Abstract
Many antiproliferative G-quadruplexes (G4s) arise from the folding of GT-rich strands. Among these, the Thrombin Binding Aptamer (TBA), as a rare example, adopts a monomolecular well-defined G4 structure. Nevertheless, the potential anticancer properties of TBA are severely hampered by its anticoagulant action and, consequently, no related studies have appeared so far in the literature. We wish to report here that suitable chemical modifications in the TBA sequence can preserve its antiproliferative over anticoagulant activity. Particularly, we replaced one residue of the TT or TGT loops with a dibenzyl linker to develop seven new quadruplex-forming TBA based sequences (TBA-bs), which were studied for their structural (CD, CD melting, 1D NMR) and biological (fibrinogen, PT and MTT assays) properties. The three-dimensional structures of the TBA-bs modified at T13 (TBA-bs13) or T12 (TBA-bs12), the former endowed with selective antiproliferative activity, and the latter acting as potently as TBA in both coagulation and MTT assays, were further studied by 2D NMR restrained molecular mechanics. The comparative structural analyses indicated that neither the stability, nor the topology of the G4s, but the different localization of the two benzene rings of the linker was responsible for the loss of the antithrombin activity for TBA-bs13.
INTRODUCTION
Aptamers are short, structured, single-stranded RNA or DNA ligands characterized by high affinity for their target (1). Thanks to their elevated potential in diagnostic and in therapeutic applications (2,3), as well as in bioengineered constructs (4–6), aptamers are, currently, one of the most investigated class of protein ligands. Many reported aptamers are G-Rich Oligonucleotides (GROs) that bind their targets adopting G-quadruplex (G4) structures (7–9). Very importantly, cell-based screening showed that some GROs selectively act as antiproliferative agents (10–13), including AS1411 (12,14–18) (also named AGRO 100, T3G2TG2TG2TG2TG2T2GTG2TG2TG2); T40214 (19,20) (G3CG3CG3CG3C); TG-ONs (21) [Telomeric G-tail Oligonucleotides, (T2AG3)n]; T-oligo (22,23) (5′-pG2T2AGTAG2T3); enriched deoxyguanosine T-oligo (24) (G2T2)4; Pur27 (25) (TG4AG3TG4AG3TG4A2G2); T-22AG (26) (G3A2GAG3A2GAG5AG2). From the analysis of the literature data, it emerges that the antiproliferative activity is usually associated with GROs incorporating GT-rich or GA-rich sequences (11).
Experimental evidence correlate the antiproliferative action of GROs with the ability to engage a common pathway of cancer cells, triggering the events that lead to cell death (11,21). It is also inferred that GROs can target a specific protein of cancer cells. For example, AS1411 binds with high affinity to nucleolin, a nucleolar protein over expressed in cancer cells (16,18,27), whereas T40214 binds to Stat-3/DNA complex (19,20).
Recently published data further stimulate the debate about the exact mechanism of action of antiproliferative GROs. Indeed, Zhang et al. (28) found that the antiproliferative activity of GROs, including AS1411 that is currently being tested in phase II clinical trials (29), might be contributed by the cytotoxicity of their guanosine-based degradation products. However, some new emerging mechanistic studies strongly suggest that GROs act, at least in part, as antiproliferative agents through their GRO sequences and, consequently, through their ability to fold into G4 structures (30,31).
While it is reasonable to suppose that GROs can target cancer cells through different pathways, poor information, if any, about the structure-activity relationships (SARs) between G-quadruplex typologies and protein interactions are available. Indeed, almost all anticancer GROs are characterized by a high grade of structural polymorphism (32–34). They form in solution several G4 structures differing for the molecularity and/or loop arrangement (35). Therefore, the possibility to fully understand the SAR of each oligodeoxynucleotide (ODN) is definitely subject to the availability of biologically active sequences forming single and well-defined G-quadruplex structures.
Interestingly, a cell-based screening of several GROs demonstrated that the GT-rich 15-mer ODN G2T2G2TGTG2T2G2 (36–44) (TBA, thrombin binding aptamer, an ODN binding and inhibiting thrombin), possesses a significant antiproliferative activity (45). Moreover, an electrophoretic mobility shift assay showed that TBA, like AS1411, is able to bind to a nucleolar protein, thought to be nucleolin (45). A powerful correlation between the stability of the TBA tertiary structure and the two related functional effects has been shown as well. Both the anticoagulant (46) and the antiproliferative efficiencies (45) of TBA are enhanced in the presence of K+ ions, which are known to promote and stabilize the structuration of TBA into a monomolecular chair-type antiparallel G-quadruplex.
Additionally, recently reported data suggested that antiparallel G-quadruplex forming ONs, including TBA, accumulate into mitochondria of CL1–0 living cells (47). Into this cellular district, G4 structures could occur to regulate some essential events (48–51). In particular, it has been shown that NOA1, a large GTP binding protein (52), binds some parallel and antiparallel G-quadruplex forming ONs (51).
Unfortunately, the potential of the TBA G4 as a molecular tool in cancer research is strongly limited by its high affinity to thrombin and to most of the thrombin precursors (53). Still, since no connection between anticoagulant and antiproliferative activities is apparent, different TBA regions and/or different binding mode with its biological targets are most likely involved, thus giving the opportunity to perform SAR studies of the two biological effects separately. Therefore, in the quest for new TBA analogues endowed with increased antiproliferative activity, it is not unreasonable to start investigating chemical modifications of TBA sequence that, safeguarding the G4 tertiary structure, selectively quench its anti-thrombin activity. In the light of this reasoning and of the literature data concerning the TBA/thrombin interaction (38–43) and the related SARs, (54–65) we have synthesized a pool of seven new 15-mer TBA analogues (TBA-bs), each of them containing a dibenzyl linker replacing one nucleoside in the loop regions. The new TBA analogues were investigated for their structural and biological properties. CD and NMR measurements indicated that most of the new analogues were able to fold into monomolecular ‘chair-like’ G4s very similar to that adopted by the TBA. Biological results obtained by performing the fibrinogen, prothrombin time (PT) and Methylthiazol Tetrazolium (MTT) tests, indicated that two out of the seven derivatives (TBA-bs4 and TBA-bs13) lost the antithrombin and anticoagulant activities but preserved (TBA-bs4) or enhanced (TBA-bs13) the antiproliferative activity with respect to the TBA. The NMR based molecular model of the G4 formed by TBA-bs13 suggested that the dibenzyl linker could obstruct the binding of TBA-bs13 to thrombin, by steric hindrance in the inner region delimited by the TT loops, thus determining the exclusive switch of the biological activity toward the antiproliferative one.
