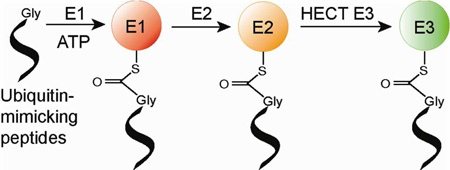
Abstract
Short heptapeptides were identified to function as ubiquitin (UB) mimics that are activated by E1 and form thioester conjugates with E1, E2 and HECT type E3 enzymes. The activities (kcat/K1/2) of E1 with the UB-mimicking peptides are 100–300 fold higher than the equally long peptide with the native C-terminal sequence of UB. By forming covalent conjugates with E1, E2 and E3 enzymes, the UB-mimicking peptides can block the transfer of native UB through the cascade.
UB is covalently attached to cellular proteins through the tandem action of E1, E2 and E3 enzymes to regulate many key biological processes including protein trafficking and degradation, complex formation, enzyme catalysis, and gene activation.1 E1 catalyzes the condensation reaction between the C-terminal carboxylate of UB and ATP to form a UB-AMP intermediate that subsequently reacts with a catalytic Cys residue of E1 to afford a UB~E1 conjugate (“~”designates the thioester linkage) (Supporting Information, Figure S1).2 UB bound to E1 is then transferred to the catalytic Cys residue of the E2 enzymes to form UB~E2 conjugates.3 In the last step, E3s recruit cellular proteins to bridge the transfer of UB from E2 to the modification targets.4 Similarly, UB-like proteins (UBLs) such as Nedd8 and SUMO are transferred by their own sets of E1-E2-E3 cascades to cellular protein targets.5 Together UB and UBLs carry a broad band of signals for the regulation of most aspects of cell biology.6
The enzymatic cascades for protein modification by UB or UBLs have been the intense focus of drug discovery efforts.7 The identification of the compound MLN4924 as a potent inhibitor of the Nedd8 E1 demonstrates the druggability of the E1 enzymes.8 MLN4924 has shown promising anticancer activities and is currently in clinical trials for the treatment of myeloma and lymphoma.9 Several inhibitors of the E1 enzyme specific for UB have been developed that inactivate the catalytic Cys residue of E1 or disrupt UB binding to E1.10 These inhibitors can attenuate the growth of cancer cells. E2 and E3 enzymes downstream of the UB transfer cascades are also valid targets for inhibitor design. Recent screening efforts have identified compounds that bind to E2 or E3 and block UB transfer.11 Here we report the identification of short peptides that mimic UB and form covalent conjugates with the E1-E2-E3 cascade (Table 1). Once these peptides are charged to the cascade enzymes, they can effectively block UB transfer through the cascade. The development of these UB-mimicking peptides provides a new way to inhibit protein modification by UB.
Table 1.
Kinetic parameters of ATP-PPi exchange catalyzed by Ube1 with the UB-mimicking peptides.
| K1/2 (µM) |
kcat (min−1) |
kcat/K1/2 (µM−1 min−1) |
|
|---|---|---|---|
| wtUB (full length) | 1.4 ± 0.5 | 88 ± 17 | 60 |
| C-terminal peptides of wtUB and variants | |||
| P1 (70VLRLRGG76) | -a | -a | 7.7 × 10−5 |
| P2 (70VWRFHGG76) | 342 ± 17 | 3.5 ± 0.6 | 1.0 × 10−2 |
| P3 (70VQRYWGG76) | 426 ± 11 | 9.7 ± 1.4 | 2.3 × 10−2 |
| P4 (70VYRFYGG76) | 141 ± 5 | 15 ± 2.7 | 1.1 × 10−2 |
K1/2 and kcat could not be determined for P1 due to its low activity.
