Abstract
Tongue squamous cell carcinoma (TSCC) is most aggressive head and neck cancer often associated with HR-HPV infection. The role of AP-1 which is an essential regulator of HPV oncogene expression and tumorigenesis is not reported in tongue cancer. One hundred tongue tissue biopsies comprising precancer, cancer and adjacent controls including two tongue cancer cell lines were employed to study the role of HPV infection and AP-1 family proteins. An exclusive prevalence (28%) of HR-HPV type 16 was observed mainly in well differentiated tongue carcinomas (78.5%). A higher expression and DNA binding activity of AP-1 was observed in tongue tumors and cancer cell lines with c-Fos and Fra-2 as the major binding partners forming the functional AP-1 complex but c-Jun participated only in HPV negative and poorly differentiated carcinoma. Knocking down of Fra-2 responsible for aggressive tongue tumorigenesis led to significant reduction in c-Fos, c-Jun, MMP-9 and HPVE6/E7 expression but Fra-1 and p53 were upregulated. The binding and expression of c-Fos/Fra-2 increased as a function of severity of tongue lesions, yet selective participation of c-Jun appears to promote poor differentiation and aggressive tumorigenesis only in HPV negative cases while HPV infection leads to well differentiation and better prognosis preferably in nonsmokers.
Head and neck squamous cell carcinoma (HNSCC) is an extremely heterogeneous group of cancers arising from different subsites such as tongue, lips, larynx and other intra-oral locations1,2. This clinical heterogeneity in terms of the site of origin also correlates with specific risk-factors, symptoms, tendency to local and distant metastasis, sensitivity to chemo-radiotherapy and the disease prognosis3,4.
Among HNSCCs, tongue squamous cell carcinoma (TSCC) is one of the most common but highly aggressive cancer particularly in younger patients and is associated with a higher rate of metastasis with poor prognosis5,6,7. In India, the incidence of TSCC is second highest in the world8. While smoking, tobacco, betel nut chewing and alcoholism are primary risk-factors, studies have reported that infection of HPV particularly high-risk HPV (HR-HPV) type 16 may also act as an independent risk-factor in inducing a substantial proportion of tongue cancer6,7,9. It has been demonstrated that unlike cervical cancer, TSCCs including oral squamous cell carcinoma and other HNSCCs with HR-HPV infection show better prognosis6,7,9,10,11,12,13,14. This has been further shown to be due to selective participation of NF-κB p65 that induces well differentiation of tumors leading to better prognosis11.
It is well established that transcription factor activator protein-1 (AP-1) formed by homo or hetero-dimerization between Jun (c-Jun, Jun-B, Jun-D) and Fos (c-Fos, FosB, Fra-1, Fra-2) family proteins15 plays a central role in HPV oncogene expression and tumorigenesis16,17. A high DNA binding activity and differential overexpression of AP-1 family proteins have been reported in many cancers, suggesting a pivotal role of AP-1 in tumor progression and metastasis14,18,19,20,21,22. AP-1 activation is known to further upregulate various downstream target genes such as cyclin Dl, c-myc, Bcl-xl, MMP-9, EGFR, and specific miRNAs etc., that are acitively involved in progression, metastasis and aggressive phenotype of various tumors14,19,20,21.
Several studies have demonstrated differential expression and high DNA binding activity of specific members of AP-1, particularly c-Fos, c-Jun and JunB during development of variety of carcinomas including oral carcinoma18,19,20,22. In contrast, Fos-related antigen 1 (Fra-1) has been shown to overexpress only in normal but absent in cancer cases (except breast cancer) indicating its possible tumor suppressor activity in these tumors20,22,23 while the Fos-related antigen 2 (Fra-2) has been found to be often highly upregulated in many cancers which show aggressive tumor phenotype and metastasis19,24,25. Although, aberrant activation and differential expression pattern of AP-1 family proteins have been reported in many cancers including oral cancer18,19,20,22, to date, there is no study that defines the role of AP-1 and its family proteins during tongue carcinogenesis. Therefore, the present study has been carried out, to investigate the role of AP-1 and its family proteins in different stages of tongue cancer including its precancer lesions to understand the contribution of specific AP-1 family proteins in presence or absence of HPV infection and their crosstalk in aggressive tongue carcinogenesis. The results demonstrated a selective interaction of c-Jun with Fra-2/c-Fos in absence of HPV promotes aggressive and invasive tongue tumorigenesis and poor prognosis while HPV infection facilitates well differentiation and better prognosis.
Results
A total of one hundred prospectively collected fresh tongue tissue biopsy specimens comprising precancer (n = 20), cancer (n = 50) and adjacent normal controls (n = 30) and two tongue cancer cell lines (UPCI:SCC090 and AW13516) were employed for the detection of HPV infection, HPV genotypes and analysis of expression and activation of AP-1 family proteins. The Clinico-epidemiological and demographical details along with status of HPV infection are presented in Table 1. The majority of tongue cancer patients (84%; 42/50) were smokers and males (n = 40) with a mean age of 40.48 ± 12.46 years.
Table 1. Clinico-pathological and demographic characteristics and their correlation with HPV16 infection in tongue cancer patients.
| Characteristics | No. of cases (%) | No. of HPV positive | No. of HPV Negative | p-value | |
|---|---|---|---|---|---|
| Adjacent normal controls | 30 | nil | — | — | |
| Precancer (n = 20) | 36.7 ± 5.99 | Mean age ± SD | nil | — | |
| Precancer (n = 20) | Leukoplakia/Erthroplakia | 11 (55%) | nil | — | — |
| Dysplasia | 9 (45%) | nil | — | — | |
| Cancer (n = 50) | 40.48 ± 12.46 | Mean age ± SD | 14 (28%) | 36 (72%) | |
| Age range | <35 | 30 (60%) | 10 (71.4%) | 20 (55.6%) | 0.3 (ns) |
| >35 | 20 (40%) | 4 (28.6%) | 16 (44.4%) | ||
| Gender | Male | 40 (80%) | 6 (42.9%) | 34(94.4%) | 0.0002 |
| Female | 10 (20%) | 8 (57.1%) | 2(5.6%) | ||
| Religion | Hindu | 40 (80%) | 13 (92.9%) | 27 (75%) | 0.2 (ns) |
| Muslim | 10 (20%) | 1 (7.1%) | 9 (25%) | ||
| Addiction habit (smoking & chewing) | Smokers | 42 (84%) | 8 (57.1%) | 34(94.4%) | 0.003 |
| Non-smokers | 8 (16%) | 6 (42.9%) | 2(5.6%) | ||
| Tumor site | Base of tongue | 22 (44%) | 12 (85.7%) | 10 (27.8%) | 0.0003 |
| Mobile tongue & other sites of tongue | 28 (56%) | 2 (14.3%) | 26 (72.2%) | ||
| Differentiation status | WDSCC | 15 (30%) | 11 (78.6%) | 4(11.2%) | 0.0001 |
| MDSCC | 8 (16%) | 2 (14.3%) | 6 (16.7%) | ||
| PDSCC | 27 (54%) | 1 (7.1%) | 26 (72.2%) | ||
| Tumor status | T1-T2 | 20 (40%) | 11 (78.6%) | 9 (25%) | 0.0009 |
| T3-T4 | 30 (60%) | 3 (21.4%) | 27 (75%) | ||
| Node Status | N0-N1 | 39 (78%) | 13 (92.9%) | 26 (72.2%) | 0.2 (ns) |
| N2-N3 | 11(22%) | 1 (7.1%) | 10 (27.8%) | ||
| Clinical Staging | Stage I–II | 14 (28%) | 10 (71.4%) | 4 (11.1%) | 0.0001 |
| Stage III–IV | 36 (72%) | 4 (28.6%) | 32 (88.9%) | ||
#n: number of patients. T: primary tumor. N: regional lymph node. Subjects comprised normal controls, precancer and HPV−ve and HPV16+ve cancer patients. P-values were obtained by probability Fisher’s (exact) test using Graph pad Prism 6.0. P-values in bold (≤0.05) are considered as statistically significant.
Exclusive prevalence of HR-HPV type 16 mainly in well differentiated tongue squamous cell carcinoma
The analysis of HPV infection and its genotype distribution in tongue cancer by HPV type-specific PCR and reverse line blot (RLB) assay demonstrated only 14 (28%) HPV+ve cases out of 50 TSCCs studied and all of them were positive exclusively for HR-HPV type 16 and no other known HPV type could be detected (supplementary Fig. 1). All precancer cases and normal adjacent controls were also found to be negative for HPV (Table 1). Of twenty precancer cases, 55% (11/20) were leukoplakia/eyrthoplakia while 45% (9/20) were dysplasia. Out of all HPV+ve TSCCs (n = 14), majority (57.2%; 8/14) of them were females. The majority (78.6%, p ≤ 0.01) of HPV infection was found in well differentiated tumors (WDSCCs) in comparison to MDSCCs and PDSCCs (see Table 1) and belonged to early stage tumors (stage I and II; 71.4%, p ≤ 0.01). Interestingly, a very low frequency of HPV infection was observed in TSCC patients who were smokers (19%; 8/42) when compared to nonsmokers (75%, 6/8; p ≤ 0.01), the majority of whom were HPV+ve (Table 1).