Taken together, the presented results provide the first evidence that the biological activities of the G4 tertiary structure of the TBA can be ‘controlled’ by a single site-specific chemical modification.
MATERIALS AND METHODS
Chemicals and anhydrous solvents were purchased from Fluka-Sigma-Aldrich. TLCs were run on Merck silica gel 60 F254 plates. Silica gel chromatography was performed using Merck silica gel 60 (0.063–0.200 mm). The API 2000 (Applied Biosystems) mass spectrometer was used to perform the analyses of the intermediates and the monomer. NMR data were collected on Varian Mercury Plus400 and UNITYINOVA 500 MHz spectrometers equipped with a broadband inverse probe with z-field gradient, and on a Varian UNITYINOVA 700 MHz spectrometer equipped with a triple resonance cryoprobe. The data were processed using the Varian VNMR and the iNMR (http://www.inmr.net) software packages. Reagents and phosphoramidites for DNA syntheses were purchased from Glenn Research. ON syntheses were performed on a PerSeptive Biosystem Expedite DNA synthesizer. HPLC purifications and analyses were carried out using a JASCO PU-2089 Plus HPLC pump equipped with a JASCO BS-997–01 UV detector. CD experiments were performed on a JASCO 715 spectropolarimeter equipped with a PTC-348 temperature controller. The fibrinogen assay was performed using a JASCO 530 UV spectrophotometer equipped with the PTC-348 temperature controller.
Synthesis of monomer phosphoramidite building block 3
Compound 2. 2,2-dibenzylpropane-1,3-diol (1.0 g, 4.1 mmol), 4,4′-dimethoxytrityl chloride (696.0 mg, 2.0 mmol) and 4-dimethylaminopyridine (25 mg, 0.21 mmol) were dissolved in dry pyridine (20 ml) and dry CH3CN (10 ml). The resulting solution was stirred at room temperature (r.t.) under argon for 1.5 h. Dry methanol (200 μl) was then added to quench the reaction. After 30 min under stirring, the solution was concentrated under reduced pressure and the residue purified by column chromatography on silica gel (eluted with 90:10:0.1 DCM/MeOH/Et3N) to give monodimethoxytritylated 2 as a clear white solid (40% yield from 1; Rf 0.7 in DCM/MeOH 90:10 v/v).
1H NMR (400 MHz, CDCl3) δ ppm 7.26–7.00 (19H); 6.90 (4H); 3.78 (s, 6H); 3.72 (s, 2H); 3.45 (s, 2H); 2.89 (s, 2H); 2.47 (s, 2H).
13C NMR (100 MHz, CDCl3) δ ppm 156.1; 145.5; 139.9; 137.2; 130.0; 129.8; 129.2; 128.2; 127.4; 127.0; 126.6; 126.4; 124.2; 113.9; 89.1; 69.9; 60.7; 55.8; 40.5; 39.4.
ESI mass (positive mode) calculated 558.28; found 559.3 [M + H]+, 581.3 [M + Na]+.
Compound 3. 2 (425 mg, 0.76 mmol) was dried in vacuo overnight before being dissolved in anhydrous DCM (8 ml) and diisopropylethylamine (530 μl, 3.0 mmol) under argon. 268 μl of β-cyanoethyl diisopropylchlorophosphoramidite was then added (1.2 mmol). After 40 min, the reaction was diluted with ethyl acetate (15 ml), and finally washed with 10% sodium carbonate solution (15 ml) and brine (15 ml). The organic layer was dried on magnesium sulphate and concentrated in vacuo. The residue was purified by silica gel chromatography eluted with DCM, ethyl acetate and triethylamine (80:10:10). The fractions containing the product were collected and concentrated under vacuum, yielding 3 as white foam (85% yield; Rf 0.65 in CHCl3/MeOH/Et3N 97:3:0.05 v/v/v).
1H NMR (400 MHz, CDCl3) δ 7.90–6.85 (23H); 3.90 (2H); 3.76 (6H); 3.65 (2H); 3.60 (2H); 3.45 (2H); 3.39−3.02 (2H); 2.60−2.45 (4H); 1.09 (6H); 1.04 (6H).
13C NMR (175 MHz, DMSO-d6) δ 158.6; 151.6; 147.3; 139.4; 135.1; 130.3; 129.1; 127.8; 127.7; 127.1; 126.4; 126.1; 124.2; 113.1, 90.5; 71.2; 64.4; 60.4; 55.2; 47.3; 33.6; 29.7; 27.6; 20.9; 19.3; 17.3.
ESI mass (positive mode) calculated 758.3; found 759.9 [M + H]+, 781.9 [M + Na]+.
Synthesis of oligomers
TBA and TBA-bs were synthesized using standard solid phase DNA chemistry on a controlled pore glass (CPG) support following the β-cyanoethyl phosphoramidite method (66,67). The coupling time for the modified monomer was prolonged from 2 min to 10 min. The oligomers were detached from the solid support and deprotected by treatment with an aqueous ammonia solution (33%) at 55°C overnight. The combined filtrates and washings were concentrated in vacuo, dissolved in H2O, and purified by HPLC using an anionic exchange column eluted with a linear gradient (from 0% to 100% B in 30 min) of phosphate buffer at pH 7.0 (A: 20 mM NaH2PO4 aqueous solution containing 20% CH3CN; B: 1.0 M NaCl, 20 mM NaH2PO4 aqueous solution containing 20% CH3CN, elution time 18.9 min). The oligomers were successively desalted by molecular exclusion chromatography on Biogel P-2 Fine. The purity (95%) was checked on HPLC using an analytical reverse phase column (Phenomenex, Clarity Oligo-RP, 3 μm, 2 × 10 mm) eluted with a gradient of CH3CN in triethylamine acetate (pH = 7.4, CH3CN from 0% to 100% in 40 min). The concentrations of the samples used in CD and UV experiments were determined by measuring the absorbance at 260 nm at 80°C and using the open access program available at http://basic.northwestern.edu/biotools/OligoCalc.html (68).
