Abstract
The spread of farming out of the Balkans and into the rest of Europe followed two distinct routes: An initial expansion represented by the Impressa and Cardial traditions, which followed the Northern Mediterranean coastline; and another expansion represented by the LBK (Linearbandkeramik) tradition, which followed the Danube River into Central Europe. Although genomic data now exist from samples representing the second migration, such data have yet to be successfully generated from the initial Mediterranean migration. To address this, we generated the complete genome of a 7,400-year-old Cardial individual (CB13) from Cova Bonica in Vallirana (Barcelona), as well as partial nuclear data from five others excavated from different sites in Spain and Portugal. CB13 clusters with all previously sequenced early European farmers and modern-day Sardinians. Furthermore, our analyses suggest that both Cardial and LBK peoples derived from a common ancient population located in or around the Balkan Peninsula. The Iberian Cardial genome also carries a discernible hunter–gatherer genetic signature that likely was not acquired by admixture with local Iberian foragers. Our results indicate that retrieving ancient genomes from similarly warm Mediterranean environments such as the Near East is technically feasible.
Keywords: Neolithic, paleogenomics, Cardial ware, migration
Introduction
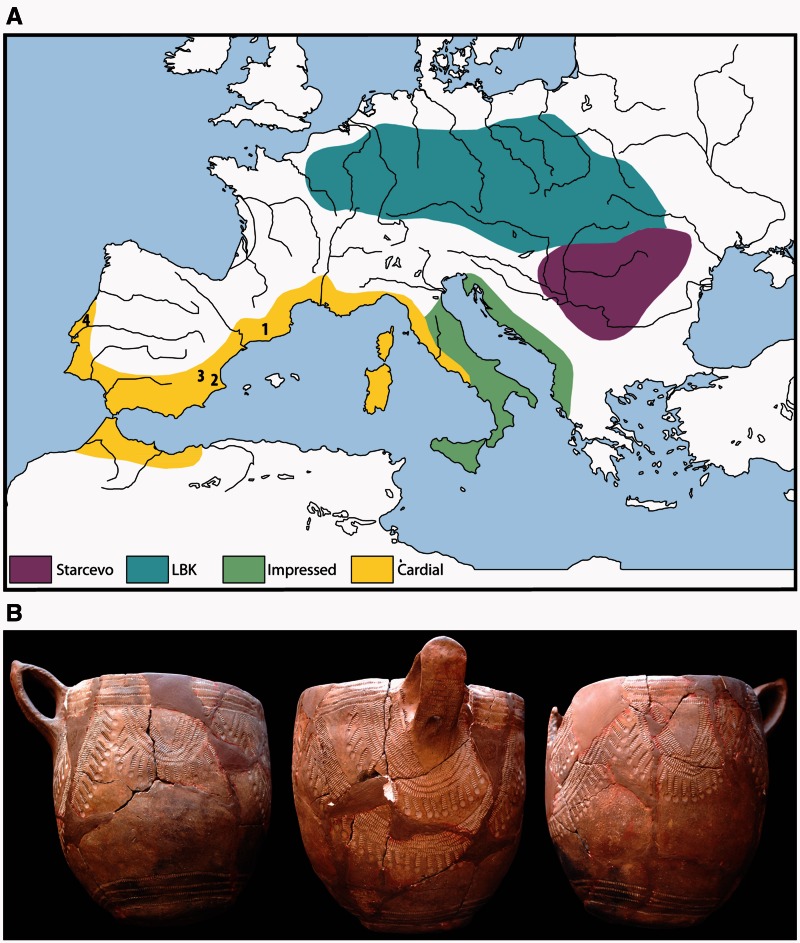
The introduction of farming into Europe around 8,000 years ago was a major demographic transition. It involved a substantial replacement of the preexisting hunter–gatherer populations by migrants of ultimate Near Eastern origin as well as new adaptive challenges (Sánchez-Quinto et al. 2012; Skoglund et al. 2012, 2014; Gamba et al. 2014; Lazaridis et al. 2014; Olalde et al. 2014; Allentoft et al. 2015; Haak et al. 2015). Initially these early farmers settled in the Balkan Peninsula, developing what today is referred to as the Starčevo–Kőrös–Criş culture (Whittle 1996) (fig. 1A). Archaeological evidence suggests that these farmers spread subsequently throughout Europe along at least two distinctive routes. Expansion along the first route commenced approximately 5,900 years BCE, and is represented by the distinct Impressa culture that spread along the central and Western Mediterranean basin. A later aspect of this culture, named Cardial for the use of the serrated edge of cockle shells in pottery decoration (fig. 1B), reached the Iberian Peninsula no later than 5,500 years BCE (Before Common Era) (fig. 1A) (Martins et al. 2015); however, the inundation of the ancient Neolithic coastline hampers our understanding of the origins and dynamics of this first expansion. The second expansion occurred in parallel with the Cardial into large areas of Central Europe along the Danube River (fig. 1A), by a culture today referred to as the Linearbandkeramik (or LBK) for the banded decoration patterns found in their pottery. From an archaeological point of view, the spread of the Cardial culture, with a distinctive package of pottery, polished stone axes and domesticates, seems to be a migration process similar to that represented by the LBK expansion (Zilhão 1993). However, the rapid expansion of the Cardial culture along the Iberian coast, as suggested by radiocarbon dates and its restricted littoral distribution, has been interpreted as the result of maritime pioneer colonization (Zilhão 2001).
Fig. 1.
Early Neolithic Cardial culture. (A) Main cultural horizons associated with the earliest Neolithic of Central and Western Europe approximately 6,000–5,500 cal BCE. 1, Cova Bonica; 2, Cova de la Sarsa; 3, Cova de l‘Or; 4, Galeria da Cisterna-Almonda. (B) Cardial ceramics from Cova de la Sarsa. The impressed decoration is characteristically made with the serrated edge of cockle shells.
Palaeogenomics represents a powerful tool for refining our understanding of such past events, and thanks to the advent of next-generation sequencing (NGS) technologies, to date 37 complete (>1×) genome drafts are available from prehistoric Eurasians, spanning from the Upper Paleolithic to the Bronze and Iron Ages (Allentoft et al. 2015; Olalde and Lalueza-Fox 2015). In addition, many more specimens have been genotyped for about 400,000 polymorphisms (Haak et al. 2015). All these ancient data have been used to reconstruct past population movements as well as adaptive challenges and selective events that arose during European prehistory.
Given its geographic location on the far western edge of Europe, the Iberian Peninsula is a critical place for estimating the final impact of the substantial population dispersals that originated in the continent’s Eastern periphery (among which are included the initial Neolithic migration and the Late Neolithic/Bronze age steppe migration).
