Abstract
The tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone (NNK, 1) is a potent lung carcinogen in laboratory animals and is believed to play a key role in the development of lung cancer in smokers. Metabolic activation of NNK leads to the formation of pyridyloxobutyl DNA adducts, a critical step in its mechanism of carcinogenesis. In addition to DNA nucleobase adducts, DNA phosphate adducts can be formed by pyridyloxobutylation of the oxygen atoms of the internucleotidic phosphodiester linkages. We report the use of a liquid chromatography–nanoelectrospray ionization–high-resolution tandem mass spectrometry technique to characterize 30 novel pyridyloxobutyl DNA phosphate adducts in calf thymus DNA (CT-DNA) treated with 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone (NNKOAc, 2), a regiochemically activated form of NNK. A 15N3-labeled internal standard was synthesized for one of the most abundant phosphate adducts, dCp[4-oxo-4-(3-pyridyl)butyl]dC (CpopC), and this standard was used to quantify CpopC and to estimate the levels of other adducts in the NNKOAc-treated CT-DNA. Formation of DNA phosphate adducts by NNK in vivo was further investigated in rats treated with NNK acutely (0.1 mmol/kg once daily for 4 days by subcutaneous injection) and chronically (5 ppm in drinking water for 10, 30, 50, and 70 weeks). This study provides the first comprehensive structural identification and quantitation of a panel of DNA phosphate adducts of a structurally complex carcinogen and chemical support for future mechanistic studies of tobacco carcinogenesis in humans.
Introduction
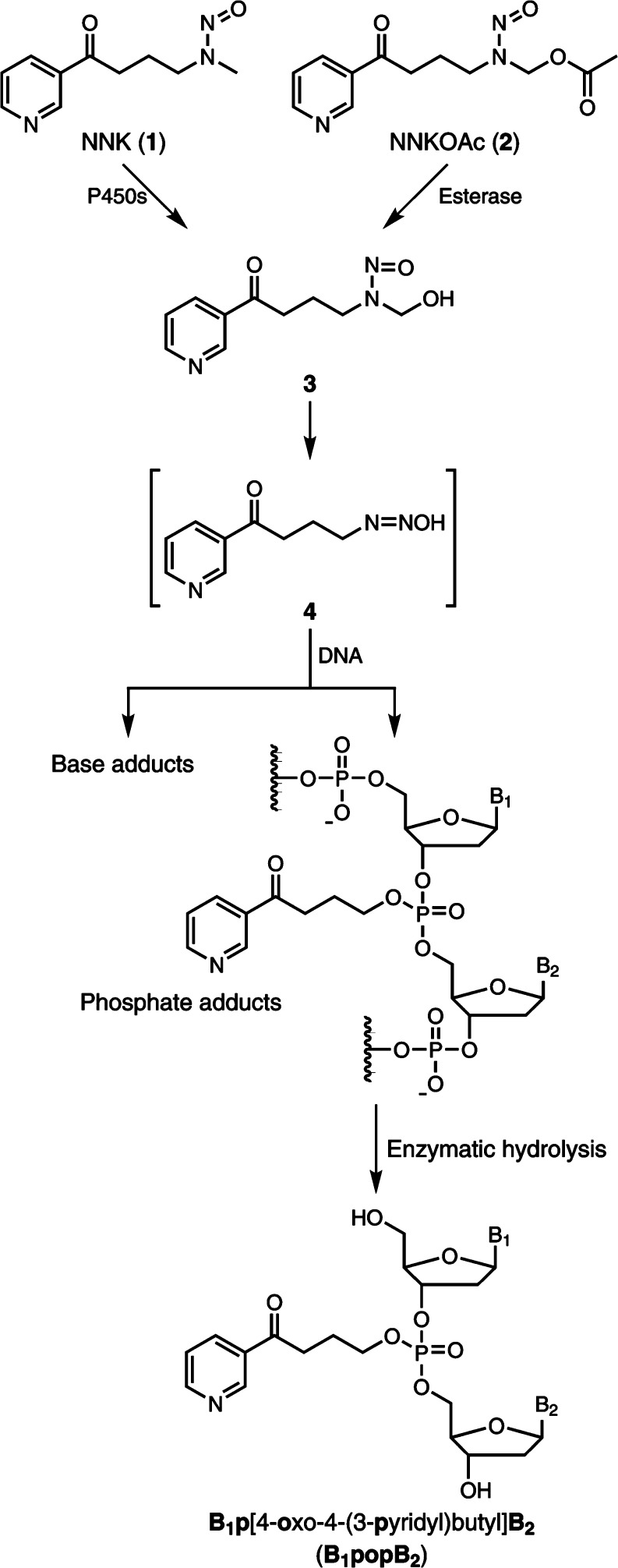
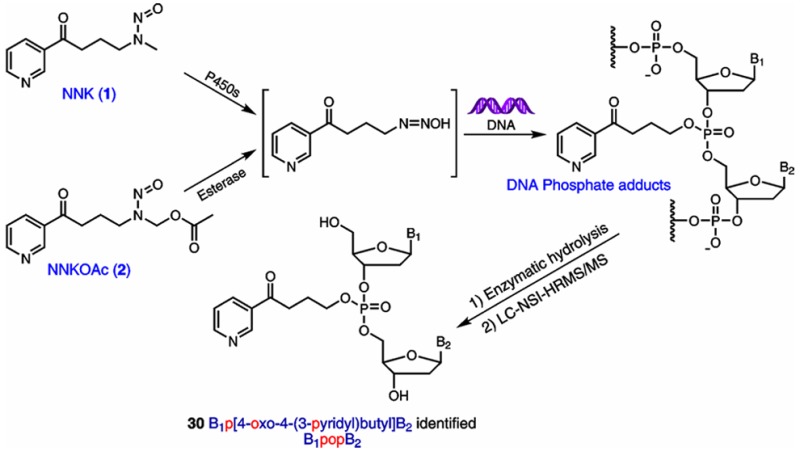
The tobacco-specific nitrosamine 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone [NNK, 1 (Figure 1)] is a powerful lung carcinogen in animal models.1 NNK and the related compound N′-nitrosonornicotine are considered “carcinogenic to humans” by the International Agency for Research on Cancer.2 NNK requires metabolic activation by cytochrome P450 enzymes to form pro-mutagenic DNA adducts, an initial key step in the multistage process of carcinogenesis. Consequently, the characterization and measurement of such adducts potentially can be used to investigate an individual’s NNK exposure and cancer risk due to tobacco use. One pathway of metabolic activation of NNK proceeds via α-hydroxylation of its methyl group to produce the unstable intermediate 3. This intermediate spontaneously yields the alkylating agent 4, which reacts with DNA to form pyridyloxobutyl (POB) DNA adducts due to DNA base alkylation.1 The major POB DNA adducts have been well-characterized, and their levels in NNK-treated animals have been quantified in our laboratory.3−5
Figure 1.