Higher DNA binding activity of AP-1 formed by heterodimerization of c-Fos/Fra-2 and selective participation of c-Jun
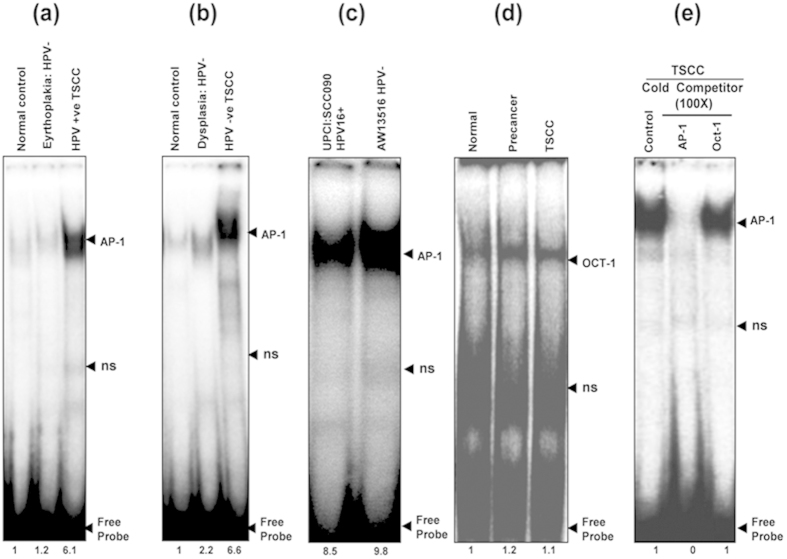
The DNA binding activity of AP-1 was analyzed by band shift assay in nuclear extracts derived from fresh tongue tissue biopsies of precancer (n = 20), cancer (n = 50) and normal adjacent control tissues (n = 30) as well as in two tongue cancer cell lines (UPCI:SCC090; HPV+ve and AW13516; HPV−ve) using γ32P-labeled probe harboring AP-1 consensus sequence. As presented in Fig. 1a–c, a higher AP-1 DNA binding activity was detected in all tongue cancer cases and cell lines, whereas it was low or nil in normal adjacent controls but a moderate binding activity was observed in precancer lesions (Fig. 1a,b). EMSA showed a gradual increase in AP-1 binding activity with the increasing severity of the lesion but relatively a higher DNA binding was observed in HPV negative tumors (Fig. 1b). The specificity of binding activity was confirmed by performing cold competition assay with 100-fold molar excess of homologous AP-1 and a heterologous Oct-1 cold probe (Fig. 1d,e).
Figure 1.
(a–e): Constitutive activation and higher DNA binding activity of AP-1 in HPV+ve and HPV−ve tongue tumor tissues and cell lines. EMSA showing DNA binding activity of AP-1 in nuclear extracts (10 μg) of different grades of tongue tissues (a,b) and cell lines (c) using γ32P- ATP-radiolabeled oligonucleotide harboring an AP-1 consensus sequence. Increasing AP-1 binding activity was observed as the severity of tongue lesions progressed from normal to precancer to invasive cancer in both HPV16+ve (a) and HPV−ve (b) cases. Both HPV16+ve UPCI:SCC090 and HPV−ve AW13516 (c) cell lines showed higher binding activity of AP-1. (d) EMSA with labeled Oct-1 probe showed uniform DNA binding in all grades of tongue tissues. Binding specificity was evidenced in a competition assay with nuclear extracts of TSCCs incubated with unlabelled 100 molar excess of specific competitor AP-1 probe and nonspecific competitor Oct-1 probe (e). Fold change in the band intensities of AP-1 are indicated in each lane. Ns: non-specific binding.
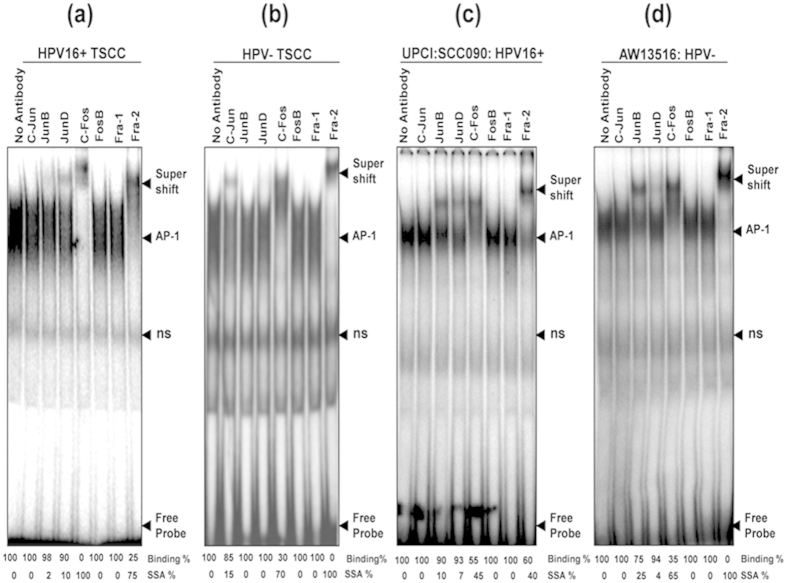
The composition of functional AP-1 complex in both HPV+ve and HPV−ve tumors was further dissected in band supershift assay with specific antibodies to identify the extent of participation of each AP-1 family proteins. The band shift assay demonstrated, c-Fos and Fra-2 as the major DNA binding partners involved in the formation of functional AP-1 complex with minor involvement of JunD/JunB in HPV+ve well differentiated tumors (64.3%, 9/14) (Fig. 2a & Table 2). In contrast, majority (80.6%, 29/36) of poorly differentiated HPV−ve cases showed minor but consistent involvement of c-Jun along with c-Fos and Fra-2 (Fig. 2b & Table 2). In almost all cases, about 90% of supershifted band was formed by c-Fos and Fra-2 which showed a total supershift making them the chief constituents of functional AP-1 in tongue cancer. Both tongue cancer cell lines also showed similar functional AP-1 complex formation mainly by c-Fos and Fra-2 along with JunB and JunD though they differ in their extent of involvement (Fig. 2c,d). HPV+ve cells however, showed only about 40% involvement of Fra-2 (Fig. 2c) and no involvement of c-Jun in HPV−ve cells (Fig. 2d).
Figure 2.
(a–d): Altered composition of functional AP-1 complex in HPV+ve and HPV−ve tongue tumor tissues and cell lines. Band supershift assay using specific antibodies (Abs) against all AP-1 family members (c-Jun, JunB, JunD, c-Fos, FosB, Fra-1, Fra-2). Panel a, b, c and d showing differential binding activity of AP-1 members in HPV+ve (a) and HPV−ve (b) TSCCs and tumor cell lines; UPCI:SCC090;HPV16+ve(c) and AW13516; HPV−ve(d). In all panels (a–d) significantly a higher (~90) binding of c-Fos and Fra-2 forms the functional AP-1 complex. HPV+ve TSCC (panel a) shows a minor participation of JunD and JunB while c-Jun selectively participates in HPV−ve tumors (panel b). The arrowhead indicates the supershifted bands. The intensities of super-shifted bands indicated and quantified in densitometric analysis. NS; non-specific binding.
Table 2. Cumulative data of activation of AP-1 family proteins in HPV16+ve & HPV−ve tongue cancer cases as observed by band supershift assay.
| AP-1 family proteins(↓) | TSCC (n = 50) |
p-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HPV16 positive TSCC (n = 14) |
HPV negative TSCC (n = 36) |
||||||||
| Nil - | Weak + | Moderate ++ | Strong ++++ | Nil - | Weak + | Moderate ++ | Strong ++++ | ||
| c-Jun | 14 | — | — | — | 7 | 9 | 20 | — | 0.0001 |
| JunB | 9 | 5 | — | — | 36 | — | — | — | 1.0 (ns) |
| JunD | 5 | 3 | 6 | — | 36 | — | — | — | 0.0001 |
| c-Fos | — | — | 2 | 12 | — | 1 | 6 | 29 | 1.0 (ns) |
| FosB | 14 | — | — | — | 36 | — | — | — | 1.0 (ns) |
| Fra-1 | 14 | — | — | — | 36 | — | — | — | 1.0 (ns) |
| Fra-2 | — | 2 | 3 | 9 | — | 2 | 2 | 32 | 0.31 (ns) |
#Data represent DNA binding activity of different AP-1 family proteins in >90% of cases with minor changes in few cases. Arbitrary level of DNA binding in band supershift assay: Nil/not detectable (0%, −); Weak (~25%, +); Moderate (~50%, ++); Strong (>80%, ++++). P-value obtained by probability Fisher’s (exact) test using Graph pad Instat (6.0) comparing the DNA binding of AP-1 proteins (nil+weak vs moderate+strong) among HPV16+ve cancer vs. HPV−ve cancer. P-values in bold (≤0.05) are considered as statistically significant. ns = not significant.
