Abstract
Although millions of RNA editing events have been reported to modify hereditary information across the primate transcriptome, evidence for their functional significance remains largely elusive, particularly for the vast majority of editing sites in noncoding regions. Here, we report a new mechanism for the functionality of RNA editing—a crosstalk with PIWI-interacting RNA (piRNA) biogenesis. Exploiting rhesus macaque as an emerging model organism closely related to human, in combination with extensive genome and transcriptome sequencing in seven tissues of the same animal, we deciphered accurate RNA editome across both long transcripts and the piRNA species. Superimposing and comparing these two distinct RNA editome profiles revealed 4,170 editing-bearing piRNA variants, or epiRNAs, that primarily derived from edited long transcripts. These epiRNAs represent distinct entities that evidence an intersection between RNA editing regulations and piRNA biogenesis. Population genetics analyses in a macaque population of 31 independent animals further demonstrated that the epiRNA-associated RNA editing is maintained by purifying selection, lending support to the functional significance of this crosstalk in rhesus macaque. Correspondingly, these findings are consistent in human, supporting the conservation of this mechanism during the primate evolution. Overall, our study reports the earliest lines of evidence for a crosstalk between selectively constrained RNA editing regulation and piRNA biogenesis, and further illustrates that such an interaction may contribute substantially to the diversification of the piRNA repertoire in primates.
Keywords: RNA editing, piRNA biogenesis, rhesus macaque, population genetics, whole-genome sequencing, RNA-Seq
Introduction
RNA editing is a core cotranscriptional process through which nucleotides are modified to generate transcript sequence different from that encoded by the genomic DNA. In the past few years, studies of the RNA editing regulation have been accelerated dramatically by the development of next generation sequencing (NGS) technology, which facilitates genome-wide determination and comparison of DNA and RNA sequences in a precise and cost-effective manner (Li et al. 2009; Ju et al. 2011; Bahn et al. 2012; Peng et al. 2012; Bazak et al. 2014; Chen et al. 2014). Despite the consequent revelation of millions of new RNA editing sites in mammals (largely A-to-I editing), only dozens of editing sites with recoding potential are known to have functional implications (Li et al. 2009). It remains controversial presently as to whether the immensely larger number of editing sites in noncoding regions (>99.9%) represents functional entity or is merely neutral transcriptional noise (Gommans et al. 2009). To this end, our recent comparative genomics study revealed that these noncoding RNA (ncRNA) editing sites are under evolutionary constraints, lending support to the functional significance of at least a proportion of these sites (Chen et al. 2014). However, the exact biological relevance of these conserved editing events in the noncoding regions remains largely unknown.
Intriguingly, considering the widespread occurrence of RNA editing in repetitive regions (e.g., >95% on primate-specific Alu elements), as well as the testis-biased expression profile of ADAR1 (a member of the adenosine deaminases acting on RNA, or ADAR, family) (Zhang et al. 2013, 2014; Chen et al. 2014), a crosstalk between RNA editing and the germ line-specific, transposons-targeting PIWI-interacting RNA (piRNA) remains a formal but as yet unexplored possibility. piRNAs are a family of small RNA species that was first identified by virtue of association with the PIWI clade of the Argonaute (AGO) proteins (Aravin, Sachidanandam, et al. 2007; Brennecke et al. 2007; Thomson and Lin 2009; Siomi et al. 2011; Luteijn and Ketting 2013). Unlike microRNAs (miRNAs) and other endogenous small interfering RNAs (siRNAs), these regulatory RNA molecules exhibit enormous sequence diversity, a predominantly gonadal expression, a strong bias for uridine at position 1 (1U bias), and unique congregation into genomic regions called piRNA clusters (Aravin et al. 2006). Moreover, no particular secondary structures, such as the stem-loop configuration in miRNA precursors, are detected in regions surrounding mature piRNAs (Seto et al. 2007). piRNAs are further distinct from other cellular small RNAs with respect to their biogenesis pathways—the primary piRNAs are first generated by a Dicer-independent processing from long, single-stranded transcripts transcribed from piRNA clusters, and may subsequently be amplified by a secondary, or “ping-pong,” cycle. In the latter mechanism, transcripts complementary to the primary piRNA sequences are cleaved by the Slicer activity of PIWI proteins, producing new secondary piRNAs that have strong bias for adenosine at the tenth nucleotide (10A bias) and further serve as guides for piRNA amplification (Brennecke et al. 2007; Aravin et al. 2008). Functionally, piRNAs and the PIWI proteins form active piRNA-induced silencing complex, a highly conserved mechanism that targets mobile transposable elements in the germ line. This protective function thus provides defense against genome instability and critically underlies gonad development and organism fertility (Vourekas et al. 2012).
Despite the seemingly straightforward connection between RNA editing and piRNAs, issues such as the restricted expression of these pathways, their distinct association with primate-specific Alu elements, the stringent requirements for high-quality tissue samples across different tissues and individuals, as well as the computational challenges in accurately identifying and verifying these events hamper further understanding of any possible mechanistic interaction between the two regulatory levels in primates.
In this study, we performed this interrogation in rhesus macaque, a close evolutionary relative of human. By combining transcriptome sequencing of multiple tissues from the same animal and its whole-genome sequencing, we deciphered accurate RNA editome across both long transcripts and the piRNA species, and further uncovered editing-bearing piRNA variants (epiRNAs). These epiRNAs are primarily processed from edited long transcripts, representing the regions where the RNA editing regulations potentially intersect piRNA biogenesis and diversify the piRNA repertoire in primates. Our population genetics analyses in human and rhesus macaque populations further showed that these epiRNA-associated RNA editing events are under selective constraints, providing the earliest clues for the functionality of such an editing–piRNA crosstalk in primates.
Results
Accurate and Quantitative Catalogs of RNA Editome and piRNAome in Primates
Considering the widespread occurrence of RNA editing in repetitive regions and the testis-enriched expression profile of ADAR1 (Chen et al. 2014), we speculated a link of RNA editing to the germ cell-specific piRNA regulation. To consider this possibility, we first profiled an accurate and more comprehensive RNA editome in rhesus macaque, by refining our previously reported RNA editing calling pipeline (Chen et al. 2014) and applying it on the seven-tissue (prefrontal cortex, cerebellum, heart, kidney, lung, muscle, and testis), poly(A)-positive RNA-Seq data of an rhesus macaque animal (100MGP-001) and its whole-genome resequencing data (tables 1 and 2, fig. 1, and see Materials and Methods). In total, 274,054 candidate editing sites were identified by this transcriptome-wide approach (http://www.rhesusbase.org/download/RNAedit/rna_edit_info.xlsx, last accessed September 12, 2015). Seventy-three of the 78 randomly selected candidate sites (93.6%) were experimentally verified by polymerase chain reaction (PCR) amplification and Sanger sequencing of both DNA and the corresponding cDNA (supplementary fig. S1, Supplementary Material online). The high validation rate suggested that most of the sites identified by the refined identification pipeline are verifiable (supplementary fig. S1, Supplementary Material online). In addition, multiple features of these candidate sites further supported that they represent bona fide RNA editing events mediated by ADARs (Ramaswami et al. 2012; Chen et al. 2014): 1) Predominant representation of the A-to-G conversion (98.2%, or 269,087 editing sites) (fig. 2A), 2) prevalent association with the Alu repeat elements (270,985 of 274,054, or 98.9%) (http://www.rhesusbase.org/download/RNAedit/rna_edit_info.xlsx, last accessed September 12, 2015), 3) a conserved local sequence context (fig. 2B), and 4) quantitative correspondence of the tissue-biased profile of the RNA editome to the expression of ADARs (fig. 2C, and see Materials and Methods) (Li and Church 2013; Chen et al. 2014).
Table 1.
Statistics of the RNA-Seq Data Used in This Study.
| Sample | Total Reads (M) | Length | Q20 (%) | Mapped (uniquely mapped [%]) Reads (%) | Reference |
|---|---|---|---|---|---|
| Small RNA-Seq | |||||
| 100MGP-001 Testis | 94.0 | 49 bp | 100 | 85 (69) | This study |
| 100MGP-001 Prefrontal cortex | 58.2 | 49 bp | 100 | 83 (17) | This study |
| 100MGP-001 Cerebellum | 62.2 | 49 bp | 100 | 92 (16) | This study |
| 100MGP-001 Heart | 61.1 | 49 bp | 100 | 81 (11) | This study |
| 100MGP-001 Kidney | 62.6 | 49 bp | 100 | 91 (19) | This study |
| 100MGP-001 Lung | 57.8 | 49 bp | 100 | 83 (18) | This study |
| 100MGP-001 Muscle | 81.8 | 49 bp | 100 | 85 (18) | This study |
| 100MGP-002 Testis | 96.2 | 49 bp | 100 | 82 (80) | This study |
| 100MGP-003 Testis | 86.9 | 49 bp | 100 | 80 (70) | This study |
| 100MGP-004 Testis | 86.3 | 49 bp | 100 | 85 (58) | This study |
| Human (A) Testis | 94.8 | 49 bp | 100 | 92 (54) | This study |
| Human (A) Prefrontal cortex | 62.6 | 49 bp | 100 | 93 (22) | This study |
| Human (A) Cerebellum | 57.0 | 49 bp | 100 | 93 (23) | This study |
| Human (A) Heart | 70.1 | 49 bp | 100 | 91 (23) | This study |
| Human (A) Kidney | 71.2 | 49 bp | 100 | 92 (25) | This study |
| Human (A) Lung | 74.6 | 49 bp | 100 | 94 (15) | This study |
| Human (A) Muscle | 80.0 | 49 bp | 100 | 93 (17) | This study |
| Human (B) Testis | 74.3 | 52 bpa | 100 | 83 (71) | This study |
| 1411H | 68.7 | 51 bp | 98 | 88 (24) | This study |
| Poly(A)-positive RNA-Seq | |||||
| 100MGP-001 Prefrontal cortex | 142.1 | 90 bp × 2 | 97 | 86 (90) | Chen et al. (2014) |
| 100MGP-001 Cerebellum | 129.0 | 90 bp × 2 | 96 | 87 (93) | Chen et al. (2014) |
| 100MGP-001 Heart | 123.7 | 90 bp × 2 | 97 | 79 (80) | Chen et al. (2014) |
| 100MGP-001 Kidney | 95.7 | 90 bp × 2 | 97 | 84 (86) | Chen et al. (2014) |
| 100MGP-001 Lung | 113.6 | 90 bp × 2 | 97 | 89 (96) | Chen et al. (2014) |
| 100MGP-001 Muscle | 120.0 | 90 bp × 2 | 97 | 82 (91) | Chen et al. (2014) |
| 100MGP-001 Testis | 100.7 | 90 bp × 2 | 97 | 87 (93) | Chen et al. (2014) |
| 100MGP-002 Testis | 128.4 | 100 bp × 2 | 98 | 88 (85) | This study |
| 100MGP-003 Testis | 109.6 | 100 bp × 2 | 98 | 87 (83) | This study |
| 100MGP-004 Testis | 104.6 | 100 bp × 2 | 100 | 84 (80) | This study |
| 1411H | 61.9 | 51 bp | 99 | 75 (80) | This study |
| 1411H siADARb | 47.1 | 51 bp | 99 | 75 (85) | This study |
| 1411H Mockc | 54.7 | 51 bp | 99 | 80 (83) | This study |
aMedian length of reads by Ion Torrent sequencing.
b1411H cells transfected with ADAR1 siRNAs.
c1411H cells transfected with siRNA negative control.