Previous studies on ancient Cardial specimens have been restricted to the analysis of uniparental markers—especially mitochondrial DNA (mtDNA)—by a traditional polymerase chain reaction (PCR) approach (Lacan, Keyser, Ricaut, Brucato, Tarrús, et al. 2011; Gamba et al. 2012). Although the resolution of these genetic markers is limited, the mtDNA haplogroup composition of the few available Cardial individuals pointed to a Near East connection and suggested a pioneer colonization from that region (Lacan, Keyser, Ricaut, Brucato, Tarrús, et al. 2011; Gamba et al. 2012).
Cardial specimens have yet to be analyzed using NGS techniques, although later Neolithic samples from Central and Northeastern Iberia have yielded genotype data that cluster them with other early European farmers (Haak et al. 2015). The lack of data from the Cardial Neolithic can be explained by the scarcity of associated human remains, as well as the warm climatic conditions of the Mediterranean, which are largely unfavorable for DNA preservation (García-Garcerà et al. 2011; Hofreiter et al. 2014). Therefore, the genomic affinities of the Western Mediterranean’s first farmers have remained unknown until now.
To clarify the origin and population affinities of this early Mediterranean migration, we analyzed six individuals from some of the oldest securely dated Cardial Iberian sites: Two from Cova Bonica (Barcelona) (supplementary fig. S1, Supplementary Material online), one from Cova de l’Or (Alicante), one from Cova de la Sarsa (Valencia), and two from the Galeria da Cisterna locus of the Almonda karst system (Portugal) (table 1). All calibrated radiocarbon dates range between approximately 5,470 and 5,220 years BCE (supplementary table S1, Supplementary Material online). As expected, the endogenous content of all samples was low. Therefore, we combined shotgun sequencing with a recently validated commercially available human whole-genome capture assay (Ávila-Arcos et al. 2015; Schroeder et al. 2015) to generate the complete genome for one of the Cova Bonica specimens (CB13) and partial genome data for the remaining samples.
Table 1.
Summary Statistics of the Sequenced Cardial Specimens.
| Sample | Site | Genome Coverage | Human DNA Shotgun (%) | Human DNA Postcapture (%) | Radiocarbon Age (cal BCE, 1σ) | Sex | mtDNA Haplogroup | mtDNA Coverage | Contamination (%) |
|---|---|---|---|---|---|---|---|---|---|
| CB13 | Cova Bonica | 1.1085 | 5.68 | 28.23 | 5,470–5,360 | F | K1a2a | 353.29 | 0.11 |
| CB14 | Cova Bonica | 0.0003 | 0.25 | — | —a | F | X2c | 4.1 | 1.35 |
| F19 | Almonda cave | 0.0129 | 0.58 | 9.66 | 5,310–5,220 | F | H4a1a | 33.79 | 29.13 |
| G21 | Almonda cave | 0.0039 | 0.09 | 1.27 | 5,330–5,230 | M | H3 | 63.55 | 1.97 |
| H3C6 | Cova de l’Or | 0.0011 | 0.06 | 1.01 | 5,360–5,310 | M | H4a1a | 4.45 | 3.96 |
| CS7675 | Cova de la Sarsa | 0.0012 | 0.08 | 1.14 | 5,321–5,227 | Mb | K1a4a1 | 0.69 | — |
aCB14 is not directly radiocarbon dated, although it comes from the same stratigraphic layer than CB13.
bCS7675 is determined to be a male from morphology.
Results and Discussion
Whole-genome capture enriched the endogenous DNA in our libraries by an average of 5- to 15-fold, consistent with performance on similarly degraded materials (Carpenter et al. 2013; Ávila-Arcos et al. 2015; Schroeder et al. 2015). However, low DNA endogenous content (below 1% in all samples except CB13) and low complexity only allowed us to retrieve one complete genome (1.1 × coverage) from the female Cova Bonica CB13 sample (table 1 and supplementary fig. S2, Supplementary Material online). For the remaining samples, we obtained mtDNA genomes at 0.7–64 × coverage and limited nuclear data (between 0.0003×and 0.0129×) (table 1 and supplementary table S2, Supplementary Material online).
We estimated very low levels (0.11%) of modern contamination at the mtDNA level for CB13 (supplementary tables S3 and S4, Supplementary Material online). The remaining samples also showed low mtDNA contamination levels (<5%) with the exception of one of the samples from Almonda (F19), which had an estimated 29% of contamination of unknown origin (supplementary table S3, Supplementary Material online). Both pre- and postcapture sequences showed the typical ancient DNA deamination pattern at the end of the reads (Brotherton et al. 2007), with deamination percentages over 20%, with the exception of F19 (supplementary fig. S3, Supplementary Material online).
We were able to determine the mtDNA haplogroups from the six Cardial individuals. The presence among our samples of haplogroups K1a, H3, and H4 (supplementary table S5, Supplementary Material online) is consistent with previous results obtained for 20 specimens including 10 Cardials from four Iberian Early Neolithic sites (Lacan, Keyser, Ricaut, Brucato, Tarrús, et al. 2011; Gamba et al. 2012). In these samples the authors describe N*, K (K1a), H (H3), U5, T2b, and X1 lineages, all of which are also present in other early Neolithic samples from the LBK culture of Central Europe (Brandt et al. 2013; Gamba et al. 2014). The haplotype K1a2a found in CB13 has been previously described in an Epicardial individual from Els Trocs (Spain) dated to 5,177–5,069 years cal BCE (Haak et al. 2015). On the other hand, the haplotype X2c from CB14 is quite rare in modern Europeans, but it has been found in a few Neolithic samples from France (Deguilloux et al. 2011; Lacan, Keyser, Ricaut, Brucato, Duranthon, et al. 2011) and Germany (Lee et al. 2012). Unfortunately, information on the Y-chromosome could not be obtained due to the low genomic coverage of the male samples.
At the phenotypic level, CB13 has derived alleles for the SLC24A5 pigmentation gene (supplementary tables S6–S8, Supplementary Material online) and appears heterozygous for the SLC45A2 skin pigmentation gene (supplementary tables S6 and S7, Supplementary Material online), both associated with light skin in Europeans. The same, light skin-related genotypic combination is also seen in several Early, Middle, and Late Neolithic individuals from Hungary (Gamba et al. 2014). Despite uncertainties associated with the low coverage, the Hirisplex pigmentation prediction (Walsh et al. 2013) yields the highest probability of this individual having dark hair (0.679). Due to a combination of missing (including the critical rs1281382 single nucleotide polymorphism [SNP] for blue eyes) and heterozygous sites at the OCA2/HERC2 haplotype (supplementary table S9, Supplementary Material online), the color of the iris could not be conclusively determined. At the rs4988235 site (which has a regulatory effect on the LCT gene), CB13 shows the ancestral variant associated with the inability to digest milk during adulthood (Itan et al. 2009), sharing this trait with all Neolithic individuals analyzed to date (Gamba et al. 2014; Lazaridis et al. 2014).
