Abstract
Background:
The effects of the vascular endothelial growth factor (VEGF) gene −2578C/A polymorphism on colorectal cancer (CRC) risk have been investigated in some studies; however, the results of these studies were conflicting and ambiguous. Therefore, we aimed to do a meta-analysis to investigate the association of VEGF −2578C/A polymorphisms with CRC risk from all eligible case-control studies published to date.
Materials and Methods:
An electronic search of the PubMed, Embase and Medline was performed. Retrieve terms were utilized as following: (“VEGF a” [MeSH Terms]) and (“polymorphism, genetic” [MeSH Terms]) and (“colorectal neoplasms” [MeSH Terms]). The association between VEGF −2578C/A polymorphisms with CRC risk was calculated with odds ratios (ORs) and their corresponding 95% of confidence intervals (CIs), and stratified analysis was also conducted with respect to ethnicity.
Results:
A comprehensive meta-analysis of eight studies, including 2312 cases and 2308 controls was performed in this work. Combined analysis revealed that a significant association between the VEGF −2578C/A polymorphism with CRC risk was identified in three comparison models including C allele versus A allele (OR = 0.85, 95% CI 0.75-0.97, P = 0.02), AA versus CA + CC (OR = 1.28, 95% CI 1.09-1.51, P = 0.003), and AA versus CC (OR = 0.77, 95% CI 0.64-0.93, P = 0.006). Moreover, a similar result was obtained in the subgroup analysis that comparison models of C allele versus. A allele (OR = 0.85, 95% CI 0.76-0.95, P = 0.004), AA versus CA + CC (OR = 1.31, 95% CI 1.09-1.57, P = 0.004), and AA versus CC (OR = 0.73, 95% CI 0.59-0.90, P = 0.004) was confirmed to be associated with CRC risk in Caucasian.
Conclusion:
It has been proved that the C allele versus A allele, AA versus CA + CC, and AA versus CC comparison models of VEGF −2578C/A polymorphism might be risk factors for CRC, but further studies with larger sample sizes are required to make a better assessment of above association.
Keywords: Colorectal cancer, meta-analysis, polymorphism, vascular endothelial growth factor
INTRODUCTION
Colorectal cancer (CRC) is a leading cause of cancer-related death all over the world.[1] However, most patients have a poor prognosis for the regional or distant spread of tumor cells at the time of diagnosis.[2] Accordingly, a growing interest is focused on an assessment of biomarkers as potential predictors of prognosis or response to therapy in CRC, which will most likely contribute to the individualized management of patients.[3]
As a vascular permeability factor, vascular endothelial growth factor (VEGF) is involved in a series of pathologic processes including tumor growth and metastasis.[4] In addition, some experiments have demonstrated prognostic significance of the VEGF expression in patients with cancer.[5,6] In particular, the expression of VEGF has been reported to be intimately correlated with the prognosis of CRC.[7,8] The VEGF gene comprises eight exons and seven introns, and several single nucleotide polymorphisms have been identified in the VEGF gene, some of which were reported to play an important role in the differential expression of VEGF in vitro.[9,10] Therefore, genetic polymorphisms of the VEGF gene are suggested to increase cancers risk and even work as a candidate marker in the prognosis of patients with cancer.[11]
VEGF −2578C/A is one of the most common VEGF polymorphisms. Numerous studies have evaluated the association between the VEGF −2578C/A polymorphism with CRC risk.[12,13,14,15,16,17,18,19] However, the results of these studies were inconclusive, probably because the sample size enclosed in any single study is so small that it lacked inadequate evidence to make a positive or negative conclusion. Furthermore, meta-analysis is a powerful means to synthesize information from varied investigations on the same issue.[20] Therefore, a meta-analysis is essential to investigate the association of VEGF −2578C/A polymorphisms with CRC risk from all eligible case-control studies published to date. Although two related meta-analysis have been performed before, they did not consisted of all related research work, and one of them failed to make a subgroup analysis to assess the effect of ethnicity on the association of −2578C/A polymorphism with CRC risk due to the paucity of eligible studies in Asian population.[21,22] And for all we know, there were some new studies published in recent years, some of which were carried out in both Asian and Caucasian population. Accordingly, a latest meta-analysis was performed in this work, in order to provide a more accurate conclusion about the association between −2578C/A polymorphism and CRC risk.
