Abstract
North American populations of Drosophila melanogaster derive from both European and African source populations, but despite their importance for genetic research, patterns of ancestry along their genomes are largely undocumented. Here, I infer geographic ancestry along genomes of the Drosophila Genetic Reference Panel (DGRP) and the D. melanogaster reference genome, which may have implications for reference alignment, association mapping, and population genomic studies in Drosophila. Overall, the proportion of African ancestry was estimated to be 20% for the DGRP and 9% for the reference genome. Combining my estimate of admixture timing with historical records, I provide the first estimate of natural generation time for this species (approximately 15 generations per year). Ancestry levels were found to vary strikingly across the genome, with less African introgression on the X chromosome, in regions of high recombination, and at genes involved in specific processes (e.g., circadian rhythm). An important role for natural selection during the admixture process was further supported by evidence that many unlinked pairs of loci showed a deficiency of Africa–Europe allele combinations between them. Numerous epistatic fitness interactions may therefore exist between African and European genotypes, leading to ongoing selection against incompatible variants. By focusing on hubs in this network of fitness interactions, I identified a set of interacting loci that include genes with roles in sensation and neuropeptide/hormone reception. These findings suggest that admixed D. melanogaster samples could become an important study system for the genetics of early-stage isolation between populations.
Keywords: Drosophila melanogaster, Drosophila Genetic Reference Panel, admixture, population ancestry, linkage disequilibrium, reference alignment
Introduction
North American populations of Drosophila melanogaster have had a disproportionate role in classical and modern Drosophila genetics (Kohler 1994). They gave rise to many of the commonly used laboratory strains that came from the T. H. Morgan lab and elsewhere. More recently, the Drosophila Genetic Reference Panel (DGRP; Mackay et al. 2012; Huang et al. 2014) introduced a set of 205 sequenced genomes from independent inbred lines collected from Raleigh, NC. The DGRP has become a widely used resource for analyses of genomic variation and its relation to phenotype. Understanding the demographic history of the DGRP and other North American populations is important for maximizing the scientific value of these genetic resources. However, D. melanogaster is not native to the western hemisphere, and the recently arrived New World populations of this species appear to have complex origins.
Drosophila melanogaster originated from sub-Saharan Africa (Lachaise et al. 1988) and probably from southern-central Africa in particular (Pool et al. 2012), where at some unknown time it became associated with human settlement. The species began to expand its geographic range, initially occupying more diverse environments within sub-Saharan Africa. On the order of 10,000 years ago, D. melanogaster managed to cross the Saharan region and expand into northern Africa and Eurasia (Lachaise et al. 1988; Baudry et al. 2004; Thornton and Andolfatto 2006). That expansion entailed a significant loss of genetic diversity, perhaps as a result of founder event population bottlenecks, with the consequence that populations from outside sub-Saharan Africa hold only a subset of the variation observed in the ancestral range (Pool et al. 2012).
The expansion of D. melanogaster from tropical Africa into temperate Old World regions appears to have had important consequences regarding adaptation to novel environments and the restriction of migration between African and non-African populations (note that here and below, “African” refers to sub-Saharan populations specifically). Tropical and temperate populations have a range of morphological differences (Capy et al. 1993), and the genomic search for loci that may encode adaptive differences between these populations has been a topic of significant interest (e.g., Kauer et al. 2002; Pool et al. 2012). Partial sexual isolation has also been reported between African and non-African strains: Female flies from African strains were found to discriminate strongly against non-African males (Wu et al. 1995; Hollocher et al. 1997; Yukilevich and True 2008).
North American populations of D. melanogaster are thought to derive from both European and African source populations. Initial evidence for this dual ancestry came from three population genetic observations from North American versus European populations. First, North American populations appeared more genetically similar to sub-Saharan populations than Eurasian populations were (Caracristi and Schlötterer 2003; Baudry et al. 2004; Haddrill et al. 2005; Nunes et al. 2008). Second, these same three studies all indicated that North American populations have higher genetic diversity than European populations, an observation that seems incompatible with a simple European origin for North American populations. And third, a North American population was found to have elevated linkage disequilibrium relative to a European population (Haddrill et al. 2005), which is likewise consistent with recent admixture in North America. Concordantly, Duchen et al. (2013) compared DGRP genomes against multilocus sequence data from African and European populations, and estimated the DGRP population to contain 15% African ancestry.
Recent population genomic analysis has confirmed the above evidence. For example, simple calculations based on genome-wide nucleotide diversity and genetic distance among African, European, and North American populations (Lack et al. 2015) suggest appreciable African ancestry levels in the DGRP population (supplementary table S1, Supplementary Material online). Furthermore, the recent studies of Kao et al. (2015) and Bergland et al. (2015) estimated ancestry proportions in multiple New World populations. Their findings support the existence of a latitudinal cline of ancestry, with African ancestry levels being relatively low in the northeast United States and intermediate in the southeast United States and Caribbean. Such a pattern may have resulted from a history in which European populations initially colonized the northeast United States, whereas African populations first reached the Caribbean (David and Capy 1988; Keller 2007), with subsequent geographic expansion and interbreeding leading to admixed populations.
This study began with the simple aim of evaluating and reporting population ancestry along DGRP genomes to bolster the interpretation of genetic and phenotypic variation in this widely studied population. Genomic patterns of ancestry suggested that natural selection had pervasively influenced the admixture process in this North American population.
In particular, I tested for “ancestry disequilibrium” (AD) between unlinked loci and detected a genome-wide influence of epistatic selection on genetic variation, consistent with ongoing selection against incompatible combinations of African and European variants (Box 1). In addition to the DGRP, I also estimated ancestry along the D. melanogaster reference genome, which may have implications for the performance of reference alignment for sequenced D. melanogaster genomes.
Box 1. Key to Abbreviations Used in This Study.
AD Ancestry disequilibrium—the correlation of population ancestry between loci. Analogous to linkage disequilibrium, but calculated using inferred ancestries rather than specific genotypes.
AD cluster A pair of genomic regions that contain one or more window pairs with strong ancestry disequilibrium.
AD hub A set of neighboring windows that overlap an unusually large number of AD clusters. These genomic regions are hotspots for ancestry disequilibrium, and may experience interlocus fitness interactions with a number of unlinked loci.
BDMI Bateson–Dobzhansky–Muller incompatibilities. Fitness may be compromised when variants from previously isolated populations are brought into contact by admixture.
IFI Interlocus fitness interaction. AD may indicate an IFI, and a BDMI is a potential explanation for an IFI.
DGRP Drosophila Genetic Reference Panel
DPGP Drosophila Population Genomics Project
DSPR Drosophila Synthetic Population Resource
Results
Genome-Wide Ancestry Proportions for DGRP and the Timing of Admixture
European versus African ancestry along North American D. melanogaster genomes was assessed using the method of Pool et al. (2012). This Hidden Markov Model (HMM) approach operates in genomic windows, comparing a focal genome with a European reference panel of genomes. For each window, it tests whether the focal genome’s genetic distance to the European reference panel resembles that expected from a non-African focal genome, or whether instead it matches the higher genetic distance expected from comparing an African focal genome against the European reference panel (Pool et al. 2012).
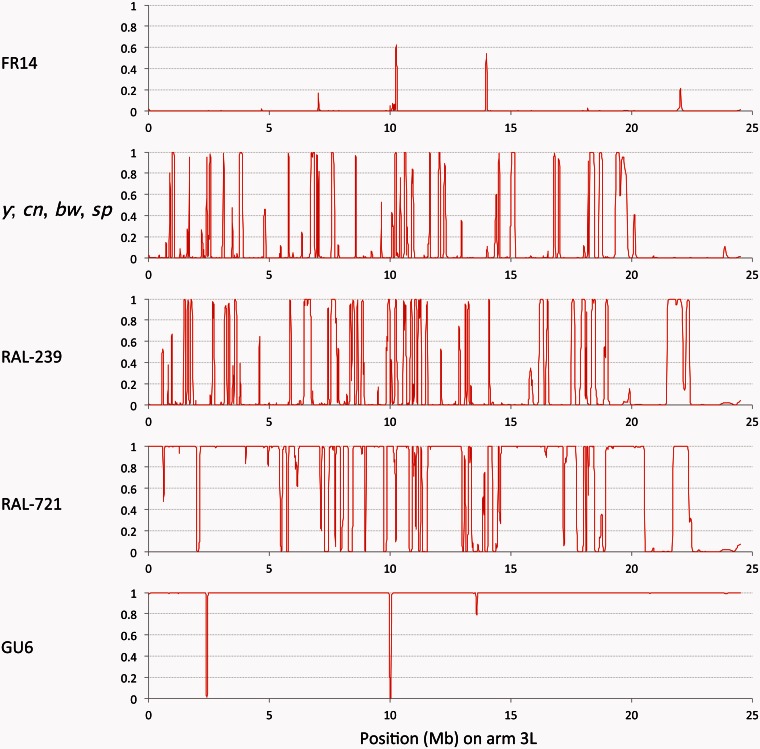
This study utilized relatively shorter windows than previously used (averaging approximately 5 kb, but scaled by local levels of diversity) to detect a somewhat less recent admixture event. However, results were highly consistent with those previously obtained from approximately 50-kb windows. Figure 1 includes representative examples of a European genome (FR14) and an African genome (GU6) that were previously estimated to contain no introgression on chromosome arm 3L (Pool et al. 2012). With approximately 5-kb windows, only a couple very narrow intervals of putative admixture were indicated. These intervals could indicate genuine introgression that was too narrow for the wider intervals to pick up, but even if they are incorrect, they suggest a very low rate of false detection of introgressed ancestry segments.