CD experiments
To perform the CD experiments on modified TBA sequences, each ON was dissolved in K+ buffer (90 mM KCl, 10 mM KH2PO4, pH 7.4) or PBS (Phosphate Buffered Saline) at the final ON concentration of 2.0 × 10−5 M and submitted to the annealing procedure (heating at 90°C for 5 min and slowly cooling at r.t.). Before each experiment the samples were equilibrated at 10°C for 30 min. CD spectra were recorded from 200 nm to 400 nm at 100 nm min−1 scanning rate, 16 s response, 2.0 nm bandwidth, cell length 0.1 cm, vol 400 μl. Each CD profile was obtained by taking the average of three scans. CD melting curves were obtained by monitoring the variation of absorbance at 295 nm from 10°C to 90°C, cell length 1 cm, vol 1400 μl, temperature scan speeds of 0.1 and 0.5°C min−1.
Furthermore, the CD melting experiments in K+ buffer were also performed at ON concentrations of 1.0 × 10−4 and 8.3 × 10−6 M. These samples were obtained by diluting a 1.0 × 10−3 M sample, properly annealed in K+ buffer. The samples at 1.0 × 10−4 M were subjected to the melting-annealing experiments (cell length 0.1 cm, vol 400 μl), whereas those at 8.3 × 10−6 M to the annealing-melting experiments (cell length 0.5 cm, vol 1200 μl). The temperature scan speed was fixed at 0.2°C min−1.
Fibrinogen clotting assay
The fibrinogen clotting times were measured spectrophotometrically following the procedure previously described (69). ONs were incubated for 1 min at 37°C in 1.0 ml of PBS containing 2.0 mg/ml of fibrinogen (fibrinogen from human plasma, F 3879, Sigma-Aldrich) in a PMMA cuvette (vol 1.5 ml, cell length 1 cm, Brand). 100 μl of human thrombin (10 NIH per ml; Sigma-Aldrich, T8885, human thrombin suitable for the thrombin time test) was then added to the solution containing the fibrinogen and the ON. For each ON, three sets of experiments at concentration of 20, 50, 100 nM were performed. The time required for fibrin polymerization was determined from the UV scattering curve, registered, as a function of time (wavelength fixed at 380 nm), in triplicate for each ON concentration. The clotting time value reported as mean ± SE was derived as the maximum of the second derivative of each scattering curve. The basal clotting time (25.6 ± 1.5 s) was referred to the clotting time value determined in absence of any ONs.
NMR experiments
NMR samples were prepared in PBS/D2O (90:10, v/v) at a single strand concentration of about 5 mM. One-dimensional NMR spectra were acquired as 16384 data points with a recycle delay of 1.0 s at 25°C. Data sets were zero filled to 32768 points prior to Fourier transformation and apodized with a shifted sine bell squared window function. Two-dimensional NMR spectra of TBA-bs12 and TBA-bs13 were acquired at 10°C using a recycle delay of 1.2 s. NOESY spectra were acquired with mixing times of 100, 200 and 300 ms. TOCSY spectra were recorded with the standard MLEV-17 spin-lock sequence and a mixing time of 80 ms. Water suppression was achieved by including a double pulsed-field gradient spin-echo (DPFGSE) module (70,71) in the pulse sequence prior to acquisition. In all 2D experiments, the time domain data consisted of 2048 complex points in t2 and 400 fids in t1 dimension. The sequence-specific resonance assignment of all proton signals of TBA-bs12 and TBA-bs13 was obtained using NOESY and TOCSY spectra following the standard procedure (72,73).
Molecular modelling
The initial structure of TBA-bs12 and TBA-bs13 was built using as the starting point the coordinates of the NMR solution structure of TBA quadruplex (PDB id. 148D) (44). The nucleoside T12 or T13, respectively, was replaced with the linker 1 (Figure 1) by using the builder module of the Insight II (2005) software package (Accelrys). The obtained molecules were parameterized according the AMBER force field and the atom of the linker 1 were selectively minimized using the steepest descent and conjugate gradient minimization methods until convergence to a RMS gradient of 0.001 Kcal mol−1 was reached. The structures were then neutralized by adding 14 sodium counterions and solvated in a square TIP3P water box (38 × 38 × 38 Å). Water molecules were minimized (RMS ≤ 0.1 Kcal mol−1) and the systems were subjected to NMR-restrained energy minimization (steepest descent and conjugate gradient methods until convergence to a RMS gradient of 0.001 Kcal mol−1) as described below. The systems were first minimized using the following restraints: 109 or 75 quadratic interproton distances (fixed-length; 30 Kcal mol−1 force constant) for TBA-bs12 or TBA-bs13, respectively; 32 flat-bottomed hydrogen bond constraints (16 hydrogen/acceptor 1.7–2.0 Å and 16 donor/acceptor 2.7–3.0 Å; 100 Kcal mol−1); 5 glycosidic torsion angles constrained to a range of –160°/–70° (flat bottomed; 200 Kcal mol−1) as required for anti Gs; 4 glycosidic torsion angles constrained to a range of 10°/100° (flat bottomed; 200 Kcal mol−1) as required for syn Gs. Next, the distance restraints were removed and the systems were subjected to a second minimization. Finally, all restraints were removed and a final energy minimization was performed. All calculations were repeated 10 times on each system until convergence at the RMS gradient ≤0.001 Kcal mol−1 was reached. For each system the average minimized structure was then obtained by using the analysis module of the Insight II 2005 software package. The average RMSD value was 0.988 Å for TBA-bs12 and 0.983 Å for TBA-bs13.
Figure 1.