DNA phosphate adduct formation by NNK and NNKOAc.
In addition to reacting with DNA base moieties, it is possible that the alkylating agent 4 can also react with the oxygen of the internucleotide phosphodiester linkages in the DNA to form DNA phosphate adducts. A wide range of alkylating agents can form phosphate adducts in DNA, including dialkylsulfates, alkyl methanesulfonates, and N-alkylnitrosoureas.6,7 Phosphate adducts comprise the major alkylated DNA lesions of some alkylating agents such as ethylnitrosourea (ENU). The proportion of the phosphate adducts in salmon sperm DNA treated with ENU was 55% of the total alkylation products.8 Studies using rodents exposed to alkylating agents have demonstrated that phosphate adducts had half-lives longer than those of any other known DNA alkylation products formed by the same compound.7,9,10 Following intraperitoneal treatment of male mice with ENU, the half-lives of the phosphate adducts formed in lung, liver, kidney, and brain were determined to be 10–15 weeks.10 Thus, the long half-lives of phosphate adducts in vivo potentially could allow one to assess chronic exposure to toxic alkylating agents. Similarly, the possible DNA phosphate adducts formed by NNK may be used to investigate exposure to NNK due to long-term cigarette smoking.
The analysis of DNA phosphate adducts is usually performed by enzymatic hydrolysis, followed by determination of the resulting phosphotriesters (PTEs), with two nucleosides and an alkyl group in the structure.6,11 Because the internucleotide bonds adjacent to a completely alkylated phosphate group are resistant to hydrolysis by nucleases,12,13 the resulting products of hydrolyzed DNA phosphate adducts are PTEs instead of monophosphate adducts. Previously, only one study has reported formation of phosphate adducts by NNK in vivo.11 A transalkylation approach was used to measure the adduct levels in mice treated with [3H]NNK. The analysis involved the use of cob(I)alamin, a strong nucleophile, to transfer the alkyl group in the phosphate adducts with the formation of an adduct–cobalamin complex and a phosphodiester. The cobalamin complex was then measured by HPLC with liquid scintillation counting. However, that study did not provide direct proof of the existence of NNK-phosphate adducts, and the structures of individual adducts were not characterized. Therefore, the formation of DNA phosphate adducts by NNK and their levels are still largely unknown.
Depending on which two nucleosides comprise the PTE, there can be 10 different combinations of the four nucleosides.6 In the case of NNK, if phosphate adducts are formed, the structures of PTEs after enzymatic hydrolysis should be B1p[4-oxo-4-(3-pyridyl)butyl]B2 (B1popB2), where B1 and B2 represent same or different nucleosides (Figure 1). Because the structure of the phosphate group in B1popB2 is tetrahedral, there can be two diastereomers for the combination with the same nucleosides, and each can be in the Rp or Sp configuration depending on which oxygen is alkylated. For the B1popB2 with different nucleosides, depending on how the two sugar moieties of the nucleosides connect to the phosphorus atom, there can be two different types of isomers, B1-5′-pop-3′-B2 and B1-3′-pop-5′-B2. Consequently, there can be 32 different PTEs formed by methyl hydroxylation of NNK (Table 1). Therefore, a specific and powerful approach is required to simultaneously characterize and measure all of the 32 combinations.
Table 1. Ten Different Combinations of NNKOAc-Derived PTEs and Their [M + H]+ Masses.
| isomer
number |
|||
|---|---|---|---|
| PTE | [M + H]+ | possibility | detected |
| ApopA | 712.2351 | 2 | 2 |
| CpopC | 664.2127 | 2 | 2 |
| GpopG | 744.2250 | 2 | 2 |
| TpopT | 694.2120 | 2 | 2 |
| ApopC | 688.2239 | 4 | 4 |
| ApopG | 728.2300 | 4 | 3 |
| ApopT | 703.2236 | 4 | 4 |
| CpopG | 704.2188 | 4 | 4 |
| CpopT | 679.2123 | 4 | 3 |
| GpopT | 719.2185 | 4 | 4 |
| total | 32 | 30 | |
In this study, we developed a novel liquid chromatography (LC)–nanoelectrospray ionization (NSI)–high-resolution tandem mass spectrometry (HRMS/MS)-based method to analyze a total of 30 NNK-derived DNA phosphate adducts (Table 1) for the first time. A mass spectrometer containing a high-field orbital trap was used with rapid, high-resolution full scan (R = 60000) detection along with MS2 product ion scan (R = 15000) detection of 10 different parent ions with unit mass quadrupole isolation. The levels of these phosphate adducts were determined in calf thymus DNA (CT-DNA) treated with 4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone [NNKOAc, 2 (Figure 1)], a regiochemically activated form of NNK, and in hepatic and pulmonary DNA of rats treated with NNK. This is the first study to provide a comprehensive analysis of phosphate adducts of DNA formed in vivo after treatment with a structurally complex carcinogen. Furthermore, this study demonstrates the first characterization and quantitation of DNA phosphate adducts of the tobacco-specific nitrosamines and provides chemical support for using phosphate adducts as potential biomarkers to investigate tobacco exposure and associated cancer risk.
Experimental Details
Caution:NNK and NNKOAc are carcinogenic. They should be handled in a well-ventilated hood with extreme care and with personal protective equipment.
Materials and Chemicals
NNK and NNKOAc were purchased from Toronto Research Chemicals. dCp[4-oxo-4-(3-pyridyl)butyl]dC (CpopC), two diastereomers, were custom-synthesized by WuXi AppTec (Hong Kong) (Scheme S1), and their structures were confirmed by one-dimensional and two-dimensional NMR in our laboratory (Figure S1). 5′-Dimethoxytrityl[15N3]-2′-deoxycytidine-3′-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite was obtained from Cambridge Isotope Laboratories (Tewksbury, MA). All other nucleoside phosphoramidites, solvents, and solid supports required for the solid phase synthesis of dinucleotide [15N3]-2′-dCpdC were acquired from Glen Research Corp. (Sterling, VA). Reagents and enzymes for DNA isolation were obtained from Qiagen Sciences (Germantown, MD). All other chemicals and solvents were purchased from Sigma-Aldrich Chemical Co. (Milwaukee, WI) or Fisher Scientific (Fairlawn, NJ).
