Abstract
Ten eleven translocation (Tet) family-mediated DNA oxidation on 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) represents a novel epigenetic modification that regulates dynamic gene expression during embryonic stem cells (ESCs) differentiation. Through the role of Tet on 5hmC regulation in stem cell development is relatively defined, how the Tet family is regulated and impacts on ESCs lineage development remains elusive. In this study, we show non-coding RNA regulation on Tet family may contribute to epigenetic regulation during ESCs differentiation, which is suggested by microRNA-29b (miR-29b) binding sites on the Tet1 3′ untranslated region (3′ UTR). We demonstrate miR-29b increases sharply after embyoid body (EB) formation, which causes Tet1 repression and reduction of cellular 5hmC level during ESCs differentiation. Importantly, we show this miR-29b/Tet1 regulatory axis promotes the mesendoderm lineage formation both in vitro and in vivo by inducing the Nodal signaling pathway and repressing the key target of the active demethylation pathway, Tdg. Taken together, our findings underscore the contribution of small non-coding RNA mediated regulation on DNA demethylation dynamics and the differential expressions of key mesendoderm regulators during ESCs lineage specification. MiR-29b could potentially be applied to enrich production of mesoderm and endoderm derivatives and be further differentiated into desired organ-specific cells.
INTRODUCTION
Understanding how embryonic stem cells (ESCs) differentiate into different functional cellular lineage is a key issue in ESCs biology (1). As an embryo develops, ESCs respond to cellular signals and differentiate to different germ layers (ectoderm, mesoderm and endoderm) followed by differentiation into various types of tissues and functional organs. This unique pluripotent property makes ESCs an ideal source for regenerative therapy. A similar process can be achieved in in vitro by inducing ESCs differentiation to specific tissue lineages through formation of embryoid bodies (EBs), which are cell aggregates that resemble the embryo at the blastocyst stage. However, a major challenge in this in vitro tissue regeneration process is inefficient differentiation toward desired therapeutic cell types due to the presence of unwanted differentiated cells of other germ layers (2). Therefore, delineating the key mechanisms in ESCs lineage development will circumvent such bottleneck in regenerative medicine.
Other than dynamic transcriptional regulations, epigenetic modifications are actively involved in ESCs development. Epigenetic modifications in form of cytosine methylation at the 5′ position (5mC) (3) in the genome have been shown to contribute to self-renewal and differentiation of ESCs (4). Recently, the novel cytosine modification known as 5-hydroxymethylcytosine (5hmC), has emerged as another significant epigenetic mark in mammalian development. 5hmC was initially identified in the T-even bacteriophage around six decades ago. Due to the recent identification of Ten-eleven translocation (Tet) family responsible for conversion of 5mC to 5hmC by oxidation (5). 5hmC is now regarded as an important intermediate in passive and active DNA demethylation pathways. Dynamic 5hmC changes have been found in many developmental processes (6). Studies document cellular 5hmC levels increases during preimplantation development and are enriched in the inner cell mass (ICM) of the blastocyst (7,8), but its level is gradually reduces during ESCs differentiation (except neural differentiation) (9). Tet1 and Tet2 are the key enzymes responsible for 5hmC maintenance in mouse ESCs and induced pluripotent stem cells (iPSCs). Both enzymes are regulated by the pluripotent transcription factor Oct4 (9). Tet1-dependent 5hmC level is responsible for loss of ESCs identity (10) and lineage differentiation potential (9). Through these studies provided solid cellular evidence about the functions of Tet1 and Tet2 in ESCs development, their molecular regulation and the regulatory network of Tet1 and Tet2 mediated 5hmC regulation in ESC development remain inconclusive. The study by Ito et al. (8) showed Tet1 repression caused overt ESCs differentiation, diminished ESCs proliferation and led to down-regulation of pluripotency factors Oct4, Sox2 and Nanog, while another report suggested that Tet1 could affect ESCs lineage differentiation through the Nodal signaling pathway and transcription factors involved in mesoderm/endoderm development (9).
During the past decade, microRNAs have been documented to be actively involved in various developmental and cellular processes, including organogenesis and differentiation (11). They represent a group of highly conserved short non-coding RNAs that suppress gene expression by binding to the 3′ untranslational region of protein coding genes (11). MicroRNAs have crucial roles in the self-renewal and differentiation of ESCs. Various studies have demonstrated microRNAs regulate ESCs development by acting on epigenetic, transcriptional and post-transcriptional levels (12,13). Specifically, the miR-29 family regulates various stem cell processes, including osteogenic stem cell differentiation (14) and induced pluripotent stem cells reprogramming (15). Emerging evidence also suggests that the miR-29 family contributes to epigenetic regulation in cancer and primordial germ cells (PGCs) development by targeting Dnmt3a/3b (16), leading to global DNA hypomethylation. Taken together, these studies implicate a possible role of miR-29 in epigenetic regulation during stem cell development. However, how microRNAs contribute to DNA demethylation and regulate ESC lineage commitment remains unclear. Since our preliminary bioinformatics analysis indicated the presence of multiple miR-29 binding sites on the 3′ UTRs of Tet mRNAs, we therefore sought to examine the regulatory role of miR-29 on demethylation pathways during ESC differentiation.
In this study, we show that miR-29b promotes in vitro ESC differentiation through the Tet1-mediated demethylation pathway. We show miR-29b reduces cellular 5hmC levels during mESCs differentiation via Tet1 repression, and regulates mesendoderm-specific differentiation of mESCs both in vitro and in vivo. Furthermore, we show that the miR-29b-Tet1 axis promotes mesendoderm lineage formation by inducing the Nodal signaling pathway and mesendoderm-related gene expression. We also find miR-29b represses the key target of the active demethylation pathway, Tdg and leads to alteration of active demethylation marks 5fC and 5caC. The findings could potentially be applied to enrich production of mesoderm and endoderm derivatives and their further differentiation into desired organ-specific cells.
MATERIALS AND METHODS
Cell culture and differentiation
Mouse E14Tg2A ES cells were maintained on 0.1% gelatin-coated culture plates in Dulbecco's Modified Eagle's Medium (DMEM, GIBCO), supplemented with 15% ES-qualified fetal bovine serum (ES-FBS, GIBCO), 55 mM β-mercaptoethanol (GIBCO), 2 mM L-glutamax (GIBCO), 0.1 mM non-essential amino acid (NEAA, GIBCO), gentamycin (GIBCO) and 1000 U/ml LIF (ESGRO, Millipore) under feeder-free condition. For regular maintenance of cell line the glycogen synthase kinase 3β inhibitor (CHIR99021) and MAPK/ERK kinase inhibitor (PD0325901) were added to a final concentration of 3 μM and 0.2 μM, respectively. Cells were passaged every 2-3 days by dissociation with recombinant trypsin (Sigma).
SiRNA transfection and miRNA overexpression
RNA interference (RNAi) experiments were performed using siRNA (GenePharm) against mouse Tet1 and Tet2. E14 cells were seeded in gelatin-coated 12-well at a density of 1 × 105 cells per well and transfected the day after (day 0) with 50 nM siRNA using Lipofectamine 2000 reagent (Invitrogen) according to the manufacturer's instructions. MiRNA mimics of miR-29b also were ordered from GenePharm, and the transfection procedure of mimics was similar to siRNA transfection. To generate miRNA stable expressing mESCs, miRNA fragment with ∼200 bp flank regions was inserted into the pcDNA3.1 (pcDNA-pre-miR-29b) for transfection in mESCs. G418 selection (1 mg/ml, Sigma) was conducted for 2 weeks to select the miR-29b stable overexpressing mESCs.
Embryoid Bodies (EBs) formation and specific induced differentiation
For LIF withdrawal and differentiation assays, cells were cultured by the hanging drop method in 10 cm petri dish and LIF was removed the day after (day 0). After 2d EB formation by the hanging drop method, EBs were transferred to petri plates for suspension cultures up to 8 days. Samples were collected at day 2, day 4, day 6 and day 8. The morphology and number of EBs were also noted. Retinoic acid (RA) or Activin A (Act A) was used to induce specific germ layer differentiation. Ectoderm was induced by Retinoic acid (10−9M, Sigma), mesendoderm was induced by different concentrations of Activin A (2.5 ng/ml for mesoderm and 50 ng/ml for endoderm inductions, R&D Systems).
RNA Extraction, cDNA synthesis and quantitative real-time polymerase chain reaction (PCR)
Total RNA was extracted by Trizol reagent (Invitrogen) according to standard protocol. The concentration and quality of all RNA samples were evaluated by Nanodrop 2000 (Thermo), and the 260/280 and 260/230 values of all samples were above 1.8 and 1.9 respectively. Reverse transcription was performed with MasterMix kit (Takara) following the standard manual. Quantitative PCR was performed using Universal SYBR Green Master mix (Applied Biosystems) on a StepOnePlus real-time PCR system (Applied Biosystems). Gene expression was normalized to Gapdh unless otherwise stated. The complete primer list is included in the supplementary information.
Fluorescence-activated cell sorting (FACS) analysis
Gata4 FACS analysis was performed according to the general protocol. mESCs were prepared at a concentration of 1 × 106cells/500 uL in rinse buffer and analyzed by Flow cytometer (BD LCSRFortessa Cell Analyzer). Both Gata4 FACS antibody and isotype control (IgG) were from Abcam.
