Summary
The evolution of sex in eukaryotes represents a paradox, given the “two-fold” fitness cost it incurs. We hypothesize that the mutational dynamics of the mitochondrial genome would have favoured the evolution of sexual reproduction. Mitochondrial DNA (mtDNA) exhibits a high mutation rate across most eukaryote taxa, and several lines of evidence suggest that this high rate is an ancestral character. This seems inexplicable given that mtDNA-encoded genes underlie the expression of life’s most salient functions, including energy conversion. We propose that negative metabolic effects linked to mitochondrial mutation accumulation would have invoked selection for sexual recombination between divergent host nuclear genomes in early eukaryote lineages. This would provide a mechanism by which recombinant host genotypes could be rapidly shuffled and screened for the presence of compensatory modifiers that offset mtDNA-induced harm. Under this hypothesis, recombination provides the genetic variation necessary for compensatory nuclear coadaptation to keep pace with mitochondrial mutation accumulation.
Introduction: The evolution of sex and the evolution of mitochondria are intrinsically linked
Widespread sexual reproduction among eukaryotes is puzzling from an evolutionary standpoint, because sex carries a “two-fold” fitness cost, and must therefore offer a benefit that surpasses this cost [1]. Several plausible theoretical models of the evolution of sex have been proposed, based on the benefits of recombination with sex, including facilitating the purging of deleterious mutations from the nuclear genome [2] or adaptation to ecological hazards such as parasites or environmental fluctuations [3]. However, while experimental and theoretical support exists for each of these classes of models, they arguably remain limited in their capacity to explain certain fundamental questions: why have all eukaryotes, but no prokaryotes, evolved life-history strategies hinged on sexual reproduction; and why has obligate sexual reproduction become so prevalent amongst certain eukaryotes (e.g., most metazoans), while other taxa (e.g., plants and certain metazoan lineages) exhibit more flexible reproductive systems based on facultative sex interspersed with clonal reproduction?
Eukaryotes are bound by two basal features. They all have – or have had during their evolutionary histories – energy-converting organelles called mitochondria [4]; and they all have – or have had – the ability to reproduce sexually [5]. We propose that the origins of this ancient association can be traced to the mutational properties of the mitochondrion’s own (mt)DNA. The mitochondria are integral to eukaryote evolution, and the ancient endosymbiosis that led to the mitochondrion provided the eukaryotes with a highly efficient form of energy conversion, presumably catalyzing the evolution of complex life [6]. However, mitochondria retain a small genome, which is destined to accumulate mutations via Muller’s ratchet as a consequence of the genome’s evolutionary constraints (e.g., uniparental inheritance and low effective population size [7]) and high mtDNA mutation rate across the eukaryote phylogeny [8,9] (with few exceptions, such as the derived slow mutation rate of many land plants [10]). Furthermore, across the eukaryote domain, de novo mutations in the mtDNA exhibit a higher fixation probability than those in the nuclear DNA [11]. This leads to a striking paradox – a genome that plays a critical role in maintaining the integrity of complex life is prone to perpetual mutational erosion across large branches of the eukaryote phylogeny.
Here, we outline a new hypothesis that can reconcile this paradox. We propose that mitochondrial genomic mutation accumulation (hereafter termed “mito-mutation accumulation”) is likely to have represented one of the original drivers of the evolution of sexual reproduction in eukaryotes. We contend that during the early evolution of eukaryotes, mito-mutation accumulation would have placed strong selection on the host genome for compensatory modifier mutations to offset the negative metabolic effects linked to deleterious mtDNA mutations. Recombination between distinct host ‘nuclear’ genotypes, achievable via sexual reproduction, would facilitate such a compensatory response, by accelerating the rate by which alleles at host nuclear loci could be shuffled and screened for compensatory function.
Below, we outline our case, which is built on a strong base of experimental studies that have elucidated how compensatory mitochondrial-nuclear (mito-nuclear) coevolution is key to maintaining organismal viability. We highlight evidence for nuclear compensatory adaptation to pathogenic mtDNA mutations, and discuss how mito-nuclear coevolution is greatly facilitated by recombination in the nuclear genome. We then identify the key assumptions on which our hypothesis rests, and present supporting evidence for each assumption. We discuss the hypothesis in the context of other evolutionary theories that have previously drawn links between mutations, mitochondria, and sex in eukaryotes. Finally, we conclude by noting that the power of our hypothesis lies in its testability: it provides a series of predictions amenable to experimental enquiry, the answers to which could provide compelling support for the contention that mito-mutation accumulation was a key driver of the evolution of sex in eukaryotes.
Experimental support for the role of compensatory nuclear adaptation to mito-mutation accumulation
Preserving the fine-scale interactions between proteins encoded by both mitochondrial and nuclear genomes is critical for maintaining oxidative phosphorylation (OXPHOS), and meeting the energy needs of the contemporary eukaryotic cell. Paradoxically, the mtDNA is prone to perpetually accumulate deleterious mutations [12], which could have dire consequences for organismal function in the absence of a compensatory nuclear response [7,13-17]. Indeed, it is now well known that mutations in the mtDNA sequence are often tied to the onset of metabolic diseases, early ageing, and infertility [18,19]. Moreover, an increased penetrance of such ailments, and a general reduction in fitness, has been observed upon artificially disrupting coevolved combinations of mito-nuclear genotypes, by expressing mtDNA haplotypes alongside evolutionary novel nuclear backgrounds [20,21]. In model systems such as Mus and Drosophila, such mito-nuclear “mismatches” have resulted in reduced metabolic functioning and lower fitness [17,22-24]. These results strongly suggest that, within any given population, mitochondrial and nuclear gene combinations are co-evolutionarily “matched” to one another. It follows that mito-nuclear coevolution may contribute to driving reproductive isolation between incipient populations, and that this might ultimately lead to speciation [14,25].
Other support for the role of mito-nuclear coevolution in maintaining organismal viability comes from studies reporting elevated substitution rates in nuclear-encoded genes that interact with mtDNA-encoded products, relative to their nuclear counterparts that interact with other nuclear-encoded genes [26-28]. These findings are important because they indicate a key role for positive selection in shaping the evolutionary rate of nuclear-encoded genes involved in mitochondrial function in order to compensate for mtDNA mutations. Given that the products of these nuclear genes are entwined in tightly regulated mito-nuclear interactions, this suggests that these nuclear genes are responding quickly and efficiently to mito-mutation accumulation, via counter-adaptations of compensatory function to ensure the integrity of metabolic function [26,29].