Synthesis of [15N3]CpopC
[15N3]-2′-dCpdC was prepared on a DNA synthesizer (ABI 394, Applied Biosystems, Foster City, CA) in accordance with standard solid phase oligodeoxynucleotide synthesis protocols. In this study, 5′-dimethoxytrityl[15N3]-2′-deoxycytidine-3′-[(2-cyanoethyl)-(N,N-diisopropyl)]phosphoramidite was manually coupled to an Ac-dC-CPG solid support, followed by standard deprotection techniques performed on the DNA synthesizer (Scheme S2). [15N3]CpopC was subsequently synthesized by reacting [15N3]-2′-dCpdC with NNKOAc in 0.01 M NaOH (37 °C) for 16 h and purified by reversed phase HPLC using an Agilent 1100 HPLC system interfaced with a DAD UV detector. The presence of two diastereomers was confirmed by comparison with their corresponding unlabeled CpopC standards via HPLC–MS (Figure S2).
In Vitro DNA Samples
CT-DNA (2 mg) was incubated with NNKOAc (2 mg, 7.54 μmol) in the presence of porcine liver esterase (4 units) in 0.1 M phosphate buffer (1 mL, pH 7.0) at 37 °C for 16–48 h. The incubation mixture was then washed three times with 2 mL of a CHCl3/isoamyl alcohol mixture (24:1). The treated DNA was precipitated via the addition of cold 2-propanol, washed with 70% EtOH and 100% EtOH sequentially, dried under a stream of N2, and stored at −20 °C until analysis.
In Vivo DNA Samples
These samples were isolated from liver and lung of male F344 rats that had been exposed to NNK in previous studies.3,4 For the acute exposure group, the rats (n = 5) were treated with 0.1 mmol of NNK/kg of body weight in 0.4 mL of saline once daily for four consecutive days by subcutaneous (sc) injection;4 for the chronic exposure group, the rats (n = 3) received 5 ppm of NNK in drinking water for 10, 30, 50, and 70 weeks.3 Liver and lung were harvested, and DNA was isolated by following the protocol described previously.3
DNA Hydrolysis and Adduct Enrichment
The DNA samples were dissolved in 0.8 mL of 10 mM sodium succinate (pH 7.4) buffer containing 5 mM CaCl2 and then mixed with 200 fmol of [15N3]CpopC-1 and [15N3]CpopC-2 as internal standards (IS), followed by the addition of deoxyribonuclease I (60 units), phosphodiesterase I (0.015 unit), and alkaline phosphatase (40 units). The solution was incubated overnight at 37 °C. The next day, 25 μL of hydrolysate was analyzed for dG by HPLC, and the amount of DNA was calculated.4 The remaining hydrolysate was filtered through 10K centrifugal filters (Ultracel YM-10, Millipore). The filtrates were loaded on 30 mg Strata X cartridges (Phenomenex) activated with 2 mL of MeOH and 2 mL of H2O. The cartridges were washed with 2 mL of H2O and 1 mL of 10% MeOH sequentially and finally eluted with 2 mL of 50% MeOH. The 50% MeOH fraction containing analytes was collected and concentrated to dryness in a centrifugal evaporator. The residue was redissolved in 10 μL of deionized H2O prior to analysis by LC–NSI–HRMS/MS.
LC–NSI–HRMS/MS
The analysis was performed on an Orbitrap Fusion Tribrid mass spectrometer (Thermo Scientific, Waltham, MA) under full scan and product ion scan modes. A trapping column (Waters nanoAcquity Symmetry C18, 5 μm, 180 μm × 20 mm) was connected to an EASY-Spray column (Thermo PepMap RSLC C18, 2 μm, 75 μm × 50 cm) mounted in an EASY-Spray ion source (Thermo Scientific). The mobile phase consisted of 5 mM NH4OAc and CH3CN with gradient elution at a flow rate of 300 nL/min. The spray voltage was 2.2 kV. The capillary temperature was 300 °C, and the S-Lens RF level was 60%. The full scan analysis was performed with a mass range of m/z 350–1000 at a resolution of 60000. The product ion scan was performed using higher-energy collisional dissociation (HCD) fragmentation with a normalized collision energy of 20 units, isolation widths of 1 Da for the possible DNA phosphate adducts, and product ion analysis performed with a mass range of m/z 100–800 at a resolution of 15000. The accurate mass tolerance used for extraction of precursor and fragment ion signals was 5 ppm.
The quantitative analysis of CpopC was performed using accurate mass-extracted ion chromatograms of m/z 326.0788 [C14H17NO6P]+ for both CpopC (parent ion at m/z 664.2) and [15N3]CpopC (parent ion at m/z 667.2) with a mass tolerance of 5 ppm. Quantitation was based on the peak area ratio of the two diastereomers of CpopC to their corresponding [15N3]CpopC, the constructed calibration curves, and the amount of IS added. A calibration curve was constructed for each diastereomer before each analysis using a series of standard solutions of CpopC and [15N3]CpopC. The calibration standard solutions contained a constant amount of [15N3]CpopC (15 fmol on-column) and varying amounts of CpopC (0.15, 0.3, 0.6, 1.5, 6, and 15 fmol on-column). The levels of other adducts were estimated on the basis of their MS signal intensities compared to CpopC in the full scan mode. All data are presented as means ± standard deviation (SD). One-way ANOVA followed by a Bonferroni post test was used for multiple comparisons. A p value of <0.05 was considered significant.
Results
Characterization of Phosphate Adducts
Our strategy for the identification of possible DNA phosphate adducts formed by NNK was based on the knowledge of phosphate adducts formed by other alkylating agents.6,11 To investigate phosphate adduct formation, DNA samples from in vitro NNKOAc treatment and in vivo NNK-treated rats were analyzed by LC–NSI–HRMS/MS. Both full and product ion spectra were recorded for structural characterization of B1popB2, the PTEs from enzymatic hydrolysis of DNA phosphate adducts.