Fig. 1.
Sample ancestry likelihood plots from chromosome arm 3L show that the proportion of sub-Saharan ancestry depends on geography and inversion status. FR14 is a European genome and shows almost no putative African ancestry. y; cn, bw, sp is the Drosophila melanogaster reference genome; it shows peaks of African ancestry probability, often on the order of 100 kb. The DGRP genome RAL-239 shows a greater density of African ancestry tracts, but these are of similar length as for the reference genome. Unlike the other genomes shown here, RAL-721 carries In(3L)P, an inversion of sub-Saharan origin; this arm shows predominantly African ancestry with mostly narrow intervals of non-African origin. GU6 is a western African genome and shows almost exclusively sub-Saharan ancestry.
By comparison, applying this ancestry HMM method to the 205 DGRP genomes revealed abundant evidence of admixture, with each chromosome arm containing a mosaic of European and African ancestry (fig. 1). From inversion-free chromosome arms, 24,034 African ancestry tracts of at least 0.05 cM were free of large-scale missing data. Based on recombination rate estimates of Comeron et al. (2012), these tracts had a median length of 0.173 cM. Using an admixture tract length simulation approach (Pool and Nielsen 2009) with appropriate ancestral population proportions, I estimate that this median tract size would be expected after approximately 1,598 generations of admixture (95% CI: 1,548–1,644). The accuracy of this estimate may be affected by demographic details, natural selection, and imprecision in recombination rate estimates. However, it is close to a previous estimate of 1,445 generations, which was based on a subset of the DGRP data (Duchen et al. 2013).
DGRP Ancestry Proportions Are Highly Variable along the Genome
Overall, levels of inferred African ancestry among the DGRP genomes averaged 19.9%, with a standard deviation of 5.5% (supplementary table S2, Supplementary Material online). Examining collective DGRP ancestry for each window, striking genome-wide variability was detected (fig. 1 and supplementary table S3, Supplementary Material online). Although the autosomes carry 23.0% African ancestry, for the X chromosome this average is reduced to 6.5%, and 37.8% of X-linked windows are completely fixed for European ancestry. These X-linked and autosomal ancestry levels mirror simple predictions based on genetic distances (supplementary table S1, Supplementary Material online). Reduced X-linked introgression was also detected by other analyses of North American populations (Kao et al. 2015; Bergland et al. 2015) and African populations (Kauer et al. 2003; Pool et al. 2012).
An outlier for higher African ancestry is chromosome arm 2L (fig. 2), which has 31.1% African ancestry (compared with 23.4%, 18.7%, and 19.8% for arms 2R, 3L, and 3R, respectively). This arm effect is largely explained by the prevalence of inversion In(2L)t (supplementary fig. S1, Supplementary Material online), the most common African-origin inversion in the DGRP (Corbett-Detig and Hartl 2012; Huang et al. 2014). In(2L)t and other inversions can have strong effects on genetic variation across whole chromosome arms (fig. 1) (Corbett-Detig and Hartl 2012; Pool et al. 2012).
Fig. 2.
The proportion of DGRP genomes that have greater than 50% probability of sub-Saharan ancestry in each genomic window is shown for the five major euchromatic chromosome arms (color-coded and labeled above). Genomes lacking at least 500 bp of called sequence within a given window were excluded, and windows with fewer than 50 genomes meeting that criterion were omitted from this plot.
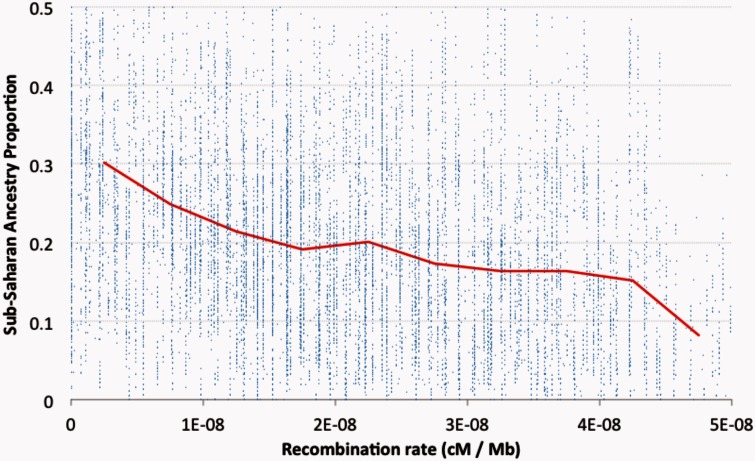
More perplexing than the between-arm ancestry differences are the strong fluctuations observed within chromosome arms, often on the scale of tens or hundreds of kilobases (fig. 2). For each chromosome arm, there is a significant negative correlation (P < 0.0001) between African ancestry and recombination rate, with Pearson r2 of 0.080 for standard autosomal arms analyzed jointly and 0.101 for the X chromosome. The mean sub-Saharan ancestry proportion is 30.2% for autosomal windows below 0.5 cM/Mb, but only 13.0% when the recombination rate is above 4 cM/Mb (fig. 3). This relationship is not expected under a neutral introgression scenario, but might result either from inefficient selection in low recombination regions against African alleles that are disadvantageous in the predominantly European gene pool and North American environment of the DGRP population, or from favored African alleles carrying longer linkage blocks in regions of low recombination.
Fig. 3.
DGRP genomes display much less sub-Saharan ancestry in windows with higher recombination rates. Here, each blue dot represents one autosomal genomic window, whereas the red line indicates the mean sub-Saharan ancestry proportion for bins of 0.5 cM/Mb. Ancestry proportions are from standard chromosome arms only. All autosomal windows with data from at least 50 standard arms were included in statistical analyses. Only windows with recombination rate up to 5 cM/Mb and sub-Saharan ancestry proportion up to 50% are shown in this plot.
Pool et al. (2012) found that admixture approximately 1,000 generations ago between African and European populations of D. melanogaster was detected reliably by the method implemented here, occasionally missing short admixture tracts but not inferring false tracts. Those simulations focused on the autosomes because X-linked admixture will be easier to detect (based on the larger X-linked diversity difference between African and European populations). Hence, lower X-linked admixture levels contradict the predictions of methodological bias. Nor is the recombination result easily explainable by such bias: Windows were scaled to contain similar numbers of polymorphisms regardless of recombination rate, and Europe/Africa diversity ratio is similar across high and low recombination regions of chromosome arms (Pool et al. 2012), so the power to detect admixture should be similar in high and low recombination regions.
Although precise neutral expectations for interlocus variance in ancestry proportion depend on unknown details of the North American colonization scenario, the dramatic and nonrandom variance observed here suggests the possibility that African and European alleles at some loci may have had unequal fitness in North American environments. To investigate which types of genes would be the most likely targets of any such selection, gene ontology (GO) enrichment analysis was performed for intervals of elevated African or European ancestry. The GO categories most enriched for European ancestry included “circadian behavior,” whereas those for African ancestry included “flight behavior” and vision-related categories (table 1 and supplementary table S4, Supplementary Material online). Importantly, this exploratory analysis does not have the power to firmly implicate any specific GO category if the number of tests is considered, but it may help generate hypotheses for downstream studies investigating the biological basis of ancestry deviations. Overall, the observed number of GO categories with raw P values below 0.01 was greater than 95% of genome-wide permutations for the analysis of elevated African ancestry (suggesting a nonrandom set of genes within outlier regions), whereas for elevated European ancestry this figure was 85%.
Table 1.
Biological Process Categories Overrepresented in Genomic Outlier Regions for Elevated African or European Ancestry.
| GO Category | Biological Process | Outlier Regions | Total Windows | P Value |
|---|---|---|---|---|
| High African ancestry | ||||
| GO:0007629 | Flight behavior | 7 | 22 | 0.00010 |
| GO:0008344 | Adult locomotory behavior | 10 | 60 | 0.00082 |
| GO:0071704 | Organic substance metabolic process | 65 | 3,456 | 0.00110 |
| GO:0009589 | Detection of UV | 3 | 5 | 0.00126 |
| GO:0009056 | Catabolic process | 27 | 459 | 0.00126 |
| GO:0042023 | DNA endoreduplication | 5 | 17 | 0.00158 |
| GO:0034059 | Response to anoxia | 3 | 6 | 0.00224 |
| GO:0007010 | Cytoskeleton organization | 27 | 449 | 0.00238 |
| GO:0008152 | Metabolic process | 67 | 4,063 | 0.00266 |
| GO:0007603 | Phototransduction, visible light | 3 | 8 | 0.00294 |
| GO:0097035 | Regulation of membrane lipid distribution | 2 | 2 | 0.00296 |
| GO:0009581 | Detection of external stimulus | 9 | 60 | 0.00356 |
| GO:0022402 | Cell cycle process | 29 | 585 | 0.00454 |
| GO:0070482 | Response to oxygen levels | 8 | 56 | 0.00480 |
| GO:0042811 | Pheromone biosynthetic process | 2 | 3 | 0.00530 |
| GO:0022607 | Cellular component assembly | 28 | 568 | 0.00612 |
| GO:0010324 | Membrane invagination | 16 | 191 | 0.00636 |
| GO:0007617 | Mating behavior | 7 | 34 | 0.00734 |
| GO:0032528 | Microvillus organization | 2 | 3 | 0.00758 |
| GO:0033993 | Response to lipid | 7 | 45 | 0.00774 |
| GO:0016044 | Cellular membrane organization | 19 | 57 | 0.00866 |
| High European ancestry | ||||
| GO:0051090 | Regulation of sequence-specific DNA binding transcription factor activity | 8 | 29 | 0.00018 |
| GO:0007623 | Circadian rhythm | 13 | 77 | 0.00064 |
| GO:0007344 | Pronuclear fusion | 3 | 6 | 0.00150 |
| GO:0045665 | Negative regulation of neuron differentiation | 4 | 10 | 0.00202 |
| GO:0003009 | Skeletal muscle contraction | 2 | 2 | 0.00288 |
| GO:0043620 | Regulation of DNA-dependent transcription in response to stress | 2 | 2 | 0.00294 |
| GO:0006633 | Cuticle hydrocarbon biosynthetic process | 2 | 3 | 0.00304 |
| GO:0006633 | Fatty acid biosynthetic process | 5 | 25 | 0.00334 |
| GO:0007281 | Germ cell development | 11 | 79 | 0.00334 |
| GO:0030178 | Negative regulation of Wnt receptor signaling pathway | 6 | 25 | 0.00350 |
| GO:0042330 | Taxis | 26 | 247 | 0.00364 |
| GO:0006333 | Chromatin assembly or disassembly | 5 | 129 | 0.00396 |
| GO:0015772 | Oligosaccharide transport | 2 | 2 | 0.00502 |
| GO:0042752 | Regulation of circadian rhythm | 8 | 44 | 0.00594 |
| GO:0048284 | Organelle fusion | 5 | 24 | 0.00700 |
| GO:0051241 | Negative regulation of multicellular organismal process | 8 | 46 | 0.00916 |
Note.—Unique biological process GO categories with raw permutation P value less than 0.01 and representation in at least two separate outlier regions are shown here (with full results in supplementary table S2, Supplementary Material online). Some of these categories could reflect targets of selection favoring African or European alleles in North America, but at present they represent hypotheses for further population genetic and functional analysis.