Differential overexpression of specific AP-1 family proteins in TSCC tissues and cell lines
Western blotting was performed to study the expression pattern of AP-1 family proteins (c-Jun, JunD, JunB, c-Fos, FosB, Fra-1 and Fra-2) in all spectrum of tongue tissue biopsies from normal, precancer to cancer including TSCC cell lines. AP-1 proteins, c-Jun (60%; 30/50, p = 0.0001), c-Fos (80%; 40/50, p = 0.0001) and Fra-2 (76%; 38/50, p = 0.0001) showed significantly a higher level of expression specifically in majority of HPV−ve tongue cancer cases as compared to precancer and normal controls (Fig. 3a, Table 3). These data very well corroborate the results of band shift assay (Fig. 2a,b). Also, JunB (p = 0.003) and JunD (p = 0.0001) proteins showed elevated expression but Fra-1 showed a low or no expression (Fig. 3a & Table 3) in tumors but it was highly expressed in normal controls (p = 0.0001) (Fig. 3a). Interestingly, despite all AP-1 proteins were gradually overexpressed with the increasing severity of lesions, only a few participated in AP-1 DNA binding activity. When, we differentiated between HPV+ve and HPV−ve tumors, we observed relatively a higher level of c-Jun, c-Fos and Fra-2 expression in HPV−ve tumors as compared to those in HPV+ve TSCCs. Though, other members except Fra-1 showed a higher expression, there was no significant difference with respect to HPV status (Fig. 3a & Table 4). Overall, gradually an increased expression of c-Jun, JunB, JunD, c-Fos and Fra-2 was observed as the lesions progressed towards malignant phenotype. Both the tongue cancer cell lines also showed comparable results but Fra-1, however showed higher expression in both cell lines (Fig. 3b).
Figure 3.
(a–e): Differential expression pattern of AP-1 family proteins and their target genes in different grades of tongue tissues and cell lines. Representative immunoblots showing overexpression of AP-1 family proteins (c-Jun, JunB, JunD, c-Fos & Fra-2) and their target genes (Bcl-2, MMP-9 and Cyclin D1) in tongue tumor biopsies (a) and cell lines (b). 40 μg cellular protein from tongue tissues and cell lines were resolved on 10% SDS-PAGE, electrotransferred and probed with specific antibodies against all AP-1 family proteins and their target genes (Bcl-2, MMP-9 and Cyclin D1). To confirm equal loading of protein, the membranes were stripped and re-probed for β-actin expression and quantitation of bands was performed by densitometric analysis (lower panel of a & b). Note that there is significantly a higher expression of c-Fos & Fra-2 in tumor tissues, while Fra-1 is completely absent. Panel c; RT-PCR showing elevated mRNA expression profile of c-Jun, c-Fos & Fra-2 transcripts in tongue tumor tissues (c). GAPDH gene was used as internal loading control (lower panel of c). (Panel d,e); showing overexpression of Bcl-2, MMP-9 and Cyclin D1 genes in HPV+ve and HPV−ve tongue tumor tissues (d) and cell lines (e). β-actin gene was used as an internal control (lower panels of d,e). Significant (p < 0.05) when compared with control vs. cases and HPV+ve vs. HPV−ve cell lines. The data in bar diagram are expressed as the mean (±SEM or ±SD) of three independent experiments. *p < 0.05 & **p < 0.01, ***p < 0.001.
Table 3. Cumulative expression pattern of AP-1 family proteins and their downstream gene products in precancer and cancer of the tongue and normal adjacent tongue tissue as observed by immunoblotting.
| Target Proteins(↓) Expression Level (→) |
Normal adjacent controls (n = 30) |
Precancer (n = 20) |
Tongue cancer (n = 50) |
p-value | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Nil (−) | Weak (+) | Moderate (++) | Strong (++++) | Nil (−) | Weak (+) | Moderate (++) | Strong (++++) | Nil (−) | Weak (+) | Moderate (++) | Strong (++++) | ||
| c-Jun | 6 | 14 | 10 | — | — | 10 | 8 | 2 | — | 4 | 16 | 30 | 0.2 (ns)1, 0.00012, 0.00023 |
| JunB | 2 | 10 | 15 | 3 | 2 | 5 | 10 | 3 | 2 | 3 | 20 | 25 | 0.7 (ns)1, 0.0032, 0.0063 |
| JunD | 5 | 15 | 10 | — | 1 | 4 | 12 | 3 | 1 | 5 | 12 | 32 | 0.0081, 0.00012, 0.4 (ns)3 |
| c-Fos | 2 | 20 | 8 | — | — | 2 | 10 | 8 | — | — | 10 | 40 | 0.00011, 0.00012, 0.073 |
| FosB | 4 | 9 | 14 | 3 | — | 5 | 10 | 5 | 2 | 8 | 10 | 30 | 0.2 (ns)1, 0.042, 0.7 (ns)3 |
| Fra-1 | — | 5 | 10 | 15 | 1 | 4 | 10 | 5 | 27 | 14 | 7 | 2 | 0.4 (ns)1, 0.00012, 0.00013 |
| Fra-2 | 5 | 20 | 5 | — | — | 2 | 10 | 8 | — | — | 12 | 38 | 0.00011,0.00012, 0.073 |
| Bcl-2 | 4 | 18 | 8 | — | — | 5 | 9 | 6 | — | 1 | 15 | 34 | 0.4 (ns)1, 0.062, 0.0063 |
| MMP-9 | 6 | 10 | 12 | 2 | — | 2 | 15 | 3 | — | — | 10 | 40 | 0.0021, 0.00012, 0.073 |
| CyclinD1 | 7 | 12 | 10 | 1 | 3 | 4 | 9 | 4 | 1 | 3 | 7 | 39 | 0.0081, 0.00012, 0.0093 |
#Arbitrary level of expression as detected by immunoblotting: Nil/not detectable (−); Weak (+); Moderate (++); Strong (++++). P-value obtained by probability Fisher’s (exact) test using Graph pad Instat 6.0 comparing the expression of proteins (nil + weak vs. moderate + strong) among 1Control vs. Pre-cancer, 2Control vs. Cancer, 3Pre-cancer vs. Cancer. P-values in bold (≤0.05) are considered as statistically significant. ns = not significant.
Table 4. Cumulative expression patterns of AP-1 family proteins and its downstream genes in HPV16+ve and HPV−ve tongue cancer cases.
| Target proteins(↓) Expression Level (→) | TSCC (n = 50) |
p-value | |||||||
|---|---|---|---|---|---|---|---|---|---|
|
HPV16 positive TSCC (n = 14) |
HPV negative TSCC (n = 36) |
||||||||
| Nil − | Weak + | Moderate ++ | Strong ++++ | Nil − | Weak + | Moderate ++ | Strong ++++ | ||
| c-Jun | — | 3 | 6 | 5 | — | 1 | 10 | 25 | 0.06 |
| JunB | 2 | 2 | 4 | 6 | — | 1 | 16 | 19 | 0.01 |
| JunD | — | 2 | 2 | 10 | 1 | 3 | 10 | 22 | 1.0 (ns) |
| c-Fos | — | — | 3 | 11 | — | — | 7 | 29 | 1.0 (ns) |
| FosB | — | 3 | 4 | 7 | 2 | 5 | 6 | 23 | 1.0 (ns) |
| Fra-1 | 6 | 7 | 1 | — | 21 | 7 | 6 | 2 | 0.4 |
| Fra-2 | — | — | 2 | 12 | — | — | 10 | 26 | 1.0 (ns) |
| Bcl-2 | — | 1 | 5 | 8 | — | — | 10 | 26 | 0.2 |
| MMP-9 | — | — | 6 | 8 | — | — | 4 | 32 | 1.0 (ns) |
| Cyclin D1 | 1 | 3 | 2 | 8 | — | — | 5 | 31 | 0.004 |
#Arbitrary level of expression as detected by immunoblotting: Nil/not detectable (−); Weak (+); Moderate (++); Strong (++++). P-value obtained by probability Fisher’s (exact) test using Graph pad Instat 6.0 comparing the expression of proteins (nil + weak vs. moderate + strong) among HPV16 + ve Cancer vs. HPV−ve Cancer. P-values in bold (≤0.05) are considered as statistically significant. ns = not significant.
Higher expression of AP-1 family gene transcripts in primary tongue tumors and cell lines
To compare AP-1 family protein expression, the gene transcripts were evaluated in total RNA isolated from tongue tissue biopsies from cancer, precancer and normal controls and tongue cancer cell lines by RT-PCR. As shown in Fig. 3c, the majority of tongue cancer cases showed higher expression of c-Jun, c-Fos and Fra-2 transcripts as compared to precancer and normal controls but the expression level of c-Fos and Fra-2 transcripts well corroborated the results of western blotting (Fig. 3a,b). We also observed similar results with tongue cancer cell lines but the expression of these genes was slightly higher in HPV−ve cells (figures not provided).
Upregulation of AP-1 target genes; MMP-9, CyclinD1 and Bcl-2 in tongue tumorigenesis
The possible factors that may contribute in potential tumor invasion and metastasis are MMP-9, Bcl-2 and Cyclin D1 genes which are known to be regulated by AP-1. We therefore, examined the expression pattern of these genes in tongue tissue biopsies and cell lines by immunoblotting. As expected significantly a higher expression of MMP-9. Bcl-2 and Cyclin D1 proteins (P < 0.05) (Fig. 3d & Table 3) was observed in the majority of cancer cases as compared to those in precancer and adjacent normal controls. As high as 80% (40/50; P = 0.0001) of tongue cancer cases showed strong expression of MMP-9, 68% (34/50; p = 0.06) for Bcl-2 and 78% (39/50; P = 0.0001) for cyclin D1 genes (Fig. 3d & Table 3). In order to examine the influence of HPV infection on these genes, we categorized them in HPV+ve and HPV−ve tumors. As depicted in the Fig. 3d and Table 4, we observed a slightly higher level of expression of all these genes in HPV−ve tongue tumors. These target genes were also found to be upregulated in both the tumor cell lines but they are more pronounced in HPV−ve cells (Fig. 3e).