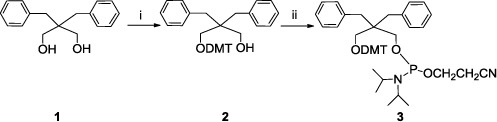
Synthesis of phosphoramidite building block. i) 2,2-dibenzylpropane-1,3-diol 1 (1.0 g, 3.9 mmol), 4,4′-dimethoxytrityl chloride (0.69 g, 3.9 mmol), 4-dimethylaminopyridine (2.5 × 10−2 g, 0.21 mmol) dry Py (10.0 mL), dry CH3CN (10.0 mL), 2.5 h, yields 40%; ii) 2 (0.425 g, 0.76 mmol), β-cyanoethyl-N,N’-diisopropyl-chloro-phosphoramidite (0.270 mL, 1.2 mmol), DIPEA (0.530 mL, 3.0 mmol), dry DCM (8 mL), 1.0 h, 85%.
Prothrombin (PT) Time
PT time was measured by using a Koagulab MJ Coagulation System with a specific kit RecombiPlas Tin HemosIL (Instrumentation Laboratories, Lexington, MA, USA). The procedure was performed according to the manufacturer's instructions. In our experimental protocol a time course (up to 15 min) of each ON or vehicle with 100 μl of plasma at 37°C was performed. In details, 2.0 μl of the corresponding ON solution (1.0×10−3 M in PBS) or vehicle (PBS) was added in the apposite microtube, to achieve the final concentration of 20 μM. Similarly, to reach the final concentration of 2 μM, 2.0 μl of 1.0 × 10−4 M ON solution was added to 100 μl of plasma at 37°C. Next, 200 μl of the kit solution containing Recombiplastin was added with consequent activation of the extrinsic pathway. The PT measurement was produced in triplicate, and the average and its standard error values were calculated. The basal clotting time was determined by measuring the clotting time in the absence of any ON (13.4 ± 0.2 s). The incubation with PBS did not modify the PT measurement.
MTT test
He-La cervical carcinoma cells were seeded in a 96-well plate at a density of 250 cells/well in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum. Briefly, 24 h after being cultured, cells were treated with the different ONs for 7 days and afterward 20 μl of MTT solution (5 mg/ml, Sigma-Aldrich) was added into each well and incubated for 4 h. The growth medium was then removed and replaced with 4 mM HCl, 0.1% Nondet P-40 (NP40) in isopropanol (150 μl/well) in order to dissolve the blue-purple crystals of formazan. The absorbance was then measured using a Microplate Reader at 595 nm. Cell viability of triplicate cultures was assessed and graphed together with the corresponding standard deviation.
RESULTS
Synthesis of ONs
The synthesis of derivative 3 (Figure 1) started from the commercially available 2,2-dibenzylpropane-1,3-diol (1), that was subjected to the standard protection of one alcoholic function with 4,4′-dimethoxytrityl group. The protected product 2 was then reacted with β-cyanoethyl-N,N’-diisopropyl-chloro-phosphoramidite to obtain 3. This monomer was used to automatically synthesize the modified ONs reported in Table 1 using the standard phosphoramidite protocol (66,67). The length of the coupling cycle using the new monomer 3 was extended from 2 min to 10 min. The yields of coupling were similar to those obtained using the standard phosphoramidite building blocks.
Table 1. TBA-based sequences (TBA-bs) containing 2,2-dibenzylpropane-1,3-diol (X) at the indicated position.
| ON | Sequence |
|---|---|
| TBA | G1G2T3T4G5G6T7G8T9G10G11T12T13G14G15 |
| TBA-bs3 | G1G2X3T4G5G6T7G8T9G10G11T12T13G14G15 |
| TBA-bs4 | G1G2T3X4G5G6T7G8T9G10G11T12T13G14G15 |
| TBA-bs7 | G1G2T3T4G5G6X7G8T9G10G11T12T13G14G15 |
| TBA-bs8 | G1G2T3T4G5G6T7X8T9G10G11T12T13G14G15 |
| TBA-bs9 | G1G2T3T4G5G6T7G8X9G10G11T12T13G14G15 |
| TBA-bs12 | G1G2T3T4G5G6T7G8T9G10G11X12T13G14G15 |
| TBA-bs13 | G1G2T3T4G5G6T7G8T9G10G11T12X13G14G15 |
X = 2,2-dibenzylpropane-1,3-diol.
CD and CD melting
The folding of TBA into the monomolecular antiparallel chair-like G-quadruplex structure causes a well known CD profile characterized by three positive bands at 212, 247 and 295 nm and a negative band around 270 nm (74–76). Indeed, CD spectroscopy is commonly used to quickly valuate the effects of sequence modifications on the general feature of TBA G-quadruplex (43,54–64,74–76). To this purpose we performed CD analyses both in PBS (Figure 2) and K+ buffer (Supplementary Figure S1). From collected data it appears that almost all new ONs in both used buffers fold into structures that display a CD profile very close to that of TBA. However, in some cases slight differences in the profile or in the intensity of bands were revealed. To analyze these effects as a function of the position occupied by dibenzyl linker along the sequence, the collected data were divided into two sets.
Figure 2.
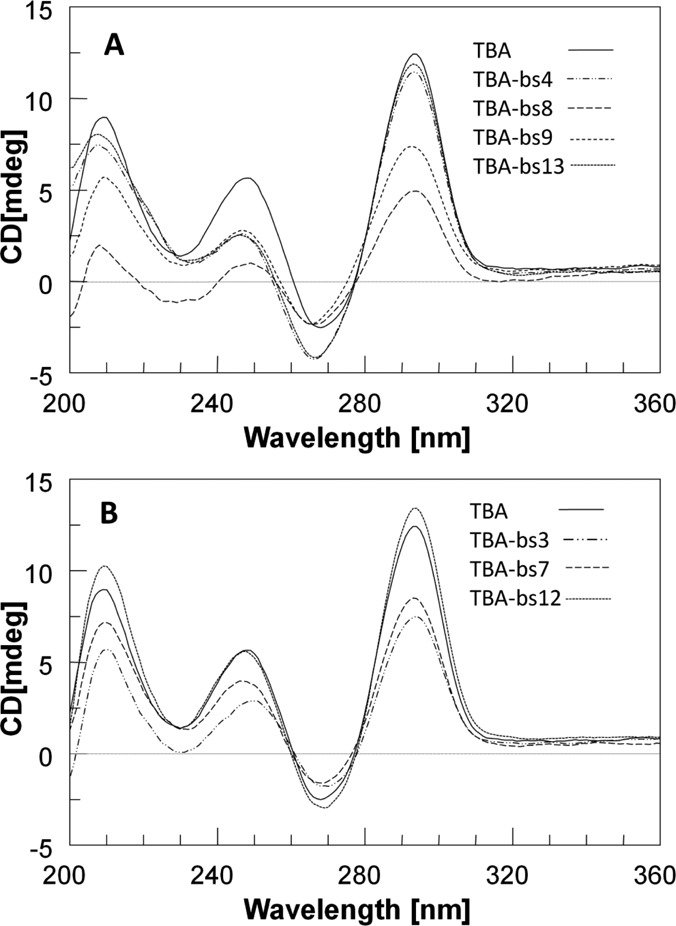
CD profiles of TBA and TBA-based sequences. Spectra were collected at 10°C, using [ON] of 2.0×10−5 M in PBS.