Teratoma formation
mESCs were trypsinized and resuspended at a concentration of 1 × 106 cells/100 μl and injected into nude mice subcutaneously. After ∼5–8 weeks, teratomas were harvested for qRT-PCR and histologic analysis when tumors exceeded 2.0 cm in diameter and were fixed overnight in 4% paraformaldehyde. Paraffin sections and H&E staining were performed according general protocol. Animal handling and maintenance were performed in accordance with institutional guidelines.
Western blot
Cells were lysed in sodium dodecyl sulphate buffer. The protein concentration was measured by BCA assay kit (Thermo). Equal amounts of cell lysates were loaded, blotted onto a PVDF membrane and probed with the following primary antibodies: anti-Tet1 (1:500, Abcam), anti-Tet2 (1:1000, Millipore), anti-Gapdh (1:4000, Abcam) anti-Lefty 1/2 (1:1000, Abcam). Gapdh was used as loading controls. After incubation with the appropriate secondary antibodies, signals were visualized by enhanced chemiluminescence (GE systems).
Luciferase reporter assay
HEK293T cells grown in 24-well plates were transfected with 50 nM miR-29b mimics (GenePharma), 100ng of pmirGLO vector (Promega) tagged with Tet1, Tet2 and Tdg 3′ UTRs that includes miR-29b binding sites or empty pmirGLO plasmid by using Lipofectamine 2000 (Invitrogen). The Firefly and Renilla luciferase activities in the cell lysates were assayed with a Dual-Luciferase Reporter Assay System (Promega) at 48h post-transfection. To generate the mutant variants, point mutations in the binding sites of miR-29b in 3′ UTRs of Tet1, Tet2 and Tdg were introduced by PCR according to in vitro mutagenesis protocol from Cold Spring Harbor (17).
5hmC Dot blot
Genomic DNA samples were prepared with 2-fold serial dilutions in TE buffer and then denatured in 0.4 M NaOH at 72°C for 10 min. Denatured DNA samples were spotted on a PVDF membrane. The membrane was baked at 80°C for 10 min and crosslinked by UV for 10 min. Then the membrane was blocked with 5% blocking buffer for 1 h, incubated with 5-hmC primary antibody (1:5000, Active Motif) 1h and after incubation with the HRP-conjugated rabbit secondary antibodies (1:10000, GE systems), signals were visualized by enhanced chemiluminescence.
Immunofluorescence
1 × 105 cells were cultured on a cover glass in a 12-well plate with 700 ul of medium. The cells were allowed to grow to desired morphology and density before staining procedure. To stain the cells, cells were first washed once with phosphate buffered saline (PBS) and fixed by 4% paraformaldehyde/4%sucrose in PBS at room temp, followed by permeabilization and DNA denaturation by 0.2% TritonX-100 in 4M HCl. After that, the cells were washed with PBS and blocked in 80 μl BSA (3%). The cells were incubated with rabbit anti-5hmC antibody (1:500, Active Motif) in BSA (3%) at 4°C overnight, and then conjugated with RED-X-conjugated mouse anti-rabbit monoclonal antibody (1:500, Santa Cruz) and DAPI (1:1000, Santa Cruz). The glass slides were mounted with a cover slip before imaging.
DNA extraction and GLIB-PCR
Genomic DNA was extracted using a genomic DNA extraction kit (Invitrogen). The concentration and quality of genomic DNA were evaluated by Nanodrop 2000 (Thermo), all samples fit the criteria for following assay. GLIB (Glucosylation, periodate oxidation and biotinylation) was precipitated using a Hydroxymethyl Collector (Active Motif) according to the manufacturers’ protocol. PCR primers for regional 5hmC level see supplementary information.
Generation of stable knockdown ES cell lines
Tet1 and Tet2 shRNA constructs were designed using the on-line design program from MIT (http://sirna.wi.mit.edu/home.php). The 19-nucleotide hairpin-type shRNAs with a 9-nucleotide loop were cloned into pSUPER-puromycin (OligoEngine) according to manufacturers’ protocol. mESCs were transfected by lipofectamine 2000 with pSuper-puro shRNA constructs and selected for 14 days in media containing 1 μg/ml puromycin. Clones were picked and screened by qRT-PCR for knockdown of target RNA expression. Established stable clones were subsequently maintained in the absence of puromycin without loss of knockdown assessed after up to 10 serial passages. shRNA oligo sequences see supplementary information.
Alkaline phosphatase (ALP), CCK-8 and propidium iodide (PI) staining assays
ALP activity detection was carried out using the Blue-color AP staining Kit (SBI) according to the manufacturer's protocol. Cell proliferation rate was measured using cell counting kit-8 (DOJINDO) according to the manufacturer's protocol. Cell cycle regulation was determined by PI (Sigma) staining assay that according to general protocol.
Statistical analyses
The error bars represent the Standard Error of Mean (SEM) of three independent experiments. *, ** and *** indicate P < 0.05, P < 0.01 and P < 0.0001, respectively (Student's t-test).
RESULTS
MiR-29b is significantly up-regulated during mESCs differentiation and is negatively associated with stemness of mESCs
The mouse miR-29 family contains three members, miR-29a, miR-29b and miR-29c, which are encoded from two intragenic clusters on chromosome 1 (miR-29a and miR-29b-1) and chromosome 6 (miR-29b-2 and miR-29c) respectively. Mature miR-29b sequence is derived from both pre-miR-29b-1/b-2 sequences. To examine the role of miR-29 in mESCs differentiation, we first examined the expression profiles of miR-29 precursors (pre-miR-29a, pre-miR-29b-1, pre-miR-29b-2 and pre-miR-29c) in mESCs (Figure 1A). Pre-miR29b-1/2 expression contributed majority of miR-29 cluster expression in mESCs (Figure 1A), suggesting miR-29b is the key miR-29 family member in mESCs. MiR-29b showed association with differentiation state as supported by higher expression in somatic cells, mouse embryonic fibroblasts (MEF) (Figure 1B). Using established mESCs differentiation model, mature miR-29b showed robust up-regulation (>20-fold) during embryonic bodies (EB) formation from day 2 to day 8 (Figure 1C), suggesting miR-29b's may play a role in mESCs differentiation.
Figure 1.
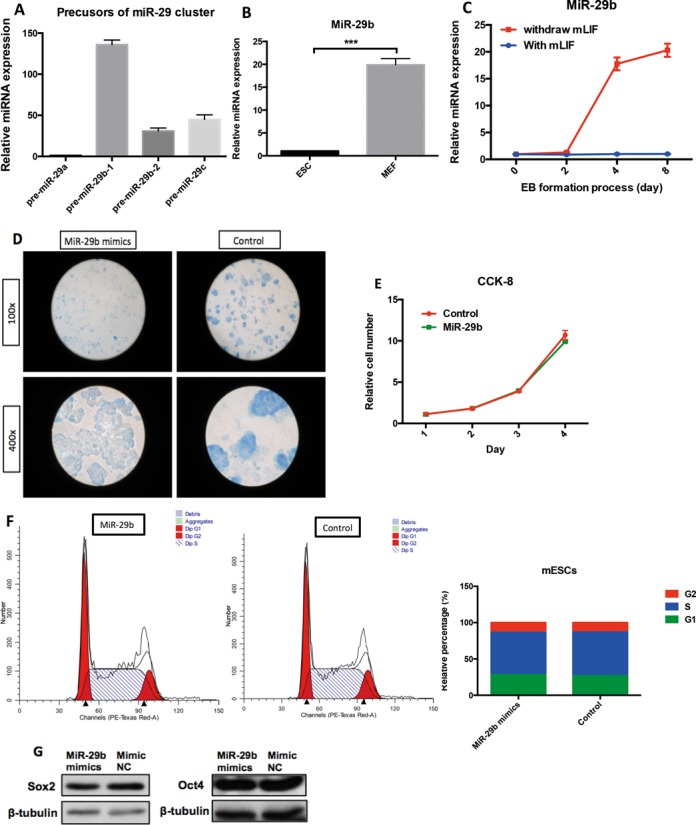
Expression pattern of miR-29 cluster and role of miR-29b in mESCs. (A) MiR-29 cluster precursors (pre-miR-29a, pre-miR-29b and pre-miRNA29c) expression in mESCs by quantitative real-time PCR. (B) MiR-29b expression pattern in the presence and absence of mLIF during EB formation. Shown here are 0-day, 2-day, 4-day and 8-day interval by quantitative real-time PCR. (C) Comparison of miR-29b expression in mESCs to MEF control by quantitative real-time PCR. (D) The effect of miR-29b on alkaline phosphotase activity of mESCs, which was carried out using the Blue-color AP staining Kit (SBI). (E) The effect of miR-29b on proliferation of mESCs. (F) The effect of miR-29b on cell-cycle of mESCs. (G) The effect of miR-29b on two pluripotent transcriptional factors (Sox2 and Oct4) by western blot in mESCs. Each bar in the figure represents the mean ± SEM of triplicates. *P < 0.05, **P < 0.01 and ***P < 0.0001.