Fra-2 silencing reduced expression of c-Jun, c-Fos, MMP-9 and HPV16 E6/E7 but upregulated p53 in tongue cancer cells
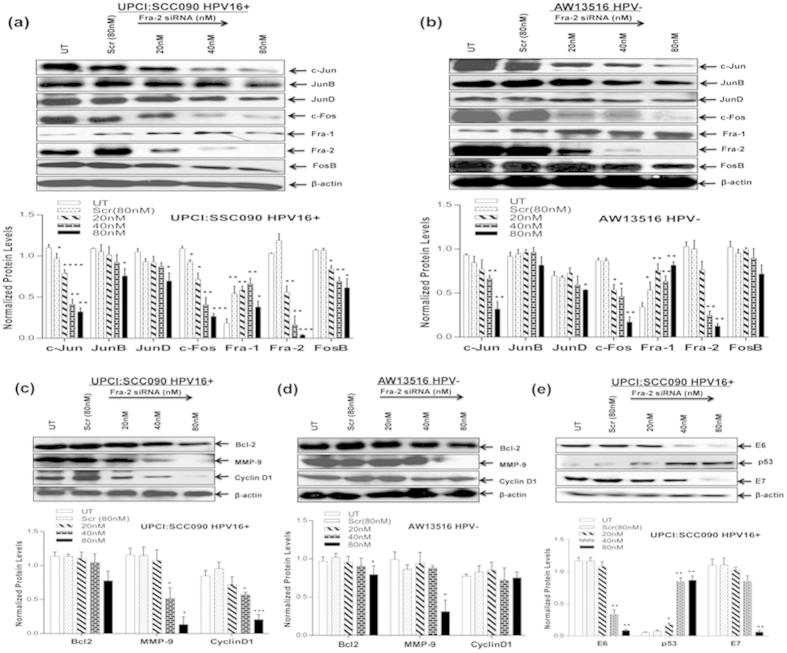
Since we observed significantly a higher expression and transactivation of Fra-2 particularly in HPV−ve aggressive tongue cancer tissues and cell lines, we examined the extent of functional contribution of Fra-2 in aggressiveness of the disease by silencing Fra-2 using Fra-2 specific siRNA in both HPV +ve and HPV−ve tongue cancer cell lines. As presented in the Fig. 4a,b, transfection of these cells by increasing concentration (20 nM to 80 nM) of Fra-2 siRNA resulted in a dose-dependent decline in the expression of Fra-2, while, it remained unchanged in scrambled siRNA treated cells even at the highest concentration (80 nM) used. Inhibition of Fra-2 expression was discernible even at 20 nM and a complete silencing was observed at 80 nM in both the cancer cells (Fig. 4a,b).
Figure 4.
(a–e): Effect of Fra-2 silencing by specific siRNA in HPV+ve and HPV−ve TSCC cells. Representative immunoblots (Panel a,b) showing complete inhibition of Fra-2 expression at the concentration of 80 nM in both UPCI:SCC090;HPV16+ve (a) and AW13516;HPV−ve cells (b). 40 μg cellular proteins isolated from both the cell lines following treatment with Fra-2-siRNA (20 nM-80 nM) and scrambled control siRNA (80 nM) for 48 hours were examined for its effects on AP-1 family proteins (a,b), their target genes; Bcl-2, MMP-9 and CyclinD1 (c,d) and HPV16 E6/E7 (e). To ensure equal loading of protein, β-actin was used as an internal control. Quantitation of bands was performed by densitometric analysis. Significant (p < 0.05) when compared with untreated control vs. treated group. The data in bar diagram are expressed as the mean ± SD of three independent experiments. *p < 0.05 & **p < 0.01.
Interestingly, Fra-2 downregulation was accompanied by a signaling that led to concomitant reduction of specific AP-1 proteins c-Fos and c-Jun expression which were also upregulated and considered important for aggressive tongue tumorigenesis (Fig. 4a,b). Curiously enough, Fra-1 which was completely absent in TSCCs showed an elevated expression as the concentration of siRNA increased in both the cell lines though it was less pronounced in HPV+ve cells (Fig. 4a,b). Most interestingly, silencing of Fra-2 led to a significant (~95%) downregulation of viral oncogenes E6/E7 expression at 80 nM concentration and a dose-dependent accumulation of p53 level in HPV+ve cells (Fig. 4e). p53 expression was detectable in cells treated with as low as 20 nM and maximum accumulation of p53 was observed at 80 nM. In contrast, untreated/scrambled siRNA treated cells showed very low or negligible expression of p53 (Fig. 4e).
In order to examine the effect of Fra-2 silencing on AP-1 target genes associated with aggressive tumorigenesis, we checked the expression of Bcl-2, MMP-9 and cyclin D1 in siRNA treated tongue cancer cell lines. As shown in Fig. 4c,d, Fra-2 siRNA at the 80 nM concentration resulted in >90% loss of specifically MMP-9 expression while it remained unchanged in cells treated with equi-molar concentration of scrambled siRNA. The expression of cyclin D1 and Bcl-2 remained almost unremarkable in HPV−ve cells (Fig. 4d) but cyclin D1 expression reduced >70% mainly in HPV+ve cells (Fig. 4c). The cancer cells transfected with Fra-2-siRNA (20–80 nM) at 48 hr or prior to harvesting, resulted in only about 30% reduction in cell proliferation (Supplementary Fig. 2a & b).
Fra-2 silencing reduced invasion and migration of tongue cancer cells in vitro
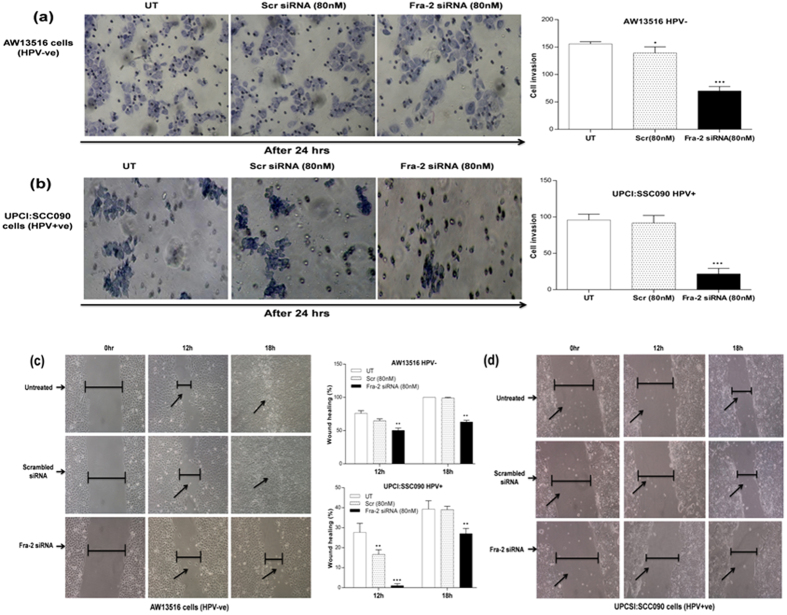
For evaluation of the potential effects of Fra-2 silencing on the invasive properties of both HPV+ve and HPV−ve tongue cancer cell lines, we have employed Matrigel invasion assay (Fig. 5a,b) using standard procedure24. The staining of invading cells through the polycarbonate membranes demonstrated that the invasion of cells was reduced after Fra-2 siRNA silencing when compared with invasion of untreated or scrambled siRNA treated cells (Fig. 5a,b). Transfection with Fra-2 siRNA resulted in a remarkable decrease in the number of invading cells in both HPV+ve and HPV−ve cells. Interestingly, Fra-2 silencing was more effective in HPV+ve cells to prevent cell invasion through the Matrigel as compared with HPV−ve cells. Quantitation of the invading HPV−ve cells showed only 45% (±70 cells, p = 0.001) invasion after treatment with Fra-2 siRNA when compared to untreated (±155.6 cells; 100%) or scrambled siRNA treated cells (±139.5 cells; 90%) (Fig. 5a). In HPV16+ve cells, we observed only 28% (±21.66 cells, p = 0.008) invasion after siRNA treatment as compared to untreated (±95.6 cells) or scrambled treated groups (±91.6 cells; 91%) (Fig. 5b). The untreated groups served as negative control and scrambled siRNA treated was considered as treated controls for both the cell lines. To determine the effects of Fra-2 on migratory property of TSCC cells and its ability to accelerate the closure of a wound, scratch assay was performed. Fra-2 knockdown in AW13516 (HPV−ve) cells for 18 hours showed an ~63% migration rate while untreated or scrambled treated control cells showed ~100% migration (Fig. 5c). Interestingly, in HPV16+ve cells following Fra-2 siRNA treatment for 18 hours showed only ~27% migration while ~40% and ~39% migration was observed in untreated or scrambled siRNA treated cells respectively (Fig. 5d). These results together correlate well with the significant decrease in proinvasive factor MMP-9 expression and the reduction in cell invasion and migration after knocking down Fra-2 in both HPV+ve and HPV−ve tongue cancer cells.
Figure 5.