MATERIALS AND METHODS
Literature search
An electronic search of the PubMed, Embase and Medline was performed to retrieve studies assessing the associations of VEGF −2578C/A polymorphism and CRC risk. Retrieve terms were utilized as following: (“VEGF a”[MeSH Terms]) AND (“polymorphism, genetic”[MeSH Terms]) AND (“colorectal neoplasms”[MeSH Terms]). Other potentially eligible studies were also found by manually searching from the reference lists of relevant reviews and included studies. All documents were updated to September 2014. The language was limited to English.
Inclusion and exclusion criteria
To be eligible for the inclusion in the meta-analysis, the following criteria were used:
Case — control studies comparing CRC cases with controls;
Studies assessing the association between VEGF −2578C/A polymorphisms and CRC risk;
Sufficient genotype data of VEGF −2578C/A polymorphisms were provided.
Studies were excluded when satisfied the following criteria:
Studies lack information about genotype frequencies or alleles;
Not case-control studies;
Studies were reviewed, letters, case reports, and editorial articles;
Family-based design.
Data extraction
Data extraction was performed independently by two reviewers. Inter-researcher disagreements were resolved by consensus or by a third investigator. The following data were extracted: First author, publication year, country, ethnicity, source of cases, source of controls, study design, number of cases, number of controls, and genotype data of VEGF +936C/T polymorphisms. Authors of the identified studies were mailed if some details were required.
Statistical analysis
The meta-analysis was carried out with two kinds of software including the STATA 11.0 (Stata Corporation, College Station, TX, USA) and Review Manager 4.2 (provided by the Cochrane Collaboration). Odds ratio (OR) and 95% confidence interval (95% CI) were used to evaluate the associations of VEGF −2578C/A polymorphism with CRC risk, and a statistical significance of OR was ascertained with the P value of Z-test <0.05. Four contrasts for the VEGF −2578C/A polymorphism were evaluated: Comparison of T allele with C allele; comparison of TT + CT versus CC; comparison of TT versus CC + CT; comparison of TT versus CC. An application to the effects models depended on the degree of between-study heterogeneity, which was estimated by Cochran's Q-test and I2 test in this meta-analysis. The heterogeneity across studies was identified by a significant Q test (P < 0.10) or I2 >50%, thus the random effects model was selected for the evaluation of each investigation with combined ORs. On the contrary, the fixed effects model was used for P value of Q-test bigger than 0.01 or I2 <50%. Hardy–Weinberg equilibrium (HWE) in the controls was appraised by a χ2 test prior to estimating the associations of VEGF −2578C/A polymorphism with CRC risk.[23] Subgroup analysis was conducted with respect to ethnicity. Sensitivity analysis was mainly performed by a sequential omission of individual studies to assess the stability of the outcomes.[24] The potential publication bias was evaluated with Egger's test and Begg's funnel plot.[25] All P values in the meta-analysis were two-sided, and statistical significance was considered when the P < 0.05.
RESULTS
Literature search
Initial search for PubMed, Embase and Medline databases of the literature yielded 69 papers, and 4 additional relevant references quoted in searched articles were also selected. There were 70 potentially relevant papers after duplicates removed. 57 irrelevant papers were excluded on the basis of title and abstract, including 42 not including −2578C/A polymorphism, 8 not control-case study, and 7 not including genotype frequencies or alleles data. Finally, 13 full-text articles were evaluated for eligibility, and 8 were included in the meta-analysis after 5 full-text reviews were excluded for lacking detailed data about assessing the association between VEGF −2578C/A polymorphisms and CRC risk. Study selection is demonstrated in Figure 1. These eight case-control studies included 2312 cases and 2308 controls reporting the relationship between VEGF −2578C/A polymorphism and CRC risk. When stratified by ethnicity, two essays involving Asians included 741 patients and 805 controls, and the other five articles were respected to Caucasian containing 1571 cases and 1503 controls. The publication year of the included studies ranged from 2007 to 2013. Eight articles provided sufficient information including the numbers of allele C and allele A in both CRC cases and controls, and selected characteristics of each study are listed in Table 1.