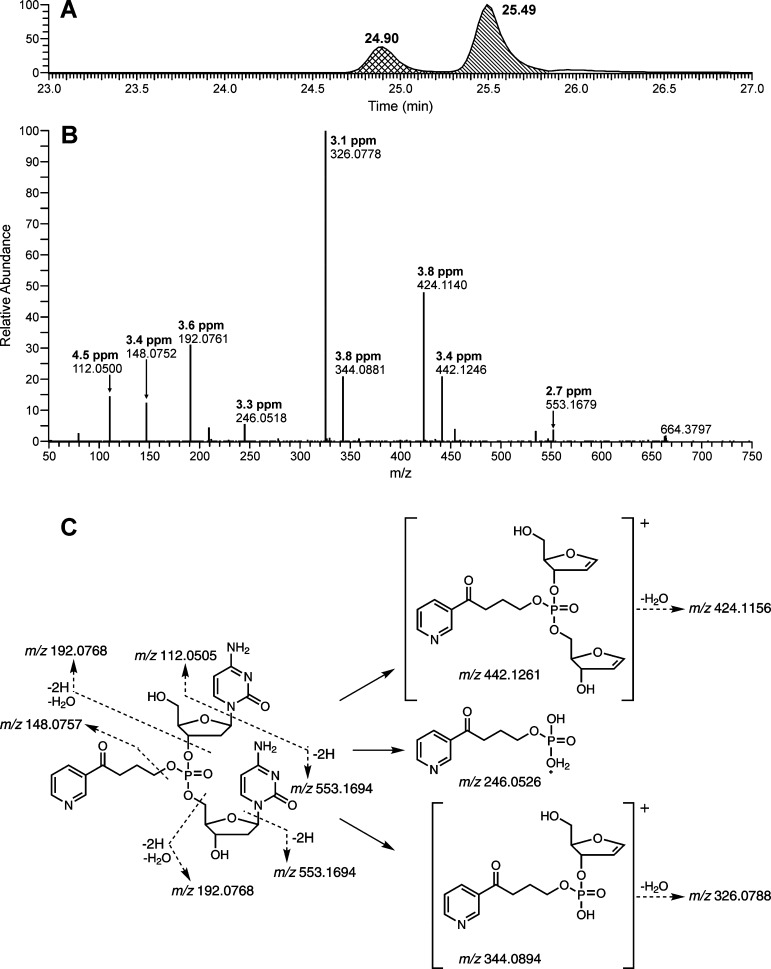
The detection and characterization of B1popB2 were first conducted in the NNKOAc-treated CT-DNA samples. The theoretical accurate masses of B1popB2 are summarized in Table 1. By analyzing the data obtained from both full and product ion scans, we observed all 10 combinations of B1popB2, and a total of 30 out of 32 possible PTEs were detected (Table 1). Their structures were characterized on the basis of the accurate masses of the precursor ions and the corresponding fragment ions. The two diastereomers of CpopC, in which both B1 and B2 are cytosine, were identified by comparison of their LC–MS retention time, full scan spectra, and product ion scan spectra to those of the standards. Two peaks were detected when the exact mass of the precursor ion of CpopC ([M + H]+, m/z 664.2127) was extracted from the full scan data (Figure 2A), indicating the presence of the two diastereomers. The product ion spectra of the two peaks showed similar fragmentation patterns (Figure 2B). The proposed structures of the major fragment ions are shown in Figure 2C. For example, the most abundant fragment ion of m/z 326.0788 was formed from loss of one nucleotide and one base plus one H2O from the precursor ion. The measured masses of all the major fragment ions were within 5 ppm of the theoretical masses of the proposed fragment ions.
Figure 2.
(A) LC–MS chromatogram, (B) product ion spectrum of the peak at 25.49 min in panel A, and (C) fragmentation pattern of CpopC. The measured masses (B) of all the major fragment ions are within 5 ppm of the theoretical masses (C) of the proposed fragment ions.
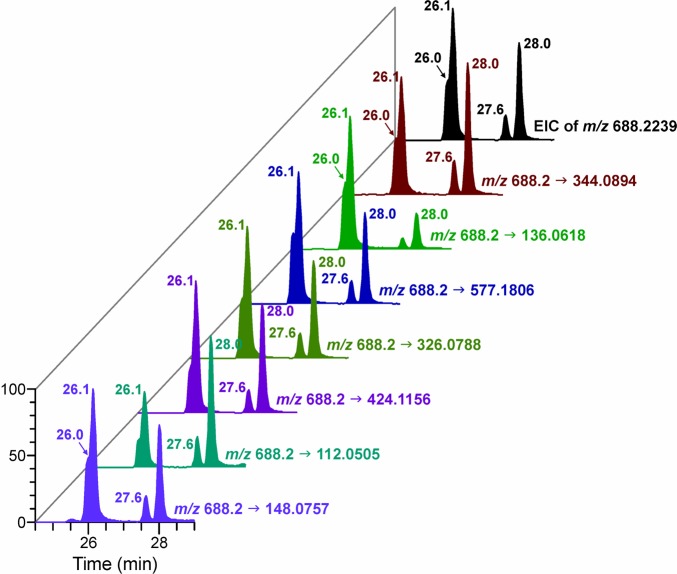
The structures of all the other B1popB2 PTEs were characterized by following the same strategy used for CpopC. With ApopC as an example, the exact mass of its precursor ion of m/z 688.2239 was extracted from the full scan data, and four peaks were detected (Figure 3), indicating the presence of four isomers. All the isomers had similar fragmentation patterns but with different abundances (Figure S3). The chromatographic traces of the major fragment ions of the four isomers matched each other (Figure 3), indicating their presence in the sample. The standards of ApopC are currently unavailable, and further work needs to be done to assign the configuration of each isomer. The extracted ion chromatograms and fragmentation patterns for characterization of all the other PTEs are available in the Supporting Information (Figure S4).
Figure 3.
LC–MS extracted precursor and fragment ion chromatograms corresponding to ApopC in an NNKOAc-treated CT-DNA sample. EIC, extracted ion chromatogram.
In the process of NNK metabolism, a diazonium intermediate may be formed by loss of hydroxide from metabolic intermediate 4 (Figure 1), and the carbonyl may displace N2 to form a cyclic oxonium ion, which may also react with the oxygen in the internucleotide linkages to form another type of phosphate adduct. If this occurs, the structures of PTEs formed after enzymatic hydrolysis should be B1p[2-(3-pyridyl)-2,3,4,5-tetrahydrofuranyl]B2 (Scheme S3), which would have the same [M + H]+ as the corresponding B1popB2. To test whether such adducts were present, the NNKOAc-treated CT-DNA sample was hydrolyzed and treated with NaBH4, which can reduce the ketone in B1popB2 to a hydroxyl group (Scheme S3), but would not change the structure of B1p[2-(3-pyridyl)-2,3,4,5-tetrahydrofuranyl]B2. The NaBH4-treated sample was analyzed by LC–NSI–HRMS/MS, and all the peaks of B1popB2 disappeared; peaks with 2 mass units higher than the corresponding B1popB2 were detected (data not shown), indicating the presence of B1popB2 instead of B1p[2-(3-pyridyl)-2,3,4,5-tetrahydrofuranyl]B2. These results indicate that the tetrahydrofuranyl adducts did not form.