(a–d): Effect of Fra-2 silencing on cell invasion and migration of HPV+ve and HPV−ve TSCC cells. Fra-2 knockdown by specific siRNA leads to alterations in invasion in both HPV−ve (a) and HPV+ve (b) TSCC cells. Matrigel invasion assay of untreated, scrambled siRNA and Fra-2 siRNA treated in TSCC cells. Equal number of cells (1 × 105) were seeded in the Matrigel coated upper chamber. After 24 hours of incubation at 37 °C and 5% CO2, the cells that invaded through the membrane were fixed and stained with Giemsa, and images were captured using inverted microscope. Representative fields of view for each well are shown. Cell invasion was quantified by counting cells in six random fields. (c,d) Scratch assay of Fra-2 silencing demonstrated that cell migration was dramatically decreased in experimental groups after Fra-2 transfection compared with untreated and scrambled treated control groups. Inverted microscope images of wound closure at 0 hours, 12 hours and 18 hours in AW13516 (c) and UPCI:SCC90 (d) cells as indicated. Migration rate was calculated by image J software. The data in bar diagram are expressed as the mean ± SD of three independent experiments. *p < 0.05, **p < 0.01, ***p < 0.001 & ****p < 0.0001.
Discussion
Human oral tongue cancer is a most aggressive disease and generally associated with a very high invasive and metastatic potential. Transcription factor, AP-1 plays a critical role in regulating wide variety of cellular processes including cell differentiation, apoptosis, transformation and signal transduction pathways, specifically during progression and metastasis of several cancers including oral cancer15,19,20,22. AP-1 has therefore emerged as an important drug discovery target but its role in tongue cancer has not been studied.
Since high-risk HPV type 16 is a major oncogenic virus often associated with tongue cancer and its oncogenic expression is again controlled by AP-1 and its family proteins, we have molecularly dissected the role of AP-1 and its family proteins and HPV during tongue tumerigenesis. We have also examined the role of AP-1 downstream target genes Bcl-2, MMP-9 and Cyclin D1 in tongue cancer.
The prevalence of HPV infection in TSCCs has been reported to vary between 0 to as high as 82%, depending on the geographic regions, tumor sites and techniques applied for detection of the virus6,7,26,27,28. Till date there have been only two studies7,26 from India which showed contrasting report of HPV prevalence with 48% vs. 82% in TSCCs. In the present study, we observed 28% prevalence (14/50) of HR-HPV16 which was found to be the exclusive type (100%; 14/14) and more than 90% of them were present in an integrated form (to be published elsewhere). Our results corroborate earlier studies that showed predominance of HPV type 16 (100%) in oral tongue cancer particularly in the base of tongue (85.7%) except one study that reported a very high prevalence (82%) of HPV that too of HR-HPV type 1826. The reasons for such a contrasting result are not known. It is interesting to note that the majority of HPV16+ve cases belonged to well differentiated tumors (78.6%) which are expected to show better prognosis when treated. This was demonstrated first in oral cancer11 and later in other head and neck cancers9,29,30,31,32. It was further demonstrated that HPV infection promotes selective participation of p65 in NF-κB complex formation leading to overexpression of p21 that induces well differentiation and better prognosis of oral cancers11.
When we correlated HPV positivity with the tumor location, majority (85.7%) of HPV infection was found to be located in the base of the tongue and are most aggressive than those located in the mobile tongue or other sites of the tongue (14.3%). Our data are in good agreement with earlier studies which showed HPV16 as the most predominant type specifically found at the base of the tongue6,28,33. Observations of a very high prevalence of HPV infection specifically at the base of the tongue is possibly due to its secluded location that allows minimal exposure to tobacco insults and food intake etc. It is intriguing to note that inspite of smoking being strongly linked to HNSCC and HR-HPV being an yet another important causative factor associated with TSCC, smoking appears to be protective against oral HPV infection. This is clear from our results that out of 10 female TSCC patients, 80% (8/10) were HPV+ve and 75% (6/8) of them were nonsmokers. Similarly, out of 40 male TSCC patients, all were smokers but only 15% (6/40) of them were HPV+ve. It suggests that female patients who are generally non-smokers or mild smokers in India and are more likely to contract HPV infection leading to well differentiated tumors which would show better prognosis while males, in contrast, are often heavy smokers and are likely to have HPV−ve poorly differentiated tumors with worst prognosis. It gains support from recent studies34,35 which also demonstrated significantly a lower rate of HPV infection in smokers as compared to nonsmokers. Thus, it appears that nicotine released and oxidative stress created in mouth due to smoking may inhibit HPV infection and its persistence.
A strong DNA binding activity of AP-1 observed in tongue cancer cases as compared to precancer or normal adjacent controls corroborate several earlier studies in variety of other carcinomas including oral and esophageal carcinomas15,19,20,22,36,37. It is known that the expression of HPV16 oncoproteins E6 and E7 can induce AP-1 activity and are responsible for tumorigenic transformation but at the same time, the expression of these viral oncoproteins is dependent mainly on the availability of AP-1 which acts as a signaling epicenter for HPV-induced tumorigenesis22,38,39. The band supershift assays demonstrated an altered functional AP-1 complex formed by heterodimerization between c-Fos and Fra-2 instead of canonical partners, c-Fos and c-Jun in both HPV+ve and HPV−ve tongue carcinoma. In sharp contrast to almost all other carcinomas, preferential heterodimerization between c-Fos and Fra-2, both of which are known to have highly oncogenic potential24,25, appears to make tongue carcinoma most aggressive.
Interestingly, in majority of HPV−ve tongue cancer cases which are shown to be highly aggressive and have worst prognosis, there is a selective participation of c-Jun along with c-Fos/Fra-2 heterodimer mainly in poorly differentiated tumors (PDSCCs/MDSCCs). Furthermore, Fra-2 silencing also downregulates c-Jun. It is indicative of a crosstalk between c-Jun and Fra-2/c-Fos that makes this subtype of tongue cancer highly aggressive. It gains support from recent report of Mariani et al. (2007) who demonstrated amplification and overexpression of c-Jun can block adipocytic differentiation leading to undifferentiated highly aggressive tumors40. Zhou et al. (2013) also showed that changes in BRCA1 gene expression is regulated by c-Jun and Fra-2 that bind to the CRE element in early stages of ovarian carcinoma that suppress BRCA1 expression in sporadic tumors and promote tumor progression41. Together, these findings suggest that selective participation of c-Jun with c-Fos-Fra-2 protein complex of AP-1 plays an important role in maintaining undifferentiated tumor phenotype, aggressiveness and poor prognosis that may lead to drug resistance, treatment failure and disease relapse.
On the other hand, participation of JunD observed specifically in HPV16+ve cases appears to induce tumor differentiation as majority of the HPV+ve cases were found to have well differentiated tumors (WDSCCs) that show better prognosis. It gains support from the study by Natio et al. (2005) who showed that JunD protein plays an important role in bone formation by stimulating differentiation in osteoblasts42. Earlier, we have demonstrated a constitutively active AP-1 formed mainly by c-Fos/JunD heterodimers in majority of oral cancers20. Interestingly, JunD/JunD homodimer often observed in precancerous oral lesions appears to prevent progression to cancer, but its participation with c-Fos induced lesions to progress to invasive cancer20. It has been also suggested that the functional change of JunD from growth suppressor to a growth promoter could be possibly due to presence of mutant form of JunD43.
Both HPV+ve and HPV−ve tongue cancer cell lines also provided similar results except that it showed preferential participation of JunB instead of c-Jun observed in HPV negative tongue cancer tissues. Ohyama et al. (2008) reported that overexpression of JunB along with c-Fos can enhance 4Nitroquinoline 1-oxide-induced tongue carcinogenesis in rat indicating an important role of JunB/c-Fos crosstalk in tongue cancer44. A higher participation of JunD only in HPV16+ve cells indicates that JunD facilitates tumor differentiation leading to better prognosis.
Immunoblotting analysis revealed a very high expression of c-Jun, c-Fos and Fra-2 in almost all tongue cancer cases and cell lines and overexpression of these genes has been shown to contribute to more aggressive disease phenotype and metastasis in breast, oral and other cancers45,46,47. Overexpression of Fra-2 has been shown to induce aggressive progression and metastasis particularly in triple negative breast cancer24,25. Interestingly, Fra-1 has been demonstrated as oncogene by many authors but we have consistently observed its role as a tumor suppressor as it did not express in cervical, oral, esophageal18,20,22 and ovarian tumor tissues23. In contrast, in breast cancer specifically those were ER/PR/Her2 negative, Fra-1 showed significantly a higher expression48. Fra-1 is also highly expressed in many other cancers and plays a role in transformation, cell proliferation and metastasis49,50. Even in the present study, while Fra-1 is almost nil in fresh tongue tumor tissues, it however, shows higher expression in tongue cancer cell lines (Fig. 3a,b). Together these findings suggest that the oncogenic or tumor suppressor function of Fra-1 is dependent on the type of tumor. It is also highly interesting to note that induction of Fra-1 expression in those tumors that do not express Fra-1 can render them chemo-radio-sensitive (Das et al. unpublished observation). This gains strong support from our present findings that knocking down of Fra-2 (responsible for aggressive tumorigenesis and drug resistance) led to upregulation of p53 (Fig. 4e) and Fra-1. When we correlated AP-1 family protein expression with HPV status, HPV−ve tumors showed higher expression of c-Jun along with c-Fos and Fra-2 as compared to those in HPV+ve cases, indicating aggressive phenotype of HPV−ve tumors than HPV positives.