The empirical evidence outlined above suggests that nuclear compensatory adaptations have commonly evolved to mitigate the effects of mito-mutation accumulation and prevent mtDNA-mediated Muller’s Rachet effects from leading to widespread lineage extinctions across the eukaryote phylogeny. Recombination in the host genome would have greatly facilitated this compensatory response, by shuffling and generating unique nuclear allelic combinations every generation, providing the genetic variation required for selection to screen for nuclear adaptations that offset mitochondrial mutational erosion (Fig. 1). In early eukaryotes, while horizontal gene transfer from other host nuclear lineages might have partially alleviated the effects of mito-mutation accumulation, we contend that a more predictable and rapid mechanism of gene mixing would have been required to cope with the perpetual mutational pressure imposed by the mitochondrial genome during this time (Fig. 1). Recombination via sex could provide such a mechanism, enabling compensatory nuclear alleles to spread quickly into new genetic backgrounds [30]. Recombination also ensures that the modifiers are not placed on nuclear genomic backgrounds that are largely deleterious in performance and hence purged under background selection.
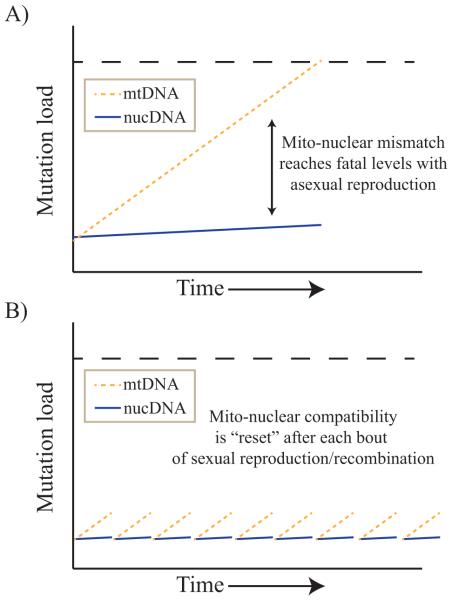
Figure 1.
Mutational meltdown in the mitochondrial DNA (mtDNA) is offset by compensatory changes in the nuclear DNA, facilitated by recombination associated with sexual reproduction, providing a mechanism for the origin and maintenance of sexual reproduction. “Mutation load” is displayed on the y-axis and denotes total number of deleterious mutations. Even though the absolute number of mutations in the mitochondrial genome remains the same in panel (a) and (b), in (b) the deleterious effects of mito-mutations are offset by nuclear compensatory adaptations in the recombined nuclear genome (hence, the mutation load is lower under sexual reproduction). A: During asexual reproduction, mito-nuclear mismatch increases over generations as mtDNA accumulates mutations faster than the nuclear genome can evolve counter-adaptations. B: With sexual reproduction, organismal viability is restored in the population by selection for compensatory genotypes in the nucDNA, facilitated by recombination. Orange/dotted lines represent mtDNA, blue/solid lines represent nucDNA, and dashed lines represent a fatal upper threshold for mito-nuclear mismatch.
Key assumptions
Our hypothesis centres on two assumptions, each of which is plausible and which has been made previously by evolutionary biologists. We outline the substantiating evidence for each below. Furthermore, we note that while there are exceptions to each of these assumptions, it is these very exceptions that provide excellent opportunities on which our hypothesis can be tested (see Predictions).
Assumption 1: The high mutation rate of the mtDNA is an ancestral condition
We assume that the high mutation rate of mtDNA, which is a hallmark of the streamlined mitochondrial genomes of metazoans [31], is a character that was shared by ancestral mitochondria during early eukaryote evolution and the progression of endosymbiosis [32]. Six lines of evidence support this assumption. First, recent studies have identified unicellular and early-diverging eukaryotic lineages (e.g., haptophyte and stramenopile algae) that similarly show elevated mtDNA mutation rates, suggesting that across eukaryotes as a whole there is a propensity for mtDNA evolutionary rates to outstrip those in the nucleus [8,9,33]. Fungi and yeast species also conform to these high mtDNA mutation rates relative to nuclear rates [34-36], indicating that elevated mtDNA mutation is not restricted to animals. Together, these data suggest that a high mutation rate is likely to have been shared by the eukaryotic ancestor, and that the low mtDNA mutation rate of many land plants is a derived condition. Indeed, studies have emerged that demonstrate low mtDNA mutation rates are not systematic of all land plants, and many such species show incredible variation in the mtDNA mutation rate, including some of the highest ever documented rates recorded in eukaryotes [37,38].
Second, other recent cellular endosymbionts likewise exhibit elevated mutation rates compared with the genomes of their hosts, suggesting that during endosymbiosis the ancestral mitochondria would have also experienced increased mutation rates [39,40]. Third, rates of substitution appear to be faster in bacteria -- which gave rise to the mitochondrial genome -- than in archaea, which gave rise to a majority of the nuclear genome [41]. Fourth, the well-documented presence of a “long-branch” leading from the bacterial ancestor to the mtDNA of modern eukaryotes (including plants) suggests a rapid increase in mutation rate in the mtDNA of early eukaryotes following endosymbiosis [42], further supporting the conclusion that the slow-evolving mtDNA of plants is a derived characteristic. Fifth, we note that the mtDNA resides within the mitochondria – a highly mutagenic site that is the major source of reactive oxygen species in the cell due to its redox activity. Reactive oxygen species production also increases during ageing and is correlated with compromised mitochondrial function [43,44]. Finally, given that mtDNA replicates more often than the nuclear DNA per cell cycle [45], and in particular undergoes a massive spike in replication during embryogenesis [46], replication errors should be more prevalent in the mtDNA, leading to an increased mutation rate [19] across the eukaryote phylogeny.