Figure 1.
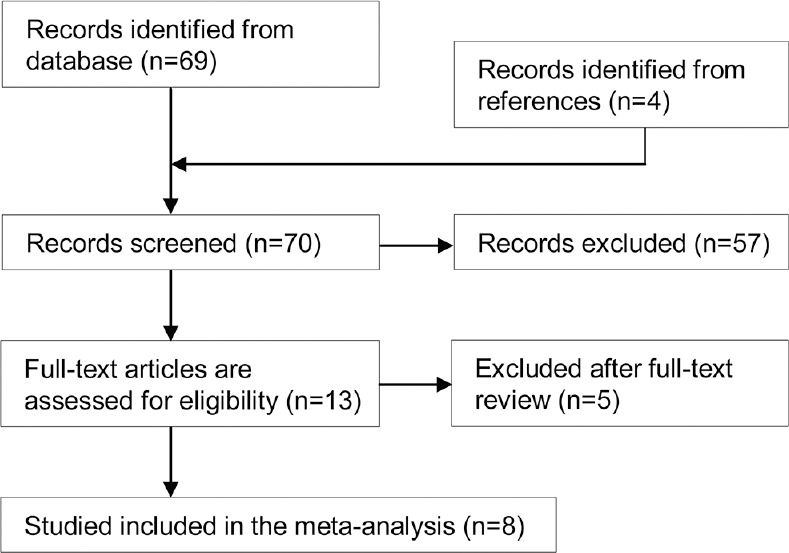
Flow diagram of included/excluded studies
Table 1.
Characteristics of the studies and populations included in the meta-analysis
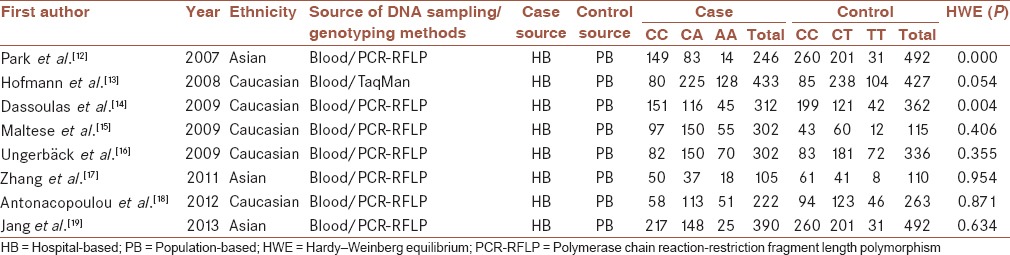
VEGF −2578C/A polymorphisms in meta-analysis
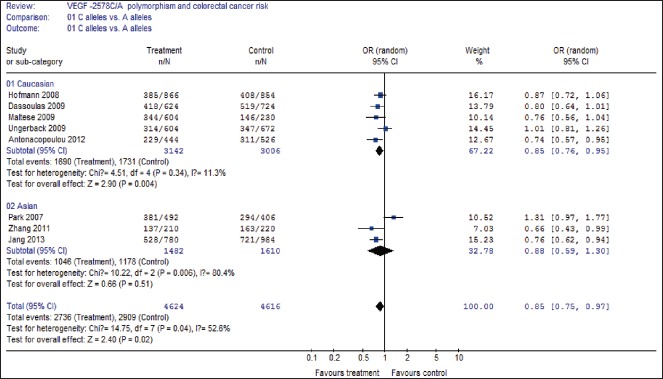
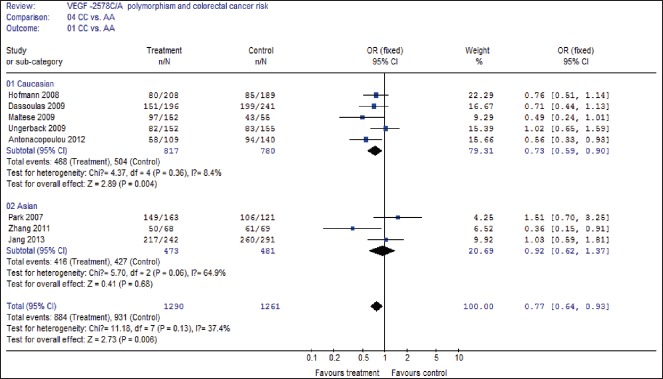
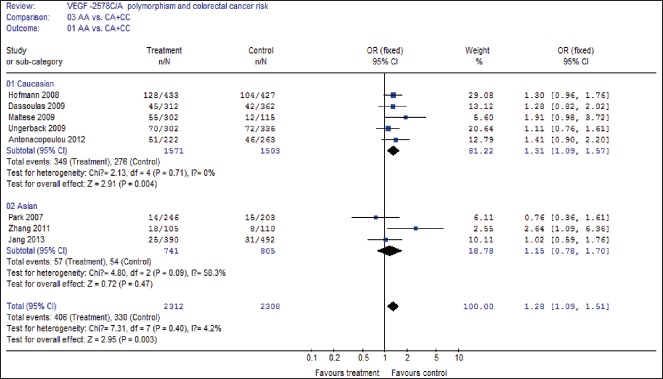
A summary of the meta-analysis results of the association between the VEGF −2578C/A polymorphisms and CRC risk is demonstrated in Table 2. In overall analysis, there was apparently between-study heterogeneity under the two comparison models of C allele versus A allele (I2 = 52.6%, Pheterogeneity = 0.04) and AA + CA versus CC (I2 = 50.4%, Pheterogeneity = 0.05). Therefore, these two genetic models used random-effects model. However, there was no evidence of heterogeneity under the other two comparison models of AA versus CC + CA (I2 = 4.2%, Pheterogeneity = 0.40), and AA versus CC (I2 = 37.4%, Pheterogeneity = 0.13), so that a fixed-effects model was applied to these two genetic models.
Table 2.
Meta-analysis of the association between the –2578C/A polymorphism and CRC risk
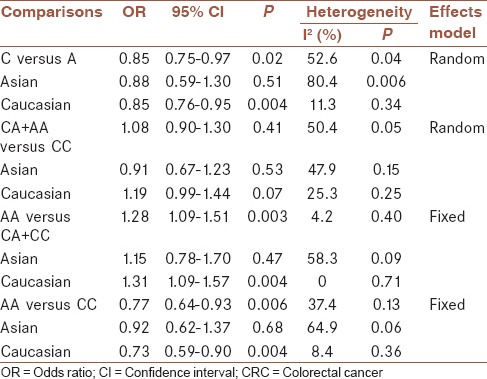