The aggressiveness of TSCCs is primarily dependent on its ability to invade adjacent tissues and metastasize to distant organ sites and the most known factors responsible are MMP-9, Cyclin D1 and Bcl-2 which also have an AP-1 binding site in their promoters. We observed significantly a higher expression of MMP-9, cyclin D1 and Bcl-2 genes in majority of tongue cancer cases and cancer cell lines particularly in HPV−ve cases that show aggressive tumor and worst prognosis. It corroborates several earlier findings51,52,53. It has been also reported that overexpression of c-Jun induces expression of MMP-9 and ECM invasion in tumor cells54. It is interesting to note that EGFR which is generally overexpressed in head and neck cancer can induce expression of MMP-9 and leads to the activation of the Raf-MEK-ERK signal transduction pathways which in turn can activate the c-Jun transcription factor. This may also contribute in regulating other critical genes, which are implicated in cell proliferation, differentiation and apoptosis15,55.
In order to understand the role of Fra-2 in tongue tumorigenesis and its possible cross talk with other critical genes such as c-Jun, c-Fos and MMP-9 that appears to contribute to the aggressiveness of the disease, Fra-2 was silenced by specific siRNA. The results demonstrated complete abolition of Fra-2 expression but expression of all the three critical genes eg. c-Fos, c-Jun and MMP-9 were also downregulated along with Fra-2. Also, HPV E6/E7 showed significant down regulation. Most interestingly, p53 as well as Fra-1 showed an elevated expression. Our observations thus suggest that complete silencing of Fra-2 also inhibits expression of c-Jun, c-Fos and MMP-9 responsible for aggressive tumor behavior but induces expression of p53 and Fra-1 which appears to function as tumor suppressors and may chemo-radio sensitize tongue tumor cells. It provides a clue for development of a novel therapeutic approach for effective targeting of chemo-radio resistant tongue cancer stem cells that cause cancer relapse. Our functional in vitro cell invasion and migration assays demonstrated Fra-2 knockdown can significantly reduce TSCC cell invasion and migration capability of both HPV+ve and HPV−ve tongue cancer cells in vitro. It has previously been reported that Fra-2 silencing significantly decreased cell migration and invasion in breast cancer cells24. These results together suggest potential effects of Fra-2 silencing on inhibiting invasion, migration and chemo-radio sensitization of TSCC cells.
In conclusion, our findings suggest that HR-HPV type 16 predominantly infects base of the tongue and persists preferably in nonsmoking individuals and is responsible for a major proportion of well differentiated tongue cancers that show better prognosis when treated. Higher DNA binding and overexpression of c-Fos and Fra-2 which form the functional AP-1 complex increased as a function of severity of tongue lesions, yet selective participation of c-Jun promotes poor differentiation, aggressive tumorigenesis and worst prognosis only in HPV−ve cases. Nevertheless, Fra-2 appears to play a critical role in tongue tumorigenesis since selective silencing of Fra-2 led to downregulation of tumorigenic genes; c-Fos, c-Jun, MMP-9 and HPV E6/E7 but upregulation of p53 and Fra-1 that may chemo-radio-sensitize tongue cancer cells leading to better treatment outcome.
Methods
Study subjects and collection of clinical samples
A total of 100 fresh tongue tissue biopsies of different histopathological grades comprising precancer (n = 20), cancer (n = 50) and adjacent normal controls (n = 30) were collected from the Department of ENT Surgery of Dr. Ram Manohar Lohia Hospital, New Delhi. None of these patients received any pre-operative radiation and chemotherapy. The clinico-pathological and epidemiological details of the patients were collected using standard proforma. Biopsy samples obtained immediately after surgery in sterile vials containing cold phosphate buffer saline (PBS), transported to lab on ice and stored in −80 °C deep freezer till further analysis.
The study was approved by the Institutional Ethics Committee of Dr. B. R. Ambedker Center for Biomedical Research (ACBR), University of Delhi, Delhi (India) and Dr. Ram Manohar Lohia Hospital, New Delhi. Written informed consent was obtained from all the subjects prior to their inclusion in the study. The study was carried out in accordance with the guidelines and principles of the Helsinki Declaration.
Cell lines and cell culture
The two tongue cancer cell lines; one HPV16 positive, UPCI:SCC090 (a kind gift from Dr. Susanne M. Gollin, University of Pittsburgh, Pittsburgh, PA) was derived from the base of tongue squamous cell carcinoma56 and the other HPV negative cell line, AW13516 (a kind gift from Dr. M. M. Vaidya, ACTREC, Tata Memorial Hospital, Navi Mumbai, India) derived from the squamous cell carcinoma of human tongue57, were maintained in MEM and IMDM medium respectively, supplemented with 10% FBS (Gibco, Invitrogen, CA) and antibiotics (penicillin and streptomycin) at 37 °C and 5% CO2.
DNA extraction and HPV genotyping
High molecular-weight genomic DNA was isolated from precancer, cancer and adjacent normal tissues as controls and two tongue cancer cell lines by standard procedure of proteinase K digestion, phenol-chloroform extraction and ethanol precipitation as followed in the lab. The initial HPV diagnosis was performed by using a pair of consensus degenerate primers (MY09 and MY11) derived from the highly conserved L1 region of HPV genome (Supplementary Table 1). PCR was performed in a 25 μl reaction mix containing 100 ng DNA, 10 mM Tris-HCl (pH 8.4), 50 mM KCl, 1.5 mM MgCl2, 125 μM of each dNTP (dATP, dCTP, dGTP and dTTP), 5 pmol of each oligonucleotide primer and 0.5 U Taq DNA polymerase (Bangalore Genei, Bangalore, India). β-globin gene was used as an internal control. HPV positive samples were subjected to comprehensive HPV genotyping assay using type-specific PCR and PGMY-reverse line blot assay (RLB) as per recommended protocol provided under the WHO HPV LabNet program58. Although the HPV+ve cell lines have previously been characterized, we reconfirmed the HPV status in these cell lines by PCR.
Preparation of protein extract and electrophoretic mobility shift assay (EMSA)
Nuclear extracts from all spectrum of tongue tissue biopsies and cell lines (UPCI:SCC90 and AW13516) were prepared by the method of Dignam59 with minor modification as described earlier11. The concentration of nuclear protein extracts was determined spectrophotometrically using standard Bradford method (Bio-Rad laboratories, Inc. CA) and stored at −80 °C till further use. AP-1-specific DNA binding activity in the nuclear extracts from fresh tongue tissues and cell lines was examined by EMSA using consensus oligonucleotide of AP-1 (5′-CGCTTGATGACTCAGCCGGAA-3′) as described earlier20. Oct-1 consensus oligonucleotide (5′-TGTCGAATGCAAATCACTAGAA-3′) was used as a control. The above oligonucleotides were annealed and labelled with [γ32P] ATP (3000 Ci/mmol; Jonaki, Hyderabad, India) by T4 polynucleotide kinase and gel purified in a 15% polyacrylamide gel17. Briefly, the binding reaction was performed in a 25 μl reaction volume containing 50% (v/v) glycerol, 60 mM HEPES (pH 7.9), 20 mM Tris-HCl (pH 7.9), 300 mM KCl, 5 mM EDTA, 5 mM DTT, 100 μg of BSA per ml, 2.5 μg of poly-dI-dC and 10 μg of nuclear extract. After 5 min, 10,000 cpm of the [γ32P] ATP 5′-end labelled double-stranded AP-1 oligonucleotide probe was added and the incubation was continued for additional 25 min at room temperature and the DNA-protein complex was resolved in a 4.5% non-denaturing polyacrylamide gel. The gel was then dried and visualized by Phosphorimager (Fujifilm FLA-5100) using Multi Gauge-ver 3.X anlaysis software. For monitoring composition of AP-1 and Oct-1, following antibodies from Santa Cruz Biotechnology (Santa Cruz, CA) were used: c-Jun (sc-45), JunB (sc-73), JunD (sc-74), c-Fos (sc-253), FosB (sc-48), Fra-1 (sc-605), Fra-2 (sc-171). The quantitative densitometric analysis was performed using Alpha Ease FC version 4.1 (Alpha Innotech Corporation, IL).
Western blotting
Protein extracted from tumor tissues and cell lines (40 μg/lane) were separated on 8–12% SDS-PAGE, electrotransferred to PVDF membrane and immunodetected using respective antibodies as described earlier20. The membrane was blocked with 10% fat free milk and incubated overnight in PBS with 5% milk, 0.05% Tween-20 and probed with polyclonal rabbit primary antibodies of respective AP-1 family members at 4 °C. These blots were washed, incubated with HRP- tagged anti-rabbit IgG secondary antibodies and visualized by Luminol detection kit (Santa Cruz Biotech, USA). The membranes were re-probed for β-actin expression as an internal control. The quantitative analysis of the bands was done using Alpha Ease FC version 4.1 (Alpha Innotech Corporation, IL). The level of expression of proteins was quantitated on an arbitrary scale with respect to β-actin expression as strong (++++) = >50%; Medium (++) = 10–50%; Weak (+) = <10% of β-actin expression and Nil/not detectable (−) = <1 as described earlier20.