Assumption 2: Fundamental evolutionary constraints limit the mtDNA itself from evolving biparental transmission
Across eukaryotes, uniparental inheritance of the mitochondria remains a ubiquitous pattern, with only a few exceptions [47]. Our hypothesis assumes that the mtDNA was under strong evolutionary constraints to avoid biparental transmission, during the transition of its host to sexual reproduction with two sexes. We acknowledge that there is considerable flexibility in mtDNA recombination rates across eukaryotes, particularly in land plants and in fungi. However, intra-individual heteroplasmy of divergent mtDNA molecules in plants is rarely reported to be caused by paternal leakage (as a consequence of sexual reproduction), and uniparental transmission of mtDNA remains the typical pattern in these lineages [48] (although, we acknowledge that comprehensive studies of heteroplasmy in plant mtDNA are lacking; see [49] for a review). This general trend is similarly the case for fungi [50], which show heteroplasmy in early life stages, but revert to homoplasmy as they develop [48]. Even in these taxa in which recombination of mtDNA has been recorded [48], they maintain uniparental inheritance of the mtDNA, which would presumably reduce the scope by which recombination could act to purge poorly performing mtDNA molecules. Genetically effective recombination between mtDNA molecules requires biparental transmission. In the absence of biparental transmission, any two mtDNA molecules would share near-identical sequences, and so too would the products of their recombination [45].
The assumption that fundamental evolutionary constraints prevented the evolution of biparental mitochondria transmission has received robust support from population genetic theory [51-54], as well as empirical work [55,56]. Population genetic models have demonstrated that biparental transmission would lead to divergent mtDNA molecules competing within the same host, and promote selection for selfish molecules that replicate faster at the expense of host metabolic efficiency [51-54].Other models have shown that negative selection against mtDNA heteroplasmy alone would suffice to lead to the evolution of mitochondrial uniparental inheritance [57]. Yet, other recent models have challenged the inherent simplicity of the evolution of uniparental inheritance, suggesting that the fitness benefits of uniparental inheritance might well be frequency dependent, which might act to limit the spread and fixation of a mutation conferring uniparental inheritance in a population [59]. At any rate, empirical work has substantiated the prediction that uniparental inheritance should be upheld to mitigate heteroplasmy-induced conflict. Studies in yeast and mice have shown that heteroplasmy leads to competition between divergent mtDNA genomes and a breakdown in fundamental metabolic functions such as respiration, nutrient intake, and even cognitive function [55,56]. Additionally, maintaining uniparental inheritance presumably enables the mtDNA to be sequestered within a metabolically quiescent germ-line, protecting the resulting offspring from inheriting a damaged mtDNA genome associated with a metabolically active cell (e.g., the sperm cell) [47,58,59].
Finally, while prokaryotes rely on horizontal gene exchange as a facilitator of adaptation, we assume this level of gene-mixing would not have sufficed for the evolving eukaryote. Transitioning to life as a highly complex cell, and harnessing the efficient energy conversion provided by mitochondria, is predicted to have led to a rapid expansion in nuclear genome size [6]. As the nuclear genome expanded, however, horizontal gene transfer would have affected a smaller and smaller fraction of the nuclear genome, diminishing the capacity of this process to reliably provide the raw genetic variation needed to screen for compensatory nuclear modifiers. Indeed, only one asexual eukaryotic lineage, the bdelloid rotifers, is believed to experience a large amount of horizontal gene transfer as a possible substitute for sex [60]. Yet rotifers also have high rates of gene conversion and expanded numbers of genes for oxidative resistance [61], suggesting that the lack of recombination in eukaryotes is exceedingly rare because multiple mechanisms, and a very specific genetic architecture, are required to supplement horizontal gene transfer.
Previous links between mutations, mitochondria, and the evolution of sex
Theories linking mutation accumulation with sex have received considerable attention since their inception. Asexual populations lack recombination, constraining the capacity by which selection can eliminate deleterious mutations from the DNA sequence without sending an entire genetic lineage to extinction. This should lead to the cumulative increase in number of mutations per genome within the population. Mutation accumulation should be particularly prevalent in small asexual populations, since the efficacy of selection in removing poorly performing lineages will be reduced, leading to mean decreases in population fitness, thus further reductions in population size, thus augmenting accumulation of further mutations. These interactions between population size and the rate of mutation accumulation are predicted to lead to mutational meltdown [62-64]. Theoretical models that underpin the Deterministic Mutation Hypothesis further show that the evolution of sex can be favoured even in the absence of small population sizes, under conditions in which the majority of mutations are slightly deleterious, their effects positively epistatic, and the mutation rate high [64-67]. Other models indicate that population demographics and the specific properties of mutations will affect the dynamics of mutation accumulation within asexual populations, and thus alter the conditions under which the evolution of sex will be favoured [68-71]. Empirical studies have generally substantiated these mutation-based theories of sex (although see [72] as an example of an exception). For example, in a recent comparative transcriptomic analysis across 29 species of the plant genus Oenothora, Hollister et al. [73] showed that functional asexual species accumulate a greater number of deleterious mutations than sexual species. Furthermore, in the fungus Aspergillus nidulans, mutation accumulation is slower in lineages propagated through sexual spores relative to lineages that were propagated through asexual spores [74].
A link between mitochondria and sex [6,53,54,75-77] has been suggested in previous studies, but these have typically assumed that gamete fusion (i.e, sex) was already in place at the time of acquisition of the mitochondrion. Here, the evolution of two different sexes – one that transmits the mitochondria (the female), and one that does not (the male) – mitigates the selfish conflict between divergent mtDNA molecules of different parents, by maintaining uniparental mtDNA transmission [53,54,76,77] (see Assumption 2 above). None of these studies envisioned that acquisition of the mitochondrion, in early eukaryote evolution, was a driving force behind the evolution of sex per se. Only one other viewpoint has previously proposed a link between recombination and mitochondria, albeit with a different process mediating the evolution of sex [75]. Lane [75] suggested the early mitochondrion in the evolving eukaryote would provide an unending source of foreign DNA that would have contaminated the nuclear genome, and this contamination of foreign DNA would have imposed selection for recombination between host genotypes to preserve nuclear chromosomes with beneficial mutations, while purging deleterious ones. This view is supported by evidence that introns in the nuclear genome are of mitochondrial origin [78]. Lane and Martin [6,79] also hypothesized that the expansion of nuclear genome size, facilitated by the mitochondrion, would have favoured a shift from unreliable lateral gene transfer to the more robust sexual recombination [6].