We identified the UB-mimicking peptides in a study profiling the specificity of the E1 enzymes with the C-terminal sequence of UB by phage display.12 We constructed a UB library with randomized C-terminal residues covering residues 71–75 of UB ending with 71LRLRGG76. Phage selection based on the reactivity of the UB clones to form thioester conjugates with Ube1, the human E1, identified UB variants with quite different C-terminal sequences from the wild type (wt) UB (Supporting Information, Figure S2). Interestingly these UB variants share similar reactivities as the wt UB with Ube1.12 As shown by the sequence alignments of the phage selected UB clones, except for Arg72 and Gly75 that have a strong preference for wt residues, positions 71 and 73 in the UB variants are predominantely occupied with bulky aromatic side chains such as Phe, Tyr and Trp, quite different from the Leu residues in the wt sequence (Figure S2). Position 74 of the UB variants may also have aromatic or positively charged His side chains to replace the wt Arg residue. It has previously been reported that short peptides corresponding to the C-terminal sequences of the wt UB can be activated by the E1 enzymes and transferred through the E1-E2-E3 cascade for protein modification.13 We were thus interested in assaying if the C-terminal peptides of the UB variants from phage selection are more reactive with the E1 enzyme than the C-terminal peptide of wt UB.
We synthesized peptides corresponding to the C-terminal sequences of wtUB (P1, VLRLRGG), and UB variants e27 (P2, VWRFHGG), e40 (P3, VQRYWGG) and e25 (P4, VYRFYGG), and measured the ATP-PPi exchange kinetics of the peptides catalyzed by Ube1 (Figure S2 and Table 1).2a The P1 peptide with the sequence of wt UB could not saturate Ube1 at a concentration as high as 500 µM in the ATP-PPi exchange reaction so only the kcat/K1/2 value could be measured. In contrast, the P2–P4 peptides displayed a much higher affinity for Ube1 than P1 and yielded K1/2 values of 141–426 µM. These peptides were 130–300 fold more active than the P1 peptide in the ATP/PPi exchange reactions based on the kcat/K1/2 values (Table 1). Despite the higher activities of the phage selected peptides with Ube1, P2–P4 were still 2,600–6,000 fold less active than full length UB, largely due to the much lower K1/2 of UB with Ube1 (1.4 µM) (Table 1). The high affinity of UB with Ube1 can be attributed to the multiple binding interfaces between UB and E1. The crystal structure of UB in complex with Uba1, the yeast E1, shows that E1 not only binds to the C-terminal peptide of UB, but also interacts with the hydrophobic patch of UB composed of residues Leu8, Ile44 and Val70, and a polar surface of UB involving Lys11, Thr12, Gln31 and Asp32 (Supporting Information, Figure S3A).14 We also synthesized 7-mer peptides with the C-terminal sequences of the UB variants e6, e19, e26, e46 and e47 from phage selection (Figure S2), and found that none of the peptides were reactive with Ube1 based on ATP-PPi exchange. These peptides have the second to last Gly (Gly75 of wt UB) replaced with Ser, Asp, Asn or Gln residues that may disrupt peptide binding to the E1 enzyme. Since the peptides P2–P4 can be activated by Ube1 like wt UB, we refer to them as “UB-mimicking peptides”.
We modeled structures of the UB-mimicking peptides bound to Uba1 based on the crystal structures of UB-Uba1 complex,14 and used the Protein Interfaces, Surfaces and Assemblies (PISA) server of the European Bioinformatics Institute (EBI) to analyze peptide binding with Uba1 (Figure S3).15 PISA calculations suggested that the wt P1 peptide and the P2, P3 and P4 peptides have similar interface areas with E1, but the binding energy of P1 is 1.5–2 kcal/mol less than the P2–P4 peptides with E1 Supporting Information, Table S1). The differences in the binding energies of the peptides with E1 calculated by PISA match the differences in ΔG calculated based on the K1/2 values of the peptides in the ATP-PPi exchange reactions surprisingly well (Table S1).