Admixture in the Reference Genome
Using the same methods as described for the DGRP genomes, the D. melanogaster reference genome was estimated to have 9.4% African ancestry (fig. 1 and supplementary table S5, Supplementary Material online). The relatively lesser degree of admixture in this genome, relative to the North Carolina DGRP population, might be expected if 1) this species’ initial colonization of the New World involved an African founder population in the Caribbean and an European founder population in the northeastern United States (David and Capy 1988; Keller 2007), and 2) the reference strain descends primarily from laboratory stocks obtained by the T. H. Morgan laboratory in the northeastern United States (Kennison JA, personal communication).
The reference genome’s segments of African ancestry are correlated with those found in the DGRP (table 2). And like the DGRP, the reference genome is more likely to carry African ancestry in low recombination regions (table 2). Hence, many of the demographic and selective events that molded complex patterns of ancestry in the DGRP may have affected other North American populations as well.
Table 2.
Relationship between Reference Genome Ancestry and DGRP Ancestry or Recombination Rate.
| Chromosome | Reference | Reference |
|---|---|---|
| Arm | Afr. Windows | Eur. Windows |
| DGRP African ancestry proportion | ||
| X | 0.118 | 0.046 |
| 2L | 0.355 | 0.216 |
| 2R | 0.329 | 0.170 |
| 3L | 0.212 | 0.176 |
| 3R | 0.419 | 0.149 |
| Recombination rate (cM/Mb) | ||
| X | 2.75 | 3.39 |
| 2L | 1.79 | 3.00 |
| 2R | 3.06 | 3.28 |
| 3L | 2.12 | 2.29 |
| 3R | 1.59 | 2.48 |
Note.—For each chromosome arm, windows called as either African or European in the reference genome are compared in two respects. First, it is shown that windows called as African in the reference genome have much higher levels of African ancestry in the DGRP. Second, as observed for the DGRP, the reference genome’s African windows have lower recombination rates (based on the estimates of Comeron et al. 2012).
The mosaic ancestry of the D. melanogaster reference genome could potentially impact the performance of reference alignment. Non-African D. melanogaster have essentially a subset of the genetic diversity present in sub-Saharan Africa. Thus, a pair of non-African genomes will have fewer sequence differences than a pair of sub-Saharan genomes or a comparison between these groups. During reference alignment, too many single nucleotide polymorphism (SNP) or indel differences from the reference genome may cause reads not to map. Thus, when the reference genome carries a European allele, European DGRP alleles might have a higher probability of mapping (and therefore a higher depth of mapped sequence reads) than African DGRP alleles. Examining the results of a single round of reference alignment, there is support for a modest effect of ancestry on aligned sequence depth. The normalized depth ratio of African versus European DGRP alleles is 1% lower in reference-European versus reference-African regions for autosomal arms, and 2% lower for the X chromosome (table 3). This ancestry effect was reduced by adding a second round of mapping to a reference sequence modified to account for a genome’s called SNPs and indels, mirroring the pipeline used by the Drosophila Genome Nexus (Lack et al. 2015). Whether ancestry affects called sequence data may thus depend on the alignment methods and options used.
Table 3.
Reference Genome Ancestry Affects the Outcomes of Reference Alignment.
| DGRP Afr./Eur. Depth Ratios | ||||||
|---|---|---|---|---|---|---|
| 1 Round of Mapping |
2 Rounds of Mapping |
|||||
| Arm | Ref. Afr. | Ref. Eur. | M-W P | Ref. Afr. | Ref. Eur. | M-W P |
| X | 1.0139 | 0.9896 | 9.6 × 10−5 | 1.0039 | 0.9934 | 0.11 |
| 2L | 1.0105 | 0.9945 | 6.3 × 10−17 | 1.0033 | 0.9966 | 1.1 × 10−4 |
| 2R | 1.0099 | 0.9996 | 7.1 × 10−7 | 1.0045 | 1.0014 | 0.056 |
| 3L | 1.0100 | 0.9987 | 1.8 × 10−12 | 1.0052 | 1.0006 | 3.6 × 10−3 |
| 3R | 1.0069 | 0.9970 | 5.3 × 10−4 | 1.0006 | 0.9994 | 0.81 |
Note.—Average mapped sequencing depth was calculated for each DGRP genome in each window, then rescaled relative to that genome’s mean depth across the chromosome arm. Ratios of rescaled depths between DGRP genomes with African versus European ancestry were then compiled separately for windows where the reference genome has either African or European ancestry (“Ref. Afr.” and “Ref. Eur.” below). DGRP alleles matching the reference genome’s local ancestry appeared to map with slightly greater success. The ancestry effect was highly significant if a single round of alignment was conducted (see Mann–Whitney P values below), but partially mitigated after a second round of mapping against a reference sequence that was modified based on SNPs and indels discovered in round 1.
Evidence for Widespread Epistatic Fitness Interactions in the DGRP
As indicated above, one general explanation for the observed relationship between recombination rate and ancestry (fig. 3) is that certain African alleles are disfavored in the primarily European gene pool of the DGRP population. In light of previous evidence (Lachance and True 2010), at least some of these loci may be involved in interlocus fitness interactions (IFIs), in which having African alleles at one locus and European alleles at another may lead to reduced survival or fecundity. If natural selection against introgressing African alleles is ongoing today, a signal of linkage disequilibrium might be observed between pairs of loci responsible for such IFIs. Such analyses have been performed for crosses between populations or species to identify pairs of loci that may constitute Bateson–Dobzhansky–Muller incompatibilities (BDMIs) of potential relevance to speciation (Gardner et al. 2000; Payseur and Hoekstra 2005; Harrison and Edmands 2006; Hohenlohe et al. 2012; Schumer et al. 2014). For the DGRP case, no admixture-driven disequilibrium is expected between unlinked loci in the absence of fitness interaction, as this would decay within a small handful of generations due to independent assortment (Wilson and Rannala 2003).
Here, I test for disequilibrium between loci on different chromosomes, not by using individual SNP genotypes, but instead based on the ancestry calls made for each genome in each of the 24,417 genomic windows. This focus on AD makes genome-scale pairwise testing computationally plausible. Fo each interchromosomal pair of windows, I calculated a Fisher’s Exact Test (FET) P value, with low one-tailed P values reflecting the preferential occurrence of Africa–Africa and Europe–Europe ancestry combinations at the two windows. Only homozygous intervals were analyzed, so each genome has just one allele per locus, and inverted chromosome arms were excluded. Results from the true data were then compared against randomly permuted data sets, in which individual labels for the second window were shifted (thus maintaining the true data’s population ancestry frequencies at each window, as well as patterns of linkage between neighboring windows). Across the genome, a notable excess of interchromosomal window pairs with low FET P values was observed (fig. 4), indicating a genome-wide signal of AD. At very low P values, the enrichment was more pronounced for X-autosome window pairs than for pairs split between the two major autosomes (fig. 4B). A much smaller enrichment may exist for negative ancestry associations (e.g., P > 0.99 in fig. 4A), but this relatively subtle genomic signal is not a focus of this study. The window pairs yielding the lowest FET P values for positive AD are shown in table 4.
Fig. 4.
A genome-wide signal of interchromosomal AD is depicted. Here, fold-enrichment (the ratio of P-value bin counts in the observed data relative to permuted data sets) is plotted for FET P values, which are low when alleles of the same ancestry tend to occur in the same genomes at unlinked loci. All comparisons between X-linked and autosomal windows, and between chromosomes 2 and 3, are plotted in separate series. (A) Enrichment is plotted for each 0.01-wide P-value bin. (B) Cumulative enrichment for all P values below a given threshold is indicated.
Table 4.
