. The patient was discharged on a 10-day course of cefpodoxime.
This is the first report of W. chitiniclastica isolated from a human in the United States. Traditional bacterial identification based on phenotypic tests, including broth microdilution and biochemical patterns (12), led to misidentification of the isolate. Careful attention to the Gram stain and MALDI-TOF MS results is essential for proper identification of this emerging pathogen.
ACKNOWLEDGMENT
We are indebted to Flora Samaroo for all her help in the microbiology laboratory.
REFERENCES
- 1.Kallstrom G, Chang T, Albertson M, Morilla D, Fisher MA, Eberly B. 2011. Recovery of a catalase-negative Staphylococcus epidermidis strain in blood and urine cultures from a patient with pyelonephritis. J Clin Microbiol 49:4018–4019. doi: 10.1128/JCM.01031-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chari A, Oakeson KF, Enomoto S, Jackson DG, Fisher MA, Dale C. 2015. Phenotypic characterization of Sodalis praecaptivus sp. nov., a close non-insect associated member of the Sodalis-allied lineage of insect endosymbionts. Int J Syst Evol Microbiol 65(Pt 5):1400–1405. doi: 10.1099/ijs.0.000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tóth EM, Schumann P, Borsodi AK, Kéki Z, Kovács AL, Márialigeti K. 2008. Wohlfahrtiimonas chitiniclastica gen. nov., sp. nov., a new gammaproteobacterium isolated from Wohlfahrtia magnifica (Diptera: Sarcophagidae). Int J Syst Evol Microbiol 58:976–981. doi: 10.1099/ijs.0.65324-0. [DOI] [PubMed] [Google Scholar]
- 5.Maurin M, Delbano JN, Mackaya L, Colomb H, Guier C, Mandjee A, Recule C, Croize J. 2007. Human infection with Schineria larvae. Emerg Infect Dis 13:657–659. doi: 10.3201/eid1304.061151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roudiere L, Jean-Pierre H, Comte C, Zorgniotti I, Marchandin H, Jumas-Bilak E. 2007. Isolation of Schineria sp. from a man. Emerg Infect Dis 13:659–661. doi: 10.3201/eid1304.061255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barker HS, Snyder JW, Hicks AB, Yanoviak SP, Southern P, Dhakal BK, Ghimire GR, Couturier MR. 2014. First case reports of Ignatzschineria (Schineria) indica associated with myiasis. J Clin Microbiol 52:4432–4434. doi: 10.1128/JCM.02183-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thaiwong T, Kettler NM, Lim A, Dirkse H, Kiupel M. 2014. First report of emerging zoonotic pathogen Wohlfahrtiimonas chitiniclastica in the United States. J Clin Microbiol 52:2245–2247. doi: 10.1128/JCM.00382-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rebaudet S, Genot S, Renvoise A, Fournier PE, Stein A. 2009. Wohlfahrtiimonas chitiniclastica bacteremia in homeless woman. Emerg Infect Dis 15:985–987. doi: 10.3201/eid1506.080232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kõljalg S, Telling K, Huik K, Murruste M, Saarevet V, Pauskar M, Lutsar I. 2015. First report of Wohlfahrtiimonas chitiniclastica from soft tissue and bone infection at an unusually high northern latitude. Folia Microbiol (Praha) 60:155–158. doi: 10.1007/s12223-014-0355-x. [DOI] [PubMed] [Google Scholar]
- 11.Almuzara MN, Palombarani S, Tuduri A, Figueroa S, Gianecini A, Sabater L, Ramirez MS, Vay CA. 2011. First case of fulminant sepsis due to Wohlfahrtiimonas chitiniclastica. J Clin Microbiol 49:2333–2335. doi: 10.1128/JCM.00001-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Senga P, Drancourta M, Gouriet F, La Scola B, Fournier PE, Rolain JM, Raoult D. 2009. Ongoing revolution in bacteriology: routine identification of bacteria by matrix-assisted laser desorption ionization time-of-flight mass spectrometry. Clin Infect Dis 49:543–551. doi: 10.1086/600885. [DOI] [PubMed] [Google Scholar]