As demonstrated in Table 2, a significant association between the VEGF −2578C/A polymorphism and CRC risk was identified in three comparison models including C allele versus A allele (OR = 0.85, 95% CI 0.75-0.97, P = 0.02), AA versus CA + CC (OR = 1.28, 95% CI 1.09-1.51, P = 0.003), and AA versus CC (OR = 0.77, 95% CI 0.64-0.93, P = 0.006), but CA + AA versus CC (OR = 1.08, 95% CI 0.90-1.30, P = 0.41) did not show association with CRC risk.
Then subgroup analysis was made to assess the potential ethnic differences, and the subjects of all included studies were divided into Asian and Caucasian populations. Results of subgroup analysis demonstrated that all comparison models of the Asians were similar to the overall populations [Figures 2–4]. A significant association with CRC risk in Caucasian populations was confirmed in comparison models of C allele versus A allele (OR = 0.85, 95% CI 0.76-0.95, P = 0.004), AA versus CA + CC (OR = 1.31, 95% CI 1.09-1.57, P = 0.004), and AA versus CC (OR = 0.73, 95% CI 0.59-0.90, P = 0.004). However, no significant association with CRC risk was found in CA + AA versus CC comparison models in Caucasian populations. Moreover, there was also no significant association of all comparison models with CRC risk in Asian populations.
Figure 2.
Forest plot of C allele versus A allele comparison model for overall comparison
Figure 4.
Forest plot of CC versus AA comparison model for overall comparison
Figure 3.