To analyze the effect of miR-29b on pluripotency of mESCs, the endogenous alkaline phosphatase (ALP) activity (indicator of ESCs stemness) was examined in miR-29b overexpressing mESCs (MiRNA mimics overexpression efficiency sees Supplementary Figure S1). mESCs transfected with miR-29b mimics showed weaker ALP staining (Figure 1D), indicating that miR-29b could repress mESCs pluripotency. Cell proliferation of mESCs was not affected by miR-29b overexpression (Figure 1E), which excluded the possibility that the repression of ALP activity was due to the anti-proliferative effect of miR-29b overexpression. In addition, cell cycle analysis showed that no difference between miR-29b mimics and control group in mESCs (Figure 1F). To our surprise, miR-29b did not affect the expression of the essential pluripotency markers, Oct4 and Sox2 (Figure 1G). These results suggested that miR-29b is a negative regulator of mESCs pluripotency, which may involve in alternative molecular pathways independent of Oct4 or Sox2.
mESCs overexpressing miR-29b exhibit increased mesendodermal and decreased ectodermal propensities
To better understand the role of miR-29b in mESCs development, we generated an ES cell line stably over-expressing (OE) miR-29b by cloning pre-miR-29b sequence and its flank regions in an expression plasmid, followed by G418 selection. Interestingly, as early as day 5 of selection, we observed the first alteration of morphological differentiation where the cells started to flatten-out. Subsequently, most colonies lost their typical undifferentiated mESCs appearance (homogeneous morphology, round and monolayer colony) and aggregated to form three-dimensional colonies (Figure 2A).
Figure 2.
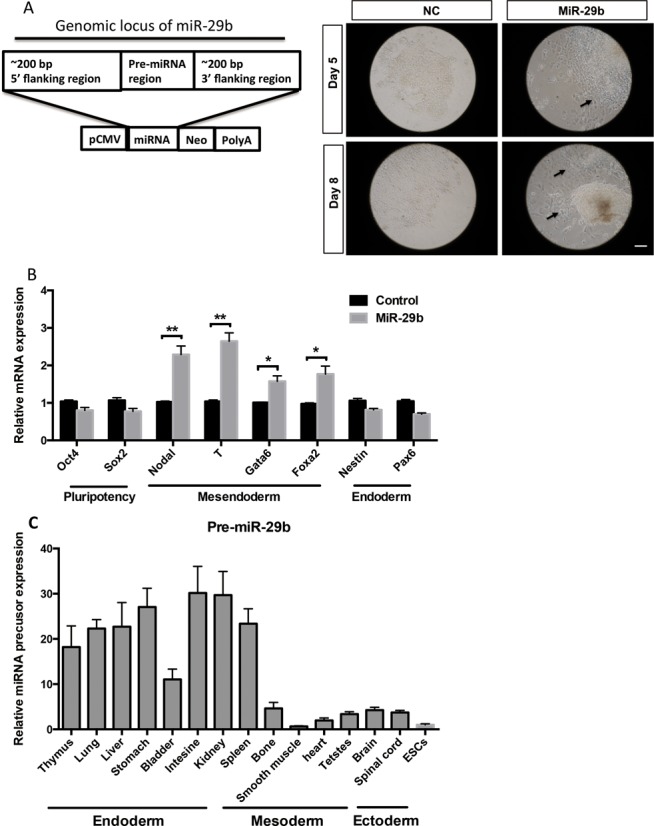
The effect of miR-29b on mESCs differentiation. (A) Scheme of the constructs used to express miRNA of interest. Representative bright-field images from pcDNA-pre-miR-29b mESCs after five or eight days of G418 induction. Scale bars: 50 μm. (B) The effect of miR-29b on pluripotent, ectoderm and mesendoderm markers expression in mESCs by qPCR. (C) Expression of miR-29b precusor in 15 mouse tissues was measured by qRT-PCR and normalized to Gapdh mRNA levels. Each bar in the figure represents the mean ± SEM of triplicates.
To identify the properties of colonies, we examined the expression of the different germ layer markers. We observed mesendoderm makers, including Nodal, T, Gata6 and Foxa2 were strongly up-regulated in miR-29b OE mESCs compared to the control (Figure 2B), while ectodermal markers, such as Nestin and Pax6, were considerably suppressed. Notably, miR-29b precusor was highly expressed in the inner organs (most of all differentiated from mesoderm and endoderm), moderately expressed in the ectoderm-derivative tissues, such as brain and spinal cord (Figure 2C). Taken together, constitutive miR-29b expression predisposes undifferentiated mESCs to express lower levels of ectoderm genes and higher mesendoderm markers, combined with expression level of miR-29b precursor in differentiated organs, suggesting its possible role in contributing a differentiation propensity on subsequent lineages.
MiR-29b promotes differentiation of embryoid bodies toward mesendoderm lineage
To evaluate whether miR-29b is involved in promoting the differentiation propensity of mESCs, we studied embryoid bodies (EBs) formation by removing mLIF. The sizes of miR-29b overexpressed embryonic bodies were significantly larger than the ones in control group (Figure 3A). All mesendoderm markers were significantly up-regulated in miR-29b OE EBs, whereas ectoderm markers showed lower expression (Figure 3B). The gradually increased levels of miR-29b also suggest progressive involvement of this miRNA in the transition from pluripotency toward acquisition of lineage-specific fates. To sum, the results indicate miR-29b may preferentially drive mesendoderm, but not ectoderm, differentiation in EBs.
Figure 3.
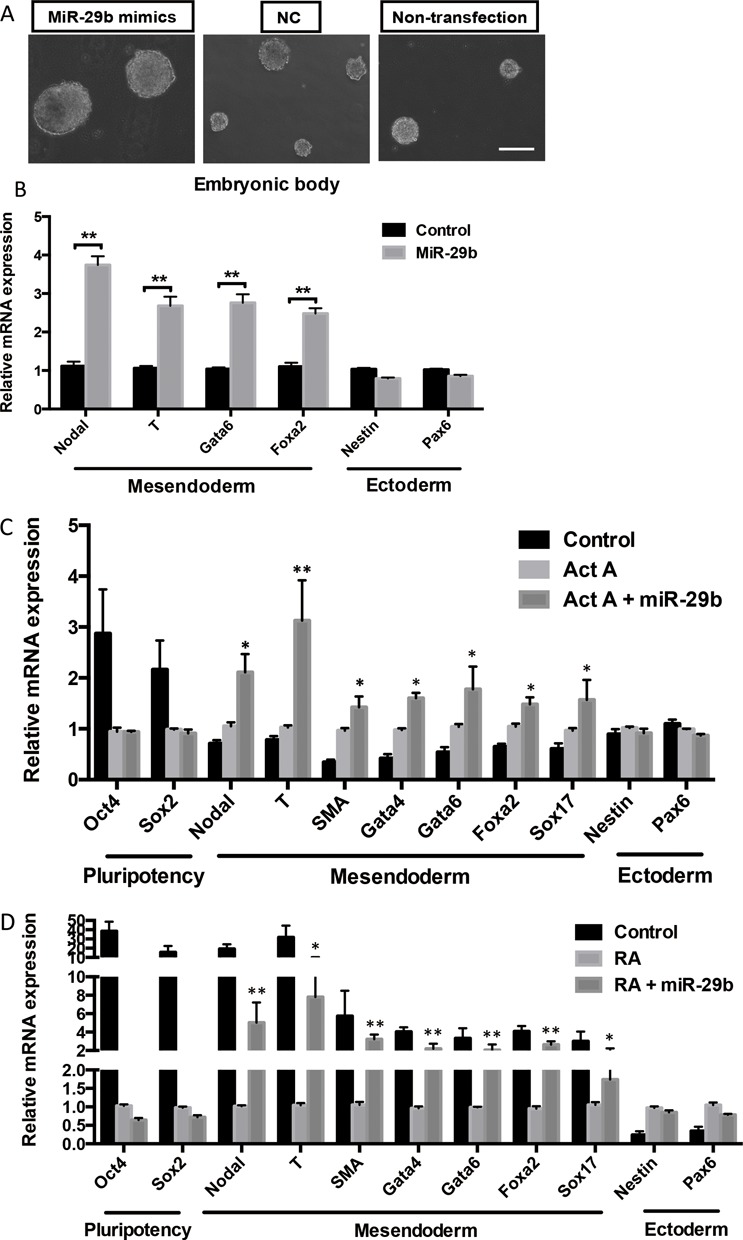
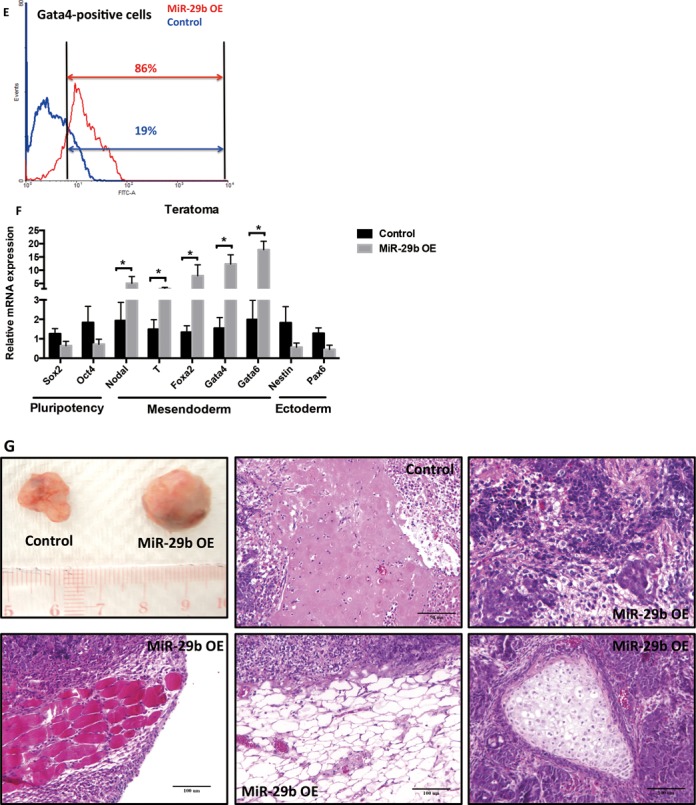
The effect of miR-29b on EB formation and differentiation by in vitro and in vivo assyas. (A) Representative bright-field images of EB formation induced by miR-29b at day 3. Scale bars: 100 μm. (B) The effects of miR-29b on different lineage markers expression in EB by qPCR. (C and D) The effects of miR-29b on different lineage markers expression of RA or Act A-induced mESCs differentiation model by qPCR. (E) Flow cytometry analysis of Gata4-postive cells in miR-29b OE mESCs. (F) Expression of differentiation markers in mIR-29b OE mESCs-formed teratoma tissues. (G) Hematoxylin/eosin (H&E) staining results of miR-29b OE-teratoma and control tissues. Each bar in the figure represents the mean ± SEM of triplicates. *P < 0.05, **P < 0.01 and ***P < 0.0001.