The Most Statistically Significant Contingency Tables for AD.
| Arm | Window Bounds (bp) | A:A | A:E | E:A | E:E | FET P | Nearest Gene |
|---|---|---|---|---|---|---|---|
| 2R | 11,353,691–11,359,516 | 26 | 13 | 24 | 102 | 6 × 10−8 | CG43729 |
| 3L | 11,079,088–11,085,304 | (11.8) | (27.2) | (38.2) | (87.8) | JIL-1 | |
| X | 20,815,287–20,820,196 | 17 | 58 | 0 | 92 | 4 × 10−7 | Npc1b (also near shakB) |
| 3L | 4,416,058–4,419,369 | (7.6) | (67.4) | (9.4) | (82.6) | DOR | |
| 2R | 9,090,338–9,096,171 | 21 | 22 | 14 | 116 | 5 × 10−7 | latheos, CG31345, FLASH |
| 3L | 6,514,568–6,520,094 | (8.7) | (34.3) | (26.3) | (103.7) | sulfateless | |
| X | 2,581,360–2,585,575 | 9 | 5 | 7 | 108 | 9 × 10−7 | period |
| 3R | 21,627,751–21,631,330 | (1.7) | (12.3) | (14.3) | (100.7) | BG642167 |
Note.—From FETs performed on all pairs of unlinked genomic windows, the listed window pairs exhibited the lowest FET P values. On the first row for each window pair, the observed two locus ancestry counts are given (e.g., the A:E column refers to the number of genomes with African ancestry for window 1 and European ancestry for window 2). The numbers in parentheses on the second row of each window pair are the expected counts under the null model of no association between the ancestries of unlinked loci. Note that JIL-1, Ncp1b/shakB, DOR, and BG642167 are within “hubs” of AD that have at least seven putative fitness interactions with unlinked loci. Genomic coordinates refer to release 5 of the Drosophila melanogaster reference genome.
To avoid treating neighboring window pairs as independent, nearby outlier P values were merged into two-dimensional “clusters” of AD, and these clusters were extended from each focal window until pairs with P < 0.05 were no longer observed with appreciable frequency. Although the merging criteria were necessarily somewhat arbitrary (see Materials and Methods), they were designed to extend clusters generously in an attempt to fully account for their effect on the genomic distribution of FET P values. Examining the chromosomal distribution of these pairwise clusters, there is little evidence that adjacent clusters are failing to be appropriately merged (supplementary fig. S2, Supplementary Material online). This procedure resulted in 676 AD clusters with no pairwise overlap. The 183 X-autosome clusters contained 47,948 window pairs with FET P < 0.05, accounting for just 33% of the genome-wide excess of significant X-autosome P values. The 493 autosome–autosome clusters contained 210,291 window pairs with FET P < 0.05, accounting for 58% of the genome-wide excess of autosomal P < 0.05 values. As excluding hundreds of the strongest AD signals still fails to fully account for the genome-wide signal of AD, it seems possible that the true number of pairwise IFIs between African and European alleles in the DGRP genomes is surprisingly large. Still, further study is needed to accurately estimate the number and strength of ancestry-associated IFIs.
Potential Genetic Targets of IFIs
The vast number of pairwise comparisons involved in genome-wide disequilibrium testing entails a multiple testing problem, with the consequence that no pairwise P value from a single hybrid or admixed population is likely to be statistically significant in a genome-wide context (Schumer et al. 2014). In this study, I tested over 28 million pairwise window combinations for X-autosome combinations (lowest P = 6 × 10−8), and did more than 73 million tests between the two major autosomes (lowest P = 4 × 10−7). Thus, no window pair had a P value low enough to remain significant after Bonferroni correction, and additional evidence is needed to draw any specific conclusions about genes causing AD in the DGRP population.
With the goal of identifying a more confident set of AD clusters, I hypothesized that some true positive loci might participate in a greater number of pairwise interactions than expected by chance. Although a plurality of all genomic windows overlapped zero AD clusters and most windows overlapped three or fewer, a smaller subset of windows overlapped several—up to a maximum of 13 pairwise between-chromosome clusters. Comparing the total “cluster counts” of windows in the real data against those from permuted data sets, I confirmed that windows overlapping multiple pairwise clusters were observed much more frequently than expected randomly (supplementary fig. S3, Supplementary Material online). For example, windows overlapping seven or more AD clusters were 3.7× more common in the real data (implying a true positive probability of 73% that at least some of a window’s pairwise clusters are genuine), and “cluster counts” of at least 7 were observed in 59 distinct genomic regions. Windows overlapping 11 or more AD clusters were enriched by a factor of 5.2×, indicating a true positive probability of 81%. Hence, a subset of windows constituting “AD hubs” have fairly strong confidence of holding genuine IFIs, even though data are limited to just one admixed population. Below, I specifically label as “AD hubs” a set of nearby windows that overlap seven or more AD clusters, and are separated by windows overlapping no fewer than five AD clusters.
AD in North American D. melanogaster could indicate IFIs resulting from adaptive functional differences between the African and European source populations. For a gene where natural selection had acted differently between those two populations, we might expect locally elevated FST values between European and West African populations. Consistent with this hypothesis, windows overlapping the largest number of AD clusters were somewhat more likely to have high FST values between European and West African populations (supplementary fig. S4, Supplementary Material online). Importantly, FST peaks are typically narrower than AD hubs, so their co-occurrence may help to localize the genetic targets of IFIs.
A thorough analysis of genes likely to underlie IFIs in North American D. melanogaster could encompass one or more follow-up studies. Still, a preliminary examination of the genes and pairwise combinations involved in AD hubs may motivate hypotheses for further genomic and functional testing, regarding the biological nature of putative incompatibilities between African and European D. melanogaster. I therefore highlight a few of the most notable genes and categories indicated by these AD hubs below.
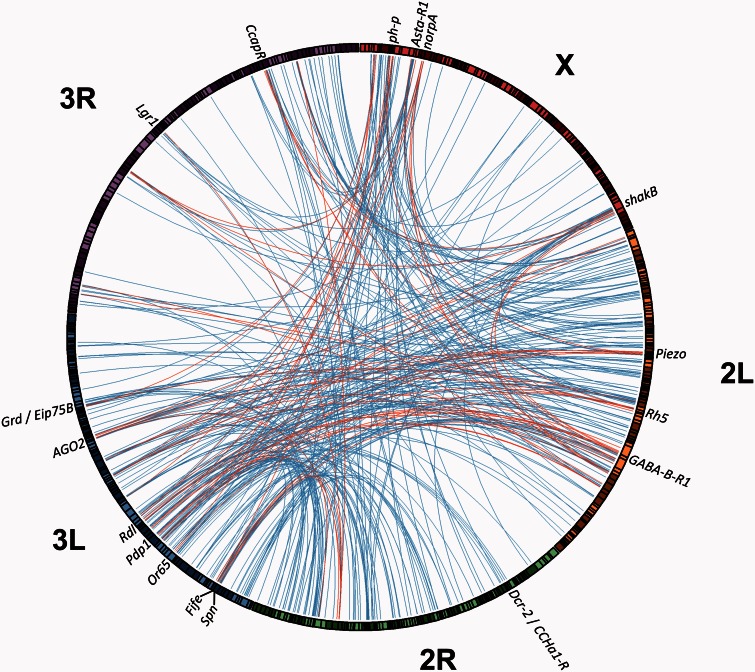
Figure 5 illustrates the pairwise components of AD hubs with at least seven pairwise interchromosomal interactions. The most extreme AD hub, overlapping 13 clusters, was centered on the gene Argonaute 2 (fig. 6). An RNA interference gene, AGO2 is involved in the loading of siRNA onto the RISC complex, and its known functions include antiviral response, chromatin silencing, and autophagy. Along with a second AD hub including Dicer-2, this result is consistent with the previous finding that RNAi genes are frequent targets of positive selection in Drosophila (Obbard et al. 2006). Another previously implicated target of selection is polyhomeotic-proximal (ph-p), which sits within one of just two AD hubs that overlap 12 clusters. This polycomb group gene, which has roles in gene silencing, nervous system development, and ecdysone response, was previously shown to have experienced a selective sweep in the African ancestral range (Beisswanger and Stephan 2008). Voigt et al. (2015) recently suggested that a second sweep may have occurred in Europe, and showed temperature-dependent expression differences between African and European ph-p alleles.
Fig. 5.
Interactions involving AD hubs that contain elevated Africa–Europe FST (see Materials and Methods) were plotted using Circos (Krzywinski et al. 2009). AD clusters linking two unlinked AD hubs are shown in red, whereas those involving a single AD hub are in blue. Locations of genes mentioned in the text are shown; these genes were indicated by patterns of cluster overlap and FST, but further research is needed to assess their potential involvement in IFIs. Chromosome arms 2 L and 3L appear to contribute strongly to AD hub interactions. Note that arm-wide African ancestry levels on these two arms are uncorrelated among DGRP genomes (Pearson r2 = 0.0016; P = 0.31).