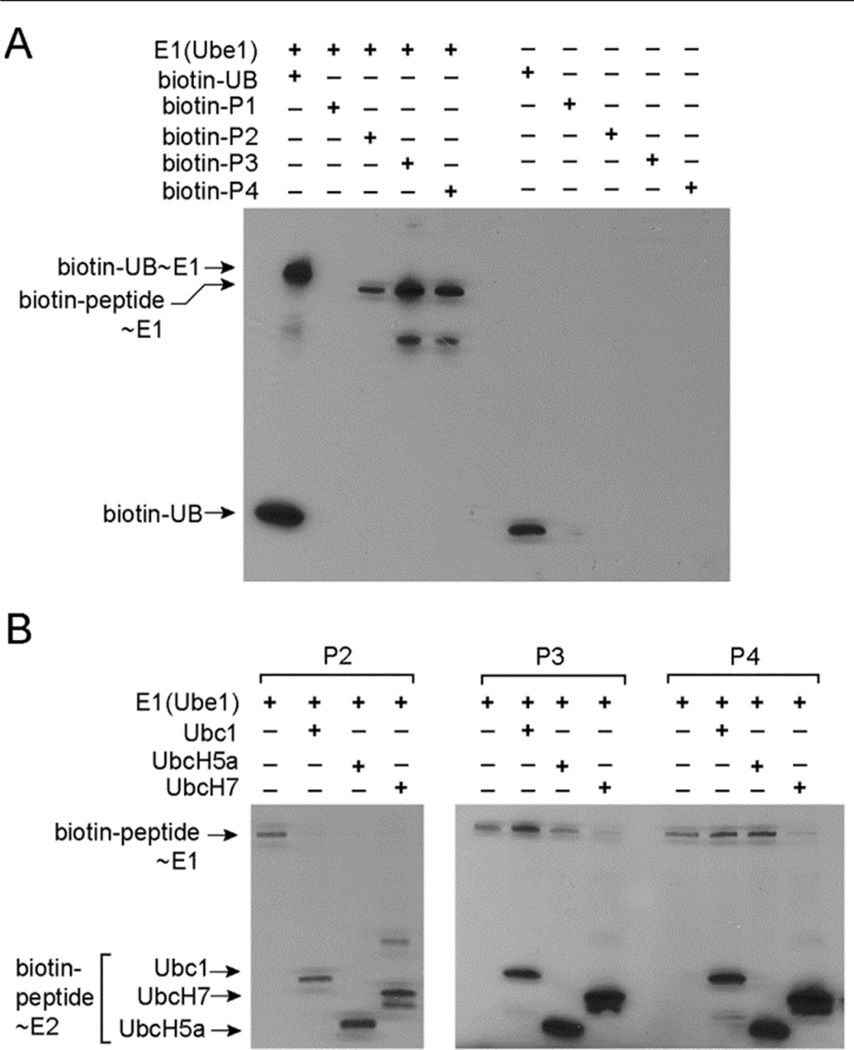
We next tested if the peptides P2–P4 can form thioester conjugates with E1 (Figure 1). We linked biotin to the N-terminal amino groups of the peptides and assayed the transfer of the peptides to Ube1 by Western blot probed with a streptavidin-horse radish peroxidase (HRP) conjugate. We found that biotin-labeled P2, P3 and P4 peptides are all transferred to E1 to form biotin-peptide~ E1 thioesters (Figure 1A). By contrast biotin labeled peptide P1 with the sequence of wtUB cannot be transferred to E1 at any detectable level. This result matches the ATP-PPi assay and suggests that P2–P4 are better substrates of the E1 enzyme than the P1 peptide with the wt sequence. We also found that P2–P4 can be transferred to E2 enzymes such as Ubc1, UbcH5a and UbcH7 to form peptide~E2 conjugates (Figure 1B).
Figure 1.
Reactivities of UB-mimicking peptides with E1 and E2 enzymes. (A) Transfer of biotin-labeled peptides to the E1 enzyme Ube1. Biotin-labeled UB was used as a control in the reactions. (B) Transfer of biotin-labeled peptides P2, P3 and P4 from E1 to the E2 enzymes Ubc1, UbcH5a and UbcH7.