Table 2.
Statistics of the Whole-Genome Sequencing Data Used in This Study.
| Sample | Total Reads (M) | Length | Total Bases | Q30 (%) | Mapped (uniquely mapped [%]) Reads (%) | Reference |
|---|---|---|---|---|---|---|
| 100MGP-001 Prefrontal cortex | 2,718.4 | 90 bp × 2 | 244.7G | 90 | 94 (82) | This study and Chen et al. (2014) |
| 100MGP-002 Blood | 905.5 | 151 bp × 2 | 135.8G | 90 | 91 (81) | This study |
| 100MGP-003 Blood | 786.3 | 151 bp × 2 | 117.9G | 89 | 91 (81) | This study |
| 100MGP-004 Blood | 1,542.3 | 151 bp × 2 | 230.5G | 90 | 91 (81) | This study |
| 100MGP-005 Blood | 1,643.5 | 151 bp × 2 | 246.5G | 89 | 91 (81) | This study |
| 100MGP-006 Blood | 1,450.3 | 151 bp × 2 | 215.7G | 92 | 91 (82) | This study |
| 100MGP-007 Blood | 1,704.2 | 151 bp × 2 | 255.4G | 89 | 91 (81) | This study |
| 100MGP-008 Blood | 1,303.5 | 151 bp × 2 | 194.2G | 90 | 91 (81) | This study |
| 100MGP-009 Blood | 711.8 | 151 bp × 2 | 106.8G | 90 | 91 (81) | This study |
| 100MGP-010 Blood | 627.6 | 151 bp × 2 | 94.1G | 90 | 92 (82) | This study |
| 100MGP-011 Blood | 695.5 | 151 bp × 2 | 104.3G | 90 | 91 (81) | This study |
| 100MGP-012 Blood | 614.5 | 151 bp × 2 | 92.2G | 89 | 91 (82) | This study |
| 100MGP-013 Blood | 736.2 | 151 bp × 2 | 110.4G | 90 | 91 (81) | This study |
| 100MGP-014 Blood | 704.4 | 151 bp × 2 | 105.7G | 89 | 91 (81) | This study |
| 100MGP-015 Blood | 679.2 | 151 bp × 2 | 101.9G | 90 | 91 (81) | This study |
| 100MGP-016 Blood | 697.1 | 151 bp × 2 | 104.6G | 90 | 91 (81) | This study |
| 100MGP-017 Blood | 706.3 | 151 bp × 2 | 105.9G | 89 | 91 (81) | This study |
| 100MGP-018 Blood | 826.2 | 151 bp × 2 | 123.9G | 90 | 91 (81) | This study |
| 100MGP-019 Blood | 892.8 | 151 bp × 2 | 133.9G | 90 | 90 (81) | This study |
| 100MGP-020 Blood | 728.3 | 151 bp × 2 | 109.2G | 89 | 90 (81) | This study |
| 100MGP-021 Blood | 855.2 | 151 bp × 2 | 128.0G | 90 | 91 (82) | This study |
| 100MGP-022 Blood | 676.5 | 151 bp x 2 | 101.5G | 89 | 91 (81) | This study |
| 100MGP-023 Blood | 706.0 | 151 bp × 2 | 105.9G | 90 | 90 (80) | This study |
| 100MGP-024 Blood | 750.9 | 151 bp × 2 | 112.6G | 90 | 91 (82) | This study |
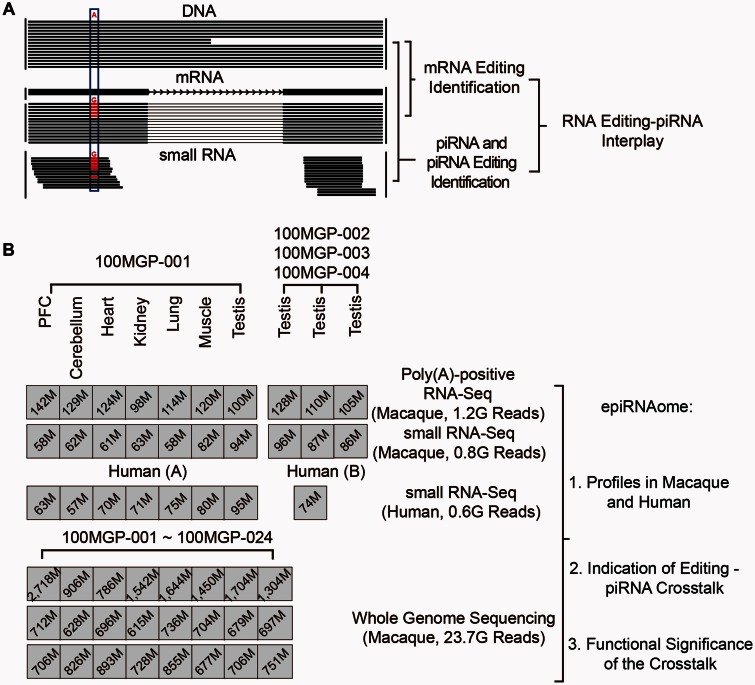
Fig. 1.
Genome-wide investigation of the crosstalk between RNA editing and piRNA regulation. (A) A schematic diagram of the principles and experimental design of this study, aimed to interrogate the RNA editing–piRNA crosstalk in primates. The blue box highlights the detection of editing site. (B) NGS data sets of rhesus macaque and human used in this study are summarized and shown with the respective numbers of total deep sequencing reads.
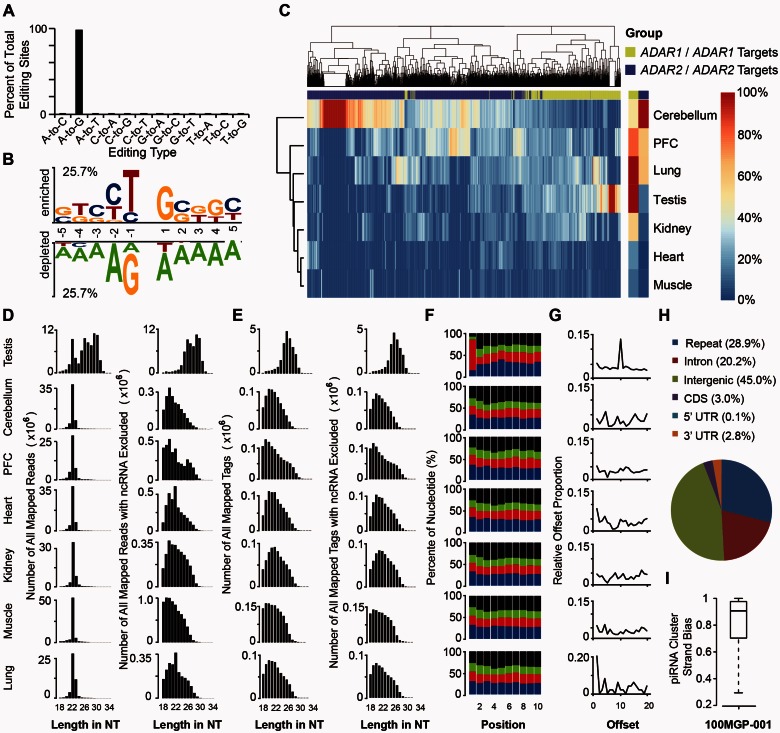
Fig. 2.
Accurate catalogs of RNA-editing sites and piRNAome profile in rhesus macaque. (A) Relative representation of RNA editing types in the macaque transcriptome. (B) The enriched (upper) and depleted (lower) nucleotide sequences flanking the focal editing sites are shown by TwoSample Logo, with the level of preference or depletion presented in height proportional to the scale. (C) Hierarchical clustering of RNA editing levels of ADARs-associated editing sites across different tissues from the same animal. RNA editing levels, as well as the expression levels of ADARs, were estimated on the basis of the poly(A)-positive RNA-Seq data, and the relative levels are shown in colors in relation to the color scale (right). Editing sites were further categorized into ADAR1-associated (green) or ADAR2-associated (purple), according to the tissue distributions of editing levels (see Materials and Methods). PFC, prefrontal cortex. The histograms show the length distribution of reads or tags in different macaque tissue types, before (D) or after (E) the exclusion of annotated ncRNA sequences. PFC, prefrontal cortex. (F) Nucleotide distribution (%) of the first 10 nt at the 5′-end of the candidate piRNAs is plotted for small RNAs in different macaque tissue types. A, U, C, and G are shown in blue, red, green, and black, respectively. (G) For each head-to-head overlapping piRNA pair, the length of sequence complementarity (Offset) was calculated. The distributions of the “Offset” values in all seven tissues are shown. (H) Pie chart showing the genomic distribution of piRNAs in different sequence regions. (I) The proportion of piRNAs exhibiting the same strand orientation as the corresponding piRNA cluster, based on the 100MGP-001 data set.