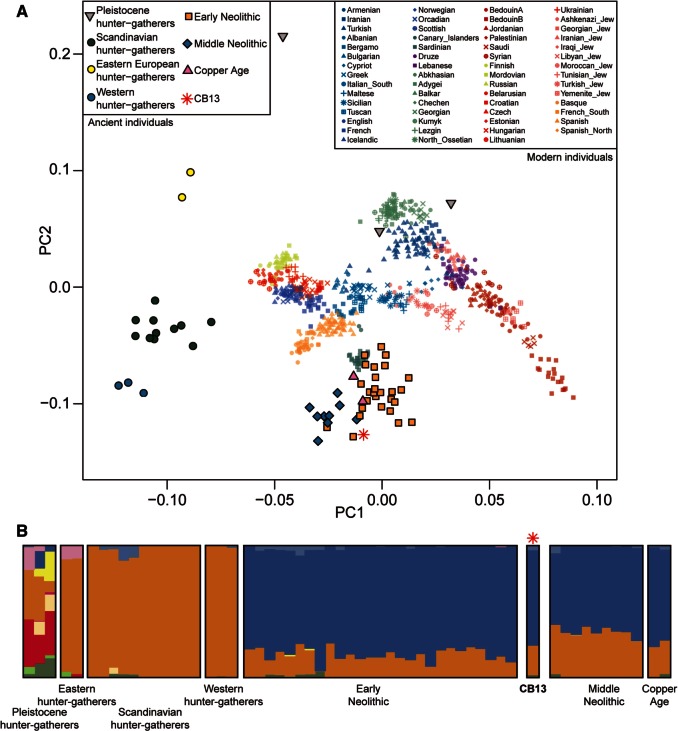
To infer the general ancestry of CB13 we performed Principal Component Analysis (PCA) with a large data set of present-day Europeans and Near Eastern individuals (Lazaridis et al. 2014), as well as on ancient individuals from different studies (Keller et al. 2012; Fu et al. 2014; Gamba et al. 2014; Lazaridis et al. 2014; Olalde et al. 2014; Raghavan et al. 2014; Seguin-Orlando et al. 2014; Skoglund et al. 2014; Haak et al. 2015) (fig. 2A and supplementary fig. S4, Supplementary Material online). Our Cardial individual clusters with other Neolithic samples, including LBK individuals from Germany (Stuttgart) and Hungary (NE1) and early Neolithic samples from Iberia, as well as later Middle Neolithic and Copper age individuals (Keller et al. 2012; Gamba et al. 2014; Lazaridis et al. 2014; Haak et al. 2015). As previously noticed (Keller et al. 2012; Gamba et al. 2014; Lazaridis et al. 2014; Skoglund et al. 2014), all these prehistoric farmers also plot close to present-day southern Europeans, in particular to Sardinians. Interestingly, these individuals and CB13 plot in between extant Near Easterners and prehistoric European hunter–gatherers, suggesting that they also share some ancestry with the latter. This pattern is consistent with previous observations (Lazaridis et al. 2014; Skoglund et al. 2014) and with our ADMIXTURE (Alexander et al. 2009) analysis (fig. 2B), where part of CB13 female’s ancestry is assigned to a component characteristic of hunter–gatherer populations.
Fig. 2.
Genetic affinities of CB13. (A) Procrustes PCA of hunter–gatherers, Early Neolithic, Middle Neolithic, and Copper Age farmers. The PCA was performed using only transversions (to avoid confounding effects related to postmortem damage). (B) Ancestry proportions assuming 11 ancestral components, as inferred by ADMIXTURE analysis.
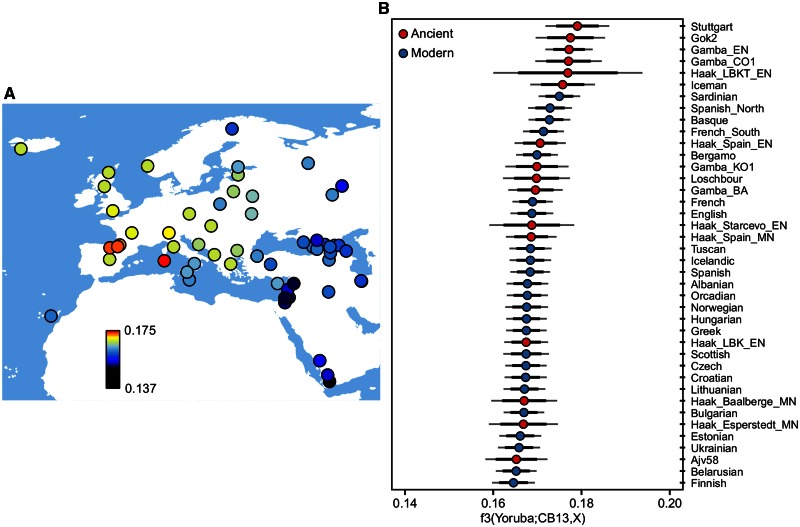
To examine which modern-day and ancient populations show the greatest shared genetic drift with the Cardial genome, we used outgroup f3-statistics (Reich et al. 2009). Among extant populations, we found the highest scores of shared genetic drift for Sardinians and, to a lesser extent, for Basques (fig. 3A). Among ancient populations/individuals, CB13 shows the highest shared genetic drift with other Neolithic individuals from different parts of Europe (fig. 3B).
Fig. 3.
Outgroup f3-statistic analysis of CB13 Cardial genome. (A) Shared genetic drift between CB13 and present-day Western Eurasian populations. (B) Top 40 populations/individuals (modern and ancient) showing the highest genetic drift with CB13. Black and gray error bars represent two and three standard errors, respectively.
The Basques have been traditionally considered one of the oldest human groups in Europe, inhabiting a marginal area in the Pyrenean mountain range and exhibiting genetic continuity since pre-Neolithic times (Cavalli-Sforza et al. 1994). The fact that modern Basque peoples speak the sole surviving relict of a pre-Indo-European language in Western Europe (the Euskera or Basque language) could have also contributed to their isolation (Renfrew and Bahn 1991). The existence of some autochthonous mtDNA subhaplogroups (Cardoso et al. 2013) has been used to further support the Basque singularity among Europeans, although this has been recently questioned by genome-wide data (Laayouni et al. 2010).
Our analysis suggests that the geographic isolation of Sardinians and Basques partially preserved the originally widespread early Neolithic population component, more than any other populations in Europe. Considering that Basques speak a pre-Indo-European language, this finding also indicates that the expansion of Indo-European languages is unlikely to have taken place during the early Neolithic. This is in agreement with the recently characterized genetic influx from the steppes in the Late Neolithic/Chalcolithic, which has been associated with the spread of Indo-European languages into Western Europe (Allentoft et al. 2015; Haak et al. 2015).