To test our hypothesis, we further examined how miR-29b would act on defined lineage differentiation conditions by inducing mesendoderm and ectoderm formation through administration of Activin A (Act A) and retinoic acid (RA) respectively. As expected, Act A treatment repressed pluripotent markers expression and induced expression for all selected mesendoderm markers, but not ectoderm markers in mESCs (Figure 3C). Transfection of miR-29b mimics further promoted mesendoderm maker expression in the presence of Act A. Similarly, miR-29b rescued mesendoderm marker expression in the cultural environment permissive for ectoderm differentiation (in the presence of RA), no significant expression was observed for ectoderm markers (Figure 3D). Flow cytometry was performed to evaluate the differentiation efficiency of mesendoderm lineage in miR-29b OE mESCs by calculating the Gata4-positve cell population. MiR-29b overexpressing mESCs led to the emergence of subpopulations expressing Gata4 as early as day 5, while control mESCs remained Gata4 negative during the same condition (Figure 3E). In addition, teratoma formation was used to explore miR-29b in modulating germ layer differentiation of mESCs in vivo. MiR-29b OE mESCs and control mESCs were injected into nude mice. After ∼ 5–8 weeks, miR-29b OE mESCs formed large aggressive tumors while control mESCs formed smaller benign teratoma (Figure 3G). Consistent with in vitro results, mesendoderm markers were induced and ectoderm markers were repressed in miR-29b OE mESCs-formed teratoma tissues (Figure 3F). By Histological analysis, all three primary germ layer lineages could be shown in control mESCs-fomed teratoma; while miR-29b OE teratoma showed many mature mesoderm and endoderm tissues and highly proliferative state (Figure 3G). These results further support that miR-29b increases mesendodermal commitment in vivo.
MiR-29b targets Tet1 and Tet2 during mESC differentiation
As the Tet family was associated with mESCs differentiation, we examined the how Tet family contributed to the dynamics of 5hmC regulation during differentiation. We first examined all three Tet family members (Tet1, Tet2 and Tet3) expression patterns during EB formation (Figure 4A). Tet1 and Tet2 were maintained at relatively high expression levels similar to other critical pluripotency factors (Oct4, Sox2 Klf4 and Nanog) in the presence of mLIF (Supplementary Figure S3B). The expression of Tet1 and Tet2 was also more abundant in MEF compared to undifferentiated mESCs (Figure 4B). Intriguingly, the responses of Tet1 and Tet2 to mLIF were very similar. In contrast to miR-29b expression profile, Tet1 and Tet2 expression decreased progressively after mLIF withdrawal (Figure 4A), suggesting both could be involved in similar mode of regulation (data not shown). Tet3, on the other hand, demonstrated a totally opposite response when compared to Tet1/2, implicating a different mode of regulation.
Figure 4.
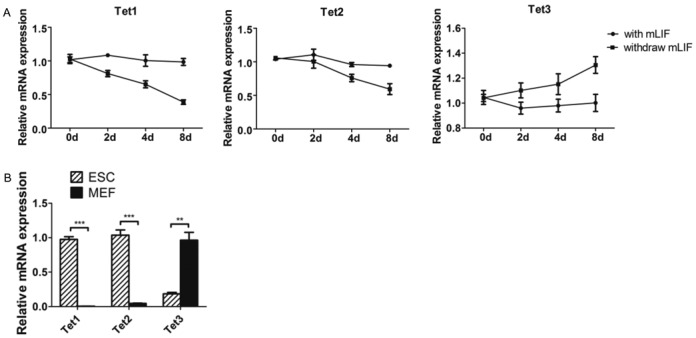
Expression pattern of Tet family in undifferentiated mESCs, differentiating EB and differentiated MEF. (A) The expression pattern of Tet family during EB formation. (B) The expression level of Tet family in mESCs and MEF. Each bar in the figure represents the mean ± SEM of triplicates. *P < 0.05, **P < 0.01 and ***P < 0.0001.
As suggested by the inverse expression pattern of Tet1/2 and miR-29b during EB differentiation, and the bioinformatic prediction that Tet1 and Tet2 are potential targets of miR-29b (Figure 5A), we hypothesized that miR-29b could repress Tet1 and Tet2 and subsequently affect active demethylation processes through alteration of global 5hmC level. This was first validated by transfection of miR-29b mimics in mESCs, which caused significant repression of Tet1 and Tet2 in mESCs at both transcriptional and post-transcriptional levels (Figure 5B and C). To show the direct interaction of miR-29b and Tet1/2, we cloned mouse Tet1 and Tet2 3′ UTRs that contains predicted miR-29 binding sites to a luciferase gene reporter (pmirGLO-Tet1/2-3′ UTRs) and determined luciferase activity in HEK293T cells transfected with miR-29b mimics. MiR-29b significantly repressed the luciferase activity, confirming both Tet1/2 are the direct binding targets of miR-29b (Figure 5D). Notably, miR-29b failed to alter luciferase reporter plasmid activity when we mutated the binding sites of miR-29 at the 3′ UTRs of Tet1/2 (2 or 3 nucleotides in ‘seed sequence’ of miR-29b in 3′ UTRs of Tet1 and Tet2 were mutated), which further proved the direct interactions of miR-29b and Tet1/2.
Figure 5.
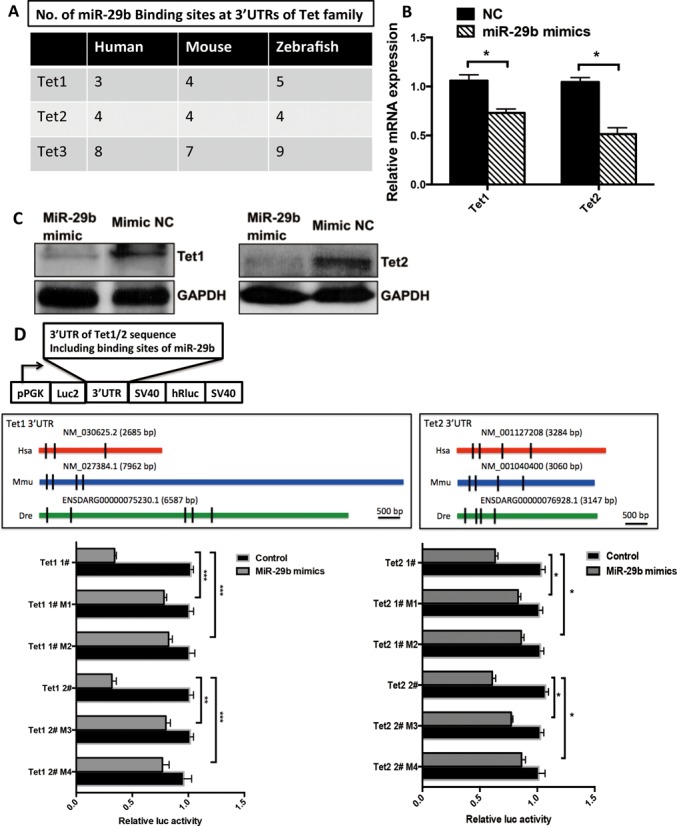
MiR-29b directly targets Tet1/2 in mESCs. (A) No. of miR-29b binding sites at 3′ UTRs of Tet family by bioinformatics prediction software. (B and C) Tet1/2 expression was repressed by miR-29b on both RNA and protein level in mESCs by real-time qPCR and western blot analysis. (D) The structure of pmirGLO dual luciferase vector and detection of direct binding between miR-29b and Tet1/2 3′ UTRs by luciferase reporter assay. Shown here are the site locations for the Tet1/2 inserts on pmirGLO Dual-Luciferase miRNA Target Expression Vector. Decrease in luciferase activity indicates positive binding. MiR-29 mimics and the luciferases reporter were co-transfected by transient transfection using HEK293T cells. A combination of controls were employed, including mutant vector and negative control of miR-29 mimics. Each bar in the figure represents the mean ± SEM of triplicates. *P < 0.05, **P < 0.01 and ***P < 0.0001.