We further set out to identify and characterize the piRNA repertoire in rhesus macaque, by performing high-quality small RNA deep sequencing on the corresponding tissues of the same animal (MGP-001) (fig. 1 and table 1). After excluding small RNAs mapped to the annotated ncRNAs in rhesus macaque (see Materials and Methods), a class of small RNAs with length ranging from 24 to 32 bp was observed specifically in testis (fig. 2D and E), represented by 58,571,712 reads (or 24,121,526 unique tags; see Materials and Methods). These small RNAs verified known features of piRNAs in mammals, including testis-exclusive tissue distribution (fig. 2D and E), 5′ uridine bias for the nucleotide composition (fig. 2F), the signature of the ping-pong biogenesis mechanism (fig. 2G) (Aravin, Sachidanandam, et al. 2007; Brennecke et al. 2007; Yan, Hu, et al. 2011), an overrepresentation in intergenic regions (fig. 2H) (Vourekas et al. 2012), as well as the clustered distribution of the small RNAs with identical transcriptional orientation as the long transcripts across the region (fig. 2I and supplementary table S1, Supplementary Material online) (Girard et al. 2006).
To facilitate cross-species comparative analyses, we also performed small RNA-Seq for the corresponding seven tissues from human. piRNAs and piRNA clusters with similar features were identified accordingly (table 1 and supplementary table S1 and fig. S2, Supplementary Material online). Together, the informative editome and piRNA profiles established across multiple tissues from the same animal, as well as the corresponding data in human, constitute the basis for systematic investigation of the relationship between the two layers of RNA regulation in the context of primate evolution.
epiRNAs: A New Class of piRNAs Processed from Edited Long Transcripts
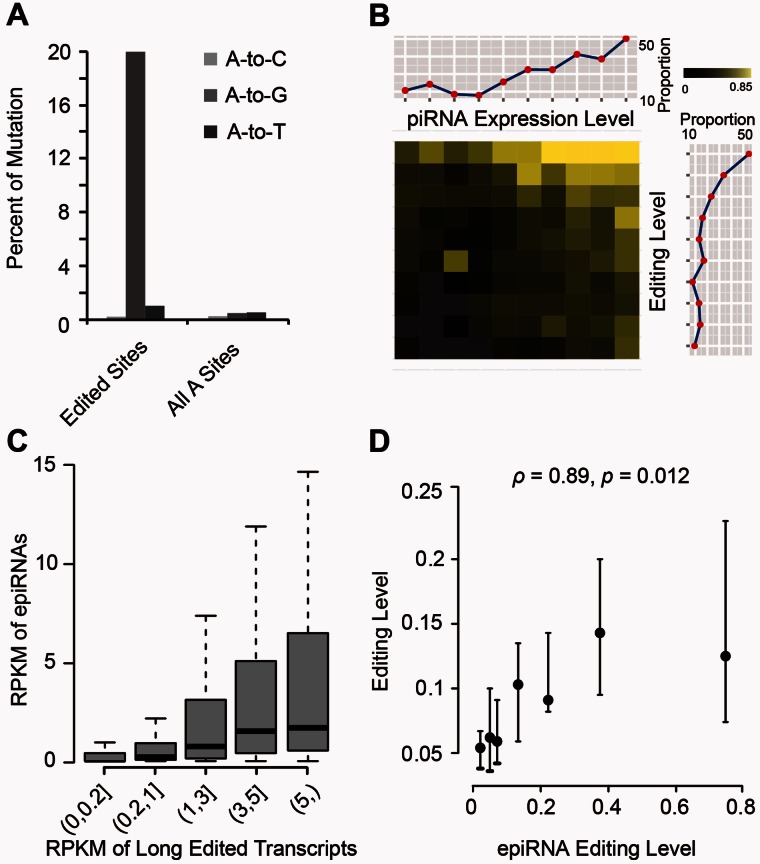
To assess whether RNA editing is associated with piRNAs, we first examined their positional overlap by comparing the genomic locations of both the poly(A)-positive RNA-associated editing sites and the uniquely mapped piRNAs, which presumably represent the loci of piRNA origin. Interestingly, 7,758 A-to-G mRNA editing sites were found to reside in the origin loci of piRNAs, of which 6,357 sites (81.9%) are transcribed in the same strand as piRNAs. Among these sites, small RNA-Seq reads further corroborated A-to-G variation on piRNAs at 1,243 positions, with a total of 3,038 piRNA reads (or 2,150 piRNA tags) harboring these variations (supplementary table S2, Supplementary Material online). Across these piRNA sequences, overrepresentation of the A-to-G variation was evident specifically at the mRNA-edited positions, but not the nonedited positions, suggesting that these variations were derived from A-to-G editing rather than technical errors (fig. 3A).
Fig. 3.
Evidence for the derivation of epiRNAs from edited long transcripts. (A) Using mRNA-associated editing sites as the reference, nucleotide variants were identified for the piRNAs. Prevalence of the indicated mutation types at the editing location (Edited Sites) and at any “A” sites on piRNAs (All A Sites) is shown. (B) The probability of detecting RNA editing sites on piRNAs was assessed in relation to other quantitative sequence features. Macaque piRNAs whose locations overlap with mRNA-associated editing sites were selected and divided into groups on the basis of two attributes—the expression levels of piRNA and the editing levels of corresponding sites identified by poly(A)-positive RNA-Seq; their distributions accordingly are shown in the dot line plots on top and left, respectively. The percentage of piRNAs in each bin that were found to harbor editing sites was calculated and represented in the central heatmap. The degree of detection probability is shown in colors in correspondence to the color scale, with missing data in gray. (C) The expression levels of piRNAs (in RPKM) are depicted in boxplots, as binned by the expression levels of the corresponding long transcripts (also in RPKM). (D) piRNAs were binned according to editing levels as estimated by piRNA reads, and plotted against the median editing levels of the corresponding events identified by poly(A)-positive RNA-Seq, with bars representing the 95% confidence interval estimated by 1,000 bootstrap samplings. For the correlation in editing frequencies between the two data sets, the Spearman’s rank correlation coefficient and P value were calculated.
Although a total of 3,038 uniquely mapped epiRNAs were identified by this initial approach, the size of epiRNA repertoire was likely to be underestimated due to bias related to reads mapping. For instance, considering the pervasive clustering editing (multiple sites in the vicinity), identification of derived epiRNAs with ≥2 editing sites might be limited due to the excess of mismatches in short reads alignment (see Materials and Methods). To address this issue, we adapted a “bisulfite-seq-mapping”-like approach as previously reported (Porath et al. 2014; Zhao et al. 2015) to remap all piRNA reads against a customized macaque genome—with macaque genome sequences being selectively modified to G at the RNA-Seq-supported editing positions and incorporated with all the possible combinations of the edited sites within clusters (see Materials and Methods)—and subsequently recovered 1,132 previously unmappable reads (or 707 piRNA tags) with unique alignment on the genome. In sum, on top of the 3,038 uniquely mapped epiRNAs (termed Group 1), at least 1,132 additional small RNAs are likely candidate epiRNAs (Group 2). We thus combined the two groups of epiRNAs for the subsequent analyses. Nevertheless, due to technical challenges in variant calling from short reads, this collection may still not sufficiently represent the true assembly of cellular epiRNAs (see Discussion).
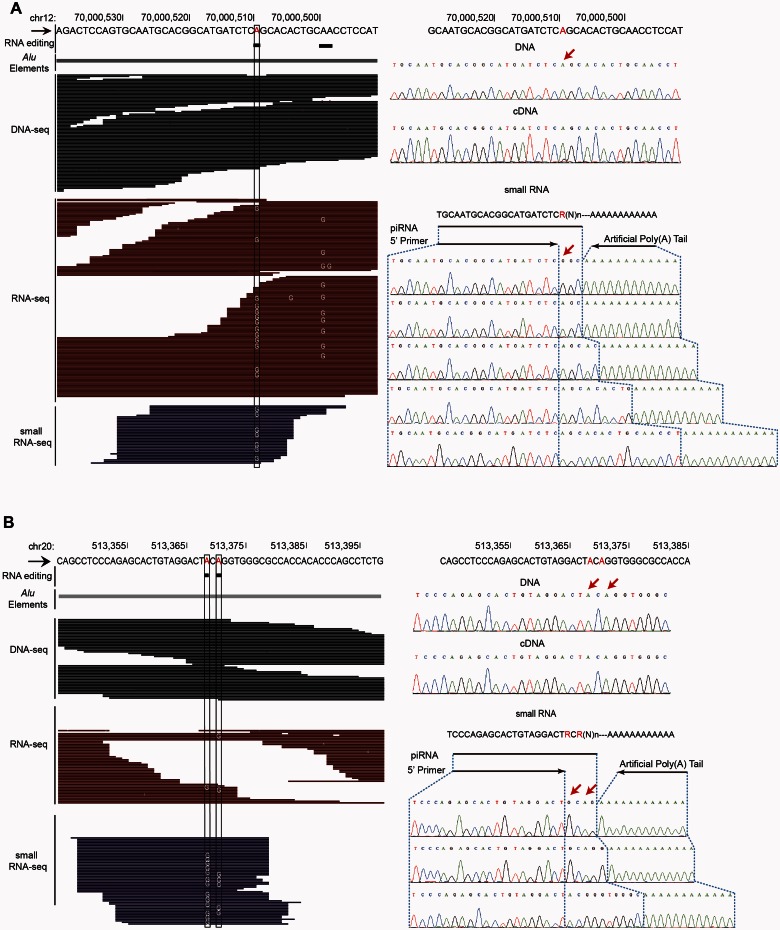
As a proof of concept, we performed independent experimental verification of the epiRNAs by amplifying and sequencing randomly selected epiRNAs and their corresponding gDNA and cDNA regions in macaque samples. For example, editing position on chr12:70000511 (rheMac2) was verified to be homozygous A allele in the gDNA sample, whereas in the corresponding cDNA sample it was heterozygous with a G allele at 21.1% frequency. This was in close agreement with the genome sequencing and poly(A)-positive RNA-Seq data (fig. 4A). Small RNAs were also amplified and cloned, and further subjected to sequencing according to a small RNA-specific verification approach (see Materials and Methods). Our data subsequently supported the existence of both epiRNAs and the corresponding wild-type piRNA (fig. 4A). We also confirmed the existence of another epiRNA spanning clustered editing sites (fig. 4B).