Our hypothesis differs from those of predecessors by proposing that the selection for sex and nuclear recombination in eukaryotes came from mutational meltdown within the mitochondrial genome itself. By recognizing that mitochondrial mutations are particularly consequential because they are more often fixed than nuclear mutations [11], and because they affect one of eukaryote life’s core functions – energy conversion, we extend the Fisher-Muller model of recombination to include the fixation of beneficial modifiers that together act to offset the effects of mito-mutation accumulation [80]. Moreover, we note that parasite-mediated theories of sexual reproduction [3] have some parallels (but also differences, e.g. in transmission mode, effective population size) to our hypothesis, given that the ancestral mitochondria can reasonably be envisaged as endosymbiotic parasites. As with any complex inter-species interaction [81], early eukaryotic-mitochondrial symbiosis may have been marked by pervasive inter-genomic conflict, signatures of which are still manifest in contemporary eukaryote lineages (e.g. cytoplasmic male sterility in plants [82]). Thus, in addition to the rapid accumulation of mito-mutations, early eukaryotic-mitochondrial conflict might conceivably have selected for mutations that benefited the mitochondria, at the expense of host fitness, further promoting the evolution of recombination in the host nuclear genome to enable an efficient response to the evolving mtDNA. As such, we believe recent theoretical advances on the evolution of sex would benefit from extension to incorporate the effects of mito-mutation accumulation, and assess whether the current hypothesis stands on rigorous footing.
Although formation of chimeric OXPHOS complexes between mitochondrial and nuclear proteins would have greatly spurred the need for exquisite mito-nuclear compatibility [7,17,83], we believe that the evolutionary transition to sexual reproduction in order to maintain mito-nuclear compatibility is very likely to have pre-dated the origin of OXHPOS complexes. Parts of the metabolic machinery of eukaryotes would have required intimate inter-genomic coordination to achieve mutual endosymbiosis long before OXPHOS-encoding genes translocated over to the nuclear genome. For example, the nuclear-encoded genes involved in importing raw materials from outside of the host and into the mitochondria would presumably have needed to co-evolve with mtDNA-encoded genes involved in converting and utilizing such materials. Indeed, we contend that it was the transition to host nuclear recombination and sex that would have provided the large selective advantage for translocated mtDNA genes to move over en masse and remain in the nucleus (Fig. 2), resulting in the greatly streamlined mito-genomes of eukaryotes compared with their bacterial ancestors.
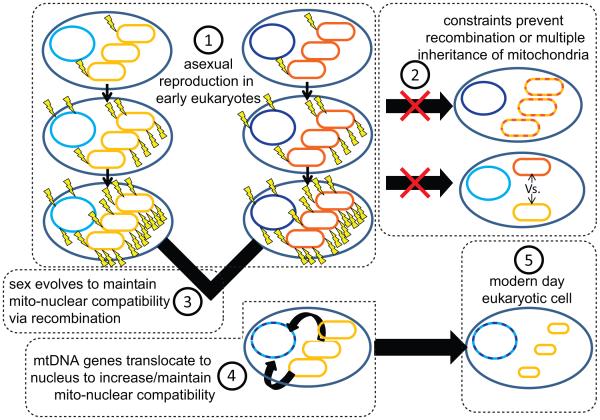
Figure 2.
A model for early eukaryotic evolution. Under our hypothesis, the evolution of recombination via sexual reproduction stems from the need to shuffle nuclear genes, as an inevitable consequence of having mitochondria. We envisage that this was driven by high mtDNA mutation rates in the early endosymbiotic bacterium that evolved into the mitochondrion. Following acquisition of the bacterial endosymbiont, the early eukaryote would have transmitted its endosymbiont vertically (number 1), without recombination, leading to mutational erosion in the mtDNA. Numbers indicate the general temporal order of other events, including the evolution of sex, based on our hypothesis. Lightning bolts denote DNA mutations, orange/blue denotes mtDNA and nuclear DNA, respectively, and different shading represents DNA from different lineages. Circles denote host nucleus and associated genome, and the eclipses denote mitochondria and their mtDNA.
Finally, empirical support for a role of mitochondrial mutation accumulation in the evolution of sex comes from a series of intriguing studies by Nedelcu and colleagues [84-86]. They showed that exposure to antioxidants inhibited sexual induction in the facultatively sexual green alga, Volvox carteri [86]; and that a two-fold increase in reactive oxygen species (ROS), induced by heat stress, resulted in an activation of the sex genes, hence invoking sexual reproduction [85]. The authors contended that the core need to cope with the deleterious effects of oxidative stress would have underpinned the evolution of sexual reproduction. We note that their hypothesis is entirely consistent with the one presented here based on mitochondrial mutation accumulation. They provide a mechanistic process that would underpin our genetic hypothesis. Mitochondria are the major source of ROS [44,87], and mutation accumulation within the mtDNA should in theory increase ROS production, invoking selection for the evolution of sex.
Testable predictions and future experiments
There are several testable predictions that stem from our hypothesis. First, we predict that eukaryote taxa with lower mtDNA mutation rates should exhibit lower propensities for sexual reproduction. Although mtDNA mutates quickly in most animal lineages, some (e.g., Cnidaria) have slowly-evolving mtDNA [88], and angiosperms in general also have slowly-evolving mtDNA [9]. Under our hypothesis, the magnitude of mtDNA-mediated selection for compensatory nuclear modifiers should be lower in these lineages. Cursory evidence suggests obligate sexual reproduction and outcrossing are relatively rare in both lineages [89,90], supporting the prediction that lineages with low mtDNA mutation rates should not have to rely as heavily on sexual reproduction to offset mtDNA-induced harm. Similarly, the protist Giardia, which has lost its mitochondria, contains meiosis-specific genes and thus the core genetic machinery required for sexual recombination [5], but it has never been directly observed to engage in sexual reproduction, suggesting sexual reproduction may be rare in eukaryotes that lack mtDNA, as we would predict. This overall correlation between propensity for sex and the tendency to accumulate mtDNA mutations should extend across diverse eukaryotic lineages, although there are obviously known exceptions to this general trend, including elevated mtDNA mutation rates in Silene plants which are known to undergo frequent selfing [38]. It is key to note that, based on our hypothesis, we assume that it is the mtDNA mutation rate that shapes the rate of sexual reproduction. We, however, acknowledge an alternative interpretation that taxa constrained to asexual reproduction may have evolved mechanisms that lower the mtDNA mutation rate. We believe some of the experiments we propose below – i.e. testing our third prediction below, by evaluating the effects of mito-nuclear mismatch on the propensity for sex - could help to disentangle these alternatives.