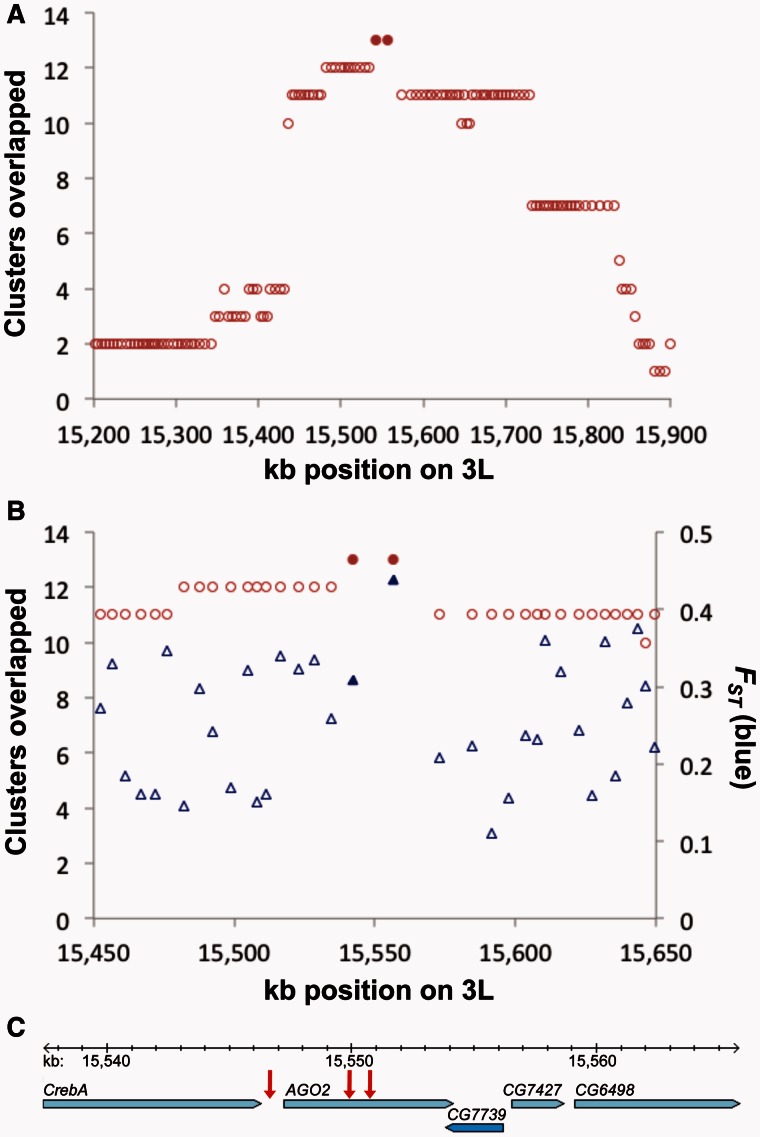
Fig. 6.
The co-occurrence of a strong AD hub and elevated FST at the gene Argonaute 2. (A) The two windows overlapping AGO2 are shaded; these represent the largest number of pairwise AD clusters overlapping any windows in the genome. (B) The same cluster overlap statistic is plotted across a narrower region, alongside window FST between European and western African populations. High values of FST could indicate adaptive functional differences between the two source populations of the North American DGRP population (the median FST on 3L is 0.19). Nearly all of the AGO2 transcript is located within the right-hand shaded window with elevated FST. (C) The two windows that overlap the most AD clusters contain five genes. Three fixed differences between European and western African genomes are located within and immediately upstream of the AGO2 gene. Genomic coordinates refer to release 5 of the Drosophila melanogaster reference genome.
AD hubs displaying the greatest number of interactions (≥11) and elevated FST values also included the genes Allatostatin A receptor 1 (AstA-R1; neuropeptide signaling), Fife (a recently described regulator of synaptic transmission; Bruckner et al. 2012), and shaking B (shakB; a synaptic gap junction protein involved in phototransduction). Other genes in AD hubs (≥7 clusters) with elevated Africa–Europe FST values included additional neuropeptide and hormone receptors (e.g., the unlinked GABA receptors Rdl, GABA-B-R1, and Grd, along with Eip75B, CCAP-R, CCHa1-R, and Lgr1), plus other genes involved in phototransduction and/or circadian rhythm (norpA, Pdp1, Rh5), as well as other genes involved in olfaction and sensory behavior (Piezo, Spn, the Or65 cluster). A full description of AD hubs and their associated interactions is given in supplementary table S6, Supplementary Material online.
Enriched GO categories for windows in AD hubs with elevated FST (see Materials and Methods) echoed many of these same themes (supplementary table S7, Supplementary Material online). These categories included “detection of chemical stimulus involved in sensory perception” (which had the lowest P value among GO categories represented by at least five AD hubs), “cellular response to stimulus,” “signal transducer activity,” “cell surface receptor signaling pathway,” “intrinsic to membrane,” aspects of transmembrane transport, and GABA and allatostatin receptor activities. Windows from 35 AD hubs met the FST criteria for this analysis, encompassing a median span of just 10 kb per hub. For 27 of these hubs, the window(s) with elevated FST included at least one gene from the GO categories mentioned above. As with the previous GO enrichment of elevated African ancestry, the analysis of AD hubs with elevated FST yielded more GO categories with raw P < 0.01 than 95% of randomized genomic permutations. However, the same qualifications apply to this exploratory analysis: It can not conclusively point to the genes and processes underlying putative incompatibilities in the DGRP, but it does suggest hypotheses for downstream molecular and genomic studies.
Less than a third of pairwise clusters involving AD hubs linked one hub to another (fig. 5). Although some of these two-hub interactions could make sense based on related known functions (e.g., a cluster that links shakB and Pdp1), other interactions are less functionally obvious (e.g., AGO2 with Piezo, and with Rh5). The fitness interactions implied by AD need not involve direct molecular interactions; they could instead stem from higher-order phenotypes. Still, AD analysis may be fairly unique among evolutionary genomic methods in its potential to identify novel functional relationships between genes. This signal could complement other genomic searches for interactors, such as correlated rates of protein evolution among species (Clark et al. 2009). But notably, AD can operate on a shorter time scale, it only requires data from a single species, and is not confined to interactions between protein-coding sequences.
Discussion
Evolutionary History and Genetic Composition of North American D. melanogaster
North American strains of D. melanogaster, including the reference genome strain and the DGRP, have taken on great importance in a wide range of genetic studies. Previous studies had shown that New World populations have an admixed history, descending from source populations both in Europe and in the African ancestral range (David and Capy 1988; Caracristi and Schlötterer 2003; Duchen et al. 2013; Kao et al. 2015; Bergland et al. 2015). However, this complex history and dual ancestry, along with its significance for Drosophila research, have not been broadly appreciated in the literature, and efforts to study the genetic composition of important fly strains have been lacking.
In this study, I estimated population ancestry (European or African) along the genomes of the reference strain and the DGRP. Results strongly support the hypothesis of admixture in North America. Greater African ancestry in the mid-Atlantic DGRP (20%) versus the northeastern reference genome (9%) is consistent with an ancestry gradient among US populations, with the highest European ancestry in the north and relatively higher African ancestry in the south (Kao et al. 2015; Bergland et al. 2015), possibly resulting from secondary contact after two separate colonizations.
An Empirical Estimate for the Generation Time of D. melanogaster
By combining our estimate of admixture onset (1,598 generations, based on the recombination-mediated shortening of ancestry tracts over time) with historical records, I can obtain a rough estimate of the DGRP population’s average generation time. In light of the hypothesized colonization of the New World by European strains through the northeast US and by African strains through the Caribbean (David and Capy 1988; Keller 2007), one plausible site of initial secondary contact would be in the far southeastern United States (Kao et al. 2015). Based on the first observation of D. melanogaster from the southern United States (1894 in Florida; Keller 2007), 109 years would have elapsed before the collection and inbreeding of the DGRP strains in 2003. Thus, an estimate of the number of generations per year is 1,598/109 = 14.7. The estimate of 1,598 generations carries important uncertainties (see Results), as does the timing of secondary contact. Generation time may also vary geographically and temporally based on climate and other factors. Temperate populations undergo reproductive diapause in winter (Saunders et al. 1989; Schmidt et al. 2005). Raleigh’s climate would seem unfavorable for outdoor reproduction of D. melanogaster for roughly 3 months of the year, whereas other populations may have longer or shorter reproductive seasons based on temperature, rainfall, resource availability, and other factors. In spite of these caveats, this estimate may be preferable to the commonly used figure of ten generations per year, for which no empirical basis is typically cited. Improving our estimation of this quantity is important for relating DNA variation and evolution to historical time, whether one is studying changes on the scale of months or millions of years.
Genome-Wide Evidence for Natural Selection Shaping Patterns of Admixture
Three patterns suggest that natural selection has powerfully influenced patterns of ancestry along DGRP genomes. First, levels of European and African ancestry vary strikingly within and between chromosome arms (fig. 2). Second, the degree of African introgression is greatly reduced in regions with higher recombination rates (fig. 3). Third, many unlinked loci have deficiencies of Africa–Europe allele combinations between them (fig. 4).
Regarding the first point, the X chromosome shows strikingly reduced African introgression relative to the autosomes. This result agrees with other recent studies (Kao et al. 2015; Bergland et al. 2015) and mirrors the situation in sub-Saharan Africa, where admixture from outside Africa is lowest on the X chromosome (Kauer et al. 2003; Pool et al. 2012). X chromosomes may thus be inhibited from introgressing between African and non-African populations in either direction. Qualitatively similar patterns have been reported from cases of hybridization involving mice, Neanderthals, and other taxa (Coyne and Orr 1989; Tucker et al. 1992; Sankararaman et al. 2014). Although my results concern the admixture of two conspecific populations, they are compatible with Haldane’s Rule (Haldane 1922), and with the findings of Lachance and True (2010), who reported a substantial rate of epistatic fitness interactions between X-linked and autosomal loci in crosses between Canadian and Caribbean strains. Haldane’s Rule suggests an elevated contribution of the X chromosome to reproductive isolation, potentially involving BDMIs in which between-locus combinations of alleles that had never coexisted arose after population divergence. A greater effect of BDMIs involving X-linked loci could be explained by recessive BDMIs (which are readily exposed in hemizygous males), or by a greater density of functional differences between African and European populations on the X chromosome (Presgraves 2008).