Fig. 4.
Experimental verification of epiRNAs. (A, B) For two selected epiRNA candidates, the deep sequencing raw data as well as the Sanger sequencing results corresponding to the genome, mRNA and small RNA in the macaque animal (100MGP-001) testis sample are shown. The detected editing sites are highlighted by black box (in deep sequencing) or red arrows (in Sanger sequencing).
As mature piRNAs are structurally unsuitable for ADARs binding, editing detected on these epiRNAs is most likely transmitted from the precursor transcripts that are targeted by ADARs prior to processing. Several lines of evidence further supported this notion: 1) The possibility of observing the edited allele “G” on piRNAs is largely accounted for (67.8%) by using features of the piRNA abundance and the editing levels on the long edited transcripts, as estimated by the poly(A)-positive RNA-Seq reads (fig. 3B and see Materials and Methods); 2) the abundance of epiRNAs were in accordance with the expression levels of the corresponding long transcripts (fig. 3C), and the editing levels estimated by short piRNA reads were closely commensurate with those of the corresponding long transcripts, as estimated by the poly(A)-positive RNA-Seq reads (fig. 3D); and 3) for long transcripts with clustered editing sites (multiple editing within a 32-bp window), similar combinatorial distributions of editing were detected on the corresponding piRNA reads, an observation that also implies an editing-elicited diversification of piRNA sequences (supplementary table S3, Supplementary Material online).
The above findings are also correspondingly consistent in human (supplementary fig. S3 and table S2, Supplementary Material online). Taken together, our studies for the first time verified experimentally the existence of epiRNAs in primates, and further showed that they may represent piRNAs generated from previously edited long transcripts. These findings, although not unexpected given the pervasive distribution of A-to-I RNA editing on long RNA transcripts, are actually nontrivial due to multiple technical challenges intrinsic to this type of analyses in primates (see Discussion). Considering the relatively small number of identified epiRNAs, it is then essential to discriminate next whether these epiRNAs represent infrequently degradation fragments of the edited long transcripts, or a functional crosstalk between RNA editing and piRNA biogenesis during the primate evolution.
Existence of epiRNAs Signifies a Regulatory Crosstalk between RNA Editing and piRNA Biogenesis
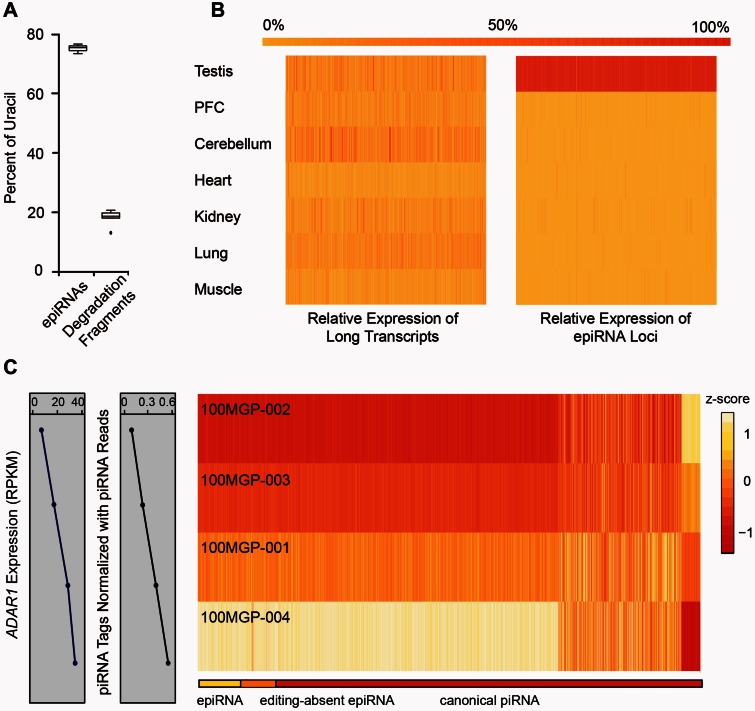
As these epiRNAs are mainly transmitted from the precursor transcripts that are targeted by ADARs prior to processing, they may actually be derived from degradation fragments of the edited long transcripts. To rule out the possibility, we further characterized them in comparison with small RNA-Seq reads detected in other somatic tissues with no definite ncRNA annotations, which potentially are degradation products. These small RNA-Seq reads did not exhibit 5′ uridine bias but showed a strong correlation in tissue expression profile with the corresponding parental transcripts (fig. 5A and supplementary fig. S4, Supplementary Material online, and see Materials and Methods). Conversely, the epiRNAs had strong sequence preference of uridine on the 5′ end (78.0%) and a largely testis-specific presence, irrespective of the tissue expression profiles of the corresponding long transcripts (fig. 5A and B). These observations thereby confirmed that epiRNAs are generated specifically rather than through nonselective degradation.
Fig. 5.
Interaction of RNA editing and piRNA biogenesis. (A) The plot shows the percentages of epiRNAs and mRNA degradation fragments with the nucleotide uridine at the 5′-end of the respective sequences. (B) Heatmaps showing the relative expression levels across seven different macaque tissues for long transcripts (left) and small RNAs (right) corresponding to the epiRNA-associated regions, with reference to the color scale on top. (C) The differences in ADAR expression levels and normalized piRNA tag types across four different animals are shown in linear graphs on the left. The heatmap on the right depicts the relative expression levels of piRNAs in each piRNA cluster of the four animals, organized and scaled in rows. These piRNA clusters were categorized into three groups: epiRNAs-expressing clusters (yellow), “editing-absent” epiRNA clusters (overlapping with editing sites on the long transcripts but lack detectable epiRNAs; orange), and canonical piRNA clusters (red), as indicated by the color bar on top.
Upon establishing the piRNA nature of these epiRNAs, we then set out to investigate whether such a crosstalk between RNA editing and piRNA biogenesis in primates could have functional implications at a global level. To this end, we performed whole-genome sequencing, mRNA-Seq and small RNA-Seq for three other macaque testis samples (table 1 and supplementary tables S1 and S2 and figs. S2 and S3, Supplementary Material online), and subsequently compared the piRNAomes and RNA editomes among the four macaque animals. Of note, for the epiRNA-associated RNA editing sites in the 100MGP-002 animal, 75.5% of the sites were also detectable on piRNAs in the other three macaque animals. This interindividual conservation of epiRNAs thus provided additional independent confirmation for the existence of these epiRNAs in vivo. Furthermore, we noted that the overall type of piRNA tags and piRNA abundance (at comparable sequencing depths) largely corresponded to individual differences in the expression levels of ADAR1 (fig. 5C), which represents the major adenosine deaminase in primate testis as revealed by the tissue expression profiles in primates (fig. 2C) (Zhang et al. 2013, 2014), by the strong quantitative correspondence between RNA editing level and ADAR1 expression level across individuals (supplementary fig. S5, Supplementary Material online), as well as by the ADAR1 knockdown assay in a testis-origin cell line (supplementary fig. S6 and table S4, Supplementary Material online). In fact, the type of piRNA tags in animal with the highest ADAR1 expression was 4.5-fold higher than that with the lowest ADAR1 expression (at comparable sequencing depths) (fig. 5C). In line with this finding, for 87% of these piRNA clusters, the relative expression levels were correlated with ADAR1 expression (Spearman Correlation Coefficient > 0.5).
Given the small representation of epiRNAs in the total pool (<0.1%), the proportionally increased abundance of the piRNA pools may not be contributed mainly by the epiRNAs themselves. Interestingly however, the epiRNAs exhibited a larger margin of variation in piRNA tag types in parallel with increased ADAR1 expression (13.4-fold vs. 4.5-fold). In addition, a larger proportion of the clusters with epiRNAs exhibited abundance closely commensurate with ADAR1 expression than that of the total piRNA clusters (98% vs. 87%, fig. 5C). Even for a class of “editing-absent” piRNAs, which have positional overlap with the editing sites on the long transcripts but lack detectable RNA editing on piRNA due to limited sensitivity of the current sequencing depth (fig. 3B and supplementary fig. S7, Supplementary Material online), the majority also showed high correlation between the piRNA expression and the RNA editing activity (95% vs. 87%, fig. 5C). These correlations provide the initial clues that RNA editing may be a causal mechanism for regulating and diversifying the piRNA repertoire in rhesus macaque (see Discussion).
Considering the difficulty in performing ADAR1 knockdown assay in primate models with piRNA regulation (see Discussion), we then focused our analysis on the public small RNA-Seq data from adar mutant C. elegans to further corroborate the causal link between RNA editing and piRNA biogenesis (Warf et al. 2012). Similar with the original report, we noted that the total piRNA reads were decreased by approximately 20% and approximately 40% in adar-1(−/−) and adar-1(−/−);adar-2(−/−) strains as compared with the wild type (supplementary fig. S8A, Supplementary Material online). For majority of piRNA loci (68.7%), the expression levels decreased in adar mutants compared with wild-type strains (supplementary fig. S8B and C, Supplementary Material online). In addition, we identified seven differentially expressed epiRNAs, all of which were downregulated in adar mutant worms (supplementary table S5, Supplementary Material online). These findings thus hinted at a direct crosstalk between ADARs and piRNA biogenesis in worms, as well as its conservation between evolutionarily divergent species.
epiRNA-Associated RNA Editing Events Are under Selective Constraints in Primates
Given the restricted expression of these regulatory pathways, as well as their distinct association with primate-specific Alu elements, direct elucidation of the functional implications of the link between RNA editing and piRNA biogenesis remains technically challenging. Alternatively, a population genetics approach, that is, characterizing polymorphisms in the epiRNA-associated genomic regions and comparing the pattern to that of the nearby regions as negative control, could potentially provide evolutionary clues to the functional significance of this crosstalk. Our previous study has proposed that for functional editing sites, the regions nearby the focal editing sites should be under selective constraints, owing to the requisite formation of local secondary structures for ADARs recognition and the evolutionary necessity to maintain such a structure for a functional editing event (Chen et al. 2014). Based on this notion, we then performed population genetics analyses to assess whether selective constraints are applied to these epiRNA-associated RNA editing events in the populations of rhesus macaque and human.