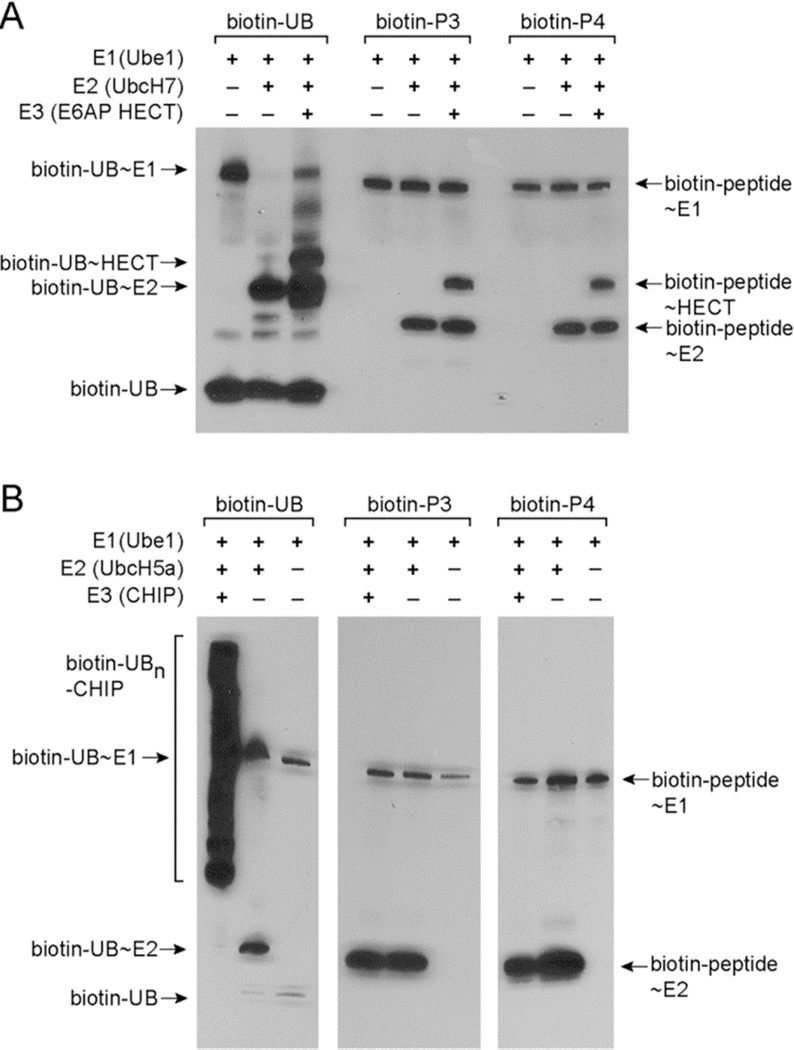
Since peptides P3 and P4 showed strong activities with the E2 enzymes, we assayed the transfer of these peptides from E2 to E3. E3s are divided into two main classes based on the mechanisms of UB transfer reactions they catalyze (Figure S1).1b The HECT type E3s have catalytic Cys residues to attack the thioester bond of the UB~E2 conjugates to form UB~HECT conjugates before passing the UB to the modified proteins. In contrast, the RING or U-box E3s bind both the UB~E2 conjugate and target proteins to facilitate the direct transfer of UB to the modified proteins. In the absence of substrate proteins, the RING and U-box E3s can be autoubiquitinated at Lys residues of the E3 enzymes. Western blots of the peptide transfer reactions showed that biotinylated P3 and P4 peptides can be transferred from UbcH7 to the HECT domain of E6AP to form peptide~HECT conjugates (Figure 2A).16 However, P3 and P4 cannot be transferred from E2 to CHIP, a U-box E3,17 for self-modification by the peptides (Figure 2B). It was previously found that the Cys thiol is more reactive with UB~E2 conjugates than the amino side chain of Lys since the thiol group is a better nucleophile.18 We thus rationalized that the HECT E3 is more reactive than the U-box E3 in the peptide transfer reaction because the catalytic Cys residue of HECT is more reactive than the Lys residues of the U-box E3 to attack the peptide~E2 conjugates.
Figure 2.
Transfer of UB-mimicking peptides P3 and P4 to the E3 enzymes. (A) Biotin-labeled P3 and P4 can be transferred to the HECT domain of E6AP. (B) Biotin-labeled P3 and P4 cannot be transferred to CHIP, a U-box E3 for self-modification.