Forest plot of AA versus CA+ CC comparison model for overall comparison
Sensitivity analysis and publication bias
A consecutive exclusion of individual studies was performed in the sensitivity analysis. The corresponding combined ORs in all individual analyses and subgroup analyses were statistically robust by deleting any single study. In addition, another sensitivity analysis was also performed by excluding studies without HWE, but the results did not changed significantly.
Publication bias was assessed by Begg's funnel plot in the meta-analysis [Figure 5]. The shape of the funnel plots showed almost symmetrical, and the P values of Egger's test and Begg's test were 0.221 and 0.138 respectively, indicating that there was no evidence of publication bias for the meta-analysis of the association between VEGF −2578C/A polymorphism and CRC risk.
Figure 5.
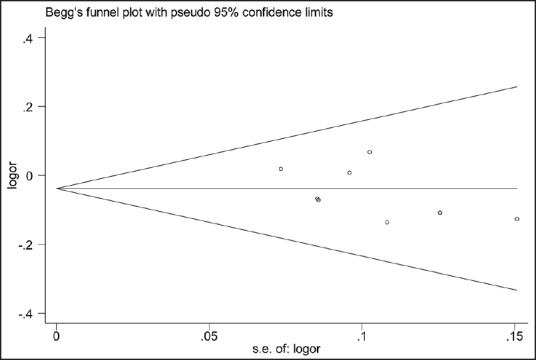
Funnel plot of C allele versus A allele, AA versus CA + CC, and CC versus AA comparison models
DISCUSSION
CRC is one of the leading causes of death all over the world, and about 1 million people have been diagnosed with CRC every year.[1] It is a huge challenge to find out an appropriate treatment to improve the poor prognosis of CRC, for the median survival in patients were still less than initially hoped for.[26]
Angiogenesis has been proved to be crucial for the progression and growth of solid cancer.[27,28] It has been identified that VEGF-mediated the angiogenesis through promoting endothelial cell growth, migration, and mitosis, and its expression is involved in the pathogenesis, progression, and metastasis of cancers.[4] Clinical studies have shown that the prognosis for various solid tumors is connected with an increased expression of VEGF.[29,30,31] In particular, the expression of VEGF has been reported to be intimately correlated with the prognosis of CRC.[7,8] Moreover, as is known, genetic polymorphisms altering the level of protein expressed are anticipated to have a substantial influence on disease activity.[32] There was growing evidence that polymorphisms in VEGF gene were associated with the production of the VEGF protein in colorectal carcinogenesis.[33,34] VEGF −2578C/A is one of the most common VEGF polymorphisms, and numerous studies have evaluated the association between the VEGF −2578C/A polymorphism and CRC risk in recent years.[12,13,14,15,16,17,18,19] However, some of those studies had used comparatively small samples, and the results remained conflicting. Therefore, meta-analysis is imperative to ensure adequate statistical power. And for all we know, this was the updated meta-analysis of the association between specific VEGF −2578C/A polymorphisms and CRC risk.
In our meta-analysis, eight case-control articles were selected for the assessment of the relationship between VEGF −2578C/A polymorphisms and CRC risk. These case-control studies selected included 2312 cases and 2308 controls. The main meta-analysis results showed that there was a significant association between the −2578C/A polymorphisms and CRC risk in comparisons of C allele versus A allele (P = 0.02), AA versus CA + CC (P = 0.003), and AA versus CC (P = 0.006), but no association was found in comparison model of CA + AA versus CC (P = 0.41). Moreover, the result of subgroup analysis confirmed a significant association with CRC risk in comparison models of C allele versus A allele (P = 0.004), AA versus CA + CC (P = 0.004), and AA versus CC (P = 0.004) in Caucasian populations.
In the subgroup analysis, a contrary conclusion was made in both Caucasian and Asian population that a significant association was found in comparisons of C allele versus A allele, AA versus CA + CC, and AA versus CC in Caucasian, but negative results were obtained in the same comparisons in Asian population. It indicated that there was a higher CRC risk for Caucasian with a VEGF −2578C/A polymorphisms compared with the Asian population.
On the basis of the above results, there was limited evidence proving that the comparison models of VEGF −2578C/A polymorphism, including C allele versus A allele, AA versus CA + CC, and AA versus CC, might be risk factors for CRC, and this conclusion was more persuading in Caucasian populations.