In Vitro Adduct Levels
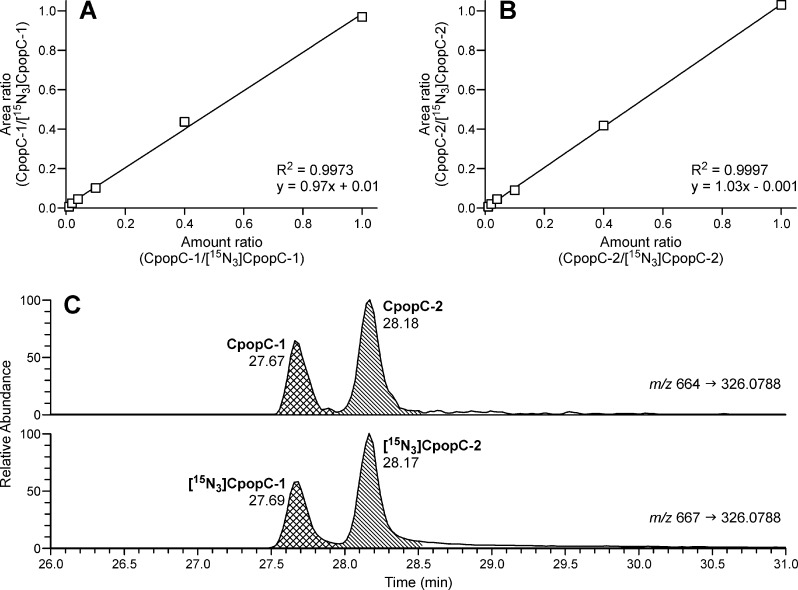
Using HCD fragmentation, the product ion spectra of CpopC and its IS [15N3]CpopC generated several major fragment ions. Because the highest signal intensities in the spectra of both CpopC and IS are the m/z 664.2 → 326.0788 and m/z 667.2 → 326.0788 transitions, respectively, they were selected for quantitative analysis. We also compared the relative signal intensities of the major fragment ions in the samples to those in the synthetic standards to confirm the identities of CpopC-1, CpopC-2, and their IS. By using this method, a limit of detection of 0.15 fmol (on-column) was obtained for both CpopC-1 and CpopC-2. The instrument response and the CpopC/[15N3]CpopC ratio were linear in the 0.15–15 fmol (on-column) range of CpopC with a typical R2 of 0.9973 for CpopC-1 (Figure 4A) and an R2 of 0.9997 for CpopC-2 (Figure 4B).
Figure 4.
Linearity of (A) CpopC-1, (B) CpopC-2, and (C) chromatograms upon analysis of CpopC in the NNKOAc-treated CT-DNA sample. The amount of CpopC was increased from 0.15 fmol to 0.3, 0.6, 1.5, 6, and 15 fmol, with a constant amount of IS (15 fmol).
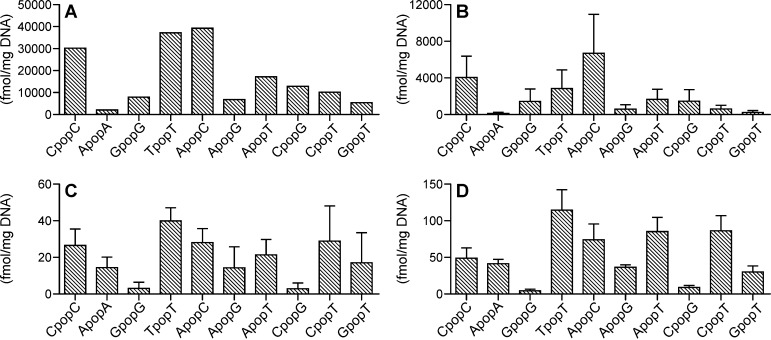
Typical chromatograms obtained upon analysis of CpopC in NNKOAc-treated CT-DNA are presented in Figure 4C. The levels of CpopC-1 and CpopC-2 were 10300 and 20200 fmol/mg DNA, or 340 and 670 adducts per 108 nucleotides, respectively. The total amount of CpopC accounted for 18% of the total phosphate adducts in the NNKOAc-treated CT-DNA (Figure 5A). The other two major PTEs, TpopT and ApopC, accounted for 22 and 23% of the total phosphate adducts, respectively (Figure 5A). We also checked the digestion efficiency by incubating the NNKOAc-treated CT-DNA with the enzymes for ≤48 h. No differences in the measured amounts of phosphate adducts were observed among the DNA samples incubated for 16 h versus 48 h. Therefore, an incubation time of 16 h was used in all the following studies.
Figure 5.
Levels of DNA phosphate adducts in (A) NNKOAc-treated CT-DNA, (B) liver DNA from NNK-treated rats (n = 5) in the acute exposure group (0.1 mmol/kg per day for 4 days), and (C) liver and (D) lung DNA of NNK-treated rats (n = 3) in the chronic exposure group (5 ppm in drinking water for 10 weeks). Values are presented as means ± SD.
In Vivo Adduct Levels
All 10 combinations of B1popB2 were detected in liver DNA from rats acutely exposed to NNK (0.1 mmol/kg per day for 4 days), and a total of 25 out of 32 PTEs were detected (Table S1). The formation pattern of DNA phosphate adducts (Figure 5B) was similar to the formation pattern in the in vitro NNKOAc-treated CT-DNA sample. The levels of CpopC-1 and CpopC-2 were 1780 ± 570 and 2290 ± 1030 fmol/mg DNA, or 59 ± 19 and 76 ± 34 adducts per 108 nucleotides, respectively. Consistent with the in vitro data, the three major PTEs were CpopC, TpopT, and ApopC, which accounted for 20, 14, and 34% of the total phosphate adducts, respectively.
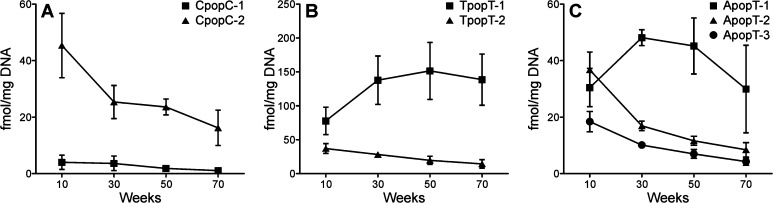
In the chronic treatment group, the phosphate adducts were measured in both liver and lung DNA. Similar to the acute group, we observed all 10 combinations of B1popB2, and a total of 23 PTEs were detected in the DNA samples (Table S1). Levels of the phosphate adducts over the course of the study are illustrated in Figure 6 and Figure S5. In general, the levels of adducts in lung DNA were higher than in liver DNA. After treatment for 10 weeks, the formation pattern of phosphate adducts in the liver (Figure 5C) was similar to the formation pattern in the lung (Figure 5D), but different from the formation pattern in the liver of the acute treatment group. Results from 30, 50, and 70 weeks also showed similar adduct formation patterns between liver and lung (Figure S5). Different levels of isomers of the same combination were observed for all 10 combinations in both liver and lung (Figure 6 and Figure S6). For example, the levels of CpopC-2 were 7–15 times higher than those of CpopC-1 in the lung throughout the study (Figure 6A).
Figure 6.
Levels of DNA phosphate adducts (A) CpopC, (B) TpopT, and (C) ApopT in lung DNA from NNK-treated rats (n = 3) in the chronic exposure group (5 ppm in drinking water for 10, 30, 50, and 70 weeks). Values are presented as means ± SD.