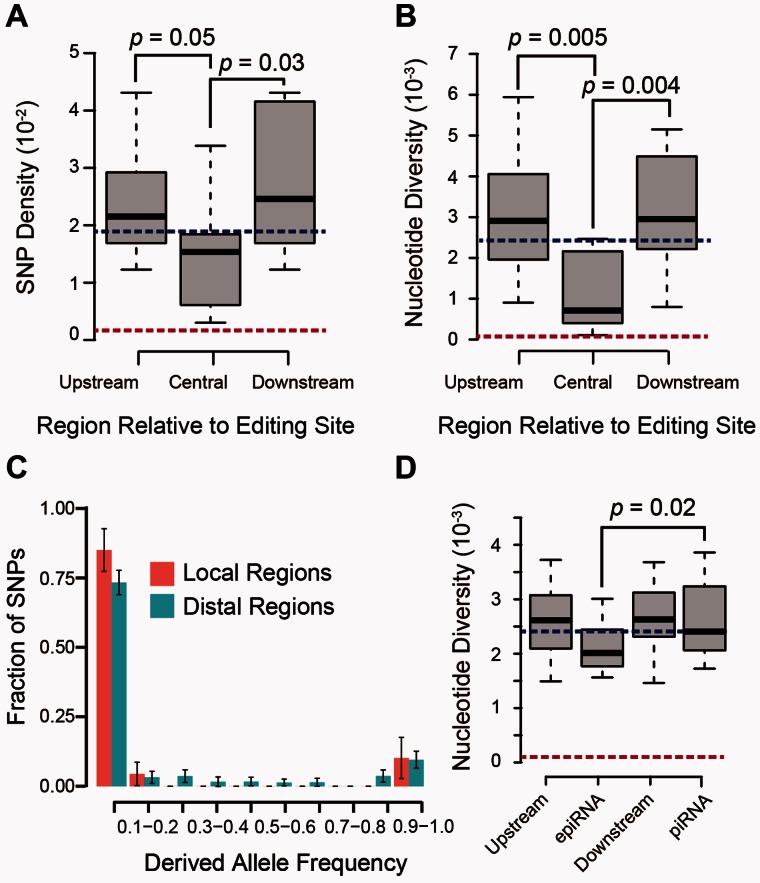
We first profiled a set of polymorphism sites in rhesus macaque populations, on the basis of whole-genome sequencing in 24 independent macaque animals from different subpopulations (see Materials and Methods). Totally, 23.7 billion paired-end reads were generated with high quality, of which 19.2 billion reads (81.2%) were uniquely mapped to the macaque genome, yielding high sequencing coverage (ranging from 26- to 70-folds) (table 2 and supplementary table S6, Supplementary Material online). Utilizing these deep sequencing data (a total sequencing output of 3,382 Gb), as well as seven public data sets for macaque genomes (Fang et al. 2011; Yan, Zhang, et al. 2011; Gokcumen et al. 2013), we profiled 54,079,575 single nucleotide polymorphic sites across the macaque genome (see Materials and Methods). To examine the functional significance of the epiRNA-associated RNA editing regulation, we then compared the distributions of macaque polymorphic sites in epiRNA regions adjacent to the focal editing sites with those in more distal regions (see Materials and Methods). As the adjacent and distal regions are both located within the same Alu element, they should have the same rate of mutation accumulation at a neutral expectation. However, our analysis evidenced substantially lower single nucleotide polymorphism (SNP) density and nucleotide diversity for regions adjoining the editing foci, thus demonstrating the presence of purifying selection on these genomic regions (supplementary fig. S9, Supplementary Material online).
However, considering the established role of piRNAs in repressing transposable elements in primates, these sequence features might be attributed to a positional effect that is potentially contributed by the underlying selectively constrained piRNAs (supplementary fig. S10, Supplementary Material online). To rule out this possibility, a subset of RNA editing sites with the 5′ and 3′ 4-bp nearby regions distributed in the central region of epiRNAs were selected for further analyses (n = 325; see Materials and Methods). Interestingly, sequences nearby the focal editing sites exhibited substantially decreased SNP density and nucleotide diversity (fig. 6A and B, Wilcoxon one-tailed test, P = 0.05, 0.03; 0.005, 0.004, respectively). The population genetics parameters of these RNA editing sites fell between those of the synonymous and nonsynonymous sites, thus indicating a moderate level of purifying selection on these sites (fig. 6A and B). Furthermore, in terms of the frequency spectra of derived allele, we discovered an excess of low-frequency mutations for editing-proximal regions in comparison with the distal piRNA regions as a control (fig. 6C).
Fig. 6.
Population genetics analysis of epiRNA-associated RNA editing events. Extent of selective constraints for epiRNA-associated editing events was evaluated as described in Materials and Methods. The distribution of SNP densities (A), SNP nucleotide diversity (B), and the frequency spectra of derived allele (C) in the local regions of epiRNA-associated focal editing sites were determined and quantitatively represented as boxplots or means ± SD, with the more distal regions (Upstream and Downstream) as the references. (D) The nucleotide diversity of epiRNAs, their flanking regions (Upstream and Downstream), and a group of 10,000 randomly selected piRNAs in piRNA clusters are shown in boxplots. Of note, the same measurements for all synonymous and nonsynonymous sites were calculated as references, with the median values denoted by the blue and red dotted lines, respectively. All P values were derived from Wilcoxon one-tailed test.
Notably, compared with canonical piRNAs (i.e., editing is absent on both piRNA and the associated mRNA), epiRNAs seemed to be under stronger selective constraints in rhesus macaque, probably due to the existence of these functional editing sites (fig. 6D, Wilcoxon one-tailed test, P = 0.02). Considering that epiRNAs with higher expression levels seemed to be under stronger selective constraints (supplementary fig. S11A, Supplementary Material online), we further investigated whether their abundance might contribute to the different strengths detected for the selection signal. However, we did not observe significant difference in the expression levels between epiRNAs and the canonical piRNAs (Wilcoxon one-tailed test, P = 0.806, supplementary fig. S11B, Supplementary Material online). More importantly, even the lowly expressed epiRNAs were under yet stronger selective constraints than the canonical piRNAs (supplementary fig. S11A, Supplementary Material online). These findings thus strengthen the notion that existence of these functional editing events, rather than the expression levels, contributes to the detection of stronger selective constraints on epiRNAs versus canonical piRNAs.
Taken together, these population genetics attributes should lend support to the functionality of these epiRNA-associated RNA editing events in rhesus macaque. Moreover, given that these regions are rarely linked to other gene regulatory processes such as splicing or gene expression, a piRNA-associated regulation represents a plausible scenario for these RNA editing events. Importantly, all of these findings are correspondingly consistent in human, further supporting the conservation of this mechanism during the primate evolution (supplementary fig. S12, Supplementary Material online). Our results thus provide the earliest evolutionary clues to the functionality of the crosstalk between RNA editing regulation and piRNA biogenesis in primates.
Discussion
An Intersection between RNA Editing Regulation and piRNA Biogenesis in Primates
Although a substantial number of editing sites have been identified across primate transcriptome, evidence for functional significance of this process is largely lacking, particularly for the overwhelmingly large group of editing sites in noncoding regions (>99.9% of total). To this end, a growing body of evidence has linked RNA editing to the small ncRNA species of miRNAs, alterations of which are known to have developmental and pathological implications (Pfeffer et al. 2005; Singh 2012; Shoshan et al. 2015). Interestingly, RNA editing regulation shares multiple spatial features with another noncoding component of the transcriptome, the piRNA-based regulation—both processes target the Alu elements, and the main enzymes for both regulatory pathways show some degree of testis-biased expression (fig. 2C and supplementary fig. S13, Supplementary Material online). Considering these shared spatial features, a link between these two regulatory mechanisms might conceivably exist. Further considering the pervasive distribution of A-to-I RNA editing in the primate genome, it is rational to speculate the existence of edited piRNAs that are derived from Alu sequences, identification of which could further substantiate the crosstalk between the two regulatory levels.
However, it is technically challenging to directly test the hypothesis, in part due to the restricted expression of these pathways, the intensive association of these regulations with primate-specific Alu elements, the stringent requirements for high-quality tissue samples across different tissues and individuals, as well as the computational challenges in accurately identifying and verifying these regulations. Exploiting the emerging primate model of rhesus macaque, our present study presents to our knowledge the first documentation of “epiRNAs” in primates, a new class of piRNA that serves as evidence for the RNA editing–piRNAs interplay.
Consistently with the reports that piRNA-like small RNAs exist in other nongermline tissues (Yan, Hu, et al. 2011), we also detected a class of small RNAs with the length of 24–32 bp in the macaque somatic tissues. However, these small RNAs only represented a minor proportion of all small RNA reads (fig. 2D), a profile that is also consistent with the previous characterization of piRNA-like small RNAs in somatic tissue (Yan, Hu, et al. 2011). Moreover, considering that these piRNA-like small RNAs do not verify the known features of canonical piRNAs in our study (fig. 2D), and that PIWI proteins were hardly expressed in these somatic tissues (supplementary fig. S13, Supplementary Material online), we focused our studies on the link of RNA editing to the germ cell piRNA regulation. Nevertheless, our present data do not fully exclude the possibility that RNA editing regulation may also crosstalk with these somatic piRNA-like small RNAs.
RNA Editing Regulation May Diversify the piRNA Repertoire in Primates
The observed correlations between the piRNAome and RNA editome in rhesus macaque, together with the additional evidence in C. elegans ADAR-KO model (supplementary fig. S8, Supplementary Material online), provide initial clues that RNA editing regulations may crosstalk with piRNA biogenesis in multiple species. However, unequivocal demonstration of this causal relationship in primates remains challenging, mainly owing to the testis-selective nature of these regulations. Due to the restricted expression of piRNA-based pathway and limited availability of corresponding cell models, further clarification of the functional implications of these epiRNAs in primates may rely on tools such as CRISPR/Cas9-based transgenic monkeys (Niu et al. 2014), which now represent effective means of genetic functional assay.