According to NMR and X-ray data, that designate the residues at position 4, 8, 9 and 13 as essential for the topology and the stability of the G4 structure (37–41,43), we grouped the CD profiles of the corresponding TBA-bs variants in Figure 2, panel A. Notably, the CD spectra of TBA-bs8 and TBA-bs9 showed a significant decreasing in the intensity of the CD band at 292 nm, compared to that of TBA. Furthermore, the spectra of the variants (Figure 2A) containing the linker 1 at position 4 or 9 or 13 were strongly similar to each other and slightly differed from that of TBA by a more negative CD signal in the range 230–270 nm.
The CD spectra of TBA-bs modified at position 3 or 7 or 12 are shown in Figure 2, panel B. All the variants in this group preserved almost unchanged the CD profile of TBA. The only difference we observed was a slight decrease in the intensity of the positive CD bands for TBA-bs3 and TBA-bs7.
To evaluate the thermal stability of the folded G-quadruplexes we performed CD melting experiments. The melting curves (Supplementary Figure S2) were registered monitoring the change of the intensity of the band at 295 nm increasing the temperature from 10°C to 90°C. The derived apparent melting temperatures (Table 2) indicated that the thermal stability of TBA-bs4 and TBA-bs13 G4s was slightly higher than that measured for TBA (ΔTm ∼3–4°C in both PBS and K+ buffer) whereas that of TBA-bs3, TBA-bs7 and TBA-bs12 was almost unchanged. In contrast, TBA-bs8 and, especially, TBA-bs9 folded into G4s considerably less stable than that of TBA in both K+ buffer (ΔTm -4.3 and -15.5°C, respectively) and PBS. Furthermore, in the case of TBA-bs9, as a result of the poor structure stability in PBS, it was not possible to calculate the melting temperature from the corresponding melting curve (Supplementary Figure S2).
Table 2. Thermal stability and biological properties of TBA and TBA-based sequences. The apparent melting temperatures (°C) were measured as minimum in the first derivative of CD melting profiles.
| Name | aTm (°C) | bΔTm | cTm (°C) | bΔTm | dw/o ± eSD 10 μM | fPT ± gSE 20 μM |
|---|---|---|---|---|---|---|
| TBA | 50.7 | 0 | 35.1 | 0 | 0.669 ± 0.054 | 49.8 ± 1.28 |
| TBA-bs3 | 50.0 | -0.7 | 35.4 | +0.3 | 0.996 ± 0.045 | 43.6 ± 2.15 |
| TBA-bs4 | 51.9 | +1.2 | 37.2 | +2.1 | 0.700 ± 0.093 | 14.1 ± 0.25 |
| TBA-bs7 | 50.0 | -0.7 | 34.5 | -0.6 | 0.400 ± 0.103 | 56.3 ± 0.97 |
| TBA-bs8 | 46.4 | -4.3 | 34.4 | -0.7 | 0.811 ± 0.047 | 25.8 ± 0.30 |
| TBA-bs9 | 35.3 | -15.4 | hn.d | hn.d. | 1.087 ± 0.1059 | 24.8 ± 0.80 |
| TBA-bs12 | 51.1 | +0.4 | 35.4 | +0.3 | 0.739 ± 0.031 | 52.0 ± 1.73 |
| TBA-bs13 | 53.3 | +2.6 | 38.9 | +3.8 | 0.498 ± 0.103 | 12.5 ± 0.55 |
aAnnealing buffer: 90 mM KCl, 10 mM potassium phosphate, pH 7.4.
bCalculated respect to TBA melting temperature.
cAnnealing buffer: PBS, pH 7,4.
dRatio between measured absorbance in MTT test in presence and in absence of ONs.
eStandard Deviation.
fMeasured Prothrombin Time in human plasma.
gStandard Error.
hnot determinable.
Finally, to probe the molecularity of the G-quadruplexes, all the CD melting experiments in K+ buffer were also performed on samples at the two additional strand concentrations of 1.0 × 10−4 and 8.3 × 10−6 M (12-fold relative dilution). For all TBAs, we did not observe significant differences between the two sets of melting profiles (Supplementary Figure S3a–S3g), thus suggesting that the modified ONs preserve the ability of TBA to fold into monomolecular G4s (77).
NMR structural characterization
The ability of all the here reported TBA-bs to fold into a TBA-like antiparallel quadruplex was also assessed by NMR spectrometry. The 1H NMR spectrum of TBA annealed in Na+ or K+ containing buffer is characterized by the presence of eight well resolved signals in the 11.5–12.5 ppm region, attributable to the exchange protected imino protons involved in the formation of two G tetrads, and by fifteen aromatic proton signals (6.8–8.5 ppm) attributable to H8 and H6 protons of guanine and thymine bases, respectively. Moreover, two peculiar upfield-shifted methyl signals of mutually H-bonded T4 and T13 are seen at around 1.0 ppm (37,40,41). Finally, the alternating syn-anti-syn-anti glycosidic bond conformation within the two stacked G-tetrads, typical of most antiparallel quadruplexes, is observed. As for the TBA, the quadruplex diagnostic imino signals were observed in the water suppressed 1H NMR spectra of the here reported TBA-bs, with the noticeable exception of TBA-bs8 (for some examples see Figure 3 and Supplementary Figure S4a–S4e). Hence, the replacement of a single T residue in one of the three loops for the dibenzyl linker did not impede the quadruplex formation, whereas the replacement of G8, the only guanosine in the sequence not engaged in the two G-tetrads, did it. In particular, as we already observed in our previous papers (54,60,64), the replacement of T3 or T4 in the TBA sequence with acyclic nucleoside analogues induces changes in the resulting NMR profiles that are very similar to those induced when the replacement occurs at T12 or T13, respectively. It is interesting to note that the similarity observed between members of each TBA-bs3/TBA-bs12 or TBA-bs4/TBA-bs13 couple was also observed by CD and by fibrinogen or prothrombin time assays (vide infra). According to the biological results, we undertook a detailed structural study of the G-quadruplexes formed by TBA-bs12 and TBA-bs13 by using the NMR-refined molecular mechanics approach.