A second prediction can be derived based on the rate of paternal leakage of mtDNA, and associated recombination, across eukaryotes. Although mtDNA is overwhelmingly transmitted through the maternal lineage in eukaryotes, rare cases have been documented of paternal mtDNA also being transmitted, suggesting recombination between mtDNA genomes to prevent mito-mutation accumulation might be possible over evolutionary timescales in some lineages [47]. Those lineages in which leakage of mtDNA from the paternal parent is prevalent should be able to resort to this mechanism in part to offset their mitochondrial mutation loads, as suggested previously [47,91]. Based on our hypothesis, it follows that species with appreciable levels of paternal mtDNA leakage should also be less likely to exhibit modes of obligate sexual reproduction.
Third, experimental disruption of coevolved mito-nuclear genotypes in facultatively sexual species should induce elevated rates of sexual reproduction in these species. Experimental mismatching of coevolved mito-nuclear genotypes, achieved by pairing the prevailing mtDNA haplotype of a population or species alongside an evolutionary novel nuclear genome sourced from a separate population/species, has been shown to decrease organismal fitness and modify patterns of gene expression [17,20-24,92,93], but has never previously been harnessed to study the propensity for sexual reproduction. Here, the assumption is that the creation of mito-nuclear mismatches will promote individuals to resort to sex to harness the benefits of recombination between divergent host nuclear genotypes, to optimize selection for counter-adaptations that restore mito-nuclear compatibility.
Finally, under the assumption that the nuclear-encoded genes that directly interact with those encoded by the mtDNA are more likely to host compensatory mutations, these nuclear-encoded genes are predicted to exhibit elevated recombination rates relative to other nuclear genes that are not involved in mito-nuclear interactions. As we noted above, there is already evidence that nuclear gene products that interact with mtDNA-encoded products evolve more quickly based on studies of animals and plants, supporting this prediction [26-28]. Recent technological developments enabling deep-sequencing to identify recombination “hot spots” across the nuclear genome [94] could be used to directly gauge recombination levels in mitochondrial interacting genes relative to those that do not interact with the mitochondria.
Conclusions
We have presented a new hypothesis for the evolution and maintenance of sexual reproduction. This hypothesis draws on a robust foundation of empirical evidence that has substantiated the evolutionary consequences of mito-mutation accumulation, and the role of mito-nuclear interactions in population coevolutionary processes. Backed by this evidence, we note that the mitochondrial genome – a universal feature of all organisms equipped for sexual reproduction – is destined to accumulate mutations. We posit that this would have invoked selection in the ancestral eukaryote for a mechanism that would facilitate the rapid screening of new combinations of nuclear genotype, for those exhibiting compensatory function to offset this mitochondrially-induced harm. We propose that this selection pressure was a catalyst for the evolution of sexual reproduction, since sex, when combined with recombination, would provide the abundant novel genetic variation necessary for selection to evaluate genotypes of compensatory effect. The power of our hypothesis lies in its testability. The predictions that we have outlined are readily amenable to scientific testing. This can be achieved through experimental studies in modern eukaryote lineages that directly evaluate whether mito-nuclear genomic mismatches can be alleviated and restored via sexual recombination, as well as by correlative studies examining mutational characteristics of mitochondrial and nuclear genomes. Our hypothesis, which links mtDNA mutation rates and mito-nuclear conflict to the propensity for sex, offers an empirically tractable solution to an enduring problem in biology.
Acknowledgements
We thank Daniel Sloan, Rachel Muller and their lab groups at Colorado State University and the Mitonuclear Ecology discussion class at Auburn University for stimulating discussion of this work. DKD/MDH were funded by the Australian Research Council.
Footnotes
The authors declare no conflicts of interest.
References
- 1.Maynard Smith J. The Evolution of Sex. Cambridge University Press; 1978. [Google Scholar]
- 2.Muller HJ. Some genetic aspects of sex. Am Nat. 1932;66:118–38. [Google Scholar]
- 3.Hamilton WD, Axelrod R, Tanese R. Sexual reproduction as an adaptation to resist parasites (a review) Proc Natl Acad Sci U S A. 1990;87:3566–73. doi: 10.1073/pnas.87.9.3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gray MW. Mitochondrial evolution. Cold Spring Harb Perspect Biol. 2012;4:a011403. doi: 10.1101/cshperspect.a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper MA, Adam RD, Worobey M, Sterling CR. Population genetics provides evidence for recombination in Giardia. Curr Biol. 2007;17:1984–8. doi: 10.1016/j.cub.2007.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Lane N. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb Perspect Biol. 2014:6. doi: 10.1101/cshperspect.a015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rand DM, Haney RA, Fry AJ. Cytonuclear coevolution: the genomics of cooperation. Trends Ecol Evol. 2004;19:645–53. doi: 10.1016/j.tree.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 8.Smith DR, Hua JM, Lee RW, Keeling PJ. Relative rates of evolution among the three genetic compartments of the red alga Porphyra differ from those of green plants and do not correlate with genome architecture. Mol Phylogenet Evol. 2012;65:339–44. doi: 10.1016/j.ympev.2012.06.017. [DOI] [PubMed] [Google Scholar]