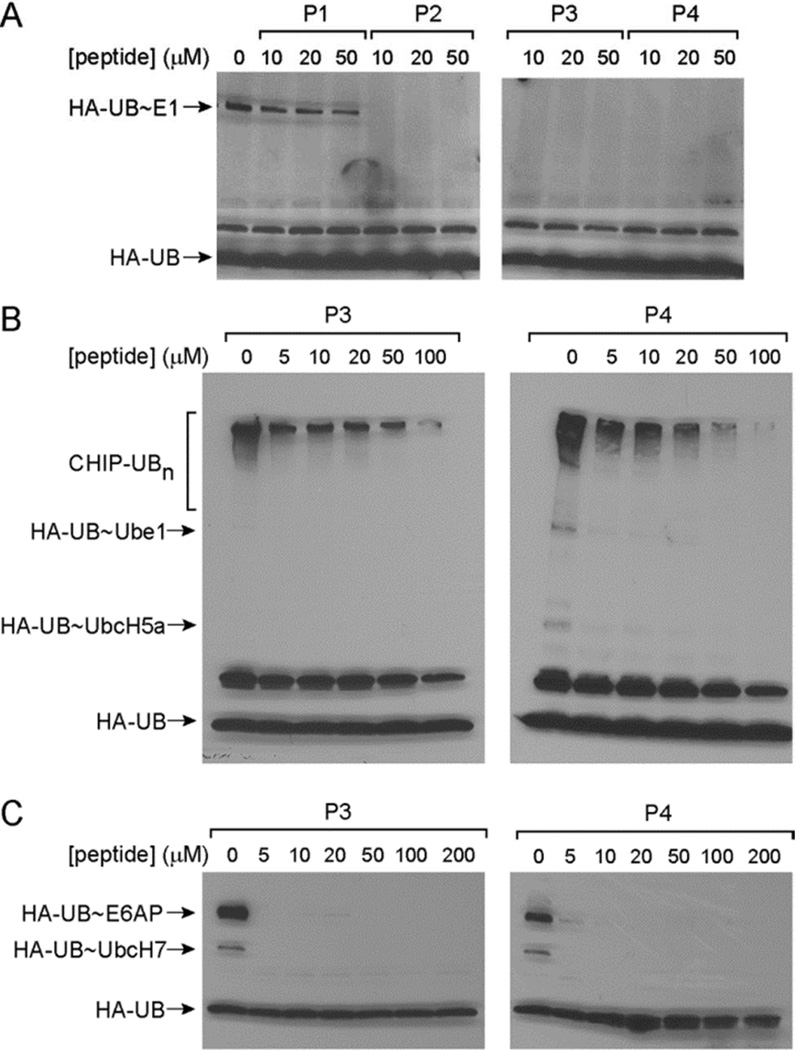
So far we have shown that peptides P2–P4 can be activated by E1 to form thioester conjugates with the E1, E2 and HECT E3s. We next analyzed whether peptide loading onto the various components of the E1-E2-E3 cascade would inhibit UB transfer through the cascade. In this way, the UB-mimicking peptides may serve as mechanism-based inhibitors that block protein ubiquitination catalyzed by enzymes of the ubiquitination cascade. To test this idea, we incubated the E1 enzyme Ube1 with peptides P1–P4 at concentrations of 10, 20 and 50 µM in the presence of ATP to allow peptide activation by E1 and the formation of peptide~E1 thioester conjugates. After a 10-minute preincubation, 5 µM UB with an N-terminal HA tag (HA-UB) was added to test if UB can still be loaded on the E1 enzyme. The reaction was allowed to proceed for another hour and was analyzed by Western blot. While P1 with the wtUB sequence cannot block the formation of UB~E1 thioester conjugates, peptides P2, P3 and P4 can completely block the formation of the UB~Ube1 conjugate at a concentration as low as 10 µM (Figure 3A). These results suggest that the formation of a peptide~E1 conjugate can inhibit UB loading onto the E1 enzyme.
Figure 3.
Inhibition of UB transfer through the E1-E2-E3 cascade by the UB-mimicking peptides. (A) Inhibition of the formation of UB~Ube1 conjugate by peptides P2, P3 and P4. (B) Inhibition of CHIP polyubiquitination by increasing concentrations of the P3 and P4 peptides. (C) Inhibition of UB transfer to the HECT domain of E6AP by increasing concentrations of P3 and P4 peptides. 5 µM HA-UB was added in each reaction after preincubation with the peptides.