We noted that the number of piRNA tag types in animal with the highest ADAR1 expression was 4.5-fold higher than that with the lowest ADAR1 expression (fig. 5C). However, based on the small number of epiRNAs detected in this study (<0.1% of the total pool), it seems plausible that these editing events may not directly contribute to the diversity and increased abundance of piRNA repertoire in correlation with ADAR1 expression (fig. 5C). There are two possible explanations to these seemingly contradictory observations. First, although 4,170 epiRNAs have been defined in this study, this number may still represent an insufficient account of the cellular epiRNA repertoire owing to the following reasons: 1) For the presumably “editing-absent” piRNA clusters that share positional overlap with RNA editing sites in the long transcripts but do not express epiRNA variants, a fraction may actually represent bona fide epiRNAs that were undetected due to the limited sensitivity provided by the current sequencing depth (fig. 3B and supplementary fig. S7, Supplementary Material online); 2) this underestimation could also be contributed by the ambiguity in small RNA sequencing reads alignment—many small RNA reads corresponding to putative epiRNAs were mapped to multiple positions across transcriptome and thus excluded initially; and 3) as calling mRNA editing events in primates is as yet a refined approach (Bazak et al. 2014), the complexity and contribution of epiRNAome may be far more extensive than expected from the current analysis. Second, the piRNA clusters that give rise to epiRNAs may represent the initial sites of priming reaction for a genome-wide regulation on piRNA biogenesis in primates. It is thus possible that these piRNAs triggered by the RNA editing regulation, including the editing-associated epiRNAs, may further activate canonical piRNA loci across the genome through the “ping-pong” biogenesis mechanism. In this regard, experimental characterization of the true genomic region distribution of piRNA targets is currently an unresolved challenge, mainly due to the difficulty in obtaining high-quality PIWI ChIP-seq data sets (Marinov et al. 2015). More efforts are thus needed to elucidate the regulatory landscape of these epiRNA. Taken together, our results are in line with the notion that piRNA-associated editing sites, which are possibly underestimated in the present study, likely contribute to the biogenesis and diversification of the total piRNA pool.
Link of RNA Editing to the Purifying Selection Signals Detected on epiRNA Regions
Our population genetics analyses showed that the epiRNA-associated RNA editing regulation is under selective constraints in primates (fig. 6 and supplementary fig. S9, Supplementary Material online), strengthening the evolutionary significance, and thus functionality, of these ADAR1-directed regulations across the primate transcriptome. Several observations further support that RNA editing per se is directly associated with these selectively constrained regulatory events. First, the population genetics analysis pinpointed the sequences immediately nearby the focal editing sites as regions with substantially decreased SNP density and nucleotide diversity—a sequence context that implies functional selection for the targeting by the catalytic domain, rather than the RNA binding domain, of the adenosine deaminase. Although it remains likely that these signals of purifying selection may actually correspond to the necessity in maintaining a stable ADAR1 binding to the substrates, we favor a simpler explanation that this selectively constrained regulation is manifested in an editing catalysis-associated functionality. Second, given that these regions with purifying selection signals are rarely linked to other functional regulations, such as splicing or gene expression regulations (Chen et al. 2014), occurrence of RNA editing may thus directly exerts functional impact on these piRNAs.
It has been well established that piRNAs are not conserved across different species, possibly a response to the quick turnover rates of their lineage-specific targets (Lu and Clark 2010). The findings that RNA editing may crosstalk with piRNA biogenesis to diversify the piRNA repertoire, as well as the fact that the crosstalk is maintained by purifying selection in the populations of human or rhesus macaque, infer that these editing–piRNAs interplay may have significant functions in controlling lineage-specific transposable elements.
Redirection of miRNA-Like piRNA Targeting by RNA Editing
Recent studies have revealed that piRNAs may inhibit mRNA expression through a similar mechanism as siRNA-mediated silencing, and that their 5′ regions are important for target recognition (Gou et al. 2014). Of note, for 1,050 epiRNAs identified in rhesus macaque, RNA editing is distributed within the first 10 bp of the piRNA 5′-end. Moreover, 89.6% of these epiRNAs constitute a significant proportion of the expression of the corresponding piRNAs loci (>5%, supplementary fig. S14, Supplementary Material online). This finding is then in line with the scenario that these piRNA-associated RNA editing events may also redirect piRNA targeting to extended Alu subfamilies through sequence pairing (Aravin, Hannon, et al. 2007; Kelleher and Barbash 2013) (supplementary fig. S14, Supplementary Material online), which is analogous to the effects of mutations on miRNA seed regions (Blow et al. 2006).
Materials and Methods
Ethics Statement
Rhesus macaque tissue samples were provided by the internationally accredited (Association for Assessment and Accreditation of Laboratory Animal Care, AAALAC) animal facility at the Institute of Molecular Medicine in Peking University. All animal studies were approved by the Institutional Animal Care and Use Committee of Peking University. Commercial human RNA samples were obtained from Clontech Laboratories, Inc. and Ambion, Inc.
Sample Preparation and Data Analysis for Whole-Genome Sequencing, RNA-Seq, and Small RNA-Seq
Strand-specific, poly(A)-positive RNA-Seq libraries were prepared from seven rhesus macaque tissues (prefrontal cortex, cerebellum, heart, kidney, lung, muscle, and testis) derived from a 5-year-old animal (100MGP-001) as previously reported (Xie et al. 2012; Chen et al. 2014), and testis tissues from three other animals (100MGP-002, 100MGP-003, and 100MGP-004) (table 1). Low molecular weight RNAs with length ranging from 15 to 40 nt were isolated from total RNA of the macaque tissue samples and from the corresponding human RNA samples ordered from Clontech Laboratories and Ambion. These small RNAs were ligated to the adapters, and amplified according to the standard Small RNA Preparation Protocol (Yan, Hu, et al. 2011). For genome resequencing, genomic DNA was obtained from prefrontal cortex of the same animal (100MGP-001), as well as from peripheral blood of 23 other macaque animals (100MGP-002–100MGP-024) (supplementary table S6, Supplementary Material online), for which sample preparation was done as previously reported (Chen et al. 2014). NGS was performed on Illumina HiSeq sequencing systems, with 90/100/151-bp paired-end reads mode, or 49-bp single-end reads mode (tables 1 and 2).
Whole-genome sequencing data in this study were combined with the published genome resequencing data from the same macaque sample (Chen et al. 2014). The genome sequencing and poly(A)-positive RNA-Seq data were then aligned to the rhesus macaque genome (rheMac2) by BWA (v0.5.9-r16), using parameters as outlined in our previous study (Chen et al. 2014). For small RNA sequencing data, reads were collapsed into one tag if they were totally identical. Small RNA tags were then aligned to the genome of human (hg19) or rhesus macaque (rheMac2) using Bowtie (V0.12.8) with one mismatch allowed, according to a pipeline described previously (Yan, Hu, et al. 2011). Deep sequencing data in this study are available at NCBI Gene Expression Omnibus and SRA under accession numbers GSE34426, GSE42857, SRP039366, and SRP049394.
Identification and Characterization of RNA Editing Sites
To identify mRNA-associated editing sites, several improvements were incorporated into our previous pipeline (Chen et al. 2014): 1) On the basis of the recent report on variant calling (Ramaswami et al. 2012), instead of using Samtools pipelines, variants on RNA molecules were directly identified by piling up poly(A)-positive RNA-Seq reads and distinguishing discrepancies between RNA sequences and the corresponding DNA sequences. The sensitivity in variant identification was thus dramatically increased. 2) To deal with potential false positives stemming from sequencing errors or mismapping, only RNA variants that satisfy specified criteria, including the minimal number of supporting reads for variant allele (2/3 for Alu/non-Alu editing sites), minimal allele frequency (0.02/0.10 for Alu/non-Alu editing sites), and required base quality (Phred score ≥ 25), were taken into consideration. 3) Candidate editing sites in non-Alu regions were further excluded if they were located in simple repeats or within a 4-bp intronic regions adjacent to the splicing junctions (Ramaswami et al. 2012). To test whether these editing sites represent bona fide editing sites, 78 candidate sites in testis with editing levels ≥ 10%, which is the approximate detection limit of Sanger sequencing, were randomly selected. PCR amplification and Sanger sequencing of both DNA and cDNA samples from testis tissues were then performed (supplementary fig. S1, Supplementary Material online).
Editing levels were estimated only for sites covered by ≥10 sequencing reads. The normalized RNA editing levels across seven tissues of the same macaque animals were clustered by pheatmap package in R (v3.0.1); and ADARs-associated sites as defined by linear regression model in our previous study (Chen et al. 2014) were used in the hierarchical clustering analyses. Overall, 32.5% and 15.3% of the A-to-G editing sites showed significantly positive correlation with ADAR2 and ADAR1 expression, respectively, in terms of tissue distribution of levels (fig. 2C).
Mapping and Definition of piRNA and piRNA Clusters
We first removed reads derived from annotated ncRNAs by filtering mappable small RNA reads against the ncRNA databases of human or rhesus macaque. Briefly, human miRNA annotated in miRBase (v20) and other ncRNAs (ribosomal RNA, transfer RNA, small nucleolar RNA, small nuclear RNA, small conditional RNA, and misc-RNA) annotated in UCSC and Ensembl were downloaded and integrated into a comprehensive data set for annotated ncRNAs in human. Subsequently, human–macaque pairwise alignment was performed to infer a list of ncRNAs in rhesus macaque, from which alignments of matched macaque sequences having >70% and <130% coverage of the query sequence were retained (Yan, Hu, et al. 2011). A small RNA-Seq read was excluded if any of its mapped locations overlaps with the regions encoding annotated ncRNAs. The remaining 24–32 nt reads were then defined as candidate piRNAs.
We next defined the piRNA clusters in the 100MGP-001 testis. Using a 3-kb-long window with 0.5-kb sliding steps, regions that encompass at least 300 uniquely mapped reads in the same orientation, with at least 60% of the reads also containing 5′ uridine sequence, were identified. Positive sliding windows were merged into a larger piRNA cluster if they located within 1 kb of each other (Yan, Hu, et al. 2011). Similar approach was used to pinpoint the piRNA clusters in other samples, with the cutoff for uniquely mapped reads being optimized according to the total sequencing depth of the respective samples.