- 9.Smith DR, Keeling PJ. Mitochondrial and plastid genome architecture: Reoccurring themes, but significant differences at the extremes. Proc Natl Acad Sci U S A. 2015 doi: 10.1073/pnas.1422049112. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolfe KH, Li WH, Sharp PM. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc Natl Acad Sci U S A. 1987;84:9054–8. doi: 10.1073/pnas.84.24.9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lynch M, Blanchard JL. Deleterious mutation accumulation in organelle genomes. Genetica. 1998;102-103:29–39. [PubMed] [Google Scholar]
- 12.Lynch M. Mutation accumulation in nuclear, organelle, and prokaryotic transfer RNA genes. Mol Biol Evol. 1997;14:914–25. doi: 10.1093/oxfordjournals.molbev.a025834. [DOI] [PubMed] [Google Scholar]
- 13.Frank SA, Hurst LD. Mitochondria and male disease. Nature. 1996;383:224. doi: 10.1038/383224a0. [DOI] [PubMed] [Google Scholar]
- 14.Burton RS, Barreto FS. A disproportionate role for mtDNA in Dobzhansky-Muller incompatibilities? Mol Ecol. 2012;21:4942–57. doi: 10.1111/mec.12006. [DOI] [PubMed] [Google Scholar]
- 15.Burton RS, Ellison CK, Harrison JS. The sorry state of F2 hybrids: consequences of rapid mitochondrial DNA evolution in allopatric populations. Am Nat. 2006;168(Suppl 6):S14–24. doi: 10.1086/509046. [DOI] [PubMed] [Google Scholar]
- 16.Burton RS, Pereira RJ, Barreto FS. Cytonuclear genomic interactions and hybrid breakdown. Ann Rev Ecol Evol Syst. 2013;44:281–302. [Google Scholar]
- 17.Wolff JN, Ladoukakis ED, Enriquez JA, Dowling DK. Mitonuclear interactions: evolutionary consequences over multiple biological scales. Philos Trans R Soc Lond B Biol Sci. 2014;369:20130443. doi: 10.1098/rstb.2013.0443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wallace DC. Diseases of the mitochondrial DNA. Annu Rev Biochem. 1992;61:1175–212. doi: 10.1146/annurev.bi.61.070192.005523. [DOI] [PubMed] [Google Scholar]
- 19.Dowling DK. Evolutionary perspectives on the links between mitochondrial genotype and disease phenotype. Biochim Biophys Acta. 2014;1840:1393–403. doi: 10.1016/j.bbagen.2013.11.013. [DOI] [PubMed] [Google Scholar]
- 20.Reinhardt K, Dowling DK, Morrow EH. Medicine. Mitochondrial replacement, evolution, and the clinic. Science. 2013;341:1345–6. doi: 10.1126/science.1237146. [DOI] [PubMed] [Google Scholar]
- 21.Morrow EH, Reinhardt K, Wolff JN, Dowling DK. Risks inherent to mitochondrial replacement. EMBO Rep. 2015;16:541–544. doi: 10.15252/embr.201439110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nagao Y, Totsuka Y, Atomi Y, Kaneda H, et al. Decreased physical performance of congenic mice with mismatch between the nuclear and the mitochondrial genome. Genes Genet Syst. 1998;73:21–7. doi: 10.1266/ggs.73.21. [DOI] [PubMed] [Google Scholar]
- 23.Yee WKW, Sutton KL, Dowling DK. In vivo male fertility is affected by naturally occurring mitochondrial haplotypes. Curr Biol. 2013;23:R55–R6. doi: 10.1016/j.cub.2012.12.002. [DOI] [PubMed] [Google Scholar]
- 24.Sackton TB, Haney RA, Rand DM. Cytonuclear coadaptation in Drosophila: Disruption of cytochrome c oxidase activity in backcross genotypes. Evolution. 2003;57:2315–25. doi: 10.1111/j.0014-3820.2003.tb00243.x. [DOI] [PubMed] [Google Scholar]
- 25.Gershoni M, Templeton AR, Mishmar D. Mitochondrial bioenergetics as a major motive force of speciation. Bioessays. 2009;31:642–50. doi: 10.1002/bies.200800139. [DOI] [PubMed] [Google Scholar]
- 26.Osada N, Akashi H. Mitochondrial-nuclear interactions and accelerated compensatory evolution: evidence from the primate cytochrome C oxidase complex. Mol Biol Evol. 2012;29:337–46. doi: 10.1093/molbev/msr211. [DOI] [PubMed] [Google Scholar]
- 27.Sloan DB, Triant DA, Wu M, Taylor DR. Cytonuclear interactions and relaxed selection accelerate sequence evolution in organelle ribosomes. Mol Biol Evol. 2014;31:673–82. doi: 10.1093/molbev/mst259. [DOI] [PubMed] [Google Scholar]
- 28.Barreto FS, Burton RS. Evidence for compensatory evolution of ribosomal proteins in response to rapid divergence of mitochondrial rRNA. Mol Biol Evol. 2013;30:310–4. doi: 10.1093/molbev/mss228. [DOI] [PubMed] [Google Scholar]
- 29.Gershoni M, Levin L, Ovadia O, Toiw Y, et al. Disrupting mitochondrial-nuclear coevolution affects OXPHOS complex I integrity and impacts human health. Genome Biol Evol. 2014;6:2665–80. doi: 10.1093/gbe/evu208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill WG, Robertson A. The effect of linkage on limits to artificial selection. Genet Res. 1966;89:311–36. doi: 10.1017/S001667230800949X. [DOI] [PubMed] [Google Scholar]
- 31.Lynch M, Koskella B, Schaack S. Mutation pressure and the evolution of organelle genomic architecture. Science. 2006;311:1727–30. doi: 10.1126/science.1118884. [DOI] [PubMed] [Google Scholar]
- 32.van der Sluis EO, Bauerschmitt H, Becker T, Mielke T, et al. Parallel structural evolution of mitochondrial ribosomes and OXPHOS complexes. Genome Biol Evol. 2015;7:1235–51. doi: 10.1093/gbe/evv061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith DR, Jackson CJ, Reyes-Prieto A. Nucleotide substitution analyses of the glaucophyte Cyanophora suggest an ancestrally lower mutation rate in plastid vs mitochondrial DNA for the Archaeplastida. Mol Phylogenet Evol. 2014;79:380–4. doi: 10.1016/j.ympev.2014.07.001. [DOI] [PubMed] [Google Scholar]
- 34.Jung PP, Friedrich A, Reisser C, Hou J, et al. Mitochondrial genome evolution in a single protoploid yeast species. G3 (Bethesda) 2012;2:1103–11. doi: 10.1534/g3.112.003152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bruns TD, Szaro TM. Rate and mode differences between nuclear and mitochondrial small-subunit rRNA genes in mushrooms. Mol Biol Evol. 1992;9:836–55. doi: 10.1093/oxfordjournals.molbev.a040760. [DOI] [PubMed] [Google Scholar]