We also assayed the competitive inhibition of UB by the peptides during UB transfer to Ube1. In this assay increasing concentrations of peptides were incubated with 1 µM UB for reaction with Ube1. The amount of UB~E1 conjugate formed in the reactions was measured by Western blot. As shown in Figure S4 in the Supporting Information, P2, P3 and P4 peptides can inhibit the formation of UB~E1 conjugate with IC50 values of 266 µM, 172 µM and 220 µM, respectively, while the P1 peptide cannot significantly inhibit UB loading on E1 at a concentration as high as 1 mM. Although the IC50 values of the P2–P4 peptides are still high for directly competing with UB in the reaction with E1, the UB-mimicking peptides can effectively block UB loading on E1 once they occupy the catalytic Cys residue of E1.
We further tested the activity of UB-mimicking peptides to inhibit UB transfer to E2 and E3 enzymes. We incubated Ube1, UbcH5a and CHIP with P3 and P4 at concentrations of 5 – 100 µM for 10 minutes to allow peptide transfer to the E1 and E2 enzymes. We then added UB to the reaction mixture and incubated the reaction for another hour. Western blot analysis of the reaction showed that P3 and P4 peptides significantly inhibited the transfer of UB to CHIP E3 at a concentration of 50 µM since the formation of polyubiquitinated CHIP was significantly decreased after preincubation of the peptides (Figure 3B). We further tested if P3 and P4 can inhibit UB transfer to the HECT domain of E6AP. After incubation of P3 and P4 with Ube1, UbcH7 and the E6AP HECT for 10 minutes, we added HA-UB to initiate UB transfer to the HECT domain. Western blot analysis of the transfer reactions showed that UB transfer to the HECT domain was blocked at a concentration as low as 5 µM of the P3 or P4 peptides (Figure 3C). The reason that the peptides showed higher activities to inhibit UB transfer to HECT E3 could be that the peptides could charge onto the catalytic Cys residue of the HECT domain and directly block UB loading on HECT (Figure 2A). However, the peptides could not be transferred to the Lys residues of the CHIP E3 to block UB conjugation (Figure 2B). Overall, our results suggest that the UB-mimicking peptides can block UB transfer through the E1-E2-E3 cascade and inhibit protein ubiquitination.
In this study we developed the UB-mimicking peptides P2–P4 that are several hundred folds more reactive towards the E1 enzyme than the P1 peptide with the C-terminal sequence of wtUB. The E1 enzyme activates the UB-mimicking peptides to form thioester conjugates with E1, E2 and HECT type E3s. The loading of the UB-mimicking peptides on enzymes of the ubiquitination cascade blocks the formation of UB thioester intermediates with these enzymes, and inhibits UB transfer through the cascade to the protein ubiquitination targets. The mechanism of the UB mimicking peptides in inhibiting the UB transfer cascade is quite unique in that they do not target a specific enzyme in the cascade, but instead block UB transfer at every step of the E1-E2-E3 cascade. In future studies, we plan to measure the inhibitory properties of the UB-mimicking peptides in cell-based assays. We expect these peptides should attenuate global protein ubiquitination in the cell rather than inhibit UB transfer through specific E2 or E3 enzymes. Furthermore since UBL proteins have their own cascade enzymes for the modification of cellular proteins, we can potentially use the same strategy to identify UBL-mimicking peptides that block UBL transfer through the cascades.
Supplementary Material
Acknowledgment
This work was supported by a lab startup grant from the University of Chicago, and a National Science Foundation CAREER award (1057092) to JY, and by the Deutsche Forschungsgemeinschaft (SCHI 425/6-1 and FZ 82) to HS. This work was also funded in part by the Chicago Biomedical Consortium with support from the Searle Funds at The Chicago Community Trust.