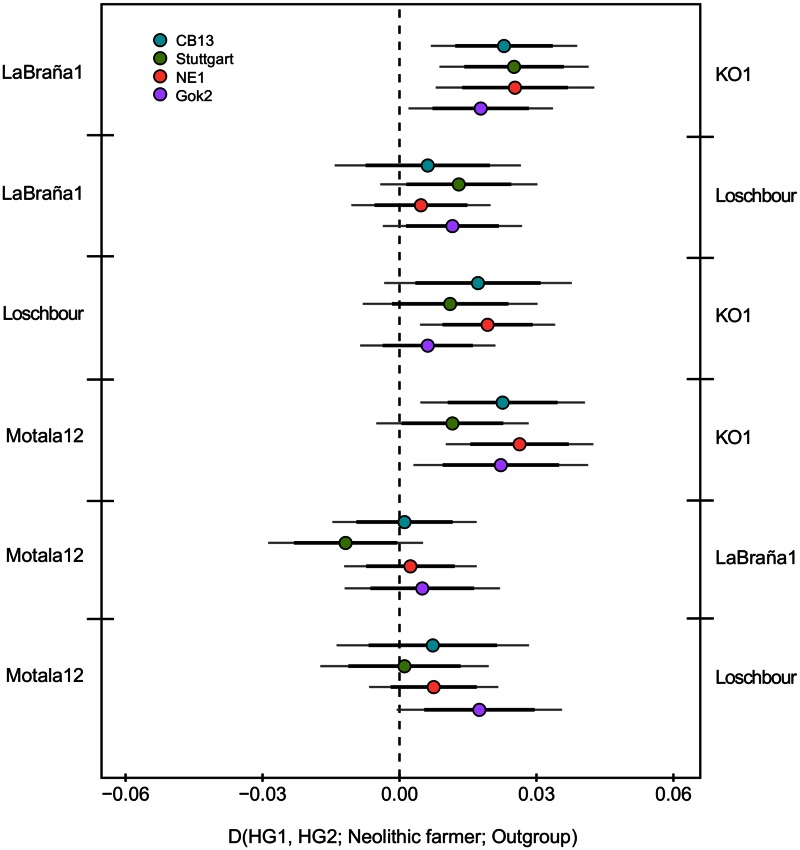
To ascertain where CB13—and other early farmers—acquired their hunter–gatherer genetic component, we computed D statistics (Durand et al. 2011) of the form D (Hunter-gatherer1, Hunter-gatherer2; Neolithic farmer, chimpanzee), testing whether a given Neolithic farmer is significantly closer to one of the two hunter–gatherers. We used La Braña 1 (Spain), Loschbour (Luxembourg), KO1 (Hungary), and Motala12 (Sweden) as hunter–gatherer references. We found that CB13 is closer to KO1 than to La Braña 1, which is only 800 km away from the Cova Bonica Cardial site (fig. 4 and supplementary table S10, Supplementary Material online). Although the number of available genomes is yet limited and KO1 is a hunter–gatherer in a farming context, these results suggest that the origin of the hunter–gatherer genomic component present in CB13 cannot be traced to the aboriginal hunter–gatherers of the Iberian Peninsula, an observation also supported by TreeMix analysis (supplementary fig. S7B, Supplementary Material online). The fact that CB13 shares more alleles with KO1 than with La Braña 1 indicates that there was some discernible East–West population structure among European hunter–gatherers, already suggested in a previous study (Haak et al. 2015). In the future, the sequencing of more ancient hunter–gatherer genomes from Greece, Italy, Southern France, and Mediterranean Iberia could potentially disentangle fine population structure patterns among these populations, allowing a further characterization of the hunter–gatherer component in early farmers.
Fig. 4.
D-statistics to determine whether CB13 and other Neolithic farmers are closer to any hunter–gatherer. Black and gray error bars represent two and three standard errors, respectively.
Conclusions
The Mediterranean region is crucial not only for understanding local cultural horizons such as the Cardial but also for unravelling potential trans-Mediterranean maritime routes and island colonization processes. Although the DNA in our samples was poorly preserved because of the warm Mediterranean climate, our results demonstrate that recovery of complete ancient genomes from areas with a similar climate (including in the Near East and North Africa) may also be possible. Cave sites in these regions clearly offer some advantages in terms of preservation.
Our analyses indicate that both the LBK and the Cardial peoples originated from a common ancient meta-population that diverged along two different migration routes, one following the Danube River (LBK) and the other one following the northern Mediterranean coastline (Impressa and Cardial). Furthermore, we detect a discernible hunter–gatherer component in the Cardial genome, which seems to derive from a population more closely related to Eastern European hunter–gatherers than to the neighboring Iberian La Braña 1 sample.
From the current genetic evidence, it seems clear that all early European farmers represent a fairly homogeneous group at both the genetic and phenotypic levels. Subsequent population movements from the Chalcolithic onwards considerably altered this scenario, and contributed to the shaping of present-day European genetic diversity (Gamba et al. 2014; Allentoft et al. 2015; Haak et al. 2015).
Materials and Methods
Sample Selection
Samples–mainly teeth–from Cardial individuals were selected based on external appearance and cave provenance. Only securely dated samples (i.e., associated with Cardial pottery remains in undisturbed deposits and/or directly dated by radiocarbon) were considered for DNA analysis. The six samples analyzed derive from four Cardial sites (supplementary materials and methods, Supplementary Material online): Cova Bonica (CB13 and CB14) in Vallirana (Barcelona), Galeria da Cisterna in Almonda (G21 and F19) (Portugal), Cova de l‘Or (H3C6) in Beniarrés (Alicante), and Cova de la Sarsa (CS7675) in Bocairent (Valencia) (table 1 and fig. 1).
DNA Extraction
The samples were extracted using a modified version of the silica-in-solution protocol described elsewhere (Rohland and Hofreiter 2007). Instead of the commonly used dentine, we targeted the cementum-rich root tip, which has been shown to contain higher levels of endogenous DNA (Adler et al. 2011; Damgaard et al. 2015). We also added a “predigestion” step to the extraction protocol that has been shown to significantly increase library efficiency (Allentoft et al. 2015; Damgaard et al. 2015). In addition, we used a recently introduced DNA binding buffer that has been shown to be more efficient at retaining short DNA fragments compared with other buffers (Allentoft et al. 2015).