However, this meta-analysis was still limited due to some deficiencies. First, the number of studies and subjects involved in researches were limited, which might provide insufficient statistical power to assess the association between VEGF −2578C/A polymorphism and CRC risk. Some connected studies with negative conclusions were quite likely to be lost. Thus, more studies were required for a more dependable consequence. Second, the sources of heterogeneity existing among studies for most polymorphisms were hard to address. Third, although there was no evident publication bias identified, potential bias might have distorted the results of the meta-analysis. Finally, due to incomplete raw data or publication limitations, relevant effect prompted by age, gender and other environmental factors could not be estimated.
Although the comparison models of VEGF −2578C/A polymorphism including C allele versus A allele, AA versus CA + CC, and AA versus CC has been shown prediction effect for CRC, some detailed information about its biological mechanism was lacked. Therefore, more researches focused on the biological mechanism should be carried out in the future on the basis of the results of this work.
Despite the above limitations, this latest meta-analysis of the association between −2578C/A polymorphism and CRC risk was statistically more persuading than any single study. It came to a conclusion that the C allele versus AA, AA versus CA + CC, and AA versus CC comparison models of VEGF −2578C/A polymorphism might be risk factors for CRC. However, in order to make a better assessment of the association between VEGF −2578C/A polymorphism and CRC risk, further studies conducted in standardized and unbiased ways are required.
Financial support and sponsorship
The work was supported by National Natural Science Foundation of China (81273585).
Conflicts of interest
There are no conflicts of interest.
AUTHOR'S CONTRIBUTIONS
ZN Cheng designed the research study. L Wang and S Ji performed an electronic search of the databases for eligible articles. L Wang and S Ji analyzed the data. L Wang wrote the paper. All authors have read and approved the content of the manuscript.
Acknowledgments
We thank all our colleagues working in the Research Institute of Drug Metabolism and Pharmacokinetics, School of Pharmaceutical Sciences, Central South University.
REFERENCES
- 1.Jemal A, Siegel R, Ward E, Hao Y, Xu J, Thun MJ. Cancer statistics, 2009. CA Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 2.Figueredo A, Coombes ME, Mukherjee S. Adjuvant therapy for completely resected stage II colon cancer. Cochrane Database Syst Rev. 2008;3:CD005390. doi: 10.1002/14651858.CD005390.pub2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Alvarez-Chaver P, Otero-Estévez O, Páez de la Cadena M, Rodríguez-Berrocal FJ, Martínez-Zorzano VS. Proteomics for discovery of candidate colorectal cancer biomarkers. World J Gastroenterol. 2014;20:3804–24. doi: 10.3748/wjg.v20.i14.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chekhonin VP, Shein SA, Korchagina AA, Gurina OI. VEGF in tumor progression and targeted therapy. Curr Cancer Drug Targets. 2013;13:423–43. doi: 10.2174/15680096113139990074. [DOI] [PubMed] [Google Scholar]
- 5.Kilic E, Schild SE, Thorns C, Bajrovic A, Rades D. Prognostic role of vascular endothelial growth factor and its receptor-1 in patients with esophageal cancer. Anticancer Res. 2014;34:5221–6. [PubMed] [Google Scholar]
- 6.Bestas R, Kaplan MA, Isikdogan A. The correlation between serum VEGF levels and known prognostic risk factors in colorectal carcinoma. Hepatogastroenterology. 2014;61:267–71. [PubMed] [Google Scholar]