- 36.Lynch M, Sung W, Morris K, Coffey N, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci U S A. 2008;105:9272–7. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sloan DB, Alverson AJ, Wu M, Palmer JD, et al. Recent acceleration of plastid sequence and structural evolution coincides with extreme mitochondrial divergence in the angiosperm genus Silene. Genome Biol Evol. 2012;4:294–306. doi: 10.1093/gbe/evs006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sloan DB, Oxelman B, Rautenberg A, Taylor DR. Phylogenetic analysis of mitochondrial substitution rate variation in the angiosperm tribe Sileneae. BMC Evol Biol. 2009:9. doi: 10.1186/1471-2148-9-260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Itoh T, Martin W, Nei M. Acceleration of genomic evolution caused by enhanced mutation rate in endocellular symbionts. Proc Natl Acad Sci U S A. 2002;99:12944–8. doi: 10.1073/pnas.192449699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marais GA, Calteau A, Tenaillon O. Mutation rate and genome reduction in endosymbiotic and free-living bacteria. Genetica. 2008;134:205–10. doi: 10.1007/s10709-007-9226-6. [DOI] [PubMed] [Google Scholar]
- 41.Sung W, Ackerman MS, Miller SF, Doak TG, et al. Drift-barrier hypothesis and mutation-rate evolution. Proc Natl Acad Sci U S A. 2012;109:18488–92. doi: 10.1073/pnas.1216223109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gray MW, Cedergren R, Abel Y, Sankoff D. On the evolutionary origin of the plant mitochondrion and its genome. Proc Natl Acad Sci U S A. 1989;86:2267–71. doi: 10.1073/pnas.86.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salmon AB, Richardson A, Perez VI. Update on the oxidative stress theory of aging: does oxidative stress play a role in aging or healthy aging? Free Radic Biol Med. 2010;48:642–55. doi: 10.1016/j.freeradbiomed.2009.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Logan A, Shabalina IG, Prime TA, Rogatti S, et al. In vivo levels of mitochondrial hydrogen peroxide increase with age in mtDNA mutator mice. Aging Cell. 2014;13:765–8. doi: 10.1111/acel.12212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lynch M. The origins of genome architecture Sunderland, Mass. Sinauer Associates; 2007. [Google Scholar]
- 46.Larsson NG. Somatic mitochondrial DNA mutations in mammalian aging. Annu Rev Biochem. 2010;79:683–706. doi: 10.1146/annurev-biochem-060408-093701. [DOI] [PubMed] [Google Scholar]
- 47.Greiner S, Sobanski J, Bock R. Why are most organelle genomes transmitted maternally? Bioessays. 2015;37:80–94. doi: 10.1002/bies.201400110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Barr CM, Neiman M, Taylor DR. Inheritance and recombination of mitochondrial genomes in plants, fungi and animals. New Phytol. 2005;168:39–50. doi: 10.1111/j.1469-8137.2005.01492.x. [DOI] [PubMed] [Google Scholar]
- 49.McCauley DE. Paternal leakage, heteroplasmy, and the evolution of plant mitochondrial genomes. New Phytol. 2013;200:966–77. doi: 10.1111/nph.12431. [DOI] [PubMed] [Google Scholar]
- 50.Hausner G. Fungal mitochondrial genomes, plasmids and introns. Appl Mycol Biotech. 2003;3:101–31. [Google Scholar]
- 51.Hoekstra RF. Evolutionary origin and consequences of uniparental mitochondrial inheritance. Hum Reprod. 2000;15(Suppl 2):102–11. doi: 10.1093/humrep/15.suppl_2.102. [DOI] [PubMed] [Google Scholar]
- 52.Hastings IM. Population genetic aspects of deleterious cytoplasmic genomes and their effect on the evolution of sexual reproduction. Genet Res. 1992;59:215–25. doi: 10.1017/s0016672300030500. [DOI] [PubMed] [Google Scholar]
- 53.Hutson V, Law R. Four steps to two sexes. Proc Biol Sci. 1993;253:43–51. doi: 10.1098/rspb.1993.0080. [DOI] [PubMed] [Google Scholar]
- 54.Hurst LD, Hamilton WD. Cytoplasmic fusion and the nature of sexes. Proc Roy Soc B Biol Sci. 1992;247:189–94. [Google Scholar]
- 55.Taylor DR, Zeyl C, Cooke E. Conflicting levels of selection in the accumulation of mitochondrial defects in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 2002;99:3690–4. doi: 10.1073/pnas.072660299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sharpley MS, Marciniak C, Eckel-Mahan K, McManus M, et al. Heteroplasmy of mouse mtDNA is genetically unstable and results in altered behavior and cognition. Cell. 2012;151:333–43. doi: 10.1016/j.cell.2012.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Christie JR, Schaerf TM, Beekman M. Selection against heteroplasmy explains the evolution of uniparental inheritance of mitochondria. PLoS Genet. 2015;11:e1005112. doi: 10.1371/journal.pgen.1005112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.de Paula WB, Agip AN, Missirlis F, Ashworth R, et al. Female and male gamete mitochondria are distinct and complementary in transcription, structure, and genome function. Genome Biol Evol. 2013;5:1969–77. doi: 10.1093/gbe/evt147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.de Paula WB, Lucas CH, Agip AN, Vizcay-Barrena G, et al. Energy, ageing, fidelity and sex: oocyte mitochondrial DNA as a protected genetic template. Philos Trans R Soc Lond B Biol Sci. 2013;368:20120263. doi: 10.1098/rstb.2012.0263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in bdelloid rotifers. Science. 2008;320:1210–3. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 61.Flot JF, Hespeels B, Li X, Noel B, et al. Genomic evidence for ameiotic evolution in the bdelloid rotifer Adineta vaga. Nature. 2013;500:453–7. doi: 10.1038/nature12326. [DOI] [PubMed] [Google Scholar]
- 62.Felsenstein J. The evolutionary advantage of recombination. Genetics. 1974;78:737–56. doi: 10.1093/genetics/78.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Muller HJ. The relation of recombination to mutational advance. Mutat Res. 1964;106:2–9. doi: 10.1016/0027-5107(64)90047-8. [DOI] [PubMed] [Google Scholar]
- 64.Lynch M, Burger R, Butcher D, Gabriel W. The mutational meltdown in asexual populations. J Hered. 1993;84:339–44. doi: 10.1093/oxfordjournals.jhered.a111354. [DOI] [PubMed] [Google Scholar]