RNA extraction and RT-PCR
Total RNA was isolated from fresh tongue biopsies comprising pre-cancer, cancer and normal adjacent control tissues and two tongue cancer cell lines using TRI reagent as per manufacturer’s protocol (Sigma-Aldrich Inc, USA). The quality and integrity of RNA was checked spectrophotometrically in a Nanodrop (Thermo scientific,USA) and on ethidium bromide-stained 1% agarose gel. For reverse transcription-PCR (RT-PCR) analysis, the isolated RNA (2 μg) was employed to prepare cDNA using first strand cDNA synthesis kit (Fermentas, USA). RT-PCR reactions were carried out in duplicates using AP-1 specific primers. The primers used for RT-PCR are listed in Supplementary Table 2. The mRNA expression levels of AP-1 genes were quantitated and normalized by GAPDH used as an internal loading control. The PCR products were analyzed on 2% agarose gel electrophoresis.
siRNA transfection and interference assay
For Fra-2-siRNA interference assay, transient transfection of commercially available Fra-2 siRNA oligonucleotides (Santa Cruz, biotech, USA) was performed according to the manufacturer’s protocol with some minor modifications60. Briefly, the complexes of Fra-2-siRNA-RNAimax (Invitrogen, USA) of different concentrations (20 nM, 40 nM and 80 nM) or scrambled siRNA (80 nM) were prepared in 6 wells plates in triplicates, after which 1 × 105 cells/well (AW13516 & UPCI:SCC090) and 2 ml antibiotics-free normal growth medium supplemented with 10% FBS was added and incubated at 37 °C in a CO2 incubator for 24–48 hours until the cells were 40–60% confluent followed by microscopic examination and protein expression analysis by immunoblotting.
In vitro Matrigel cell invasion assay
The effect of Fra-2 silencing on invasive characteristics of tongue cancer cells was evaluated using Matrigel invasion assay. After 24 hours of Fra-2 siRNA or scramble siRNA transfection, TSCC cells were trypsinized and resuspended in FBS-free medium. A total of 1 × 105 cells were plated in upper chamber of the transwell with a matrigel-coated membrane (8.0 μm pore size; Corning Costar Corp. USA). Medium with serum was added to the lower chamber as a chemoattractant. After incubation for 24 hours, cells on the lower surface of the membrane were fixed with formaldehyde and stained with Giemsa. Non-migratated cells were mechanically removed by a cotton swab. The images of invaded cells were acquired by an inverted microscope with a magnification of 20X. The number of invaded cells was quantified by counting cells in five randomly selected fields under microscope. Comparisons were made between the untreated or scramble siRNA treated and Fra-2 knockdown wells. All experiments were performed in triplicate.
In vitro cell migration assay
To investigate the migratory capacity of TSCC cells, cell scratch assay was performed. Both HPV+ve and HPV−ve TSCC cells were transfected with 80 nM Fra-2 siRNA and scramble siRNA as control as described above and incubated under standard conditions to achieve knockdown of Fra-2. After 24 hours, a scratch was carefully made by scraping through each well using a sterile pipette tip. The cells were then washed with PBS to remove detached cells. Scratches were monitored with an inverted microscope immediately after wounding and after incubation in an incubator (37 °C, 5% CO2) at different time points. Images were taken exactly at the same position before and after the incubation and migration rate was calculated by using image J software.
Statistical Analysis
The data analysis was performed using the statistical software Graph Pad Prism (version 6.0) and image J software. The association between HPV infection and expression profile of AP-1 proteins among different histopathological grades and clinico-pathological parameters in TSCC cases was determined using Fischer’s exact test and students t-test (two tailed). The p value ≤ 0.05 was considered as statistically significant.
Additional Information
How to cite this article: Gupta, S. et al. Selective participation of c-Jun with Fra-2/c-Fos promotes aggressive tumor phenotypes and poor prognosis in tongue cancer. Sci. Rep. 5, 16811; doi: 10.1038/srep16811 (2015).
Supplementary Material
Acknowledgments
The authors thank Dr. Susanne M. Gollin (University of Pittsburgh, Pittsburgh, PA) and Dr. M.M. Vaidya, (ACTREC, Tata Memorial Hospital, Navi Mumbai, India) for kind gift of UPCI:SCC090 and AW13516 tongue cancer cell lines, respectively. We thank Dr. Laishram R. Singh (Dr. B.R. Ambedkar Centre for Biomedical Research, University of Delhi, New Delhi) for extending some laboratory help. Authors also thank Dr. Gitanjali Bugnait, Deen Dayal Upadhaya hospital, New Delhi for providing a few clinical samples. We thank Dr. Sutapa Mahata and Abhishek Tyagi (Institute of Cytology & Preventive Oncology, Noida) for helping in EMSA and cell culture experiments. We also thank Dr. Amjad Ali (National Institute of Immunology, Delhi) for helping in invasion assay experiments. This work has been supported by the research grants from Indian Council of Medical Research (ICMR) and J. C. Bose Fellowship from DST, Government of India (to BCD) and ICMR Senior Research Fellowship to SG.
Footnotes
Author Contributions There is a total of 7 authors who have contributed in this study. S.G. and P.K. jointly participated in collection of samples, carrying out experiments, statistical data analysis, interpretation and initial writing of the manuscript. H.K. helped in monitoring experiments and critical reading of the manuscript. N.S. senior ENT surgeon who took biopsy and provided biopsy samples and clinical details of the patients and critically reviewed the manuscript. D.S. monitored the study and critically reviewed the manuscript. A.C.B. monitored the study, participated in the interpretation of data, drafting and critical reviewing of the manuscript. B.C.D. conceptualized, designed and supervised the study, participated in interpretation of the results and critically reviewed, finalized and communicated the manuscript. All authors have read and approved the final manuscript.
References
- Bosch F. X. et al. Head and neck tumor sites differ in prevalence and spectrum of p53 alterations but these have limited prognostic value. Int J Cancer 111, 530–538 (2004). [DOI] [PubMed] [Google Scholar]
- Kamangar F., Dores G. M. & Anderson W. F. Patterns of cancer incidence, mortality, and prevalence across five continents: defining priorities to reduce cancer disparities in different geographic regions of the world. J Clin Oncol 24, 2137–2150 (2006). [DOI] [PubMed] [Google Scholar]
- Lim Y. C., Song M. H., Kim S. C., Kim K. M. & Choi E. C. Preserving level IIb lymph nodes in elective supraomohyoid neck dissection for oral cavity squamous cell carcinoma. Arch Otolaryngol Head Neck Surg 130, 1088–1091 (2004). [DOI] [PubMed] [Google Scholar]
- Stransky N. et al. The mutational landscape of head and neck squamous cell carcinoma. Science 333, 1157–1160 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petersen P. E. Oral cancer prevention and control–the approach of the World Health Organization. Oral Oncol 45, 454–460 (2009). [DOI] [PubMed] [Google Scholar]
- Lee S. Y. et al. Relevance of human papilloma virus (HPV) infection to carcinogenesis of oral tongue cancer. Int J Oral Maxillofac Surg 39, 678–683 (2010). [DOI] [PubMed] [Google Scholar]
- Elango K. J. et al. Role of human papilloma virus in oral tongue squamous cell carcinoma. Asian Pac J Cancer Prev 12, 889–896 (2011). [PubMed] [Google Scholar]
- Mathew Iype E. et al. Squamous cell carcinoma of the tongue among young Indian adults. Neoplasia 3, 273–277 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhry C. et al. Improved survival of patients with human papillomavirus-positive head and neck squamous cell carcinoma in a prospective clinical trial. J Natl Cancer Inst 100, 261–269 (2008). [DOI] [PubMed] [Google Scholar]
- Ringstrom E. et al. Human papillomavirus type 16 and squamous cell carcinoma of the head and neck. Clin Cancer Res 8, 3187–3192 (2002). [PubMed] [Google Scholar]
- Mishra A., Bharti A. C., Varghese P., Saluja D. & Das B. C. Differential expression and activation of NF-kappaB family proteins during oral carcinogenesis: Role of high risk human papillomavirus infection. Int J Cancer 119, 2840–2850 (2006). [DOI] [PubMed] [Google Scholar]
- Licitra L. et al. High-risk human papillomavirus affects prognosis in patients with surgically treated oropharyngeal squamous cell carcinoma. J Clin Oncol 24, 5630–5636 (2006). [DOI] [PubMed] [Google Scholar]
- Lowy D. R. & Munger K. Prognostic implications of HPV in oropharyngeal cancer. N Engl J Med 363, 82–84 (2010). [DOI] [PubMed] [Google Scholar]
- Eferl R. & Wagner E. F. AP-1: a double-edged sword in tumorigenesis. Nat Rev Cancer 3, 859–868 (2003). [DOI] [PubMed] [Google Scholar]
- Angel P. & Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim Biophys Acta 1072, 129–157 (1991). [DOI] [PubMed] [Google Scholar]