The formation of different isomers from some combinations was variable over the course of the study (Figure 6 and Figure S6). For example, the highest level of TpopT-1 in the lung was at 50 weeks, while that of TpopT-2 was at 10–30 weeks (Figure 6B). The formation of different isomers of some combinations showed differences over time. For example, the levels of ApopT-1 in the lung increased after treatment, reached a peak at 30 weeks, and decreased afterward, while the levels of the other two isomers reached a peak at 10 weeks and then decreased over time throughout the study (Figure 6C). Only three rats were used for this analysis, and adduct levels were only significantly different between certain time points; for example, the level of CpopC-2 at 10 weeks was significantly higher than at 30, 50, or 70 weeks (p < 0.05). However, other adducts were not significantly different over time, such as CpopC-1, so more replicates would need to be analyzed to investigate whether the levels of these adducts are changing over time.
To determine the relative proportion of phosphate adducts to the total adducts formed by NNKOAc/NNK, the levels of corresponding major base adducts, including O2-[4-(3-pyridyl)-4-oxobut-1-yl]thymidine (O2-POB-dThd), 7-[4-(3-pyridyl)-4-oxobut-1-yl]guanine (7-POB-Gua), and O6-[4-(3-pyridyl)-4-oxobut-1-yl]deoxyguanosine (O6-POB-dGuo), were measured using our previously developed methods (Table S1).4,5 The total amount of the three major base adducts was 3070 pmol/mg DNA in NNKOAc-treated CT-DNA, 47600 fmol/mg DNA in liver in the acute NNK treatment group, and 1140–2310 fmol/mg DNA in liver in the chronic NNK treatment group. The amount of the three base adducts reported in our previous study was 2300–5570 fmol/mg DNA in lung in the chronic NNK treatment group.3 Consequently, the phosphate adducts accounted for 5% of the total DNA adducts in NNKOAc-treated CT-DNA, 30% in the acute NNK treatment group, and 5–9% in the chronic NNK treatment group.
Discussion
The interaction of chemical carcinogens with DNA to form covalent DNA adducts is a key step in genotoxic chemical carcinogenesis. The levels of DNA adducts in vivo can be used to investigate exposure plus metabolic activation of carcinogens and potentially evaluation of cancer risk. Most studies of DNA adducts have focused on the interaction of carcinogens with DNA bases and formation of premutagenic DNA base adducts. In addition to base adducts, certain carcinogens can react with the oxygen in the internucleotide phosphate linkages to form DNA phosphate adducts, which may also be used as a biomarker of DNA damage. However, the characteristics and biological significance of the DNA phosphate adducts are still largely unknown. In this study, a panel of phosphate adducts were detected and characterized in NNKOAc-treated CT-DNA and NNK-treated rat liver and lung DNA. The levels of the phosphate adducts were assessed in these modified DNA samples. This is the first study that has directly and comprehensively characterized DNA phosphate adducts formed in vivo by NNK or any other structurally complex carcinogen. The use of state-of-the-art, high-resolution mass spectrometry was critical to the success of this study.
One reason that relatively little knowledge has been obtained on DNA phosphate adducts is a lack of appropriate analytical methods. Early efforts to detect the phosphate adducts involved alkali-induced strand breaks at the sites of the phosphate groups and quantitation of the total amount of adducts.14,15 However, this measurement is not specific because of the possible presence of other alkali-labile lesions in the DNA. The 32P postlabeling technique was also used for the detection of phosphate adducts.16,17 Before labeling, because the PTEs are not a substrate for phosphorylation by polynucleotide kinase, they have to be converted into either phosphodiester dinucleoside monophosphates or 3′-phosphate-alkylated mononucleotides by treatment with alkali. Therefore, it is not possible to measure individual PTEs by using this method. The same problem exists for the strong nucleophile-based transalkylation approach, which measures the alkyl-nucleophile complex instead of directly analyzing individual PTEs.11 Studies using mass spectrometry were able to directly detect PTEs without any hydrolysis or transalkylation procedures.13,18 Haglund et al. characterized 10 different ethyl PTEs in DNA treated with ENU using an LC–ESI–MS/MS method.13 Zhang et al. quantified a methyl PTE of thymidyl(3′–5′)thymidine in the DNA from cells treated with methylnitrosourea and methyl methanesulfonate by tandem mass spectrometry.18 These studies demonstrated that mass spectrometry was an appropriate technique for the analysis of DNA phosphate adducts. Previously, we have developed HRMS/MS-based methods for the analysis of DNA base adducts, which provided high sensitivity and selectivity for specific and accurate measurement of these adducts.19,20 The same basic approach was used here to measure phosphate adducts; however, a mass spectrometer containing a higher-field, higher-performance orbital trap was used allowing for sequential acquisition of 10 different MS2 fragmentation events, along with the full scan event, with a rate sufficient to allow for 10–15 data points across narrow chromatographic peaks. In the study presented here, we applied this technique to analyze DNA phosphate adducts and characterized and quantified 30 out of 32 possible PTEs formed by NNKOAc/NNK in the in vitro and in vivo DNA samples.
The random formation of phosphate adducts in vitro has been implied by the results obtained from CT-DNA treated with dimethylsulfate or diethylsulfate.21 Therefore, all 32 PTEs of B1popB2 could potentially be formed in the NNKOAc-treated CT-DNA. We detected 30 instead of 32 PTEs, with three instead of four isomers detected for ApopG and CpopT. This may be due to the insufficient chromatographic resolution of isomer peaks under the current chromatographic conditions. The frequencies of dA- and dC-containing PTEs were 28 and 24%, respectively, which were comparable with their normal nucleoside contents in CT-DNA,21 suggesting in vitro random formation of phosphate adducts. However, the frequency of dG-containing PTEs (8%) was much lower than its nucleoside content (23%), while the frequency of dT-containing PTEs (39%) was higher than its nucleoside content (25%). This difference is possibly caused by different MS responses of dG- and dT-containing PTEs because of the different ionization efficiency in positive ion mode, because their levels were estimated on the basis of their MS signal intensities. In addition, different recoveries of PTEs during the sample preparation may also contribute to this difference.
The in vivo formation of phosphate adducts is nonrandom in mice treated with nitrosodiethylamine, with levels of dT-containing PTEs formed higher than the levels of those containing dG.21 In our study, 24 and 23 PTEs of B1popB2 were detected in rats acutely and chronically exposed to NNK, respectively. Consistent with previous studies, the formation of these phosphate adducts was nonrandom, with levels of dT-containing PTEs higher than its content, and dG-containing PTEs lower than its content in both the acute and chronic treatment groups. In addition to MS response and recovery mentioned before, it has been proposed that the nonrandom formation is determined by two factors: the phosphate oxygen having to compete with adjacent nucleophilic sites for the alkylating electrophile and the electrophile’s inherent reactivity, with the more reactive electrophiles yielding a more random formation of PTEs.22 Differences in repair processes of different adducts may also contribute to their nonrandom distribution in DNA. Although the repair mechanisms of phosphate adducts formed by NNK are still unknown, studies of ethyl PTEs suggested that the repair of those phosphate adducts was sequence-specific.21
Although the level of phosphate adduct formation was lower than the level of base adduct formation, some adducts, such as TpopT, were persistent in vivo in the chronic NNK treatment group. While the levels of three major base adducts (O2-POB-dThd, 7-POB-Gua, and O6-POB-dGuo) decreased at the end of the chronic treatment,3 the levels of TpopT-1 were still persistently high even at 70 weeks. Such phosphate adducts could be used to reflect the amount of the potently mutagenic base adducts and might be markers for the evaluation of chronic exposure to NNK and associated cancer risk.