- 7.Galizia G, Lieto E, Ferraraccio F, Orditura M, De Vita F, Castellano P, et al. Determination of molecular marker expression can predict clinical outcome in colon carcinomas. Clin Cancer Res. 2004;10:3490–9. doi: 10.1158/1078-0432.CCR-0960-03. [DOI] [PubMed] [Google Scholar]
- 8.Kaio E, Tanaka S, Kitadai Y, Sumii M, Yoshihara M, Haruma K, et al. Clinical significance of angiogenic factor expression at the deepest invasive site of advanced colorectal carcinoma. Oncol Basel. 2003;64:61–73. doi: 10.1159/000066511. [DOI] [PubMed] [Google Scholar]
- 9.Paré-Brunet L, Glubb D, Evans P, Berenguer-Llergo A, Etheridge AS, Skol AD, et al. Discovery and functional assessment of gene variants in the vascular endothelial growth factor pathway. Hum Mutat. 2014;35:227–35. doi: 10.1002/humu.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Almawi WY, Saldanha FL, Mahmood NA, Al-Zaman I, Sater MS, Mustafa FE. Relationship between VEGFA polymorphisms and serum VEGF protein levels and recurrent spontaneous miscarriage. Hum Reprod. 2013;28:2628–35. doi: 10.1093/humrep/det308. [DOI] [PubMed] [Google Scholar]
- 11.Eng L, Azad AK, Habbous S, Pang V, Xu W, Maitland-van der Zee AH, et al. Vascular endothelial growth factor pathway polymorphisms as prognostic and pharmacogenetic factors in cancer: A systematic review and meta-analysis. Clin Cancer Res. 2012;18:4526–37. doi: 10.1158/1078-0432.CCR-12-1315. [DOI] [PubMed] [Google Scholar]
- 12.Park HM, Hong SH, Kim JW, Oh D, Hwang SG, An HJ, et al. Gender-specific association of the VEGF -2578C > A polymorphism in Korean patients with colon cancer. Anticancer Res. 2007;27:2535–9. [PubMed] [Google Scholar]
- 13.Hofmann G, Langsenlehner U, Renner W, Langsenlehner T, Yazdani-Biuki B, Clar H, et al. Common single nucleotide polymorphisms in the vascular endothelial growth factor gene and colorectal cancer risk. J Cancer Res Clin Oncol. 2008;134:591–5. doi: 10.1007/s00432-007-0322-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dassoulas K, Gazouli M, Rizos S, Theodoropoulos G, Christoni Z, Nikiteas N, et al. Common polymorphisms in the vascular endothelial growth factor gene and colorectal cancer development, prognosis, and survival. Mol Carcinog. 2009;48:563–9. doi: 10.1002/mc.20495. [DOI] [PubMed] [Google Scholar]
- 15.Maltese P, Canestrari E, Ruzzo A, Graziano F, Falcone A, Loupakis F, et al. VEGF gene polymorphisms and susceptibility to colorectal cancer disease in Italian population. Int J Colorectal Dis. 2009;24:165–70. doi: 10.1007/s00384-008-0586-x. [DOI] [PubMed] [Google Scholar]
- 16.Ungerbäck J, Elander N, Dimberg J, Söderkvist P. Analysis of VEGF polymorphisms, tumor expression of VEGF mRNA and colorectal cancer susceptibility in a Swedish population. Mol Med Rep. 2009;2:435–9. doi: 10.3892/mmr_00000118. [DOI] [PubMed] [Google Scholar]
- 17.Zhang L, Zhang G, Wang P, Gong J, Cao Y, Tang L. Association of vascular endothelial growth factor -2578C/A gene polymorphism in Chinese patients with colon cancer. Genet Test Mol Biomarkers. 2011;15:117–21. doi: 10.1089/gtmb.2010.0166. [DOI] [PubMed] [Google Scholar]
- 18.Antonacopoulou AG, Kottorou AE, Dimitrakopoulos FI, Triantafyllia V, Marousi S, Koutras A, et al. VEGF polymorphisms may be associated with susceptibility to colorectal cancer: A case-control study. Cancer Biomark. 2011;10:213–7. doi: 10.3233/CBM-2012-0249. [DOI] [PubMed] [Google Scholar]
- 19.Jang MJ, Jeon YJ, Kim JW, Cho YK, Lee SK, Hwang SG, et al. Association of VEGF and KDR single nucleotide polymorphisms with colorectal cancer susceptibility in Koreans. Mol Carcinog. 2013;52:E60–9. doi: 10.1002/mc.21980. [DOI] [PubMed] [Google Scholar]