MiR-29b represses cellular 5hmC level during mesendoderm differentiation through Tet1 inhibition
Recent studies have shown global 5hmC decreases when Tet1 and Tet2 are repressed in mESCs. We next checked whether miR-29b could lead to the similar effect. As expected, repression of Tet1 and Tet2 by siRNAs led to 5hmC reduction (Figure 6A and Supplementary Figure S2). Ectopic expression of miR-29b caused reduction of cellular 5hmC level as shown by dot blot and immunofluorescence analyses (Figure 6B and C). To further examine the effect of miR-29b on induced mesendoderm differentiation, we selected SMA and Gata4 as the representative markers of mesoderm and endoderm. Immunofluorescence staining confirmed that miR-29b could induce expression of these two markers with different concentrations of Act A (Figure 6G and H), indicating that miR-29b could promote the differentiations of specific mesoderm and endoderm. Importantly, the expression of both markers inversely correlated with cellular 5hmC level (Figure 6G and H), which suggested that miR-29b also played a role in the late stage differentiation from mesendoderm further to mesoderm and endoderm.
Figure 6.
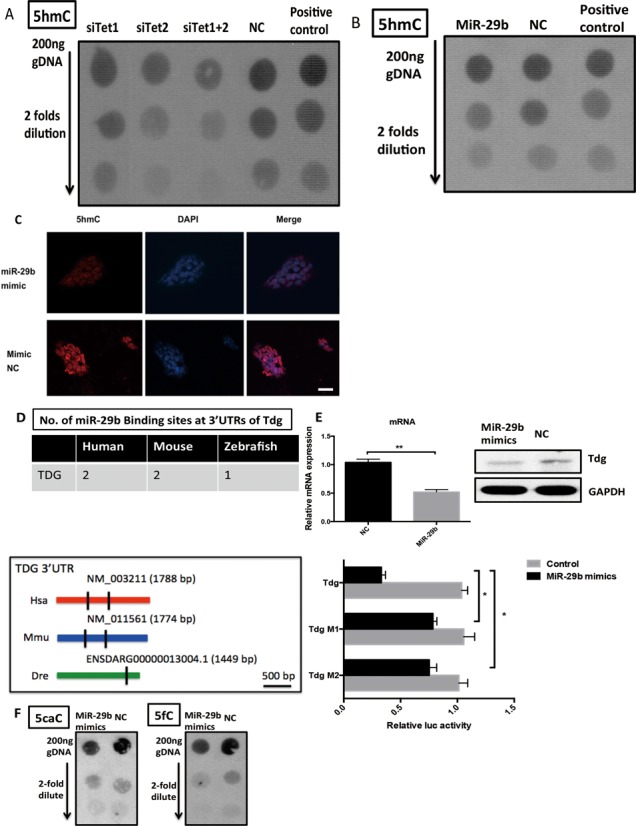
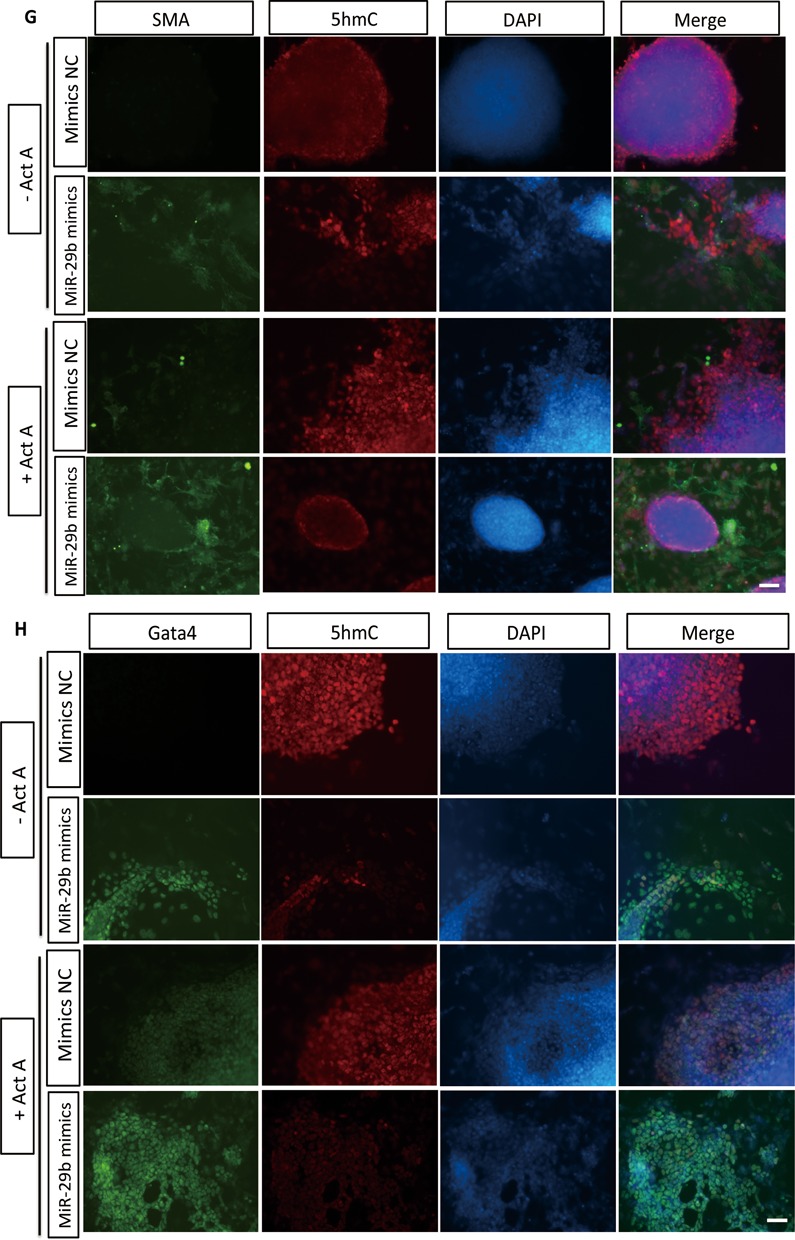
The effects of MiR-29b on global 5hmC, 5caC, 5fC levels and Tdg in mESCs. (A) The effects of Tet1/2 on 5hmC in mESCs by dot blot. Serial 2-fold dilution was applied. (B) The effects of miR-29b on global 5hmC by dot blot. Serial 2-fold dilution was applied in vertical order on mESC transfected with miR-29b mimics and control. (C) The effects of miR-29b on global 5hmC by immunofluorescence. The scale bar is 40 μM. (D) The effects of miR-29b on Tdg and global 5caC and 5fC levels in mESCs. Online prediction software was used to predict potential No. of miR-29b binding sites at Tdg 3′ UTR region. Tdg expression level was repressed via qPCR and western-blot assays. The direct binding between miR-29b and Tdg 3′ UTR was proved by dual luciferase reporter assay. (E and F) The immunostaining results of mesoderm and endoderm markers (SMA and Gata4) in miR-29b overexpressed mESCs and control group. The scale bar is 10 μM. Each bar in the figure represents the mean ± SEM of triplicates. *P < 0.05, **P < 0.01 and ***P < 0.0001.
We also evaluated the effect of Tet1 and Tet2 on the pluripotent state of mESCs by alkaline phosphatase (ALP) assay. Tet1 stable KD mESCs (Supplementary Figure S4) showed decreased ALP level in mESCs (Figure 7A) but was negative for Tet2 KD mESCs (Supplementary Figure S5A). Besides, three important pluripotent transcriptional factors (Sox2, Oct4 and Nanog) were not affected in Tet1 KD mESCs at both mRNA and protein levels (Figure 7B and C), which is consistent with the effects of miR-29b on these transcriptional factors. Then we focused the role of Tet1 in mESCs differentiation. As expected, knockdown Tet1 in mESCs also induced mesendoderm marker and repressed ectoderm marker expression (Figure 7D), indicating that miR-29b/Tet1 axis could induce specific lineage differentiation of mESCs. Immunostaining results of SMA and Gata4 further confirmed the role of Tet1 in mESCs differentiation in vitro. Importantly, the differentiation phenotype associated with Tet1 depletion positively correlated with cellular 5hmC content (Figure 7E and F). While knockdown Tet2 in mESCs did not show any obvious differentiation phenotype of mESCs (data not shown), suggesting that other regulatory mechanisms may involve in the regulation of Tet2 in mESCs.
Figure 7.
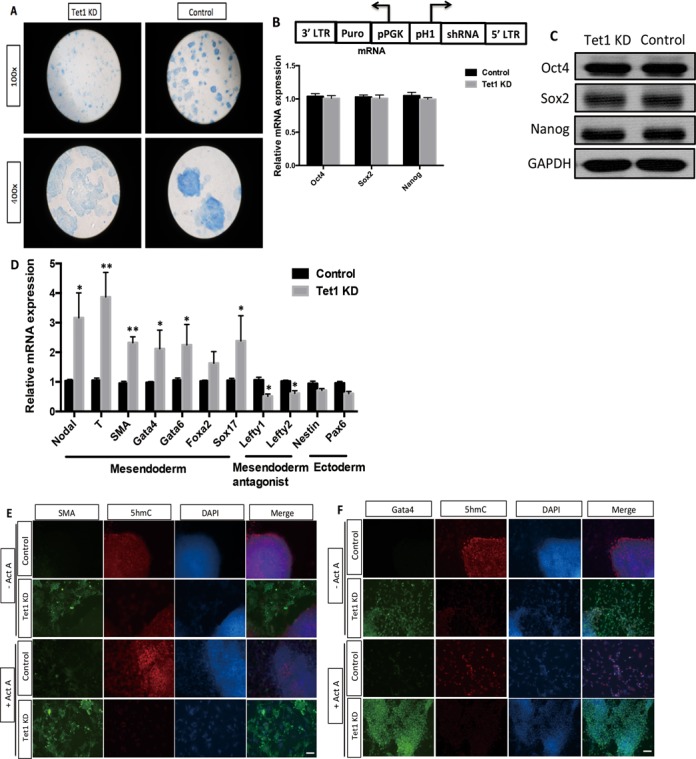
Tet1 KD mESCs inclines to mesendoderm differentiation. (A) The construct of Tet1 shRNA and the effects of Tet1 on ALP activity of mESCs by Blue-color AP staining kit from SBI. (B and C) The expression levels of pluripotent transcriptional factors (Oct4, Sox2 and Nanog) in Tet1 KD mESCs by western blot analysis. (D) The effect of Tet1 on different lineage markers expression of mESCs by qPCR. (E and F) The immunostaining results of mesoderm and endoderm markers, SMA and Gata4 in Tet1 KD mESCs. The scale bar is 10 μM. Each bar in the figure represents the mean ± SEM of triplicates.