Library Preparation
Sequencing libraries were constructed using the NEB’s NEBNext DNA Sample Prep Master Mix Set 2 (E6070) and Illumina-specific adapters, following established protocols (Meyer and Kircher 2010). Following the Bst fill-in step, the libraries were amplified and indexed in 50 µl PCRs containing 1 × KAPA HiFi HotStart Uracil + ReadyMix (KAPA Biosystems, Woburn, MA) and 200 nM of each of Illumina’s Multiplexing PCR primer in PE1.0 (5′-AATGATACGGCGACCACCGAGATCTACACTCTTT CCCTACACGAC GCTCTT CCGATCT) and a custom-designed index primer with a six nucleotide index (5′-CAAGCAGAAGACGGCATAC GAGATNNNNNNGTGACTGGAGTTC). Thermocycling conditions were as follows: 1 min at 94 °C, followed by 8–12 cycles of 15 s at 94 °C, 20 s at 60 °C, and 20 s at 72 °C, and a final extension step of 1 min at 72 °C. The optimal number of cycles was determined by quantitative PCR, as done in Meyer and Kircher (2010). The amplified libraries were then purified using Agencourt AMPure XP beads (Beckman Coulter, Krefeld, Germany) and quantified on an Agilent 2200 TapeStation (Agilent Technologies, Palo Alto, CA).
Initial Screening and Whole-Genome Capture
To establish their overall efficiency, the libraries were sequenced on an Illumina HiSeq 2000 run in 100 SR mode at the Danish National High-Throughput Sequencing Centre in Copenhagen, Denmark. Following this initial screening run, libraries were enriched using the MYbait Human Whole Genome Capture Kit from MYcroarray (Ann Arbor, MI) (Ávila-Arcos et al. 2015; Schroeder et al. 2015). The libraries were captured following the manufacturer’s instructions (http://www.mycroarray.com/pdf/MYbaits-manual-v2.pdf). The captured libraries were amplified for 10–20 cycles using primers IS5 (5′-AATGATACG GCGACCACCGA) and IS6 (5′-CAAGCAGAAGACGGCA TACGA) and the same PCR set-up conditions as above. Subsequently, libraries were purified and quantified as above, pooled in equimolar amounts, and sequenced. Base-calling was performed using the Illumina software CASAVA 1.8.2.
Cova Bonica DNA Reextraction
Several libraries were constructed from the most efficient sample, CB13, a tooth root from Cova Bonica. After the exhaustion of the original extract, the undigested tooth pellet was reextracted and a new library was built. The subsequent sequencing results showed that this second-round library had a significantly higher human DNA content and higher complexity than previous libraries. In addition, reads from this library showed a lower mean fragment length and higher deamination levels at the extremes (supplementary fig. S5, Supplementary Material online), suggesting that reextracting the pellet significantly increased endogenous DNA yields. Similar observations have been made previously and it has been proposed that reextraction triggers the release of a DNA fraction located in the deep, crystalline dentine structure and thus better protected from hydrolysis and microbial action (Orlando et al. 2011; Der Sarkissian et al. 2014). All of these observations are also in line with the proposed notion that a predigestion step can significantly improve efficiency in ancient samples (Damgaard et al. 2015).
Sequencing Data Processing
Owing to postmortem degradation, ancient DNA fragments are usually very short, resulting in sequencing of the adapter, which has been ligated during library preparation. Thus, AdapterRemoval (Lindgreen 2012) was used to remove adapter sequences from the 3′-end of the reads, and to remove stretches of consecutive bases with 0, 1, or 2 quality scores. Before mapping, we discarded sequences shorter than 30 bp. Reads were mapped with BWA-0.6.1 (Li and Durbin 2009) to the human reference genome (hg19) and to the rCRS mitochondrial genome (Andrews et al. 1999), disabling the seed and setting parameters “-n 0.01” and “-o 2.” Duplicate sequences were removed using PicardTools (http://picard.sourceforge.net/). To estimate the coverage attained for each sample, the DepthOfCoverage tool implemented in GATK (McKenna et al. 2010) was used. The sequence statistics for each sample are displayed in supplementary table S2, Supplementary Material online. Only reads with mapping quality greater than 30 were kept for further analysis.
Sex Determination
The sex of each individual was determined following the approach in Skoglund et al. (2013). This method computes the ratio of chrY reads to chrX + chrY reads. The differences in coverage between sexual and autosomal chromosomes provide direct evidence of the individual’s sex (females are expected to have an X-chromosome coverage similar to that of autosomes, whereas males show an X-chromosome with half the coverage for autosomes and also a significant presence of Y-chromosome reads).
mtDNA Haplogroup Determination
Consensus sequences were called using samtools and bcftools (Li et al. 2009), requiring a support of at least three reads and using a majority rule. Haplogroups were determined using haplogrep tool (Kloss-Brandstätter et al. 2011) (supplementary table S5, Supplementary Material online).
Ancient DNA Authenticity
To estimate the rate of modern human DNA contamination in our samples, several procedures were followed:
Misincorporation Patterns at the Ends of the Reads
It has been observed in different studies that the endogenous DNA bears a distinctive pattern at the 5′- and 3′-ends resulting from chemical processes related to depurination, fragmentation, and subsequent deamination of the DNA templates (Briggs et al. 2007). This pattern of miscoding lesions (increased ratio of C to T changes at the 5′-ends and of G to A at the 3′-ends) is typically used as a proxy for authenticity. The damage pattern at the end of the reads was determined with the mapDamage2.0 software (Jónsson et al. 2013) (supplementary fig. S3, Supplementary Material online).
mtDNA Contamination Estimates
The analysis of the reads mapped to the mtDNA allows us to check whether the majority of the reads derives from a single biological source. After calling consensus sequences, we estimated contamination in the mitochondria using contamMix-1.0.10. This software implements a Bayesian approach described in Fu et al. (2013). In the case of CB13, the high coverage allowed us to use only transversions that are not affected by postmortem damage. Contamination estimates are shown in supplementary tables S3 and S4, Supplementary Material online.
Phenotypic Traits
For the low-coverage Cova Bonica genome, it was possible to screen for phenotypic traits of interest that have been the subject of recent natural selection in Europeans, according to several studies (Beleza et al. 2013; Grossman et al. 2013). We examined the reads overlapping the SNPs and haplotypes involved in potential selective sweeps, notably those related to pigmentation and lactase persistance (supplementary tables S6–S9, Supplementary Material online). The hair color was estimated with the Hirisplex prediction model (Walsh et al. 2013, 2014).
Population Genetic Reference Data
We used two different reference data sets:
The Human Origins data set released in a previous study (Lazaridis et al. 2014), which contains 1,941 present-day individuals from worldwide populations genotyped at 594,924 sites. We next added variants from CB13 and other Eurasian ancient individuals from different studies (Fu et al. 2014; Gamba et al. 2014; Lazaridis et al. 2014; Olalde et al. 2014; Raghavan et al. 2014; Seguin-Orlando et al. 2014; Skoglund et al. 2014). For each ancient sample, we randomly sampled one read at each position, discarding alleles not present in the reference data set, and discarding reads with mapping quality lower than 30 and base quality lower than 30. Moreover, data from the study of Haak et al. (2015), containing 69 ancient Europeans, were downloaded and merged with the data set, resulting in a total of 93 ancient individuals.