- 20.Greco T, Zangrillo A, Biondi-Zoccai G, Landoni G. Meta-analysis: Pitfalls and hints. Heart Lung Vessel. 2013;5:219–25. [PMC free article] [PubMed] [Google Scholar]
- 21.Zhao Z, Ba C, Wang W, Wang X, Xue R, Wu X. Vascular endothelial growth factor (VEGF) gene polymorphisms and colorectal cancer: A meta-analysis of epidemiologic studies. Genet Test Mol Biomarkers. 2012;16:1390–4. doi: 10.1089/gtmb.2012.0266. [DOI] [PubMed] [Google Scholar]
- 22.Chen Q, Zhou Z, Shan L, Hua Y, Zeng H, Liu P, et al. Association of the vascular endothelial growth factor -2578C/A polymorphism with cancer risk: A meta-analysis update. Biomed Rep. 2014;2:823–830. doi: 10.3892/br.2014.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Munafò MR, Clark TG, Flint J. Assessing publication bias in genetic association studies: Evidence from a recent meta-analysis. Psychiatry Res. 2004;129:39–44. doi: 10.1016/j.psychres.2004.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Chootrakool H, Shi JQ, Yue R. Meta-analysis and sensitivity analysis for multi-arm trials with selection bias. Stat Med. 2011;30:1183–98. doi: 10.1002/sim.4143. [DOI] [PubMed] [Google Scholar]
- 25.Sterne JA, Egger M, Smith GD. Systematic reviews in health care: Investigating and dealing with publication and other biases in meta-analysis. BMJ. 2001;323:101–5. doi: 10.1136/bmj.323.7304.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyle P, Ferlay J. Mortality and survival in breast and colorectal cancer. Nat Clin Pract Oncol. 2005;2:424–5. doi: 10.1038/ncponc0288. [DOI] [PubMed] [Google Scholar]
- 27.Yancopoulos GD, Davis S, Gale NW, Rudge JS, Wiegand SJ, Holash J. Vascular — Specific growth factors and blood vessel formation. Nature. 2000;407:242–8. doi: 10.1038/35025215. [DOI] [PubMed] [Google Scholar]
- 28.Carmeliet P, Jain RK. Angiogenesis in cancer and other diseases. Nature. 2000;407:249–57. doi: 10.1038/35025220. [DOI] [PubMed] [Google Scholar]
- 29.Nishida N, Yano H, Komai K, Nishida T, Kamura T, Kojiro M. Vascular endothelial growth factor C and vascular endothelial growth factor receptor 2 are related closely to the prognosis of patients with ovarian carcinoma. Cancer. 2004;101:1364–74. doi: 10.1002/cncr.20449. [DOI] [PubMed] [Google Scholar]
- 30.Masuya D, Huang C, Liu D, Kameyama K, Hayashi E, Yamauchi A, et al. The intratumoral expression of vascular endothelial growth factor and interleukin-8 associated with angiogenesis in nonsmall cell lung carcinoma patients. Cancer. 2001;92:2628–38. doi: 10.1002/1097-0142(20011115)92:10<2628::aid-cncr1616>3.0.co;2-f. [DOI] [PubMed] [Google Scholar]
- 31.Fontanini G, Faviana P, Lucchi M, Boldrini L, Mussi A, Camacci T, et al. A high vascular count and overexpression of vascular endothelial growth factor are associated with unfavourable prognosis in operated small cell lung carcinoma. Br J Cancer. 2002;86:558–63. doi: 10.1038/sj.bjc.6600130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taylor JG, Choi EH, Foster CB, Chanock SJ. Using genetic variation to study human disease. Trends Mol Med. 2001;7:507–12. doi: 10.1016/s1471-4914(01)02183-9. [DOI] [PubMed] [Google Scholar]
- 33.Watson CJ, Webb NJ, Bottomley MJ, Brenchley PE. Identification of polymorphisms within the vascular endothelial growth factor (VEGF) gene: Correlation with variation in VEGF protein production. Cytokine. 2000;12:1232–5. doi: 10.1006/cyto.2000.0692. [DOI] [PubMed] [Google Scholar]
- 34.Bae SJ, Kim JW, Kang H, Hwang SG, Oh D, Kim NK. Gender-specific association between polymorphism of vascular endothelial growth factor (VEGF 936 C >T) gene and colon cancer in Korea. Anticancer Res. 2008;28:1271–6. [PubMed] [Google Scholar]