- Butz K. & Hoppe-Seyler F. Transcriptional control of human papillomavirus (HPV) oncogene expression: composition of the HPV type 18 upstream regulatory region. J Virol 67, 6476–6486 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosl F., Das B. C., Lengert M., Geletneky K. & zur Hausen H. Antioxidant-induced changes of the AP-1 transcription complex are paralleled by a selective suppression of human papillomavirus transcription. J Virol 71, 362–370 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain S. et al. Transcription factor AP-1 in esophageal squamous cell carcinoma: alterations in activity and expression during human Papillomavirus infection. BMC Cancer 9, 329 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milde-Langosch K. The Fos family of transcription factors and their role in tumourigenesis. Eur J Cancer 41, 2449–2461 (2005). [DOI] [PubMed] [Google Scholar]
- Mishra A., Bharti A. C., Saluja D. & Das B. C. Transactivation and expression patterns of Jun and Fos/AP-1 super-family proteins in human oral cancer. Int J Cancer 126, 819–829 (2009). [DOI] [PubMed] [Google Scholar]
- Ozanne B. W. et al. Transcriptional regulation of cell invasion: AP-1 regulation of a multigenic invasion programme. Eur J Cancer 36, 1640–1648 (2000). [DOI] [PubMed] [Google Scholar]
- Prusty B. K. & Das B. C. Constitutive activation of transcription factor AP-1 in cervical cancer and suppression of human papillomavirus (HPV) transcription and AP-1 activity in HeLa cells by curcumin. Int J Cancer 113, 951–960 (2005). [DOI] [PubMed] [Google Scholar]
- Hein S. et al. Expression of Jun and Fos proteins in ovarian tumors of different malignant potential and in ovarian cancer cell lines. Oncol Rep 22, 177–183 (2009). [DOI] [PubMed] [Google Scholar]
- Milde-Langosch K. et al. Role of Fra-2 in breast cancer: influence on tumor cell invasion and motility. Breast Cancer Res Treat 107, 337–347 (2008). [DOI] [PubMed] [Google Scholar]
- Schroder C. et al. The transcription factor Fra-2 promotes mammary tumour progression by changing the adhesive properties of breast cancer cells. Eur J Cancer 46, 1650–1660 (2010). [DOI] [PubMed] [Google Scholar]
- Balaram P. et al. Human papillomaviruses in 91 oral cancers from Indian betel quid chewers–high prevalence and multiplicity of infections. Int J Cancer 61, 450–454 (1995). [DOI] [PubMed] [Google Scholar]
- Honig J. F. Non radioactive in situ hybridization for detection of human papilloma virus DNA in squamous cell carcinoma of tongue. Bull Group Int Rech Sci Stomatol Odontol 35, 107–115 (1992). [PubMed] [Google Scholar]
- Machado J. et al. Low prevalence of human papillomavirus in oral cavity carcinomas. Head Neck Oncol 2, 6 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison M. L. Human papillomavirus and prognosis of oropharyngeal squamous cell carcinoma: implications for clinical research in head and neck cancers. J Clin Oncol 24, 5623–5625 (2006). [DOI] [PubMed] [Google Scholar]
- Jo S. et al. Human papillomavirus infection as a prognostic factor in oropharyngeal squamous cell carcinomas treated in a prospective phase II clinical trial. Anticancer Res 29, 1467–1474 (2009). [PMC free article] [PubMed] [Google Scholar]
- Shaw R. & Robinson M. The increasing clinical relevance of human papillomavirus type 16 (HPV-16) infection in oropharyngeal cancer. Br J Oral Maxillofac Surg 49, 423–429 (2010). [DOI] [PubMed] [Google Scholar]
- Syrjanen S. The role of human papillomavirus infection in head and neck cancers. Ann Oncol 21 Suppl 7, vii243–245 (2010). [DOI] [PubMed] [Google Scholar]
- Dahlgren L. et al. Human papillomavirus is more common in base of tongue than in mobile tongue cancer and is a favorable prognostic factor in base of tongue cancer patients. Int J Cancer 112, 1015–1019 (2004). [DOI] [PubMed] [Google Scholar]
- Zwenger S. R. Bogarting that joint might decrease oral HPV among cannabis users. Curr Oncol 16, 5–7 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoler D. L. et al. Human papillomavirus and tobacco use in tongue base cancers. Ear Nose Throat J 92, 372–380 (2013). [DOI] [PubMed] [Google Scholar]
- Battista S. et al. Increase in AP-1 activity is a general event in thyroid cell transformation in vitro and in vivo. Oncogene 17, 377–385 (1998). [DOI] [PubMed] [Google Scholar]
- Chen T. K., Smith L. M., Gebhardt D. K., Birrer M. J. & Brown P. H. Activation and inhibition of the AP-1 complex in human breast cancer cells. Mol Carcinog 15, 215–226 (1996). [DOI] [PubMed] [Google Scholar]
- Offord E. A. & Beard P. A member of the activator protein 1 family found in keratinocytes but not in fibroblasts required for transcription from a human papillomavirus type 18 promoter. J Virol 64, 4792–4798 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cripe T. P. et al. Transcriptional activation of the human papillomavirus-16 P97 promoter by an 88-nucleotide enhancer containing distinct cell-dependent and AP-1-responsive modules. New Biol 2, 450–463 (1990). [PubMed] [Google Scholar]
- Mariani O. et al. JUN oncogene amplification and overexpression block adipocytic differentiation in highly aggressive sarcomas. Cancer Cell 11, 361–374 (2007). [DOI] [PubMed] [Google Scholar]
- Zhou L. et al. Microenvironmental regulation of BRCA1 gene expression by c-Jun and Fra2 in premalignant human ovarian surface epithelial cells. Mol Cancer Res 11, 272–281 (2013). [DOI] [PubMed] [Google Scholar]
- Naito J. et al. Menin suppresses osteoblast differentiation by antagonizing the AP-1 factor, JunD. J Biol Chem 280, 4785–4791 (2005). [DOI] [PubMed] [Google Scholar]
- Agarwal S. K. et al. Transcription factor JunD, deprived of menin, switches from growth suppressor to growth promoter. Proc Natl Acad Sci USA 100, 10770–10775 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohyama M. et al. Expressions of junB and c-fos are enhanced in 4-nitroquinoline 1-oxide-induced rat tongue cancers. Pathol Int 54, 35–40 (2004). [DOI] [PubMed] [Google Scholar]
- Ritke M. K., Bergoltz V. V., Allan W. P. & Yalowich J. C. Increased c-jun/AP-1 levels in etoposide-resistant human leukemia K562 cells. Biochem Pharmacol 48, 525–533 (1994). [DOI] [PubMed] [Google Scholar]
- Turatti E., da Costa Neves A., de Magalhaes M. H. & de Sousa S. O. Assessment of c-Jun, c-Fos and cyclin D1 in premalignant and malignant oral lesions. J Oral Sci 47, 71–76 (2005). [DOI] [PubMed] [Google Scholar]
- Zhang Y. et al. c-Jun, a crucial molecule in metastasis of breast cancer and potential target for biotherapy. Oncol Rep 18, 1207–1212 (2007). [PubMed] [Google Scholar]
- Kharman-Biz A. et al. Expression of activator protein-1 (AP-1) family members in breast cancer. BMC Cancer 13, 441 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belguise K., Kersual N., Galtier F. & Chalbos D. FRA-1 expression level regulates proliferation and invasiveness of breast cancer cells. Oncogene 24, 1434–1444 (2005). [DOI] [PubMed] [Google Scholar]
- Yang S. et al. MicroRNA-34 suppresses breast cancer invasion and metastasis by directly targeting Fra-1. Oncogene 32, 4294–4303 (2012). [DOI] [PubMed] [Google Scholar]
- Mineta H. et al. Prognostic value of vascular endothelial growth factor (VEGF) in head and neck squamous cell carcinomas. Br J Cancer 83, 775–781 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- P O. C. et al. Overexpression of epidermal growth factor receptor in human head and neck squamous carcinoma cell lines correlates with matrix metalloproteinase-9 expression and in vitro invasion. Int J Cancer 86, 307–317 (2000). [DOI] [PubMed] [Google Scholar]
- Xie X., Clausen O. P., De Angelis P. & Boysen M. The prognostic value of spontaneous apoptosis, Bax, Bcl-2, and p53 in oral squamous cell carcinoma of the tongue. Cancer 86, 913–920 (1999). [DOI] [PubMed] [Google Scholar]
- Crowe D. L., Brown T. N., Kim R., Smith S. M. & Lee M. K. A c-fos/Estrogen receptor fusion protein promotes cell cycle progression and proliferation of human cancer cell lines. Mol Cell Biol Res Commun 3, 243–248 (2000). [DOI] [PubMed] [Google Scholar]
- Reddy S. P. & Mossman B. T. Role and regulation of activator protein-1 in toxicant-induced responses of the lung. Am J Physiol Lung Cell Mol Physiol 283, L1161–1178 (2002). [DOI] [PubMed] [Google Scholar]
- White J. S. et al. The influence of clinical and demographic risk factors on the establishment of head and neck squamous cell carcinoma cell lines. Oral Oncol 43, 701–712 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatake R. J. et al. Establishment and characterization of four new squamous cell carcinoma cell lines derived from oral tumors. J Cancer Res Clin Oncol 116, 179–186 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson M. et al. DNA Extraction and HPV DNA Testing. in Human Papillomavirus Laboratory Manual, 1st edn (eds Unger E. R. & Dillner J.), Ch. 5, 35–63 (World Health Organization, 2010).
- Dignam J. D. Preparation of extracts from higher eukaryotes. Methods Enzymol 182, 194–203 (1990). [DOI] [PubMed] [Google Scholar]
- Shukla S. et al. Functional regulatory role of STAT3 in HPV16-mediated cervical carcinogenesis. PLoS One 8, e67849 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.