The 1000 Genomes project (1000 Genomes Project Consortium 2012) phase 3 initial callset downloaded from ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/release/20130502/. Only SNPs found to be polymorphic in Yoruba were used for the analysis. Variants from CB13 and other Neolithic farmers and hunter–gatherers from different studies (Gamba et al. 2014; Lazaridis et al. 2014; Olalde et al. 2014; Skoglund et al. 2014) were added following the same strategy as for the Human Origins reference data set.
Principal Component Analysis
PCA was performed on a reference set of 777 modern West Eurasians and a set of 58 ancient individuals, including CB13. First, for each ancient sample, individual PCA was computed using EIGENSOFT (Patterson et al. 2006). Then, using Procrustes transformation (Skoglund et al. 2012) implemented in the vegan package (http://vegan.r-forge.r-project.org), the first two principal components were transformed to match the configuration of reference only PC1 and PC2. Finally, the average of the transformed coordinates for reference individuals was plotted, together with transformed coordinates for each ancient sample (fig. 2A and supplementary fig. S4, Supplementary Material online). To avoid possible confounding effects caused by postmortem deamination, the PCA was generated with only transversion positions.
Outgroup f3-Statistics
To study the degree of genetic relatedness between CB13 Cardial sample and different present-day and ancient populations, we computed outgroup f3-statistics (Reich et al. 2009; Raghavan et al. 2014) of the form f3 (Yoruba; CB13,X). This statistic measures the amount of genetic drift shared by CB13 and population X, after their divergence from Yoruba. Standard errors were computed using a weighted block jackknife approach (Busing et al. 1999) over 5-Mb blocks (fig. 3).
D Statistics
Using the 1000 Genomes reference data set, we computed D statistics (Durand et al. 2011) of the form D (Hunter-gatherer1, Hunter-gatherer2; Neolithic farmer; Outgroup) to test whether any Neolithic farmer, including CB13 Cardial sample, is closer to one of the two hunter–gatherers (fig. 4 and supplementary table S10, Supplementary Material online). Standard errors were computed using a weighted block jackknife approach (Busing et al. 1999) over 5-Mb blocks.
Admixture
We carried out model-based clustering analysis using ADMIXTURE (Alexander et al. 2009) on the Human Origins reference data set, including 1,941 present-day individuals and 93 ancient individuals. First, we performed LD-pruning on the data set using PLINK (Purcell et al. 2007) with the flag –indep-pairwise 200 25 0.4, resulting in 283,136 SNPs that were used for analysis. ADMIXTURE was run with the cross validation (–cv) flag considering K = 2 to K = 19, with 25 replicates for each value of K. The lowest median CV error was obtained for K = 11. In figure 2B we show the ancestry proportions for K = 11 of 58 ancient individuals, including the CB13 cardial female. CB13 displays an ancestry make up similar to other early Neolithic samples, with an orange ancestral component characteristic of hunter–gatherer populations and a blue ancestral component related to the arrival of the Neolithic.
Treemix Analysis
We applied TreeMix (Pickrell and Pritchard 2012) to the Human Origins reference data set to infer maximum-likelihood trees and admixture graphs. We included nine ancient individuals: CB13, MA1 (Raghavan et al. 2014), LaBraña1 (Olalde et al. 2014), Loschbour (Lazaridis et al. 2014), Motala12 (Lazaridis et al. 2014), KO1 (Gamba et al. 2014), NE1 (Gamba et al. 2014), Gok2 (Skoglund et al. 2014) and Stuttgart (Lazaridis et al. 2014), and also three present-day populations: Mbuti, Papuan, and Karitiana. Only 90,604 sites with information in all the ancient individuals were used. We root the graphs with Mbuti, disabled sample size correction (‐noss), performed a round of global realignments of the graph (‐global), and computed standard error of migration weights (‐se).
We considered up to four migration edges and kept for each edge the graph with the highest log-likelihood among ten replicate runs (supplementary figs. S6 and S7, Supplementary Material online). All the graphs place our CB13 Cardial sample close to other European early farmers, especially close to NE1 and Stuttgart. Interestingly, the graph with four migration edges (supplementary fig. S7B, Supplementary Material online) places the group formed by the European early farmers (CB13, NE1, Gok2, and Stuttgart) basal to hunter–gatherers, MA1 and Karitiana, consistent with them having basal Eurasian ancestry (Lazaridis et al. 2014). Furthermore, around 46% of the European Early farmer ancestry is contributed by a hunter–gatherer population most closely related to KO1.
Supplementary Material
Supplementary materials and methods, figures S1–S7, and tables S1–S10 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors thank the staff at the Danish National High Throughput Sequencing Centre for technical support and the Museu de Prehistòria de la Diputació Provincial de València, Direcció General de Cultura de la Generalitat Valenciana, and Ajuntament de Bocairent for granting permission to analyze the Cova de la Sarsa and Cova de l’Or samples. They thank Professor Jean-Jacques Hublin, Professor Michael Richards and the Max Planck Society for supporting the radiocarbon part, Professor Eske Willerslev for providing critical input, and Dr Philip Johnson for providing the contamMix software. The Centre for GeoGenetics is funded by the Danish National Research Foundation (DNRF94). Cova Bonica work is supported by Servei d’Arqueologia i Paleontologia (2014/100639), Generalitat de Catalunya (2014SGR-108), and Ministerio de Ciencia e Innovación (HAR2011-26193) projects. H.S. was supported by an ERC Synergy Grant (FP7/2007-2013/319209); C.L.-F. by a FEDER and Spanish Government Grant BFU2012-34157; and S.C. by a grant 2014 SGR 464 from Departament d’Economia i Coneixement (Generalitat de Catalunya). D.C.S-G. acknowledges support from the Generalitat Valenciana (VALi + d APOSTD/2014/123), the BBVA Foundation (I Ayudas a Investigadores, Innovadores y Creadores Culturales), and the European Union (FP7/2007-2013—MSCA-COFUND, no. 245743 via a Braudel-IFER-FMSH). I.O. was funded by a predoctoral fellowship from the Basque Government (DEUI), and M.S. and J.D. by postdoctoral grants from Fundação para a Ciência e a Tecnologia (FCT) and Juan de la Cierva Subprogram (JCI-2011-09543), respectively. Alignment data are available through the Sequence Read Archive (SRA) at the accession code SRP057056.