MiR-29b may associate with active DNA demethylation by repressing Tdg
Other than passive demethylation, 5hmC can also be converted to unmodified cytosine through active demethylation pathway by thymine DNA glycosylase (TDG)-mediated base excision repair (BER) of 5fC and 5caC. These modified bases are generated by iterative oxidation of 5hmC by Tet enzymes. Interestingly, in addition to Tet family, the bioinformatic prediction suggested Tdg was also a potential target of miR-29b, and the target sites were relatively conserved in human and mouse (Figure 6D). The finding implicated the possible association of miR-29b in the active demethylation pathway. Both RNA and protein expression assays confirmed miR-29b mimics could lead to down-regulation of Tdg (Figure 6E) by around half compared to the controls. The results were also supported by luciferase reporter assay, where miR-29b direct binds to Tdg 3′ UTR. However, only 5fC appeared to be significantly affected by miR-29b, only moderate differences in global 5caC levels were observed (Figure 6F), suggesting that regional level of 5fC/5caC at specific gene maybe more important than overall level change.
MiR-29b/Tet1 regulatory axis involves in epigenetic inhibition of Nodal signaling antagonist Lefy1/2
To further delineate the downstream targets regulated by miR-29b/Tet1 regulatory axis, we searched for established pathways required for directing mesendoderm specification. Enlighten by the results that miR-29b could further promote Act A-induced mesendoderm differentiation, we focused on Actvin/Nodal signaling pathways. The link between miR-29b/Tet1 regulatory axis and Nodal signaling has been indicated by a remarkable up-regulation of Nodal expression in response to miR-29b mimics (>2.6-fold), Activin A plus miR-29b treatment (>2.4-fold) and Tet1 KD condition (>3-fold) (Figures 2B, 3C and 7D). To further delineate the molecular mechanisms, we examined the regulatory role of Lefty1 and Lefty2. Both are the known antagonists of Nodal signaling, and lead to inhibition of mesendoderm differentiation through Activin/TGF-beta/Nodal pathway (18). They are among the earliest genes to be repressed upon ESCs differentiation, and studies have shown both gene promoters are relatively hypomethylated in stem cells and hypermethylated in differentiated cells (9). Therefore, we hypothesized that decreased active demethylation activity (i.e 5hmC levels) of the promoter regions may also contribute to down-regulation of both targets. To examine the 5hmC level on the promoters, we scanned the promoter regions rich in CG contents, four potential 5hmC sites were predicted on Lefty1 and Lefty2 promoters respectively (Figure 8A). The corresponding 5hmC level at these sites was examined in Tet1 KD and miR-29b overexpressing mESCs by GLIB-qPCR approach. The results showed 5hmC level were decreased at all predicted sites on Lefty1 and Lefty2 promoters (Figure 8B), and were concordant with decreased expression profiles (Figure 8C and D). These confirmed that miR-29b-Tet1 axis could affect Lefty1/2 expression through epigenetic repression of 5hmC level on the promoter region.
Figure 8.
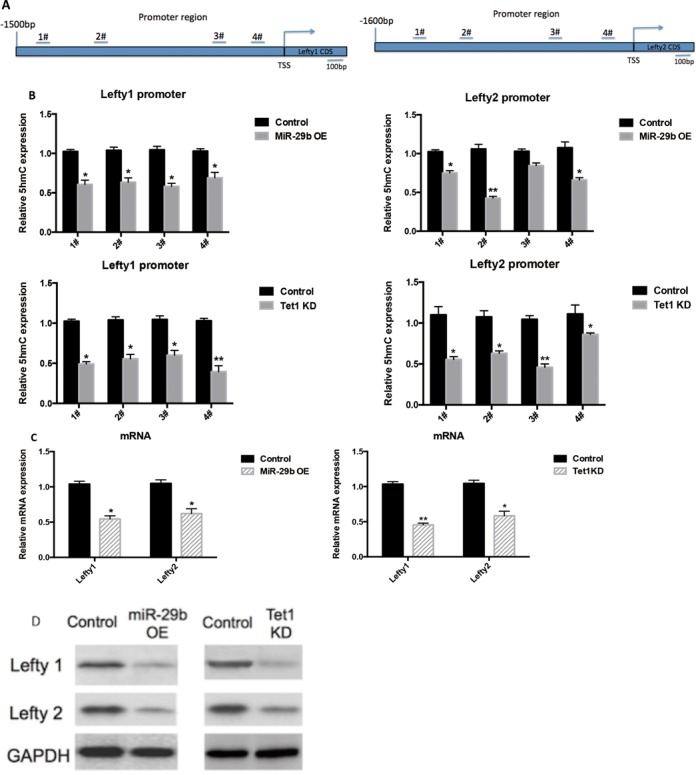
MiR-29b-Tet1 axis epigenetically regulates two key inhibitors of mesendoderm markers. (A) The selected promoter regions of Lefty1/2, the predicted. (B) Real time quantitative PCR showing 5hmC in Lefty1/2 promoters were decreased in both miR-29b OE and Tet1 KD mESCs. (C and D) Western blot showing Lefty1/2 was repressed in miR-29b OE and Tet1 KD mESCs.
DISCUSSION
In this study, we show the mesendoderm lineage specification of mESCs can be promoted by miR-29b (Supplementary Figure S6). Recent genome-wide studies have suggested dynamic DNA methylation is required in various key developmental processes, including embryonic stem differentiation, induced pluripotent cell reprogramming, germ cell and zygote development (19). The maintenance of ES cell depends on precise transcriptional control of pluripotency and lineage-specification factors, which is associated with a distinct methylation pattern. Though TETs and TDG represent the key players in DNA demethylation pathways of the pluripotency regulatory circuit, the underlying molecular mechanism on TET and TDG regulation, the associated gene targets and how to modulation of differentiation remains largely elusive. Here we demonstrate a novel miR-29b/Tet1 axis epigenetically modulates mESCs pluripotency and mesendoderm differentiation. Ectopic expression of miR-29b in mESCs significantly enhanced their potential to commit mesendoderm lineage through induction of Nodal signaling pathway. Notably, miR-29b modulates its action through demethylation pathways by targeting Tet1 and Tdg, leading to decreased 5hmC levels and up-regulation of Nodal signaling antagonists Lefty1/2 and mesoderm/endoderm transcription factors SMA and Gata4. To our knowledge, this is the first report on the modulation of DNA demethylation intermediates by microRNA in ESCs development.
It is now known that microRNA is actively involved in gene regulation of ESCs (11), and miR-29 is no exception. Despite sharing similar seed sequences and that it is conserved in mammalians, miR-29 family members are expressed differently predominantly in various subcellular locations. Expression of miR-29a is cytoplasmic, whereas miR-29b and miR-29c are mainly in the nucleus (20,21). Such difference implies the diverse biological functions among miR-29 members.
Based on recent miRNA studies (22–24), we postulate the expression difference of miR-29b in cytoplasmic and nuclear compartment could attribute to the storage effect or nuclear activities. The nucleolus may serve as a site of storage for miRNAs, which remain inactive until released by cell stress, and/or (ii) that nuclear miRNAs function in a manner distinct from the canonical cytoplasmic RISC paradigm. Given that the nucleolus is the site of ribosomal gene transcription, ribosomal RNA maturation and RNA editing, it is possible that nuclear miRNAs participate in the regulation of these processes, or are themselves subject to RNA editing. Another possibility is interacting with transcriptional factors or other nucleus factors in the nucleus, such as Klf4 (25), Mycn (26), Yy1 (27) and Sp1 (28). Previous studies have also suggested the pivot role of miR-29b during late stage of cellular differentiation in normal and pathological conditions, including skeletal myogenesis (14,27), osteoblast differentiation (29), osteoclastogenesis (30), muscle differentiation (31–33), renal and cardiovascular injury (34,35), pulmonary fibrosis (36), myoblast differentiation (37) and acute myeloid leukemia (38). An interesting phenomenon is that all of these somatic cell lineages are differentiated from mesendoderm, which indicates that miR-29b may play a key role in the late differentiation stage for mesendoderm as was confirmed by our teratoma formation assay (Figure 3). However, the role of miR-29b in the early differentiation from undifferentiated ESCs to mesendoderm is still not completely understood. Similar to other differentiation-related microRNAs in ESCs (11), miR-29b expression increases sharply during mESCs differentiation. Interestingly, such differentiation promotion is not related to repression of pluripotent factors Oct4 and Sox2, which suggests the presence of alternative regulatory pathways in ESCs differentiation. To make the picture even more complicated, we found a subset of microRNAs (miR-22, miR-290) that act on Oct4/Sox2 based on our bioinformatics prediction may also target the Tet gene family based on our bioinformatics prediction (Supplementary Figure S3A). Whether these Oct4/Sox2 related microRNAs also target Tet family members awaits further investigation. We also evaluated some other miRNAs (potential miRNAs that may target Tet family), including miR-26, miR-93, miR-291 and miR-302, during mESCs differentiation and only miR-26b showed increased trend (Supplementary Figure S3C). The bioinformatics prediction and opposite expression trend of miR-26b and Tet1/Tet2 in mESCs differentiation suggested that miR-26b may also play a role in mESCs, just like miR-29b. Therefore miRNAs, Tet family and downstream target/pathway formed a network in ESCs, which connects genetics and epigenetics, coding and non-coding RNAs. More investigation is needed to further illuminate this complicated network.