Because this is the first time DNA phosphate adducts formed by NNK have been characterized, the biological significance of those adducts is still unknown. Previous studies showed that the activities of DNA polymerases were affected by the formation of phosphate adducts. The effect of ethyl PTE modification on DNA polymerase I activity was investigated by using modified oligonucleotides, and the rates and extents of polymerization directed by the modified templates were 25 or 50% lower than control, indicating the replication rates of cellular DNA were inhibited by ethyl PTE modification.23 Another study used oligonucleotides with an isopropyl PTE as templates for in vitro DNA synthesis catalyzed by E. coli DNA polymerase I, and the results showed that the isopropyl PTE inhibited DNA chain elongation.24 Moreover, the neutralization of the negative charge in the phosphate group by the formation of phosphate adducts may interfere with the binding of DNA to other macromolecules, resulting in cellular dysfunctions. It has been shown that many of the physical characteristics of DNA were not significantly affected by the presence of a single alkyl PTE,25 while another study demonstrated long-range perturbations in duplex stability and conformation in isopropyl PTE-containing oligonucleotides.26 Because the pyridyloxobutyl moiety in the PTE of B1popB2 is larger than the isopropyl group, it is more likely to affect the stability of DNA or DNA replication processes. The potential biological effects of these phosphate adducts identified here warrant further investigation.
Conclusion
By using the developed LC–NSI–HRMS/MS method, we investigated DNA phosphate adduct formation by the tobacco-specific carcinogen NNK. We provide convincing evidence of the formation of 30 of 32 possible phosphate adducts, and their individual levels in both in vitro and in vivo DNA samples were determined. Certain phosphate adducts such as TpopT are persistent and abundant in NNK-treated rats over 70 weeks, suggesting that they could be potential biomarkers for chronic exposure to NNK. The results of this study lay the groundwork for establishing a representative array of phosphate adducts as biomarkers of tobacco carcinogen exposure in future studies.
Acknowledgments
We thank Xun Ming for his help with the mass spectrometry analysis and Dr. Silvia Balbo for providing rat lung DNA samples. We also thank Robert Carlson for editorial assistance.
Glossary
Abbreviations
- B1popB2
B1p[4-oxo-4-(3-pyridyl)butyl]B2
- CpopC
dCp[4-oxo-4-(3-pyridyl)butyl]dC
- CT-DNA
calf thymus DNA
- ENU
ethylnitrosourea
- HCD
higher-energy collisional dissociation
- IS
internal standards
- LC-NSI-HRMS/MS
liquid chromatography-nanoelectrospray ionization-high resolution tandem mass spectrometry
- NNK
4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone
- NNKOAc
4-(acetoxymethylnitrosamino)-1-(3-pyridyl)-1-butanone
- POB-DNA adduct
pyridyloxobutyl-DNA adduct
- PTEs
phosphotriesters
- SD
standard deviation
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.chemrestox.5b00318.
Synthesis and NMR data of CpopC, synthesis of [15N3]-2′-dCpdC, LC–MS chromatograms of CpopC and [15N3]CpopC, product ion spectra of ApopC, extracted ion chromatograms and fragmentation patterns of B1popB2 PTEs, structure of B1p[2-(3-pyridyl)-2,3,4,5-tetrahydrofuranyl]B2, levels of DNA phosphate adducts in liver and lung DNA of NNK-treated rats, and levels of POB adducts and total DNA phosphate adducts in NNKOAc-treated CT-DNA and in liver DNA from NNK-treated rats (PDF)
This study was supported by National Cancer Institute (NCI) Grant CA-81301. Mass spectrometry analyses were conducted in the Analytical Biochemistry Shared Resource of the Masonic Cancer Center, supported in part by NCI Grant CA-77598.
The authors declare no competing financial interest.
Supplementary Material
References
- Hecht S. S. (1998) Biochemistry, biology, and carcinogenicity of tobacco-specific N-nitrosamines. Chem. Res. Toxicol. 11, 559–603 10.1021/tx980005y. [DOI] [PubMed] [Google Scholar]
- International Agency for Research on Cancer (2012) Personal habits and indoor combustions. In IARC Monographs on the Evaluation of Carcinogenic Risks to Humans, Vol. 100E, pp 319–331, IARC, Lyon, France. [PMC free article] [PubMed] [Google Scholar]
- Balbo S.; Johnson C. S.; Kovi R. C.; James-Yi S. A.; O′Sullivan M. G.; Wang M.; Le C. T.; Khariwala S. S.; Upadhyaya P.; Hecht S. S. (2014) Carcinogenicity and DNA adduct formation of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of its metabolite 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol in F-344 rats. Carcinogenesis 35, 2798–2806 10.1093/carcin/bgu204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lao Y.; Villalta P. W.; Sturla S. J.; Wang M.; Hecht S. S. (2006) Quantitation of pyridyloxobutyl DNA adducts of tobacco-specific nitrosamines in rat tissue DNA by high-performance liquid chromatography-electrospray ionization-tandem mass spectrometry. Chem. Res. Toxicol. 19, 674–682 10.1021/tx050351x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.; Wang M.; Villalta P. W.; Lindgren B. R.; Upadhyaya P.; Lao Y.; Hecht S. S. (2009) Analysis of pyridyloxobutyl and pyridylhydroxybutyl DNA adducts in extrahepatic tissues of F344 rats treated chronically with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone and enantiomers of 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanol. Chem. Res. Toxicol. 22, 926–936 10.1021/tx900015d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G. D.; Le Pla R. C.; Farmer P. B. (2010) Phosphotriester adducts (PTEs): DNA’s overlooked lesion. Mutagenesis 25, 3–16 10.1093/mutage/gep038. [DOI] [PubMed] [Google Scholar]