Figure 3.
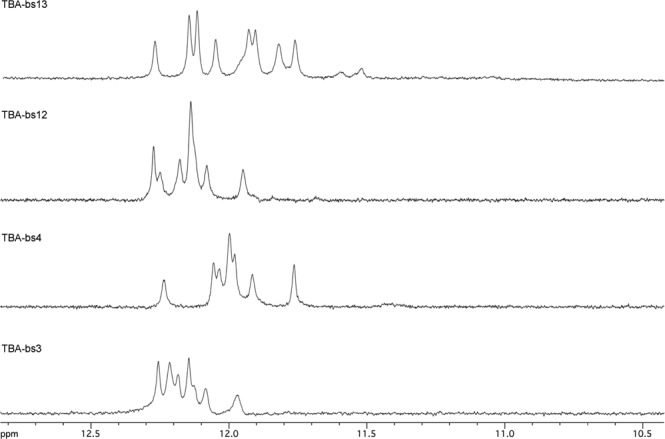
Representative 1H NMR imino proton regions of TBA-bs incorporating the 2,2-dibenzylpropane-1,3-diol at the indicated position (25°C; PBS buffer, H2O/D2O 9:1).
Structural investigation of TBA-bs12
The 1H NMR spectrum of TBA-bs12 recorded at 25°C (Supplementary Figure S4a) was characterized by the presence of eight partially overlapped signals in the 11.9–12.3 ppm region attributable to the exchange-protected imino protons involved in the formation of Hoogsteen hydrogen bonds of two G-tetrads, in close analogy with previous observations on the antiparallel quadruplex formed by the unmodified TBA (41). Because of the presence of the two benzene rings in the linker 1 at position 12, the aromatic portion of the 1H spectrum (6.8–8.5 ppm) was populated by more than the 14 signals belonging to the protons of guanines and thymines of the G4 structure. In addition, other weaker signals belonging to one or more minor conformations likely including the unfolded form were observed in this region. With the aim of minimizing the exchange between the G-quadruplex and the unfolded form, we recorded the 1H NMR spectrum of TBA-bs12 also at 10°C (Figure 4, panel A). As expected, a general simplification of the NMR spectrum was observed. In particular, at 10°C most of the weaker signals disappeared and an improved dispersion of the signals was obtained. The combined analysis of 2D NOESY and TOCSY spectra (700 MHz, 10°C) allowed us to get the nearly complete assignment of exchangeable and non-exchangeable protons using the standard procedure (Supplementary Table S1) (72,73). The TOCSY data are not shown here, but they were used to confirm the assignments based on the NOESY data. The overall analysis of NOE connectivities and the evaluation of the Δδ between TBA-bs12 and TBA (see Supplementary Table S2) confirmed that TBA-bs12 adopts a three-dimensional structure that is very similar to the TBA chair-like antiparallel quadruplex. In particular, the analysis of the intranucleotide and internucleotides H8/H6 to H1′ NOE connectivities (Figure 4, panel B) confirmed that four out of the eight guanosines involved in the two G-tetrads adopt the syn glycosidic bond conformation, as seen for TBA. The NOE connectivities between the methyl signal of T9 and four imino protons confirmed that T9 is stacked above the G1-G15-G10-G6 G-tetrad (syn Gs are underlined) (Figure 4, panel C). Finally, the very low Δδ observed for the methyl signal of T4 (δ 1.07 ppm; Δδ 0.04 ppm) disclosed that the substitution of T12 with the linker 1 did not interfere with the tertiary structure adopted by TBA-bs12, even in the close proximity of the substitution site, and suggested that the two benzene rings could not occupy the cleft delimited by the two TT loops (37,40,41,44). The latter observation is noteworthy, considering that most of the X-ray diffraction data of TBA-thrombin complexes have shown that the TBA recognizes the thrombin anion-binding exosite I (ABE I; also known as the fibrinogen exosite) by using the T3-T4 and T12-T13 loops (37,39–43,54,60,61,64). To get insights about the orientation of the benzene rings of 1 with respect to the quadruplex scaffold, and to rationalize the effects on the antiproliferative and antithrombin activities shown by TBA-bs12, we performed a NMR-restrained molecular mechanics study as described in Materials and Methods. As we expected, among the 109 NMR interproton distances used in the calculations only a few involved close contacts with the atoms of the linker 1, and the resulting averaged structure confirmed in full (Figure 5) that in the G4 structure formed by TBA-bs12, the two benzene rings are positioned aside from the G4 core, thus allowing its recognition by the thrombin ABE I.
Figure 4.
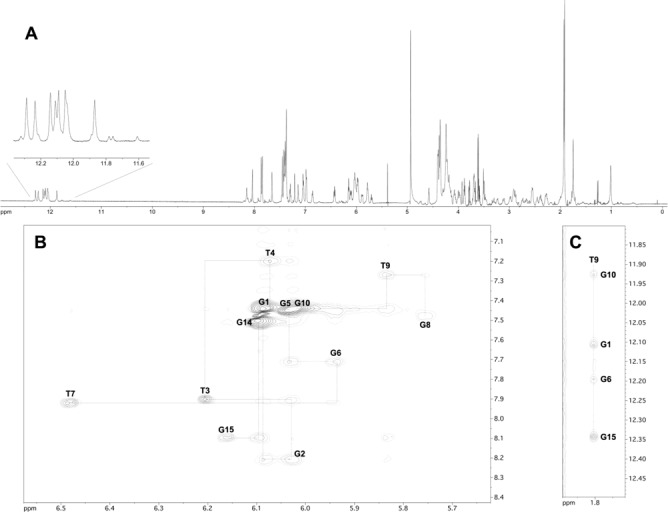
NMR study of TBA-bs12 annealed in PBS/D2O 9:1 (10°C, 700 MHz). (A) 1H NMR spectrum. The expansion of the imino proton region is shown in the inset. (B, C) Expansions of the 2D NOESY (200 ms mixing time) spectrum. The sequential H1′-H8 or H1′-H6 connectivities and the self-peaks involving H1′ protons have been drawn in B using dotted lines and labels, respectively. NOE correlations between T9 methyl protons and H8 protons of the G10-G1-G6-G15 tetrad are shown in C.