The potential involvement of miR-29b in epigenetic regulation in development has been largely reflected in DNA methylation, including modulation of Dnmt3a/b expression in primordial germ cells of female mouse embryos (39) and epigenetic modification of MMP-2/9 in cardiovascular diseases (40). Our data clearly demonstrate miR-29 involves in DNA demethylation pathways through Tet and Tdg. In contrast to the recent reports focusing on biochemical validation on miR-29/Tet interactions using fibroblast or cancer lines (31,34), our study demonstrates the functional relationship between miR-29 and Tet using a dynamic developmental model approach that is supported by phenotypic and molecular evidence. Consistent with previous findings (9), we show that Tet1, as well as Tet2, are the key enzymes responsible for the cellular of 5hmC level in mESCs and that their activity correlates closely with the pluripotent state. Although Tet1 and Tet2 demonstrate similar expression profiles in response to mLIF and get repressed by miR-29b, Tet2 fails to affect mESCs differentiation. Our observation is consistent with recent ChIP-seq data (41). Such downstream target specificity may be attributed to the structural difference between Tet1 and Tet2, where Tet1 contains an additional CXXC motif for binding to CpG island (42). The bindings are enriched at the promoter region regardless of modified or unmodified cytosine.
The formation of mesoderm or endoderm from mesendoderm lineage is a critical step in the establishment of subsequent derivatives (43). Modulation by miR-495 in mesendoderm lineage specification of ESCs has also been reported recently (12), but in an opposite way compared to miR-29b. MiR-495 acts as a negative regulator of mesendoderm differentiation through targeting Dnmt3a in mESCs. Both miR-495 and miR-29b demonstrate a sharp change on expression in the first 4 days of EB formation, suggesting they are the essential regulators for early epigenome and transcriptome dynamics. Interestingly, miR-495 appears to regulate DNA methylation only as no targets are predicted on Tet and Tdg. Coupled with miR-29b that acts on DNA demethylation pathway. A ‘yin-yang’ microRNA regulation may exist in mesendoderm lineage development. Delineating how this yin-yang event is regulated would lead to identification of key regulators in mesendoderm lineage development. It is therefore imperative to characterize the genome-wide pattern of 5hmC and other cytosine derivatives, including 5-carboxylcytosine (5caC) and 5-formylcytosine (5fC) in miR-29 induced mESCs differentiation to have a robust understanding of the epigenetic hotspots during mESCs differentiation.
Although we show miR-29b may be involved in the active demethylation pathway by repressing Tdg, the dot blot data on the 5caC and 5fC are inconclusive. We expect miR-29b would cause similar effects in Tdg-deficient cell knock out cells, leading to accumulation of 5caC and 5fC (44). A plausible explanation is that miR-29b targets not only Tdg, but also Tet. Since both enzyme families are involved in conversion of 5fC and 5caC by oxidation and BER respectively, the cellular level of 5fC and 5caC will be affected. Therefore, it is possible that both passive and active demethylation mechanisms are affected by miR-29b.
In summary, we demonstrate miR-29b directly regulates Tet and Tdg in mESCs. Ectopic miR-29 expression activates mesendoderm lineage development through epigenetic modulation of mesendoderm and ectoderm targets, and corresponding signature pathways like Nodal pathway (Supplementary Figure S6). The findings provide a better understanding of the role of microRNA in mESCs lineage specification, which will be helpful in efficient differentiation of ESCs into specific therapeutic cell types of choice without contamination of unwanted differentiated cells from other germ layers. We believe miR-29 induced mesendoderm progenitors will be of great value in regenerative medicine. It can also serve as an experimental platform for studying the molecular regulations of mesodermal and endodermal specification.
Supplementary Material
Acknowledgments
We thank Prof. Bo Feng for providing mESCs line, E14Tg2A, Prof. Kingston Mak for technical support on primary MEF isolation and Prof. Xiaohua Jiang for support on teratoma formation assay.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
General Research Fund from Hong Kong Research Grant Council [469713]; Direct Research Fund from the Faculty of Medicine, The Chinese University of Hong Kong [MD12465] and School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong [4620504]. Funding for open access charge: School of Biomedical Sciences, Faculty of Medicine, The Chinese University of Hong Kong [4620504].
Conflict of interest statement. None declared.
REFERENCES
- 1.Young R.A. Control of the embryonic stem cell state. Cell. 2011;144:940–954. doi: 10.1016/j.cell.2011.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lacoste A., Berenshteyn F., Brivanlou A.H. An efficient and reversible transposable system for gene delivery and lineage-specific differentiation in human embryonic stem cells. Cell Stem Cell. 2009;5:332–342. doi: 10.1016/j.stem.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 3.Farthing C.R., Ficz G., Ng R.K., Chan C.-F., Andrews S., Dean W., Hemberger M., Reik W. Global mapping of DNA methylation in mouse promoters reveals epigenetic reprogramming of pluripotency genes. PLoS Genet. 2008;4:e1000116. doi: 10.1371/journal.pgen.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meissner A., Mikkelsen T.S., Gu H., Wernig M., Hanna J., Sivachenko A., Zhang X., Bernstein B.E., Nusbaum C., Jaffe D.B., et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766-770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tahiliani M., Koh K.P., Shen Y., Pastor W.A., Bandukwala H., Brudno Y., Agarwal S., Iyer L.M., Liu D.R., Aravind L., et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koh K.P., Rao A. DNA methylation and methylcytosine oxidation in cell fate decisions. Curr. Opin. Cell Biol. 2013;25:152–161. doi: 10.1016/j.ceb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ruzov A., Tsenkina Y., Serio A., Dudnakova T., Fletcher J., Bai Y., Chebotareva T., Pells S., Hannoun Z., Sullivan G., et al. Lineage-specific distribution of high levels of genomic 5-hydroxymethylcytosine in mammalian development. Cell Res. 2011;21:1332–1342. doi: 10.1038/cr.2011.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ito S., D'Alessio A.C., Taranova O.V., Hong K., Sowers L.C., Zhang Y. Role of Tet proteins in 5mC to 5hmC conversion, ES-cell self-renewal and inner cell mass specification. Nature. 2010;466:1129–1133. doi: 10.1038/nature09303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koh K.P., Yabuuchi A., Rao S., Huang Y., Cunniff K., Nardone J., Laiho A., Tahiliani M., Sommer C.A., Mostoslavsky G., et al. Tet1 and Tet2 regulate 5-hydroxymethylcytosine production and cell lineage specification in mouse embryonic stem cells. Cell Stem Cell. 2011;8:200–213. doi: 10.1016/j.stem.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freudenberg J.M., Ghosh S., Lackford B.L., Yellaboina S., Zheng X., Li R., Cuddapah S., Wade P.A., Hu G., Jothi R. Acute depletion of Tet1-dependent 5-hydroxymethylcytosine levels impairs LIF/Stat3 signaling and results in loss of embryonic stem cell identity. Nucleic Acids Res. 2012;40:3364–3377. doi: 10.1093/nar/gkr1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gangaraju V.K., Lin H. MicroRNAs: key regulators of stem cells. Nat. Rev. Mol. Cell. Biol. 2009;10:116–125. doi: 10.1038/nrm2621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang D., Wang G., Zhu S., Liu Q., Wei T., Leng Y., Duan T., Kang J. MiR-495 suppresses mesendoderm differentiation of mouse embryonic stem cells via the direct targeting of Dnmt3a. Stem Cell Res. 2014;12:550–561. doi: 10.1016/j.scr.2014.01.005. [DOI] [PubMed] [Google Scholar]
- 13.Tay Y., Zhang J., Thomson A.M., Lim B., Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- 14.Suh J.S., Lee J.Y., Choi Y.S., Chong P.C., Park Y.J. Peptide-mediated intracellular delivery of miRNA-29b for osteogenic stem cell differentiation. Biomaterials. 2013;34:4347–4359. doi: 10.1016/j.biomaterials.2013.02.039. [DOI] [PubMed] [Google Scholar]