Ancestry patterns within chromosomes further suggest that European and African alleles at many loci have had unequal fitness in the DGRP population. Sharp peaks of African or European ancestry were apparent. Levels of African introgression into this primarily European population were much lower in higher recombination regions. This signal could result from African alleles favored in the DGRP population, with longer linkage blocks hitchhiking with them in regions of low recombination. Alternatively, in accord with the X-autosome contrast and the AD analysis, African alleles at many loci may have been selected against in the DGRP population (perhaps due to incompatibilities with European alleles at other loci, or else directional selection based on the North Carolina environment or the prevalent mating system). In either case, the functional differences between African and European populations that selection acted upon in North America are likely to represent products of positive selection in one or both of the source populations. The brief evolutionary time scale of African and European populations’ separation (roughly 0.1Ne generations; Thornton and Andolfatto 2006) leaves little time for mutation and drift alone to produce such differences.
Impact of Natural Selection on Ancestry Inferences
There are reasons to be skeptical of some extreme DGRP ancestry deviations. The two intervals of maximal African ancestry are near Cyp6g1 and overlapping Ace, loci with strong selective sweeps related to 20th century insecticide usage (Catania et al. 2004; Karasov et al. 2010). At these loci, sweeps that occurred after the divergence of the Raleigh population from its source populations (perhaps less than 150 years ago; Keller 2007) could result in biased ancestry inference.
Although it would be desirable to annotate each case in which very recent selective sweeps may have influenced ancestry calling, this goal may require significant methodological advances. The HMM used here should be more sensitive to cases involving very recent selection affecting the European reference panel, but such sweeps could either be global (with the same or different haplotypes fixing in each population), or shared by the European and African reference panels but not the DGRP, or specific to the European sample. These scenarios each lead to distinct predictions for variation among populations, whereas typical genome scans focused on European reference panel will mainly pick up sweeps that happened prior to American colonization (which are of less concern here).
This issue reflects a general challenge for ancestry inference. Other reference panel approaches should be subject to similar effects of recent selection. Methods that do not use reference panels may return nongeographic divisions in the data, such as clustering inverted versus standard chromosome arms (Corbett-Detig and Hartl 2012), and even if inverted arms were removed, output could prove similarly uninformative or biased in cases of recent sweeps. Hence, the ancestry inferences presented here (supplementary tables S3 and S5, Supplementary Material online) should be regarded as provisional, and should be revisited in light of future methodological developments.
For either hard sweeps or moderately soft sweeps affecting the European sample, such recent selection should increase that population’s haplotype homozygosity, since there has been very little time for mutation and recombination since the adaptive event. Although such a pattern is observed at Cyp6g1, in general the inferred peaks of African ancestry in the DGRP show no such pattern (supplementary fig. S5, Supplementary Material online). Thus, although recent selection may drive some apparent ancestry deviations, most of the genomic variance in DGRP ancestry suggested in figure 2 seems likely to be genuine.
AD and Its Possible Causes
Consistent with the hypothesis of epistatic incompatibilities or other fitness interactions between African and European alleles, I found that AD is widespread in the DGRP genomes and may involve a large number of locus pairs. The clearest explanation for AD is an incompatibility between an African allele at one locus and a European allele at another, producing an epistatic fitness interaction due to consequences for survival and/or reproduction. Positive assortative mating—for example, if flies with African alleles at certain loci mate preferentially—might also contribute to AD among wild-caught individuals. Thus, AD could stem from interactions between individuals in addition to epistasis within individuals. It is worth mentioning, however, that this study does not directly examine wild-caught flies, but instead the genomes of strains that were inbred for 20 generations, and had originated from greater than 200 independent isofemale lines. Recessive BDMIs will be unmasked by the inbreeding process. If two-locus genotypes influence the viability and fertility of sibling cross offspring, selection might act across multiple generations of inbreeding until one allelic combination is fixed. The survival of an inbred strain might also be affected by the combinations of African and European alleles that its founders possessed. Thus, inbreeding and the opportunity to study mostly homozygous genomes may amplify the signal of IFIs and aid the search for causative loci.
Recently, Corbett-Detig et al. (2013) reported interchromosomal disequilibrium from a mapping population known as the Drosophila Synthetic Population Resource (DSPR; King et al. 2012), which comprises over 1,700 recombinant inbred lines, each derived from 8 geographically diverse founder strains after 50 generations of interbreeding. In light of this study, and given the mix of cosmopolitan and sub-Saharan strains in the DSPR, it seems possible that population differentiation contributed many of these incompatibilities. Examining the 22 SNP pairs identified by that study, none corresponded to AD window pairs from my analysis (P > 0.05 in all cases where African alleles existed at both loci), and only 1 of the 44 SNPs was located within an AD hub (a window on arm 3R with Europe–West Africa FST of 0.46, upstream of the genes βTub97EF and CG4815). However, the 30 autosomal windows containing these SNPs had a median ancestry deviation statistic (see Materials and Methods) of 5.8% toward European ancestry (Mann–Whitney test P = 0.0037 comparing these SNPs’ ancestry deviations against all other autosomal windows). The modest overlap could indicate that SNPs identified by Corbett-Detig et al. (2013) are unrelated to Africa–Europe genetic differentiation. However, the DSPR loci might subject to strong epistatic selection (in order to be observed on a short laboratory time scale), and such alleles might have been purged from the DGRP population by now, whereas AD in this study may be driven by incompatibilities of more moderate effect. Further analysis of the selection coefficients that may drive DGRP ancestry deviations and AD, in light of nuances such as demographic details and dominance, is clearly warranted.
Previously, it was shown that one solution to the multiple testing problem inherent in genome-wide disequilibrium testing is to add data from a second independent hybrid/admixed population and require that both populations show a disequilibrium signal for the same locus pair (Schumer et al. 2014). Appropriate genomic data for a second admixed population are not yet available for D. melanogaster (the admixture tracts found in sub-Saharan populations are too long for locus-specific analysis; Pool et al. 2012). However, the two-population approach could become feasible if enough nonpooled genomes were sequenced from a region such as Saharan Africa, Madagascar (Baudry et al. 2004), northern Australia, or possibly South America. The suitability of a population will depend on the timing and amount of admixture, and analysis supporting an independent history of admixture relative to North America.
Here, I found that even without data from a second population, statistical power can be gained by focusing on “AD hubs.” Loci participating in multiple pairwise interactions were far more common in the real data than expected randomly, allowing the identification of a set of loci with fairly strong confidence of contributing to IFIs (e.g., 73% to 81% posterior probabilities), including those discussed above. Many of these AD hubs include genes with roles in neurotransmission and sensation. Experimentation will be needed to test whether population differences at these genes impact ecological or reproductive aspects of behavior, the maintenance of function in novel thermal environments, or other processes.
It will also be of interest to compare the genomic admixture patterns identified in the DGRP to broader latitude clines in eastern North America and elsewhere (Turner et al. 2008; Kolaczkowski et al. 2011; Fabian et al. 2012; Kao et al. 2015; Bergland et al. 2015), with the expectation that many loci subject to ancestry deviations in North Carolina may show atypical clinal patterns as well. Such analyses should be conducted based on ancestry proportions along the cline, as opposed to FST between northern and southern populations (which will vary along the genome based on the histories of the source populations).
I have not estimated the precise number of loci contributing to ongoing fitness interactions in the DGRP population, and further methodological advances toward this goal would be desirable. However, the above analyses hint that this number may be substantial. Excluding several hundred of the most extreme pairwise interactions did not erase the genomic signal of AD. I also identified 59 AD hubs, and these appear to interact with a larger number of partner loci (fig. 5). These findings, together with the pronounced genomic variance in ancestry and its correlation with recombination rate, suggest that natural selection has profoundly influenced the genomic consequences of admixture between temperate and tropical populations of D. melanogaster. This work provides an intriguing example of admixture between genetically differentiated populations, in a species in which large populations may facilitate an important role for natural selection in the genome (Sella et al. 2009; Langley et al. 2012). Importantly, putative incompatibilities in this system may be particularly amenable to functional characterization.
Significance of Mosaic Ancestry for Drosophila Research
Being the first and most completely sequenced D. melanogaster genome, the genome of the y; cn, bw, sp laboratory strain is typically the standard against which newly sequenced genomes are compared. In an evolutionary context, however, this genome is not an obvious “reference,” being the result of a complex history involving founder events and admixture. The reference genome’s mosaic ancestry could impact reference alignments and downstream analyses, presenting a possible source of bias for population genomic studies. The minor effect of ancestry on mapped sequence depth observed here was reduced by a second round of mapping (table 3), as implemented by the Drosophila Genome Nexus (Lack et al. 2015). The ancestry effect might be further minimized by accounting for known variation during reference alignment, or by using an alternative reference genome with similar genetic distances to all strains of D. melanogaster (e.g., from Zambia; Pool et al. 2012).
The mosaic ancestry of DGRP and laboratory strains may also be relevant to a range of phenotypic and genetic studies. The European and African source populations likely differed in various phenotypes (Capy et al. 1993). Some of the phenotypic diversity resulting from their admixture may persist today and contribute to the trait variation of populations such as the DGRP. As a potential example, variants at many of the AD hub genes mentioned above were found to have associations with sleep traits in the DGRP (Harbison et al. 2013), including AlstA-R1, βTub97EF, Eip75B, Grd, norpA, the Or65 cluster, Rdl, shakB, and Spn. It could be worthwhile to incorporate ancestry into similar genome-wide association studies, perhaps using window ancestries for a preliminary “admixture mapping” phase requiring fewer genome-wide tests. Ancestry-associated phenotypic variation might have longer linkage blocks flanking the causative sites, potentially making it easier to detect but more challenging to pinpoint within ancestry blocks. In light of the AD results cited above, admixture could also be a source of epistatic interactions in the DGRP and lab strains, which might impact phenotypes of interest and contribute to genetic background effects.