We then investigated whether these candidate piRNAs recapitulate the known features of piRNAs, such as 5′ uridine bias, complementary piRNAs sequences as a result of the “ping-pong” biogenesis mechanism, as well as distinctive genomic distributions. Briefly, the nucleotide compositions of the first 10 bp on 5′-end of these candidate piRNAs were determined to assess the extent of 5′ uridine bias (fig. 2F). To assess the contribution of “ping-pong” mechanism to the generation of these candidate piRNAs, we first selected candidate piRNAs with both 5′ uridine (feature of primary piRNA) and adenosine at the tenth position (feature of secondary piRNA). We then used piRNAs with 5′ uridine (presumably the primary piRNAs) as the reference to identify from the remaining candidate piRNAs any complementary piRNAs (putative secondary piRNAs). We then calculated the lengths of the complementary regions and compared the length distribution in different samples, using an approach as previously proposed (Yan, Hu, et al. 2011) (fig. 2G). These candidate piRNAs were then assigned to different types of genomic regions on the basis of RhesusBase (V2) annotations (Zhang et al. 2014). For piRNAs at positions with multiple definitions, they were given a single assignment according to the order of repeat, exon, intron, UTR, and intergenic regions (fig. 2H). For candidate piRNAs located in clusters, the proportions of piRNAs showing the same strand orientation as the corresponding piRNA cluster were also calculated (fig. 2I).
Qualitative and Quantitative Account of epiRNAs
In an effort to pinpoint editing sites associated with the piRNAs, we compared the genomic locations of RNA editing sites defined by the long transcripts and by the uniquely mapped piRNAs. Through this approach, a total of 3,038 piRNA reads (or 2,150 piRNA tags) were identified (termed Group 1). We also remapped unaligned piRNA tags against a customized macaque genome in which the mRNA Seq-supported edited sites were selectively modified to G. To this end, adjacent editing sites within 32 bp were considered to be in the same cluster. Genomic sequences for each cluster, as well as sequences in the 5′ and 3′ adjacent regions, were then retrieved. Some or all of the edited sites in these genome sequences were modified to the nucleotide G—considering all possible combinations of the edited sites within the cluster—to generate multiple sequence contigs. Previously unmapped piRNAs tags were then mapped to these contigs by Bowtie (V0.12.8) with no mismatches allowed. This second read-alignment step yielded additional 1,132 small RNAs that harbor putative RNA editing sites (Group 2).
Candidate epiRNAs from both groups were combined and subjected to further characterization. Editing levels of the identified sites were estimated based on the set of uniquely mapped small RNA-Seq reads, in which annotatable ncRNA reads were excluded (fig. 3D). Linear regression was performed to study the relationship between the probabilities of detecting RNA editing sites on piRNA, and the expression level of piRNA and/or the corresponding editing level in mRNA as estimated by poly(A)-positive RNA-Seq reads. The adjusted R-square values obtained under the regression model were then used to assess the power of predicting the existence of edited allele “G” on piRNAs, on the basis of piRNA abundance and the editing levels on the long edited transcripts (fig. 3B). The tissue expression profiles of epiRNAs and the corresponding long transcripts, calculated on the basis of RPKM values for the 300-bp regions spanning the epiRNAs, were quantitatively determined and compared (Xie et al. 2012) (figs. 3C and 5B). As negative controls, we made use of the tissue expression profiles of small RNA-Seq reads (ncRNA-derived tags excluded) from the somatic tissues (supplementary fig. S4, Supplementary Material online). Annotations for the genomic context and Alu subfamilies were downloaded from the RhesusBase (V2) and used for the definition of the genomic distributions of macaque epiRNAs (fig. 6).
Cell Culture and RNA-Seq Studies of 1411H Cell Line
To experimentally examine the role of ADAR1 on RNA editing sites in testis, and further the functional implications of epiRNAs in cell lines, RNA-Seq studies were performed on the testis-origin 1411H cell line (Public Health England, 06011805). 1411H cells were cultured in complete growth medium DMEM (Hyclone, UT), supplemented with 10% FBS, 100 U/ml penicillin, and 100 µg/ml streptomycin, at 37 °C and 5% CO2. Proliferative cultures were transfected with synthetic single-strand small RNA oligos (GenePharma, Shanghai, China): 1) siRNA negative control (50 nM) and 2) mixture of three ADAR1 siRNAs (50 nM; si-seq1, si-seq2, si-seq3) (supplementary table S4 and fig. S6, Supplementary Material online). These siRNAs were designed for ADAR1 specifically, and their sequences contain multiple mismatches relative to the ADAR2 sequence (supplementary fig. S6D, Supplementary Material online). The silencing efficiency was further assessed by quantitative PCR (qPCR) and Western blotting (supplementary fig. S6A and B, Supplementary Material online). Briefly, total RNA samples for qRT-PCR experiment were prepared using Trizol (Invitrogen), and reverse transcribed into cDNA using SuperScriptTM II Reverse Transcriptase (Invitrogen). qPCR was performed on AB Step One Plus (Applied Biosystems) with ADAR1 primers (ATCAGCGGGCTGTTAGAATATG and AAACTCTCGGCCATTGATGAC). Total protein samples were also extracted using Cell Lysis Buffer (CST) containing PMSF (sigma) and 1× protease inhibitors (Roche) for Western blotting, with antibodies for ADAR1 and GAPDH purchased from Santa Cruz Biotechnology (sc-73408) and EASYBIO (BE0023), respectively. Upon RNA extraction, poly(A)-positive RNA-Seq libraries and small RNA-Seq libraries were also prepared and sequenced on a Hiseq 2500 sequencing system with 51 single-end reads mode.
The orthologous editing sites in human were identified on the basis of pairwise genome alignment between human and rhesus macaque (Karolchik et al. 2014), and sites with the edited form encoded in the human reference genome were excluded. The editing levels were determined only for sites covered by ≥10 sequence reads in both the control and the experimental groups. The expression levels of genes were estimated in the form of RPKM, as shown in our previous study (Xie et al. 2012).
Experimental Verification of epiRNAs
Total RNAs were isolated from frozen testis tissues using Trizol reagent (Invitrogen, CA). Genomic DNA was also obtained from the same tissue samples. Editing sites on the mRNA transcripts were validated by means of PCR amplification and Sanger sequencing of both the DNA and corresponding RNA. For epiRNA verification, 1 µg of total RNA was first polyadenylated by poly(A) polymerase (NEB, MA, UK) in a 20-µl reaction mixture at 37 °C for 1 h, and subsequently isolated by phenol–chloroform extraction and ethanol precipitation. Then, 0.5 µg 3′ RACE adapter GCGAGCACAGAATTAATACGACTCACTATAGGT12VN and 200 U SuperScriptTM II Reverse Transcriptase (Invitrogen) were used to reversely transcribe the poly(A) tail-added small RNA molecules (Shi and Chiang 2005). Using the resultant cDNAs as template, specific epiRNA sequences were amplified by EasyTaq DNA polymerase (TransGen), with piRNA-specific 5′ primers (TGCAATGCACGGCATGATCTC in fig. 4A or TCCCAGAGCACTGTAGGACT in fig. 4B) and the 3′ RACE adapter outer primer (GCGAGCACAGAATTAATACGACT). PCR products were then cloned in T1-vector for Sanger sequencing.
Population Genetics Analysis
Whole-genome sequencing data of seven macaque animals (Fang et al. 2011; Yan, Zhang, et al. 2011; Gokcumen et al. 2013) downloaded from SRA (SRA023856, SRA037810, and ERP002376), together with the whole-genome sequencing data of 24 independent macaque animals generated in this study, were mapped to the macaque genome (rheMac2) by BWA (0.7.10-r789). Macaque polymorphism sites in this population of 31 animals were then profiled by the standard GATK (V3.2-2) pipelines with Unified Genotyper. To examine whether piRNAs and epiRNAs are under purifying selection, we first retrieved sequences of equal lengths that are upstream and downstream to the corresponding piRNAs/epiRNAs, and used them as the references. Both piRNA/epiRNA and the control adjacent regions were then individually divided into 15 subregions for the calculation and comparison of the SNP densities and nucleotide diversity, with any missing alleles substituted with the reference alleles. For the estimation of the frequency spectra of a given derived allele, as defined by the EPO pipeline (Paten et al. 2008), 1,000 times of bootstrap were performed to estimate the confidence intervals of the proportions of polymorphism sites. Similar population genetics analyses were performed to analyze evolutionary significance of the editing sites on epiRNAs, in terms of the SNP densities, nucleotide diversity, and the frequency spectra of derived allele for the focal regions (8-nt regions surrounding the editing sites) versus the reference regions (8-nt sequences upstream and downstream of the focal regions) (fig. 6). To investigate the functionality of epiRNAs in human, we also profiled human polymorphism data of 67 individuals with high sequencing coverage from the 1000 Genomes project (Genomes Project Consortium et al. 2012; Chen et al. 2015), and performed similar population genetics analyses (supplementary fig. S12, Supplementary Material online).