- 15.Guo X., Liu Q., Wang G., Zhu S., Gao L., Hong W., Chen Y., Wu M., Liu H., Jiang C., et al. microRNA-29b is a novel mediator of Sox2 function in the regulation of somatic cell reprogramming. Cell Res. 2012;23:142–156. doi: 10.1038/cr.2012.180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Valinluck V., Sowers L.C. Endogenous cytosine damage products alter the site selectivity of human DNA Maintenance methyltransferase DNMT1. Cancer Res. 2007;67:946–950. doi: 10.1158/0008-5472.CAN-06-3123. [DOI] [PubMed] [Google Scholar]
- 17.Sambrook J., Russell D.W. In Vitro mutagenesis using double-stranded DNA templates: selection of mutants with DpnI. CSH Protoc. 2006 doi: 10.1101/pdb.prot3813. doi:10.1101/pdb.prot3813. [DOI] [PubMed] [Google Scholar]
- 18.Whitman M. Nodal signaling in early vertebrate review embryos: themes and variations. Dev. Cell. 2001;1:605–617. doi: 10.1016/s1534-5807(01)00076-4. [DOI] [PubMed] [Google Scholar]
- 19.Borgel J., Guibert S., Li Y., Chiba H., Schübeler D., Sasaki H., Forné T., Weber M. Targets and dynamics of promoter DNA methylation during early mouse development. Nat. Genet. 2010;42:1093–1100. doi: 10.1038/ng.708. [DOI] [PubMed] [Google Scholar]
- 20.Hwang H.-W., Wentzel E.A., Mendell J.T. A hexanucleotide element directs microRNA nuclear import. Science. 2007;315:97–100. doi: 10.1126/science.1136235. [DOI] [PubMed] [Google Scholar]
- 21.Liao J.-Y., Ma L.-M., Guo Y.-H., Zhang Y.-C., Zhou H., Shao P., Chen Y.-Q., Qu L.-H. Deep sequencing of human nuclear and cytoplasmic small RNAs reveals an unexpectedly complex subcellular distribution of miRNAs and tRNA 3′ trailers. PLoS One. 2010;5:e10563. doi: 10.1371/journal.pone.0010563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Politz J.C.R., Zhang F., Pederson T. MicroRNA-206 colocalizes with ribosome-rich regions in both the nucleolus and cytoplasm of rat myogenic cells. Proc. Natl. Acad. Sci. U.S.A. 2006;103:18957–18962. doi: 10.1073/pnas.0609466103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Libert S., Zwiener J., Chu X., Vanvoorhies W., Roman G., Pletcher S.D. Regulation of drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- 24.Humphreys D.T., Hynes C.J., Patel H.R., Wei G.H., Cannon L., Fatkin D., Suter C.M., Clancy J.L., Preiss T. Complexity of murine cardiomyocyte miRNA biogenesis, sequence variant expression and function. PLoS One. 2012;7:e30933. doi: 10.1371/journal.pone.0030933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cittelly D.M., Finlay-Schultz J., Howe E.N., Spoelstra N.S., Axlund S.D., Hendricks P., Jacobsen B.M., Sartorius C.A., Richer J.K. Progestin suppression of miR-29 potentiates dedifferentiation of breast cancer cells via KLF4. Oncogene. 2012;32:2555–2564. doi: 10.1038/onc.2012.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ugalde A.P., Pamsay A.J., de la Rosa J., Varela I., Marino G., Cadinanos J., Lu J., Freije J.M.P., Lopez-Otin C. Aging and chronic DNA damage response activate a regulatory pathway involving miR-29 and p53. EMBO J. 2011;30:2219–2232. doi: 10.1038/emboj.2011.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang H., Garzon R., Sun H., Ladner K.J., Singh R., Dahlman J., Cheng A., Hall B.M., Qualman S.J., Chandler D.S., et al. NF-kappaB-YY1-miR-29 regulatory circuitry in skeletal myogenesis and rhabdomyosarcoma. Cancer Cell. 2008;14:369–381. doi: 10.1016/j.ccr.2008.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu S., Wu L.C., Pang J., Santhanam R., Schwind S., Wu Y.Z., Hickey C.J., Yu J., Becker H., Maharry K., et al. Sp1/NFkappaB/HDAC/miR-29b regulatory network in KIT-driven myeloid leukemia. Cancer Cell. 2010;17:333–347. doi: 10.1016/j.ccr.2010.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Z., Hassan M.Q., Jafferji M., Aqeilan R.I., Garzon R., Croce C.M., van Wijnen A.J., Stein J.L., Stein G.S., Lian J.B. Biological functions of miR-29b contribute to positive regulation of osteoblast differentiation. J. Biol. Chem. 2009;284:15676–15684. doi: 10.1074/jbc.M809787200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Franceschetti T., Kessler C.B., Lee S.K., Delany A.M. miR-29 promotes murine osteoclastogenesis by regulating osteoclast commitment and migration. J. Biol. Chem. 2013;288:33347–33360. doi: 10.1074/jbc.M113.484568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Morita S., Horii T., Kimura M., Ochiya T., Tajima S., Hatada I. miR-29 represses the activities of DNA methyltransferases and DNA demethylases. Int. J. Mol. Sci. 2013;14:14647–14658. doi: 10.3390/ijms140714647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Winbanks C.E., Wang B., Beyer C., Koh P., White L., Kantharidis P., Gregorevic P. TGF- regulates miR-206 and miR-29 to control myogenic differentiation through regulation of HDAC4. J. Biol. Chem. 2011;286:13805–13814. doi: 10.1074/jbc.M110.192625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang X.H., Hu Z., Klein J.D., Zhang L., Fang F., Mitch W.E. Decreased miR-29 suppresses myogenesis in CKD. J. Am. Soc. Nephrol. 2011;22:2068–2076. doi: 10.1681/ASN.2010121278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang P., Huang B., Xu X., Sessa W.C. Ten-eleven translocation (Tet) and thymine DNA glycosylase (TDG), components of the demethylation pathway, are direct targets of miRNA-29a. Biochem. Biophys. Res. Commun. 2013;437:368–373. doi: 10.1016/j.bbrc.2013.06.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kriegel A.J., Liu Y., Fang Y., Ding X., Liang M. The miR-29 family: genomics, cell biology, and relevance to renal and cardiovascular injury. Physiol. Genomics. 2012;44:237–244. doi: 10.1152/physiolgenomics.00141.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cushing L., Kuang P.P., Qian J., Shao F., Wu J., Little F., Thannickal V.J., Cardoso W.V., Lü J. miR-29 Is a major regulator of genes associated with pulmonary fibrosis. Am. J. Respir. Cell Mol. Biol. 2011;45:287–294. doi: 10.1165/rcmb.2010-0323OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei W., He H.-B., Zhang W.-Y., Zhang H.-X., Bai J.-B., Liu H.-Z., Cao J.-H., Chang K.-C., Li X.-Y., Zhao S.-H. miR-29 targets Akt3 to reduce proliferation and facilitate differentiation of myoblasts in skeletal muscle development. Cell Death Dis. 2013;4:e668. doi: 10.1038/cddis.2013.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gong J.-N., Yu J., Lin H.-S., Zhang X.-H., Yin X.-L., Xiao Z., Wang F., Wang X.-S., Su R., Shen C., et al. The role, mechanism and potentially therapeutic application of microRNA-29 family in acute myeloid leukemia. Cell Death Differ. 2013;21:100–112. doi: 10.1038/cdd.2013.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Takada S., Berezikov E., Choi Y.L., Yamashita Y., Mano H. Potential role of miR-29b in modulation of Dnmt3a and Dnmt3b expression in primordial germ cells of female mouse embryos. RNA. 2009;15:1507–1514. doi: 10.1261/rna.1418309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen K.-C., Wang Y.-S., Hu C.-Y., Chang W.-C., Liao Y.-C., Dai C.-Y., Juo S.-H.H. OxLDL up-regulates microRNA-29b, leading to epigenetic modifications of MMP-2/MMP-9 genes: a novel mechanism for cardiovascular diseases. FASEB J. 2011;25:1718–1728. doi: 10.1096/fj.10-174904. [DOI] [PubMed] [Google Scholar]
- 41.Xu Y., Wu F., Tan L., Kong L., Xiong L., Deng J., Barbera A.J., Zheng L., Zhang H., Huang S., et al. Genome-wide regulation of 5hmC, 5mC, and gene expression by Tet1 hydroxylase in mouse embryonic stem cells. Mol. Cell. 2011;42:451–464. doi: 10.1016/j.molcel.2011.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang H., Zhang X., Clark E., Mulcahey M., Huang S., Shi Y.G. TET1 is a DNA-binding protein that modulates DNA methylation and gene transcription via hydroxylation of 5-methylcytosine. Cell Res. 2010;20:1390–1393. doi: 10.1038/cr.2010.156. [DOI] [PubMed] [Google Scholar]
- 43.Tada S., Era T., Furusawa C., Sakurai H., Nishikawa S., Kinoshita M., Nakao K., Chiba T., Nishikawa S. Characterization of mesendoderm: a diverging point of the definitive endoderm and mesoderm in embryonic stem cell differentiation culture. Development. 2005;132:4363–4374. doi: 10.1242/dev.02005. [DOI] [PubMed] [Google Scholar]
- 44.Hu X., Zhang L., Mao S.-Q., Li Z., Chen J., Zhang R.-R., Wu H.-P., Gao J., Guo F., Liu W., et al. Tet and TDG mediate DNA demethylation essential for mesenchymal-to-epithelial transition in somatic cell reprogramming. Cell Stem Cell. 2014;14:512–522. doi: 10.1016/j.stem.2014.01.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