References
- 1000 Genomes Project Consortium. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adler CJ, Haak W, Donlon D, Cooper A. 2011. Survival and recovery of DNA from ancient teeth and bones. J Archaeol Sci. 38:956–964. [Google Scholar]
- Alexander DH, Novembre J, Lange K. 2009. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19:1655–1664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allentoft ME, Sikora M, Sjögren K-G, Rasmussen S, Rasmussen M, Stenderup J, Damgaard PB, Schroeder H, Ahlström T, Vinner L, et al. 2015. Population genomics of Bronze Age Eurasia. Nature 522:167–172. [DOI] [PubMed] [Google Scholar]
- Andrews RM, Kubacka I, Chinnery PF, Lightowlers RN, Turnbull DM, Howell N. 1999. Reanalysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 23:147. [DOI] [PubMed] [Google Scholar]
- Ávila-Arcos MC, Sandoval-Velasco M, Schroeder H, Carpenter ML, Malaspinas A-S, Wales N, Peñaloza F, Bustamante CD, Gilbert MTP. 2015. Comparative performance of two whole-genome capture methodologies on ancient DNA Illumina libraries. Methods Ecol Evol. 6:725–734. [Google Scholar]
- Beleza S, Santos AM, McEvoy B, Alves I, Martinho C, Cameron E, Shriver MD, Parra EJ, Rocha J. 2013. The timing of pigmentation lightening in Europeans. Mol Biol Evol. 30:24–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt G, Haak W, Adler CJ, Roth C, Szécsényi-Nagy A, Karimnia S, Möller-Rieker S, Meller H, Ganslmeier R, Friederich S, et al. 2013. Ancient DNA reveals key stages in the formation of central European mitochondrial genetic diversity. Science 342:257–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Briggs AW, Stenzel U, Johnson PLF, Green RE, Kelso J, Prüfer K, Meyer M, Krause J, Ronan MT, Lachmann M, et al. 2007. Patterns of damage in genomic DNA sequences from a Neandertal. Proc Natl Acad Sci U S A. 104:14616–14621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brotherton P, Endicott P, Sanchez JJ, Beaumont M, Barnett R, Austin J, Cooper A. 2007. Novel high-resolution characterization of ancient DNA reveals C > U-type base modification events as the sole cause of post mortem miscoding lesions. Nucleic Acids Res. 35:5717–5728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busing FMTA, Meijer E, Van Der Leeden R. 1999. Delete- m Jackknife for Unequal m. Stat Comput. 9:3–8. [Google Scholar]
- Cardoso S, Valverde L, Alfonso-Sánchez MA, Palencia-Madrid L, Elcoroaristizabal X, Algorta J, Catarino S, Arteta D, Herrera RJ, Zarrabeitia MT, et al. 2013. The expanded mtDNA phylogeny of the Franco-Cantabrian region upholds the pre-neolithic genetic substrate of Basques. PLoS One 8:e67835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter ML, Buenrostro JD, Valdiosera C, Schroeder H, Allentoft ME, Sikora M, Rasmussen M, Gravel S, Guillén S, Nekhrizov G, et al. 2013. Pulling out the 1%: Whole-genome capture for the targeted enrichment of ancient DNA sequencing libraries. Am J Hum Genet. 93:852–864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavalli-Sforza LL, Menozzi P, Piazza A. 1994. The history and geography of human genes. Princeton (NJ): Princeton University Press. [Google Scholar]
- Damgaard PB, Margaryan A, Schroeder H, Orlando L, Willerslev E, Allentoft ME. 2015. Improving access to endogenous DNA in ancient bones and teeth. Sci Rep. 5:11184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deguilloux M-F, Soler L, Pemonge M-H, Scarre C, Joussaume R, Laporte L. 2011. News from the west: ancient DNA from a French megalithic burial chamber. Am J Phys Anthropol. 144:108–118. [DOI] [PubMed] [Google Scholar]
- Der Sarkissian C, Ermini L, Jónsson H, Alekseev AN, Crubezy E, Shapiro B, Orlando L. 2014. Shotgun microbial profiling of fossil remains. Mol Ecol. 23:1780–1798. [DOI] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Li H, Moorjani P, Jay F, Slepchenko SM, Bondarev AA, Johnson PLF, Aximu-Petri A, Prüfer K, de Filippo C, et al. 2014. Genome sequence of a 45,000-year-old modern human from western Siberia. Nature 514:445–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Mittnik A, Johnson PLF, Bos K, Lari M, Bollongino R, Sun C, Giemsch L, Schmitz R, Burger J, et al. 2013. A revised timescale for human evolution based on ancient mitochondrial genomes. Curr Biol. 23:553–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamba C, Fernández E, Tirado M, Deguilloux MF, Pemonge MH, Utrilla P, Edo M, Molist M, Rasteiro R, Chikhi L, et al. 2012. Ancient DNA from an Early Neolithic Iberian population supports a pioneer colonization by first farmers. Mol Ecol. 21:45–56. [DOI] [PubMed] [Google Scholar]
- Gamba C, Jones ER, Teasdale MD, McLaughlin RL, Gonzalez-Fortes G, Mattiangeli V, Domboróczki L, Kővári I, Pap I, Anders A, et al. 2014. Genome flux and stasis in a five millennium transect of European prehistory. Nat Commun. 5:5257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Garcerà M, Gigli E, Sanchez-Quinto F, Ramirez O, Calafell F, Civit S, Lalueza-Fox C. 2011. Fragmentation of contaminant and endogenous DNA in ancient samples determined by shotgun sequencing; prospects for human palaeogenomics. PLoS One 6:e24161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, Park DJ, Griesemer D, Karlsson EK, Wong SH, et al. 2013. Identifying recent adaptations in large-scale genomic data. Cell 152:703–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haak W, Lazaridis I, Patterson N, Rohland N, Mallick S, Llamas B, Brandt G, Nordenfelt S, Harney E, Stewardson K, et al. 2015. Massive migration from the steppe was a source for Indo-European languages in Europe. Nature 522:207–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofreiter M, Paijmans JLA, Goodchild H, Speller CF, Barlow A, Fortes GG, Thomas JA, Ludwig A, Collins MJ. 2014. The future of ancient DNA: technical advances and conceptual shifts. Bioessays 37:284–293. [DOI] [PubMed] [Google Scholar]
- Itan Y, Powell A, Beaumont MA, Burger J, Thomas MG. 2009. The origins of lactase persistence in Europe. PLoS Comput Biol. 5:e1000491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jónsson H, Ginolhac A, Schubert M, Johnson PLF, Orlando L. 2013. mapDamage2.0: fast approximate Bayesian estimates of ancient DNA damage parameters. Bioinformatics 29:1682–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A, Graefen A, Ball M, Matzas M, Boisguerin V, Maixner F, Leidinger P, Backes C, Khairat R, Forster M, et al. 2012. New insights into the Tyrolean Iceman’s origin and phenotype as inferred by whole-genome sequencing. Nat Commun. 3:698. [DOI] [PubMed] [Google Scholar]