Materials and Methods
Genomes and Ancestry Inference
Aside from the D. melanogaster reference genome (release 5.57), all genomes analyzed here were originally described in the DGRP (Mackay et al. 2012; Huang et al. 2014) or the Drosophila Population Genomics Project, phase 2 (Pool et al. 2012). The alignments used in this study were generated using a common pipeline, involving a second round of mapping to a modified reference genome, as described by Lack et al. (2015).
Ancestry estimation was performed using the HMM approach originally described by Pool et al. (2012). Briefly, this method utilizes the difference in genetic distance between two types of pairwise comparisons: 1) Comparisons among “cosmopolitan” genomes from outside sub-Saharan Africa, which have reduced diversity stemming from the out-of-Africa bottleneck; and (2) comparisons between sub-Saharan and cosmopolitan genomes, which have similarly higher distances as comparisons between sub-Saharan genomes. These comparisons are evaluated with the aid of two reference panels of genomes (sub-Saharan and cosmopolitan). Distances are initially assessed in nonoverlapping windows across the genome. In each window for a focal genome being tested, its genetic distance to the cosmopolitan panel is tested to evaluate whether it more closely resembles the comparisons among cosmopolitan genomes (perhaps indicating cosmopolitan ancestry) or the comparison between the sub-Saharan and cosmopolitan genomes (favoring sub-Saharan ancestry of the tested genome). A likelihood of each ancestry type is obtained for each window for this genome, with the HMM then returning final ancestry probabilities in each case.
Following Lack et al. (2015), the sub-Saharan reference panel consisted of 27 Rwanda genomes, whereas the cosmopolitan panel included 9 France and 3 Egypt genomes. Chromosome arms with inversions were excluded from reference panels, based on evidence that inversions have recently moved between populations (Corbett-Detig and Hartl 2012; Pool et al. 2012). As in previous studies, admixed segments of the African reference panel were identified and masked prior to analyzing genomes from other populations.
Based on the relatively older admixture of North American populations (compared with the apparently very recent introgression studied in Africa), a somewhat smaller window size was used in the present analysis. Windows were scaled by genetic diversity, as defined by 100 nonsingleton SNPs in the Rwanda sample, so that all windows contained similar amounts of genetic variation. Otherwise, ancestry was assessed exactly as previously described (Pool et al. 2012; Lack et al. 2015). Regions of genomes previously inferred to contain residual heterozygosity or identity by descent with another analyzed genome were excluded from all analyses.
Ancestry Deviations and GO Enrichment
Population ancestry proportions among DGRP genomes were found to vary on both local and broader genomic scales. To analyze genes that could be responsible for local peaks of African or European ancestry, a simple “ancestry deviation” statistic was implemented. This statistic was defined as the difference between the proportion of African ancestry in the focal window and the median of that quantity in the 51st to 250th windows on each side. This procedure helped to account for the regional ancestry background while excluding windows that may deviate along with the focal window due to the same instance of natural selection. Outlier windows for ancestry deviation were defined as based on the 2.5% tails for each chromosomal arm. To avoid double-counting the same putative instance of selection, “outlier regions” grouped outlier windows with up to two nonoutlier windows between them.
The set of all genes overlapping outlier regions (including the next exon on each side of the region) was subjected to GO enrichment analysis. GO categories corresponding to the overlapping genes were counted only once per region. The locations of all outlier regions (in terms of the windows that each spanned) were randomly permuted within their original chromosome arms 50,000 times, a practice that accounts for the effects of varying gene lengths. For each GO category, the proportion of random permutations generating at least as many outliers as observed in the real data constituted a P value.
Reference Genome Ancestry and Reference Alignment
The effect of reference genome ancestry on mapped sequencing depth was also assessed. Depth was analyzed for DGRP genomes after one and two rounds of mapping through the pipeline of Lack et al. (2015). Here, the second round of mapping is to a reference sequence altered based on the SNPs and indels called after the first round of mapping, and the two round data object corresponds to that distributed within the Drosophila Genome Nexus. Average depth was summarized for each genome in each window for all called sites, then all values were rescaled by dividing by that genome’s mean depth across a chromosome arm. For each window, the ratio of rescaled depths between DGRP genomes with African versus European local ancestry was calculated (provided at least five African and five European alleles were present). For each chromosome arm, I then recorded DGRP African/European depth ratios separately for windows where the reference genome had African or else European ancestry. A Mann–Whitney test then gauged the significance of a difference in DGRP depth ratios in reference-African versus reference-European windows.
AD Testing and Analysis
Analogous to linkage disequilibrium, I tested for AD using the ancestry inferred for each genome in each window, asking whether having an African allele in one window boosted the chance of having an African allele in an unlinked window. FETs were applied to each interchromosomal pair of windows that both had at least two African-ancestry and two European-ancestry DGRP genomes. The 24,417 genomic windows used in this pairwise analysis had a median sample size of 164 genomes after the exclusion of heterozygous regions and inverted chromosome arms.
Genomic distributions of FET P values were compared between the real data and permuted data sets in which individual labels were consistently shifted for the second window in a pair (thus maintaining linkage patterns among windows). Due to the computationally intensive analysis, just ten permuted genomic data sets were assessed, but each one contains roughly 100 million P values, and consistent results were observed from one replicate to the next.
To bin multiple neighboring window pairs that could result from the same pair of interacting loci, a set of the most extreme AD window pairs were extended to form “AD clusters.” Specific criteria for selecting and extending these criteria were as follows: 1) Identify each interchromosomal window pair with a raw FET P value below 0.0001 as starting points for AD clusters. 2) While holding one member of the focal window pair constant, extend the cluster from the other window by advancing in each direction until 10 consecutive P values above 0.05 are observed. Repeat to extend bidirectionally from the first member of the window pair as well, holding the second member of the window pair constant. 3) Consider clusters to encompass the full two-dimensional range of windows between the window start and stop positions identified for each side of the pair above. Merge any clusters that have overlapping boundaries on both chromosomes, giving the merged cluster the maximal span indicated by the boundaries of its component clusters.
Windows were considered to lie within AD “hubs” if they overlapped at least seven interchromosomal AD clusters, and if their cluster count was within 1 of the local maximum (with cluster counts of 4 or below preventing the extension of AD hubs). To test whether the empirical data contained an unexpected number of AD hubs, it was compared against the permuted data described above. The enrichment of genomic windows overlapping a given number of AD clusters was noted. Enrichments were converted into true positive probabilities using the equation (e − 1)/e. Here, 1 reflects the rate of false positives for every e total positives.
GO enrichment analysis on AD hubs with elevated Africa–Europe FST (Hudson et al. 1992) was conducted as described above for African ancestry deviation, except for the specific criteria for outlier regions. To focus on loci with at least modest evidence for adaptive differences between the source populations of North American D. melanogaster, a minimum FST of 0.35 (autosomes) or 0.42 (X chromosome) was required, corresponding to roughly the upper 15% quantile of this statistic. FST was evaluated between a France sample and a panel of four small western African samples from Cameroon, Gabon, Guinea, and Nigeria (Pool et al. 2012).
Supplementary Material
Supplementary figures S1–S5 and tables S1–S7 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/)
Acknowledgments
The author thanks J.J. Emerson for assistance with the admixture detection HMM, R.B. Corbett-Detig for helpful manuscript suggestions, and J.B. Lack for bioinformatic assistance.