Footnotes
Supporting Information Available Supporting figures and detailed experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.(a) Hershko A, Ciechanover A. Annu Rev Biochem. 1998;67:425. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]; (b) Pickart CM. Annu Rev Biochem. 2001;70:503. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 2.(a) Haas AL, Rose IA. J. Biol. Chem. 1982;257:10329. [PubMed] [Google Scholar]; (b) Haas AL, Warms JV, Hershko A, Rose IA. J. Biol. Chem. 1982;257:2543. [PubMed] [Google Scholar]; (c) Schulman BA, Harper JW. Nat Rev Mol Cell Biol. 2009;10:319. doi: 10.1038/nrm2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.(a) Wenzel DM, Stoll KE, Klevit RE. Biochem J. 2011;433:31. doi: 10.1042/BJ20100985. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ye Y, Rape M. Nat Rev Mol Cell Biol. 2009;10:755. doi: 10.1038/nrm2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.(a) Deshaies RJ, Joazeiro CA. Annu Rev Biochem. 2009;78:399. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]; (b) Rotin D, Kumar S. Nat Rev Mol Cell Biol. 2009;10:398. doi: 10.1038/nrm2690. [DOI] [PubMed] [Google Scholar]
- 5.(a) Hori T, Osaka F, Chiba T, Miyamoto C, Okabayashi K, Shimbara N, Kato S, Tanaka K. Oncogene. 1999;18:6829. doi: 10.1038/sj.onc.1203093. [DOI] [PubMed] [Google Scholar]; (b) Johnson ES. Annu. Rev. Biochem. 2004;73:355. doi: 10.1146/annurev.biochem.73.011303.074118. [DOI] [PubMed] [Google Scholar]
- 6.(a) Hochstrasser M. Nature. 2009;458:422. doi: 10.1038/nature07958. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) van der Veen AG, Ploegh HL. Annu. Rev. Biochem. 2012;81:323. doi: 10.1146/annurev-biochem-093010-153308. [DOI] [PubMed] [Google Scholar]
- 7.(a) Bedford L, Lowe J, Dick LR, Mayer RJ, Brownell JE. Nat Rev Drug Discov. 2011;10:29. doi: 10.1038/nrd3321. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Hoeller D, Dikic I. Nature. 2009;458:438. doi: 10.1038/nature07960. [DOI] [PubMed] [Google Scholar]; (c) Nalepa G, Rolfe M, Harper JW. Nat Rev Drug Discov. 2006;5:596. doi: 10.1038/nrd2056. [DOI] [PubMed] [Google Scholar]
- 8.(a) Brownell JE, Sintchak MD, Gavin JM, Liao H, Bruzzese FJ, Bump NJ, Soucy TA, Milhollen MA, Yang X, Burkhardt AL, Ma J, Loke HK, Lingaraj T, Wu D, Hamman KB, Spelman JJ, Cullis CA, Langston SP, Vyskocil S, Sells TB, Mallender WD, Visiers I, Li P, Claiborne CF, Rolfe M, Bolen JB, Dick LR. Mol. Cell. 2010;37:102. doi: 10.1016/j.molcel.2009.12.024. [DOI] [PubMed] [Google Scholar]; (b) Soucy TA, Smith PG, Milhollen MA, Berger AJ, Gavin JM, Adhikari S, Brownell JE, Burke KE, Cardin DP, Critchley S, Cullis CA, Doucette A, Garnsey JJ, Gaulin JL, Gershman RE, Lublinsky AR, McDonald A, Mizutani H, Narayanan U, Olhava EJ, Peluso S, Rezaei M, Sintchak MD, Talreja T, Thomas MP, Traore T, Vyskocil S, Weatherhead GS, Yu J, Zhang J, Dick LR, Claiborne CF, Rolfe M, Bolen JB, Langston SP. Nature. 2009;458:732. doi: 10.1038/nature07884. [DOI] [PubMed] [Google Scholar]