- Kloss-Brandstätter A, Pacher D, Schönherr S, Weissensteiner H, Binna R, Specht G, Kronenberg F. 2011. HaploGrep: a fast and reliable algorithm for automatic classification of mitochondrial DNA haplogroups. Hum Mutat. 32:25–32. [DOI] [PubMed] [Google Scholar]
- Laayouni H, Calafell F, Bertranpetit J. 2010. A genome-wide survey does not show the genetic distinctiveness of Basques. Hum Genet. 127:455–458. [DOI] [PubMed] [Google Scholar]
- Lacan M, Keyser C, Ricaut F-X, Brucato N, Duranthon F, Guilaine J. 2011. Ancient DNA reveals male diffusion through the Neolithic Mediterranean route. Proc Natl Acad Sci U S A. 108:9788–9791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacan M, Keyser C, Ricaut F-X, Brucato N, Tarrús J, Bosch A, Guilaine J, Crubézy E, Ludes B. 2011. Ancient DNA suggests the leading role played by men in the Neolithic dissemination. Proc Natl Acad Sci U S A. 108:18255–18259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazaridis I, Patterson N, Mittnik A, Renaud G, Mallick S, Kirsanow K, Sudmant PH, Schraiber JG, Castellano S, Lipson M, et al. 2014. Ancient human genomes suggest three ancestral populations for present-day Europeans. Nature 513:409–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EJ, Makarewicz C, Renneberg R, Harder M, Krause-Kyora B, Müller S, Ostritz S, Fehren-Schmitz L, Schreiber S, Müller J, et al. 2012. Emerging genetic patterns of the european neolithic: perspectives from a late neolithic bell beaker burial site in Germany. Am J Phys Anthropol. 148:571–579. [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25:1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The Sequence Alignment/Map format and SAMtools. Bioinformatics 25:2078–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgreen S. 2012. AdapterRemoval: easy cleaning of next-generation sequencing reads. BMC Res Notes. 5:337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins H, Oms FX, Pereira L, Pike AWG, Rowsell K, Zilhão J. 2015. Radiocarbon dating the beginning of the Neolithic in Iberia: new results, new problems. J Mediterr Archaeol. 28:105–131. [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20:1297–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer M, Kircher M. 2010 Illumina sequencing library preparation for highly multiplexed target capture and sequencing. Cold Spring Harb Protoc. Available from: http://dx.doi.org/10.1101/prot5448.
- Olalde I, Allentoft ME, Sánchez-Quinto F, Santpere G, Chiang CWK, DeGiorgio M, Prado-Martinez J, Rodríguez JA, Rasmussen S, Quilez J, et al. 2014. Derived immune and ancestral pigmentation alleles in a 7,000-year-old Mesolithic European. Nature 507:225–228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalde I, Lalueza-Fox C. 2015. Modern humans’ paleogenomics and the new evidences on the European prehistory. Sci Technol Archaeol Res. 1:STAR20151120548. [Google Scholar]
- Orlando L, Ginolhac A, Raghavan M, Vilstrup J, Rasmussen M, Magnussen K, Steinmann KE, Kapranov P, Thompson JF, Zazula G, et al. 2011. True single-molecule DNA sequencing of a pleistocene horse bone. Genome Res. 21:1705–1719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. 2006. Population structure and eigenanalysis. PLoS Genet. 2:e190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickrell JK, Pritchard JK. 2012. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 8:e1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, Bakker PIW De, Daly MJ, et al. 2007. PLINK : a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 81:559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW, Orlando L, Metspalu E, et al. 2014. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature 505:87–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. 2009. Reconstructing Indian population history. Nature 461:489–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renfrew C, Bahn P. 1991. Archaeology theories, methods, and practice. New York: Thames and Hudson. [Google Scholar]
- Rohland N, Hofreiter M. 2007. Ancient DNA extraction from bones and teeth. Nat Protoc. 2:1756–1762. [DOI] [PubMed] [Google Scholar]
- Sánchez-Quinto F, Schroeder H, Ramirez O, Avila-Arcos MC, Pybus M, Olalde I, Velazquez AMV, Marcos MEP, Encinas JMV, Bertranpetit J, et al. 2012. Genomic affinities of two 7,000-year-old Iberian hunter-gatherers. Curr Biol. 22:1494–1499. [DOI] [PubMed] [Google Scholar]
- Schroeder H, Ávila-Arcos MC, Malaspinas A-S, Poznik GD, Sandoval-Velasco M, Carpenter ML, Moreno-Mayar JV, Sikora M, Johnson PLF, Allentoft ME, et al. 2015. Genome-wide ancestry of 17th-century enslaved Africans from the Caribbean. Proc Natl Acad Sci U S A. 112:3669–3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seguin-Orlando A, Korneliussen TS, Sikora M, Malaspinas A, Manica A, Moltke I, Westaway M, Lambert D, Khartanovich V, Wall JD, et al. 2014. Genomic structure in Europeans dating back at least 36,200 years. Science 346:1113–1118. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Malmström H, Omrak A, Raghavan M, Valdiosera C, Günther T, Hall P, Tambets K, Parik J, Karl-Göran S, et al. 2014. Genomic diversity and admixture differs for stone-age Scandinavian foragers and farmers. Science 201:786–792. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Malmström H, Raghavan M, Storå J, Hall P, Willerslev E, Gilbert MTP, Götherström A, Jakobsson M. 2012. Origins and genetic legacy of Neolithic farmers and hunter-gatherers in Europe. Science 336:466–469. [DOI] [PubMed] [Google Scholar]
- Skoglund P, Storå J, Götherström A, Jakobsson M. 2013. Accurate sex identification of ancient human remains using DNA shotgun sequencing. J Archaeol Sci. 40:4477–4482. [Google Scholar]
- Walsh S, Chaitanya L, Clarisse L, Wirken L, Draus-Barini J, Kovatsi L, Maeda H, Ishikawa T, Sijen T, de Knijff P, et al. 2014. Developmental validation of the HIrisPlex system: DNA-based eye and hair colour prediction for forensic and anthropological usage. Forensic Sci Int Genet. 9:150–161. [DOI] [PubMed] [Google Scholar]
- Walsh S, Liu F, Wollstein A, Kovatsi L, Ralf A, Kosiniak-Kamysz A, Branicki W, Kayser M. 2013. The HIrisPlex system for simultaneous prediction of hair and eye colour from DNA. Forensic Sci Int Genet. 7:98–115. [DOI] [PubMed] [Google Scholar]
- Whittle A. 1996. Europe in the Neolithic: the creation of new worlds. Cambridge: Cambridge University Press. [Google Scholar]
- Zilhão J. 1993. The spread of agro-pastoral economies across Mediterranean Europe: a view from the far west. J Mediterr Archaeol. 6:5–63. [Google Scholar]
- Zilhão J. 2001. Radiocarbon evidence for maritime pioneer colonization at the origins of farming in west Mediterranean Europe. Proc Natl Acad Sci U S A. 98:14180–14185 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.