Figure 5.
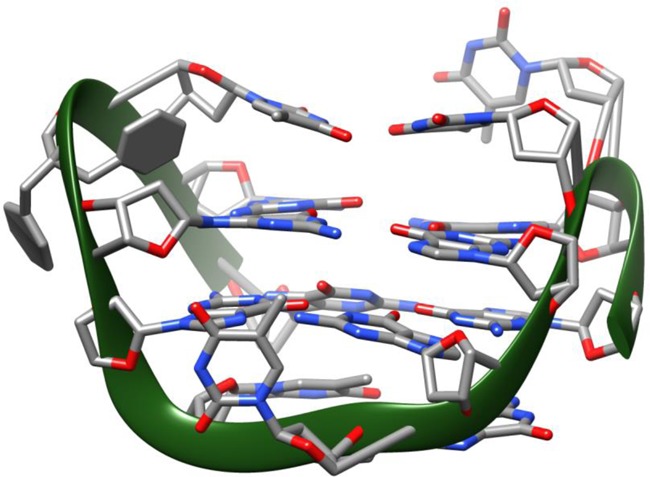
3D average structure of TBA-bs12 obtained by NMR-restrained molecular mechanics. Nucleotides are shown as sticks, the 2,2-dibenzylpropan-1,3-diol linker as sticks and filled rings and the phosphate backbone as a green solid ribbon.
Structural investigation of TBA-bs13
The NMR structural elucidation of the G4 formed by TBA-bs13 in PBS buffer was performed as described above for TBA-bs12 and in Materials and Methods. The NMR characterization confirmed the formation of a stable antiparallel chair-like G4 stabilized by two G-tetrads (8 well resolved imino proton signals; Figure 6, panel A) connected by the T3-T4, T12-X13 and T7-G8-T9 loops. Intra- and inter-nucleotide NOE connectivities confirmed that the methyl group of T9 is stacked above the G1-G15-G10-G6 tetrad (Figure 6, panel C) and that the two G-tetrads are made up of alternating syn and anti Gs (Figure 6, panel B). All together, the analysis of the 1H chemical shifts (Supplementary Table S3) and Δδ between the NMR signals of TBA-bs13 and TBA (Supplementary Table S4) indicated that the incorporation of the dibenzyl linker at position 13 of TBA does not induce major distortions in the resulting G4 scaffold. However, differently from TBA-bs12, we observed a significant up-field shifting for the imino protons of G5, G6, G10, G11, G14 and G15, and a down-field shifting for the methyl signals of T4 and T12. These NMR evidence suggested that the two benzene rings of the linker were not projected aside the G4 core as found for TBA-bs12. Accordingly, among the 75 NOE distance restraints included in the calculations, 8 involved NOE contacts between the atoms of the linker and those of nucleosides G11, T12 and G14. The resulting minimized average structure of TBA-bs13 obtained by the NMR-refined molecular mechanics calculations is shown in Figure 7. According to the simulated structure, the two benzene rings of X13 are located in the pocket delimited by the two TT loops. In particular, the ring coloured in red in Figure 7 is stacked exactly above the 6-membered ring of G11, whereas the blue ring is turned away the G2-G5-G11-G14 tetrad pointing at the T3-T4 loop. These findings are in agreement with the observed Δδ caused by the strong benzene rings current.
Figure 6.
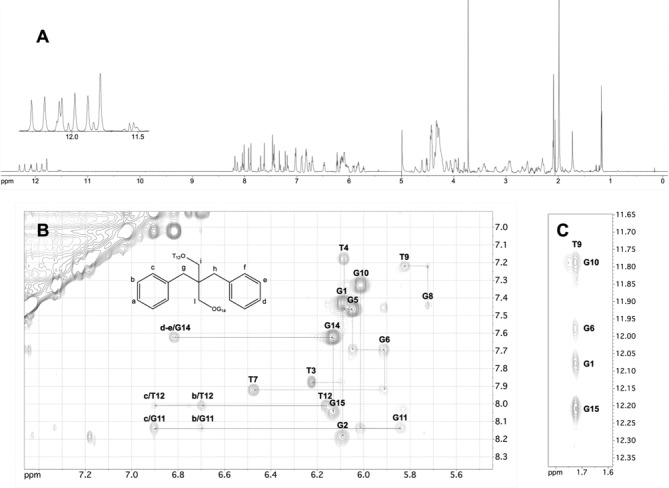
NMR study of TBA-bs13 annealed in PBS/D2O 9:1 (10°C, 700 MHz). (A) 1H NMR spectrum. The expansion of the imino proton region is shown in the inset. (B, C) Expansions of the 2D NOESY (200 ms mixing time) spectrum. The NOE connectivities between atoms of X13 and nucleotides atoms are traced and labelled in panel B. The sequential H1′-H8 or H1′-H6 connectivities and the self-peaks involving H1′ protons have been drawn in B using dotted lines and labels, respectively. NOE correlations between T9 methyl protons and H8 protons of the G10-G1-G6-G15 tetrad are shown in C.
Figure 7.