- 9.(a) Milhollen MA, Traore T, Adams-Duffy J, Thomas MP, Berger AJ, Dang L, Dick LR, Garnsey JJ, Koenig E, Langston SP, Manfredi M, Narayanan U, Rolfe M, Staudt LM, Soucy TA, Yu J, Zhang J, Bolen JB, Smith PG. Blood. 2010;116:1515. doi: 10.1182/blood-2010-03-272567. [DOI] [PubMed] [Google Scholar]; (b) Soucy TA, Dick LR, Smith PG, Milhollen MA, Brownell JE. Genes & cancer. 2010;1:708. doi: 10.1177/1947601910382898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.(a) Kitagaki J, Yang Y, Saavedra JE, Colburn NH, Keefer LK, Perantoni AO. Oncogene. 2009;28:619. doi: 10.1038/onc.2008.401. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ungermannova D, Parker SJ, Nasveschuk CG, Wang W, Quade B, Zhang G, Kuchta RD, Phillips AJ, Liu X. PLoS One. 2012;7:e29208. doi: 10.1371/journal.pone.0029208. [DOI] [PMC free article] [PubMed] [Google Scholar]; (c) Xu GW, Ali M, Wood TE, Wong D, Maclean N, Wang X, Gronda M, Skrtic M, Li X, Hurren R, Mao X, Venkatesan M, Beheshti Zavareh R, Ketela T, Reed JC, Rose D, Moffat J, Batey RA, Dhe-Paganon S, Schimmer AD. Blood. 2010;115:2251. doi: 10.1182/blood-2009-07-231191. [DOI] [PMC free article] [PubMed] [Google Scholar]; (d) Yang Y, Kitagaki J, Dai RM, Tsai YC, Lorick KL, Ludwig RL, Pierre SA, Jensen JP, Davydov IV, Oberoi P, Li CC, Kenten JH, Beutler JA, Vousden KH, Weissman AM. Cancer Res. 2007;67:9472. doi: 10.1158/0008-5472.CAN-07-0568. [DOI] [PubMed] [Google Scholar]
- 11.(a) Aghajan M, Jonai N, Flick K, Fu F, Luo M, Cai X, Ouni I, Pierce N, Tang X, Lomenick B, Damoiseaux R, Hao R, Del Moral PM, Verma R, Li Y, Li C, Houk KN, Jung ME, Zheng N, Huang L, Deshaies RJ, Kaiser P, Huang J. Nat. Biotechnol. 2010;28:738. doi: 10.1038/nbt.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]; (b) Ceccarelli DF, Tang X, Pelletier B, Orlicky S, Xie W, Plantevin V, Neculai D, Chou YC, Ogunjimi A, Al-Hakim A, Varelas X, Koszela J, Wasney GA, Vedadi M, Dhe-Paganon S, Cox S, Xu S, Lopez-Girona A, Mercurio F, Wrana J, Durocher D, Meloche S, Webb DR, Tyers M, Sicheri F. Cell. 2011;145:1075. doi: 10.1016/j.cell.2011.05.039. [DOI] [PubMed] [Google Scholar]; (c) Orlicky S, Tang X, Neduva V, Elowe N, Brown ED, Sicheri F, Tyers M. Nat. Biotechnol. 2010;28:733. doi: 10.1038/nbt.1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhao B, Bhuripanyo K, Schneider J, Zhang K, Schindelin H, Boone D, Yin J. ACS Chemical Biology. 2012 doi: 10.1021/cb300339p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.(a) Jonnalagadda S, Ecker DJ, Sternberg EJ, Butt TR, Crooke ST. J Biol Chem. 1988;263:5016. [PubMed] [Google Scholar]; (b) Madden MM, Song W, Martell PG, Ren Y, Feng J, Lin Q. Biochemistry. 2008;47:3636. doi: 10.1021/bi702078m. [DOI] [PubMed] [Google Scholar]
- 14.Lee I, Schindelin H. Cell. 2008;134:268. doi: 10.1016/j.cell.2008.05.046. [DOI] [PubMed] [Google Scholar]
- 15.Krissinel E, Henrick K. J. Mol. Biol. 2007;372:774. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 16.Huang L, Kinnucan E, Wang G, Beaudenon S, Howley PM, Huibregtse JM, Pavletich NP. Science. 1999;286:1321. doi: 10.1126/science.286.5443.1321. [DOI] [PubMed] [Google Scholar]
- 17.Jiang J, Ballinger CA, Wu Y, Dai Q, Cyr DM, Hohfeld J, Patterson C. J. Biol. Chem. 2001;276:42938. doi: 10.1074/jbc.M101968200. [DOI] [PubMed] [Google Scholar]
- 18.Wenzel DM, Lissounov A, Brzovic PS, Klevit RE. Nature. 2011;474:105. doi: 10.1038/nature09966. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.