- Singer B. (1985) In vivo formation and persistence of modified nucleosides resulting from alkylating agents. Environ. Health Perspect. 62, 41–48 10.1289/ehp.856241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek D. T.; Weis C. C.; Swenson D. H. (1980) A comprehensive quantitative analysis of methylated and ethylated DNA using high pressure liquid chromatography. Carcinogenesis 1, 595–606 10.1093/carcin/1.7.595. [DOI] [PubMed] [Google Scholar]
- Den Engelse L.; De Graaf A.; De Brij R. J.; Menkveld G. J. (1987) O2- and O4-ethylthymine and the ethylphosphotriester dTp(Et)dT are highly persistent DNA modifications in slowly dividing tissues of the ethylnitrosourea-treated rat. Carcinogenesis 8, 751–757 10.1093/carcin/8.6.751. [DOI] [PubMed] [Google Scholar]
- Shooter K. V.; Slade T. A. (1977) The stability of methyl and ethyl phosphotriesters in DNA in vivo. Chem.-Biol. Interact. 19, 353–361 10.1016/0009-2797(77)90057-6. [DOI] [PubMed] [Google Scholar]
- Haglund J.; Henderson A. P.; Golding B. T.; Tornqvist M. (2002) Evidence for phosphate adducts in DNA from mice treated with 4-(N-methyl-N-nitrosamino)-1-(3-pyridyl)-1-butanone (NNK). Chem. Res. Toxicol. 15, 773–779 10.1021/tx015542o. [DOI] [PubMed] [Google Scholar]
- Miller P. S.; Fang K. N.; Kondo N. S.; Ts’o P. O. P. (1971) Syntheses and properties of adenine and thymine nucleoside alkyl phosphotriesters, the neutral analogs of dinucleoside monophosphates. J. Am. Chem. Soc. 93, 6657–6665 10.1021/ja00753a054. [DOI] [PubMed] [Google Scholar]
- Haglund J.; Van Dongen W.; Lemiere F.; Esmans E. L. (2004) Analysis of DNA-phosphate adducts in vitro using miniaturized LC-ESI-MS/MS and column switching: Phosphotriesters and alkyl cobalamins. J. Am. Soc. Mass Spectrom. 15, 593–606 10.1016/j.jasms.2003.12.012. [DOI] [PubMed] [Google Scholar]
- Snyder R. D.; Regan J. D. (1981) Quantitative estimation of the extent of alkylation of DNA following treatment of mammalian cells with non-radioactive alkylating agents. Mutat. Res. Lett. 91, 307–314 10.1016/0165-7992(81)90006-3. [DOI] [PubMed] [Google Scholar]
- Shooter K. V.; Merrifield R. K. (1976) An assay for phosphotriester formation in the reaction of alkylating agents with deoxyribosenucleic acid in vitro and in vivo. Chem.-Biol. Interact. 13, 223–236 10.1016/0009-2797(76)90076-4. [DOI] [PubMed] [Google Scholar]
- Saris C. P.; Damman S. J.; van den Ende A. M.; Westra J. G.; den Engelse L. (1995) A 32P-postlabelling assay for the detection of alkylphosphotriesters in DNA. Carcinogenesis 16, 1543–1548 10.1093/carcin/16.7.1543. [DOI] [PubMed] [Google Scholar]
- Singh R.; Sweetman G. M.; Farmer P. B.; Shuker D. E.; Rich K. J. (1997) Detection and characterization of two major ethylated deoxyguanosine adducts by high performance liquid chromatography, electrospray mass spectrometry, and 32P-postlabeling. Development of an approach for detection of phosphotriesters. Chem. Res. Toxicol. 10, 70–77 10.1021/tx960135b. [DOI] [PubMed] [Google Scholar]
- Zhang F.; Bartels M. J.; Pottenger L. H.; Gollapudi B. B.; Schisler M. R. (2007) Quantitation of lower levels of the DNA adduct of thymidylyl(3′-5′)thymidine methyl phosphotriester by liquid chromatography/negative atmospheric pressure chemical ionization tandem mass spectrometry. Rapid Commun. Mass Spectrom. 21, 1043–1048 10.1002/rcm.2933. [DOI] [PubMed] [Google Scholar]
- Ma B.; Villalta P. W.; Balbo S.; Stepanov I. (2014) Analysis of a malondialdehyde-deoxyguanosine adduct in human leukocyte DNA by liquid chromatography nanoelectrospray-high-resolution tandem mass spectrometry. Chem. Res. Toxicol. 27, 1829–1836 10.1021/tx5002699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balbo S.; Villalta P. W.; Hecht S. S. (2011) Quantitation of 7-ethylguanine in leukocyte DNA from smokers and nonsmokers by liquid chromatography-nanoelectrospray-high resolution tandem mass spectrometry. Chem. Res. Toxicol. 24, 1729–1734 10.1021/tx200262d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guichard Y.; Jones G. D.; Farmer P. B. (2000) Detection of DNA alkylphosphotriesters by 32P postlabeling: evidence for the nonrandom manifestation of phosphotriester lesions in vivo. Cancer Res. 60, 1276–1282. [PubMed] [Google Scholar]
- Le Pla R. C.; Bowman K. J.; Farmer P. B.; Jones G. D. (2006) Phosphate alkylation in different DNA substrates: the role of local DNA sequence and electrophile character in determining the nonrandom nature of phosphotriester adduct formation. Chem. Res. Toxicol. 19, 407–413 10.1021/tx050298c. [DOI] [PubMed] [Google Scholar]
- Miller P. S.; Chandrasegaran S.; Dow D. L.; Pulford S. M.; Kan L. S. (1982) Synthesis and template properties of an ethyl phosphotriester modified decadeoxyribonucleotide. Biochemistry 21, 5468–5474 10.1021/bi00265a014. [DOI] [PubMed] [Google Scholar]
- Yashiki T.; Yamana K.; Nunota K.; Negishi K. (1992) Sequence specific block of in vitro DNA synthesis with isopropyl phosphotriesters in template oligodeoxyribonucleotides. Nucleic Acids Symp. Ser. 27, 197–198. [PubMed] [Google Scholar]
- Kan L.; Voituriez L.; Cadet J. (1988) Nuclear magnetic resonance studies of cis-syn, trans-syn, and 6–4 photodimers of thymidylyl(3′-5′)thymidine monophosphate and cis-syn photodimers of thymidylyl(3′-5′)thymidine cyanoethyl phosphotriester. Biochemistry 27, 5796–5803 10.1021/bi00415a060. [DOI] [PubMed] [Google Scholar]
- Lawrence D. P.; Chen W. Q.; Zon G.; Stec W. J.; Uznanski B.; Broido M. S. (1987) NMR studies of backbone-alkylated DNA: duplex stability, absolute stereochemistry, and chemical shift anomalies of prototypal isopropyl phosphotriester modified octanucleotides, (Rp,Rp)- and (Sp,Sp)-(d-[GGA(iPr)ATTCC])2 and -(d-[GGAA(iPr)TTCC])2. J. Biomol. Struct. Dyn. 4, 757–783 10.1080/07391102.1987.10507677. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.