- 65.Kondrashov AS. Deleterious mutations and the evolution of sexual reproduction. Nature. 1988;336:435–40. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- 66.de Visser JA, Elena SF. The evolution of sex: empirical insights into the roles of epistasis and drift. Nat Rev Genet. 2007;8:139–49. doi: 10.1038/nrg1985. [DOI] [PubMed] [Google Scholar]
- 67.Kimura M, Maruyama T. The mutational load with epistatic gene interactions in fitness. Genetics. 1966;54:1337–51. doi: 10.1093/genetics/54.6.1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Otto SP, Barton NH. Selection for recombination in small populations. Evolution. 2001;55:1921–31. doi: 10.1111/j.0014-3820.2001.tb01310.x. [DOI] [PubMed] [Google Scholar]
- 69.Jiang X, Hu S, Xu Q, Chang Y, et al. Relative effects of segregation and recombination on the evolution of sex in finite diploid populations. Heredity (Edinb) 2013;111:505–12. doi: 10.1038/hdy.2013.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Keightley PD, Otto SP. Interference among deleterious mutations favours sex and recombination in finite populations. Nature. 2006;443:89–92. doi: 10.1038/nature05049. [DOI] [PubMed] [Google Scholar]
- 71.Kaiser VB, Charlesworth B. Muller's ratchet and the degeneration of the Drosophila miranda neo-Y chromosome. Genetics. 2010;185:339–48. doi: 10.1534/genetics.109.112789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Cutter AD, Payseur BA. Rates of deleterious mutation and the evolution of sex in Caenorhabditis. J Evol Biol. 2003;16:812–22. doi: 10.1046/j.1420-9101.2003.00596.x. [DOI] [PubMed] [Google Scholar]
- 73.Hollister JD, Greiner S, Wang W, Wang J, et al. Recurrent loss of sex is associated with accumulation of deleterious mutations in Oenothera. Mol Biol Evol. 2015;32:896–905. doi: 10.1093/molbev/msu345. [DOI] [PubMed] [Google Scholar]
- 74.Bruggeman J, Debets AJ, Wijngaarden PJ, deVisser JA, et al. Sex slows down the accumulation of deleterious mutations in the homothallic fungus Aspergillus nidulans. Genetics. 2003;164:479–85. doi: 10.1093/genetics/164.2.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lane N. Life ascending : the ten great inventions of evolution. W.W. Norton; New York: 2009. [Google Scholar]
- 76.Hadjivasiliou Z, Pomiankowski A, Seymour RM, Lane N. Selection for mitonuclear co-adaptation could favour the evolution of two sexes. Proc Roy Soc B Biol Sci. 2012;279:1865–72. doi: 10.1098/rspb.2011.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lane N. Mitonuclear match: Optimizing fitness and fertility over generations drives ageing within generations. Bioessays. 2011;33:860–9. doi: 10.1002/bies.201100051. [DOI] [PubMed] [Google Scholar]
- 78.Martin W, Koonin EV. Introns and the origin of nucleus-cytosol compartmentalization. Nature. 2006;440:41–5. doi: 10.1038/nature04531. [DOI] [PubMed] [Google Scholar]
- 79.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–34. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 80.Fisher RA. The genetical theory of natural selection. Claredon Press; Oxford: 1930. [Google Scholar]
- 81.Otto SP, Nuismer SL. Species interactions and the evolution of sex. Science. 2004;304:1018–20. doi: 10.1126/science.1094072. [DOI] [PubMed] [Google Scholar]
- 82.Budar F, Touzet P, De Paepe R. The nucleo-mitochondrial conflict in cytoplasmic male sterilities revisited. Genetica. 2003;117:3–16. doi: 10.1023/a:1022381016145. [DOI] [PubMed] [Google Scholar]
- 83.Blier PU, Dufresne F, Burton RS. Natural selection and the evolution of mtDNA-encoded peptides: evidence for intergenomic co-adaptation. Trends Genet. 2001;17:400–6. doi: 10.1016/s0168-9525(01)02338-1. [DOI] [PubMed] [Google Scholar]
- 84.Nedelcu AM. Sex as a response to oxidative stress: stress genes co-opted for sex. Proc Biol Sci. 2005;272:1935–40. doi: 10.1098/rspb.2005.3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Nedelcu AM, Marcu O, Michod RE. Sex as a response to oxidative stress: a twofold increase in cellular reactive oxygen species activates sex genes. Proc Biol Sci. 2004;271:1591–6. doi: 10.1098/rspb.2004.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nedelcu AM, Michod RE. Sex as a response to oxidative stress: the effect of antioxidants on sexual induction in a facultatively sexual lineage. Proc Biol Sci. 2003;270(Suppl 2):S136–9. doi: 10.1098/rsbl.2003.0062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Dowling DK, Simmons LW. Reactive oxygen species as universal constraints in life-history evolution. Proc Biol Sci. 2009;276:1737–45. doi: 10.1098/rspb.2008.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hellberg ME. No variation and low synonymous substitution rates in coral mtDNA despite high nuclear variation. BMC Evol Biol. 2006;6:24. doi: 10.1186/1471-2148-6-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Fautin DG. Reproduction of Cnidaria. Can J Zool. 2002;80:1735–54. [Google Scholar]
- 90.Silvertown J. The evolutionary maintenance of sexual reproduction: Evidence from the ecological distribution of asexual reproduction in clonal plants. Int J Plant Sci. 2008;169:157–68. [Google Scholar]
- 91.Gemmell NJ, Metcalf VJ, Allendorf FW. Mother's curse: the effect of mtDNA on individual fitness and population viability. Trends Ecol Evol. 2004;19:238–44. doi: 10.1016/j.tree.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 92.Barreto FS, Pereira RJ, Burton RS. Hybrid dysfunction and physiological compensation in gene expression. Mol Biol Evol. 2014;32:613–22. doi: 10.1093/molbev/msu321. [DOI] [PubMed] [Google Scholar]
- 93.Innocenti P, Morrow EH, Dowling DK. Experimental evidence supports a sex-specific selective sieve in mitochondrial genome evolution. Science. 2011;332:845–8. doi: 10.1126/science.1201157. [DOI] [PubMed] [Google Scholar]
- 94.Hussin JG, Hodgkinson A, Idaghdour Y, Grenier JC, et al. Recombination affects accumulation of damaging and disease-associated mutations in human populations. Nat Genet. 2015;47:400–4. doi: 10.1038/ng.3216. [DOI] [PubMed] [Google Scholar]