Supplementary Material
Supplementary figures S1–S14 and tables S1–S6 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Supplementary Material
Acknowledgments
The authors thank Dr Heping Cheng and Dr Yangming Wang at Peking University, and Yong E. Zhang at Institute of Zoology, Chinese Academy of Science for insightful suggestions. They acknowledge Dr Jia Yu and Ms Xin Wen at Peking Union Medical College Hospital, Ms Shuxian Li and Meng Fang at Peking University for assistance in the study. This work was supported by grants from the National Key Basic Research Program of China (2013CB531202, 2012CB518004) and the National Natural Science Foundation of China (31471240, 31522032, 31171269, 31221002). This work was also supported by the Ministry of Science and Technology of Taiwan (MOST101-2320-B-182-036-MY3 and MOST103-2321-B-182-002), the National Health Research Institute of Taiwan (NHRI-EX104-10321SI), and Chang Gung Memorial Hospital (CMRPD1C0841) to B.C.-M.T. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- Aravin A, Gaidatzis D, Pfeffer S, Lagos-Quintana M, Landgraf P, Iovino N, Morris P, Brownstein MJ, Kuramochi-Miyagawa S, Nakano T, et al. 2006. A novel class of small RNAs bind to MILI protein in mouse testes. Nature 442:203–207. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Hannon GJ, Brennecke J. 2007. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 318:761–764. [DOI] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Bourc’his D, Schaefer C, Pezic D, Toth KF, Bestor T, Hannon GJ. 2008. A piRNA pathway primed by individual transposons is linked to de novo DNA methylation in mice. Mol Cell. 31:785–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aravin AA, Sachidanandam R, Girard A, Fejes-Toth K, Hannon GJ. 2007. Developmentally regulated piRNA clusters implicate MILI in transposon control. Science 316:744–747. [DOI] [PubMed] [Google Scholar]
- Bahn JH, Lee JH, Li G, Greer C, Peng G, Xiao X. 2012. Accurate identification of A-to-I RNA editing in human by transcriptome sequencing. Genome Res. 22:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazak L, Haviv A, Barak M, Jacob-Hirsch J, Deng P, Zhang R, Isaacs FJ, Rechavi G, Li JB, Eisenberg E, et al. 2014. A-to-I RNA editing occurs at over a hundred million genomic sites, located in a majority of human genes. Genome Res. 24:365–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blow MJ, Grocock RJ, van Dongen S, Enright AJ, Dicks E, Futreal PA, Wooster R, Stratton MR. 2006. RNA editing of human microRNAs. Genome Biol. 7:R27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennecke J, Aravin AA, Stark A, Dus M, Kellis M, Sachidanandam R, Hannon GJ. 2007. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 128:1089–1103. [DOI] [PubMed] [Google Scholar]
- Chen JY, Peng Z, Zhang R, Yang XZ, Tan BC, Fang H, Liu CJ, Shi M, Ye ZQ, Zhang YE, et al. 2014. RNA editome in rhesus macaque shaped by purifying selection. PLoS Genet. 10:e1004274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J-Y, Shen QS, Zhou W-Z, Peng J, He BZ, Li Y, Liu C-J, Luan X, Ding W, Li S, et al. 2015. Emergence, retention and selection: a trilogy of origination for functional de novo proteins from ancestral LncRNAs in primates. PLoS Genet. 11e:1005391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang X, Zhang Y, Zhang R, Yang L, Li M, Ye K, Guo X, Wang J, Su B. 2011. Genome sequence and global sequence variation map with 5.5 million SNPs in Chinese rhesus macaque. Genome Biol. 12:R63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genomes Project Consortium, Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. 2012. An integrated map of genetic variation from 1,092 human genomes. Nature 491:56–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Girard A, Sachidanandam R, Hannon GJ, Carmell MA. 2006. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature 442:199–202. [DOI] [PubMed] [Google Scholar]
- Gokcumen O, Tischler V, Tica J, Zhu Q, Iskow RC, Lee E, Fritz MH, Langdon A, Stutz AM, Pavlidis P, et al. 2013. Primate genome architecture influences structural variation mechanisms and functional consequences. Proc Natl Acad Sci U S A. 110:15764–15769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gommans WM, Mullen SP, Maas S. 2009. RNA editing: a driving force for adaptive evolution? Bioessays 31:1137–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gou LT, Dai P, Yang JH, Xue Y, Hu YP, Zhou Y, Kang JY, Wang X, Li H, Hua MM, et al. 2014. Pachytene piRNAs instruct massive mRNA elimination during late spermiogenesis. Cell Res. 24:680–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju YS, Kim JI, Kim S, Hong D, Park H, Shin JY, Lee S, Lee WC, Yu SB, Park SS, et al. 2011. Extensive genomic and transcriptional diversity identified through massively parallel DNA and RNA sequencing of eighteen Korean individuals. Nat Genet. 43:745–752. [DOI] [PubMed] [Google Scholar]
- Karolchik D, Barber GP, Casper J, Clawson H, Cline MS, Diekhans M, Dreszer TR, Fujita PA, Guruvadoo L, Haeussler M, et al. 2014. The UCSC Genome Browser database: 2014 update. Nucleic Acids Res. 42:D764–D770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher ES, Barbash DA. 2013. Analysis of piRNA-mediated silencing of active TEs in Drosophila melanogaster suggests limits on the evolution of host genome defense. Mol Biol Evol. 30:1816–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Church GM. 2013. Deciphering the functions and regulation of brain-enriched A-to-I RNA editing. Nat Neurosci. 16:1518–1522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li JB, Levanon EY, Yoon JK, Aach J, Xie B, Leproust E, Zhang K, Gao Y, Church GM. 2009. Genome-wide identification of human RNA editing sites by parallel DNA capturing and sequencing. Science 324:1210–1213. [DOI] [PubMed] [Google Scholar]
- Lu J, Clark AG. 2010. Population dynamics of PIWI-interacting RNAs (piRNAs) and their targets in Drosophila. Genome Res. 20:212–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luteijn MJ, Ketting RF. 2013. PIWI-interacting RNAs: from generation to transgenerational epigenetics. Nat Rev Genet. 14:523–534. [DOI] [PubMed] [Google Scholar]
- Marinov GK, Wang J, Handler D, Wold BJ, Weng Z, Hannon GJ, Aravin AA, Zamore PD, Brennecke J, Toth KF. 2015. Pitfalls of mapping high-throughput sequencing data to repetitive sequences: Piwi’s genomic targets still not identified. Dev Cell. 32:765–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. 2014. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 156:836–843. [DOI] [PubMed] [Google Scholar]
- Paten B, Herrero J, Beal K, Fitzgerald S, Birney E. 2008. Enredo and Pecan: genome-wide mammalian consistency-based multiple alignment with paralogs. Genome Res. 18:1814–1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng Z, Cheng Y, Tan BC, Kang L, Tian Z, Zhu Y, Zhang W, Liang Y, Hu X, Tan X, et al. 2012. Comprehensive analysis of RNA-Seq data reveals extensive RNA editing in a human transcriptome. Nat Biotechnol. 30:253–260. [DOI] [PubMed] [Google Scholar]
- Pfeffer S, Sewer A, Lagos-Quintana M, Sheridan R, Sander C, Grasser FA, van Dyk LF, Ho CK, Shuman S, Chien M, et al. 2005. Identification of microRNAs of the herpesvirus family. Nat Methods. 2:269–276. [DOI] [PubMed] [Google Scholar]
- Porath HT, Carmi S, Levanon EY. 2014. A genome-wide map of hyper-edited RNA reveals numerous new sites. Nat Commun. 5:4726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramaswami G, Lin W, Piskol R, Tan MH, Davis C, Li JB. 2012. Accurate identification of human Alu and non-Alu RNA editing sites. Nat Methods. 9:579–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seto AG, Kingston RE, Lau NC. 2007. The coming of age for Piwi proteins. Mol Cell. 26:603–609. [DOI] [PubMed] [Google Scholar]
- Shi R, Chiang VL. 2005. Facile means for quantifying microRNA expression by real-time PCR. Biotechniques 39:519–525. [DOI] [PubMed] [Google Scholar]
- Shoshan E, Mobley AK, Braeuer RR, Kamiya T, Huang L, Vasquez ME, Salameh A, Lee HJ, Kim SJ, Ivan C, et al. 2015. Reduced adenosine-to-inosine miR-455-5p editing promotes melanoma growth and metastasis. Nat Cell Biol. 17:311–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh M. 2012. Dysregulated A to I RNA editing and non-coding RNAs in neurodegeneration. Front Genet. 3:326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siomi MC, Sato K, Pezic D, Aravin AA. 2011. PIWI-interacting small RNAs: the vanguard of genome defence. Nat Rev Mol Cell Biol. 12:246–258. [DOI] [PubMed] [Google Scholar]
- Thomson T, Lin H. 2009. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu Rev Cell Dev Biol. 25:355–376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vourekas A, Zheng Q, Alexiou P, Maragkakis M, Kirino Y, Gregory BD, Mourelatos Z. 2012. Mili and Miwi target RNA repertoire reveals piRNA biogenesis and function of Miwi in spermiogenesis. Nat Struct Mol Biol. 19:773–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warf MB, Shepherd BA, Johnson WE, Bass BL. 2012. Effects of ADARs on small RNA processing pathways in C . elegans. Genome Res. 22:1488–1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie C, Zhang YE, Chen JY, Liu CJ, Zhou WZ, Li Y, Zhang M, Zhang R, Wei L, Li CY. 2012. Hominoid-specific de novo protein-coding genes originating from long non-coding RNAs. PLoS Genet. 8:e1002942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan G, Zhang G, Fang X, Zhang Y, Li C, Ling F, Cooper DN, Li Q, Li Y, van Gool AJ, et al. 2011. Genome sequencing and comparison of two nonhuman primate animal models, the cynomolgus and Chinese rhesus macaques. Nat Biotechnol. 29:1019–1023. [DOI] [PubMed] [Google Scholar]
- Yan Z, Hu HY, Jiang X, Maierhofer V, Neb E, He L, Hu Y, Hu H, Li N, Chen W, et al. 2011. Widespread expression of piRNA-like molecules in somatic tissues. Nucleic Acids Res. 39:6596–6607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Liu CJ, Shi M, Kong L, Chen JY, Zhou WZ, Zhu X, Yu P, Wang J, Yang X, et al. 2013. RhesusBase: a knowledgebase for the monkey research community. Nucleic Acids Res. 41:D892–D905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SJ, Liu CJ, Yu P, Zhong X, Chen JY, Yang X, Peng J, Yan S, Wang C, Zhu X, et al. 2014. Evolutionary interrogation of human biology in well-annotated genomic framework of rhesus macaque. Mol Biol Evol. 31:1309–1324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao HQ, Zhang P, Gao H, He X, Dou Y, Huang AY, Liu XM, Ye AY, Dong MQ, Wei L. 2015. Profiling the RNA editomes of wild-type C . elegans and ADAR mutants. Genome Res. 25:66–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.