References
- Baudry E, Viginier B, Veuille M. 2004. Non-African populations of Drosophila melanogaster have a unique origin. Mol Biol Evol. 21:1482-1491. [DOI] [PubMed] [Google Scholar]
- Beisswanger S, Stephan W. 2008. Evidence that strong positive selection drives neofunctionalization in the tandemly duplicated polyhomeotic genes in Drosophila. Proc Natl Acad Sci U S A. 105:5447–5452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergland AO, Tobler R, Gonzalez J, Schmidt P, Petrov D, Forthcoming 2015. Secondary contact and local adaptation contribute to genome-wide patterns of clinal variation in Drosophila melanogaster. Mol Ecol. Advance Access published November 7, 2015; doi: 10.1111/mec.13455c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruckner JJ, Gratz SJ, Slind JK, Geske RR, Cummings AM, Galindo SE, Donohue LK, O’Connor-Giles KM. 2012. Fife, a Drosophila Piccolo-RIM homolog, promotes active zone organization and neurotransmitter release. J Neurosci. 32:17048–17058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capy P, Pla E, David JR. 1993. Phenotypic and genetic variability of morphometrical traits in natural populations of Drosophila melanogaster and D. simulans. I. Geographic variations. Genet Sel Evol. 25:517–536. [Google Scholar]
- Caracristi G, Schlötterer C. 2003. Genetic differentiation between American and European Drosophila melanogaster populations could be attributed to admixture of African alleles. Mol Biol Evol. 20:792–799. [DOI] [PubMed] [Google Scholar]
- Catania F, Kauer MO, Daborn PJ, Yen JL, Ffrench-Constant RH, Schlötterer C. 2004. A world-wide survey of an Accord insertion and its association with DDT resistance in Drosophila melanogaster. Mol Ecol. 13:2491–2504. [DOI] [PubMed] [Google Scholar]
- Clark NL, Gasper J, Sekino M, Springer SA, Aquadro CF, Swanson WJ. 2009. Coevolution of interacting fertilization proteins. PLoS Genet. 5:e1000570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeron J, Ratnappan R, Bailin S. 2012. The many landscapes of recombination in Drosophila melanogaster. PLoS Genet. 8:e1002905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Hartl DL. 2012. Population genomics of inversion polymorphisms in Drosophila melanogaster. PLoS Genet. 8:e1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett-Detig RB, Zhou J, Clark AG, Hartl DL, Ayroles JF. 2013. Genetic incompatibilities are widespread within species. Nature 504:135–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne JA, Orr HA. 1989. Patterns of speciation in Drosophila. Evolution 43:362–381. [DOI] [PubMed] [Google Scholar]
- David JR, Capy P. 1988. Genetic variation of Drosophila melanogaster natural populations. Trends Genet. 4:106–111. [DOI] [PubMed] [Google Scholar]
- Duchen P, Živković D, Hutter S, Stephan W, Laurent S. 2013. Demographic inference reveals African and European admixture in the North American Drosophila melanogaster population. Genetics 193:291–301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabian DK, Kapun M, Nolte V, Kofler R, Schmidt PS, Schlötterer C, Flatt T. 2012. Genome‐wide patterns of latitudinal differentiation among populations of Drosophila melanogaster from North America. Mol Ecol. 21:4748–4769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner K, Buerkle A, Whitton J, Rieseberg L. 2000. Inferring epistasis in wild sunflower hybrid zones . In: Wolf JB, Brodie ED, III, Wade MJ, editors. Epistasis and the evolutionary process. New York: Oxford University Press; p. 264–279. [Google Scholar]
- Haddrill PR, Thornton KR, Charlesworth B, Andolfatto P. 2005. Multilocus patterns of nucleotide variability and the demographic and selection history of Drosophila melanogaster populations. Genome Res. 15:790–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldane JB. 1922. Sex ratio and unisexual sterility in hybrid animals. J Genet. 12:101–109. [Google Scholar]
- Harbison ST, McCoy LJ, Mackay TF. 2013. Genome-wide association study of sleep in Drosophila melanogaster. BMC Genomics 14:281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison JS, Edmands S. 2006. Chromosomal basis of viability differences in Tigriopus californicus interpopulation hybrids. J Evol Biol. 19:2040–2051. [DOI] [PubMed] [Google Scholar]
- Hohenlohe PA, Bassham S, Currey M, Cresko WA. 2012. Extensive linkage disequilibrium and parallel adaptive divergence across threespine stickleback genomes. Philos Trans R Soc Lond B Biol Sci. 367:395–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollocher H, Ting C-T, Pollack F, Wu C-I. 1997. Incipient speciation by sexual isolation in Drosophila melanogaster: variation in mating preference and correlation between sexes. Evolution 51:1175–1181. [DOI] [PubMed] [Google Scholar]
- Huang W, Massouras A, Inoue Y, Peiffer J, Rámia M, Tarone AM, Turlapati L, Zichner T, Zhu D, Lyman RF, et al. 2014. Natural variation in genome architecture among 205 Drosophila melanogaster Genetic Reference Panel lines. Genome Res. 24:1193–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson RR, Slatkin M, Maddison WP. 1992. Estimation of levels of gene flow from DNA sequence data. Genetics 132:583–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kao JY, Zubair A, Salomon MP, Nuzhdin SV, Campo D. 2015. Population genomic analysis uncovers African and European admixture in Drosophila melanogaster populations from the south-eastern United States and Caribbean Islands. Mol Ecol. 24:1499–1509. [DOI] [PubMed] [Google Scholar]
- Karasov T, Messer PW, Petrov DA. 2010. Evidence that adaptation in Drosophila is not limited by mutation at single sites. PLoS Genet. 6:e1000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauer M, Dieringer D, Schlötterer C. 2003. Nonneutral admixture of immigrant genotypes in African Drosophila melanogaster populations from Zimbabwe. Mol Biol Evol. 20:1329–1337. [DOI] [PubMed] [Google Scholar]
- Kauer M, Zangerl B, Dieringer D, Schlötterer C. 2002. Chromosomal patterns of microsatellite variability contrast sharply in African and non-African populations of Drosophila melanogaster. Genetics 160:247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller A. 2007. Drosophila melanogaster’s history as a human commensal. Curr Biol. 7:R77–R81. [DOI] [PubMed] [Google Scholar]
- King EG, Macdonald SJ, Long AD. 2012. Properties and power of the Drosophila Synthetic Population Resource for the routine dissection of complex traits. Genetics 191:935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohler RE. 1994. Lords of the fly: Drosophila genetics and the experimental life. Chicago (IL): University of Chicago Press. [Google Scholar]
- Kolaczkowski B, Kern AD, Holloway AK, Begun DJ. 2011. Genomic differentiation between temperate and tropical Australian populations of Drosophila melanogaster. Genetics 187:245–260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, Schein J, Birol İ, Connors J, Gascoyne R, Horsman D, Jones SJ, Marra MA. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19:1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachaise D, Cariou ML, David JR, Lemeunier F, Tsacas L, Ashburner M. 1988. Historical biogeography of the Drosophila melanogaster species subgroup. In: Hecht MK, Wallace B, Prance GT, editors. Evolutionary biology. New York: Plenum; p. 159–225. [Google Scholar]
- Lachance J, True JR. 2010. X-autosome incompatibilities in Drosophila melanogaster: tests of Haldane’s rule and geographic patterns within species . Evolution 64:3035–3046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lack JL, Cardeno CM, Crepeau MW, Taylor W, Corbett-Detig RB, Stevens KA, Langley CH, Pool JE. 2015. The Drosophila Genome Nexus: a population genomic resource of 605 Drosophila melanogaster genomes. Genetics 199:1229–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley CH, Stevens K, Cardeno C, Lee YCG, Schrider DR, Pool JE, Langley SA, Suarez C, Corbett-Detig RB, Kolaczkowski B, et al. 2012. Genomic variation in natural populations of Drosophila melanogaster. Genetics 192:533–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TFC, Richards S, Stone EA, Barbadilla A, Ayroles JF, Zhu D, Casillas S, Han Y, Magwire MM, Cridland JM, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482:173–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunes MD, Neumeier H, Schlötterer C. 2008. Contrasting patterns of natural variation in global Drosophila melanogaster populations. Mol Ecol. 17:4470–4479. [DOI] [PubMed] [Google Scholar]
- Obbard DJ, Jiggins FM, Halligan DL, Little TJ. 2006. Nature selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 16:580–585. [DOI] [PubMed] [Google Scholar]
- Payseur BA, Hoekstra HE. 2005. Signatures of reproductive isolation in patterns of single nucleotide diversity across inbred strains of mice. Genetics 171:1905–1916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Corbett-Detig RB, Sugino RP, Stevens KA, Cardeno CM, Crepeau MW, Duchen P, Emerson JJ, Saelao P, Begun DJ, et al. 2012. Population genomics of sub-Saharan Drosophila melanogaster: African diversity and non-African admixture. PLoS Genet. 8:e1003080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pool JE, Nielsen R. 2009. Inference of historical changes in migration rate from the lengths of migrant tracts. Genetics 181:711–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves DC. 2008. Sex chromosomes and speciation in Drosophila. Trends Genet. 24:336–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature 507:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saunders DS, Henrich VC, Gilbert LI. 1989. Induction of diapause in Drosophila melanogaster: photoperiodic regulation and the impact of arrhythmic clock mutations on time measurement. Proc Natl Acad Sci U S A. 86:3748–3752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt PS, Matzkin L, Ippolito M, Eanes WF. 2005. Geographic variation in diapause incidence, life-history traits, and climatic adaptation in Drosophila melanogaster. Evolution 59:1721–1732. [PubMed] [Google Scholar]
- Schumer M, Cui R, Powell D, Dresner R, Rosenthal GG, Andolfatto P. 2014. High-resolution mapping reveals hundreds of genetic incompatibilities in hybridizing fish species. eLIFE 3:e02535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella G, Petrov DA, Przeworski M, Andolfatto P. 2009. Pervasive natural selection in the Drosophila genome? PLoS Genet. 5:e1000495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton KR, Andolfatto P. 2006. Approximate Bayesian inference reveals evidence for a recent, severe bottleneck in a Netherlands population of Drosophila melanogaster. Genetics 172:1607–1619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker PK, Sage RD, Warner J, Wilson AC, Eicher EM. 1992. Abrupt cline for sex chromosomes in a hybrid zone between two species of mice. Evolution 46:1146–1163. [DOI] [PubMed] [Google Scholar]
- Turner TL, Levine MT, Eckert ML, Begun DJ. 2008. Genomic analysis of adaptive differentiation in Drosophila melanogaster. Genetics 179:455–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voigt S, Laurent S, Litovchenko M, Stephan W. 2015. Positive selection at the polyhomeotic locus led to decreased thermosensitivity of gene expression in temperate Drosophila melanogaster. Genetics 200:591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson GA, Rannala B. 2003. Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-I, Hollocher H, Begun DJ, Aquadro CF, Xu Y, Wu ML. 1995. Sexual isolation in Drosophila melanogaster: a possible case of incipient speciation. Proc Natl Acad Sci U S A. 92:2519–2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yukilevich R, True JR. 2008. Incipient sexual isolation among cosmopolitan Drosophila melanogaster populations. Evolution 62:2112–2121 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.