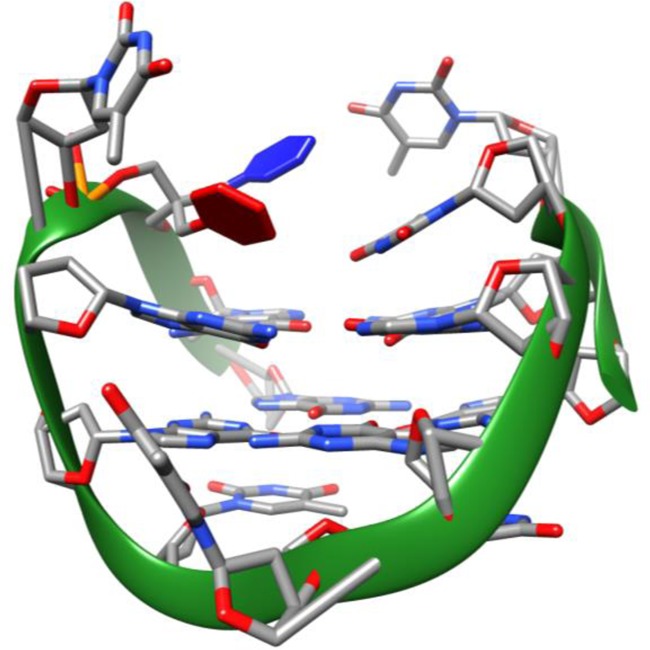
3D average structure of TBA-bs13 obtained by NMR-restrained molecular mechanics. Nucleotides are shown as sticks, the 2,2-dibenzylpropan-1,3-diol linker as sticks and red and blue filled rings and the phosphate backbone as a green solid ribbon.
Fibrinogen assay
In order to assess the effect on the enzymatic activity of thrombin and to correlate the biological effects with the structural data, the modified ONs were tested in the fibrinogen clotting assay. The thrombin-induced clotting of fibrinogen was measured spectrophotometrically, following the increase in absorbance at 380 nm as a function of time (69). The assay was performed using 2 mg per ml of fibrinogen, 1 NIH of thrombin and various concentrations of each aptamer (20, 50 and 100 nM). The comparison of resulting data (Figure 8 and Supplementary Table S5) evidenced marked differences among the modified TBA-bs in their ability to inhibit the fibrinogen clotting formation. TBA-bs12 showed an increased inhibitory effect relative to the unmodified sequence, at all explored concentrations. TBA, TBA-bs3 and TBA-bs7 prolonged the fibrinogen clotting time at almost the same extent. TBA-bs9 and, especially, TBA-bs8 showed a significant decreased inhibitory effect relatively to TBA. Most significantly, at all explored concentrations, TBA-bs4 and TBA-bs13 resulted totally unable to compete with fibrinogen for the binding to the enzyme.
Figure 8.
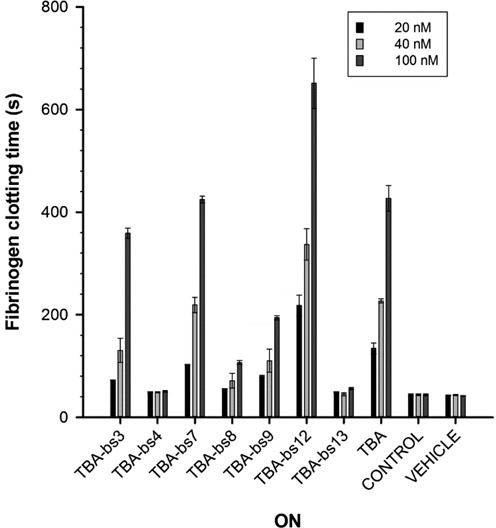
Fibrinogen clotting time (sec) in presence of each ON measured at three different concentrations (20, 40 and 100 nM). The bars of control and vehicle represent the fibrinogen clotting time values of the system in absence of any ONs and diluted with the buffer alone, respectively.
PT assay
To validate the results obtained from the fibrinogen clotting assay, we performed PT tests on human plasma (PT assay) (36,54,60,61). As shown in Figure 9 and in the Table 2 and Supplementary Table S6, the PT values pointed to a trend of the anticoagulant activities that was almost in agreement with the antithrombin activities measured in the fibrinogen assay. Major difference resulted in the behaviour of TBA-bs12, which showed in fibrinogen assay, but not in PT test, a significant increasing in the antithrombin efficiency with respect to TBA. Herein, among the tested ONs, TBA-bs7 proceeded as the best anticoagulant one. The PT values measured in presence of TBA-bs4 or TBA-bs13 confirmed that these modified sequences were wholly unable to inhibit thrombin.
Figure 9.
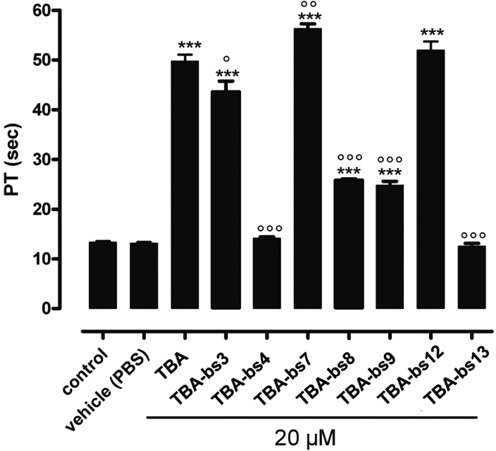
PT values obtained following 15 min of ON incubation with human plasma at concentration of 20 μM [ON]. Each measurement has been performed in triplicate and it was shown as mean ± SEM. PT values are expressed in seconds. The basal PT time is 13.4 ± 0.2 s. *** = P<0.001 vs. vehicle; °,°°, °°° = P<0.05, <0.01, <0.001 versus TBA.
MTT test
On the basis of literature data (45), all modified sequences and TBA were examined for their antiproliferative activities against He-La cervical carcinoma cell line at three different concentrations, 1 (Figure 10, panel A), 5 (Supplementary Figure S5) and 10 μM (Figure 10, panel B and Table 1). As a result, all but TBA-bs9 were judged to be active against this tumor cell line. Specifically, TBA-bs7 and TBA-bs13 had the greatest effect on the cell growth, at all tested concentrations. Notably, TBA-bs4 and TBA-bs13 preserved the antiproliferative activity of TBA against He-La cells, but, according to fibrinogen and PT data, lacked in thrombin affinity. To better explore the antiproliferative effect of TBA-bs13, dose-response experiments at concentrations between 1.0 × 10−3 and 50 μM were performed. The obtained clear dose-response curves indicated a 5-fold increased cell growth inhibition for TBA-bs13 with respect to TBA (Figure 11 and Supplementary Tables S7 and S8).
Figure 10.
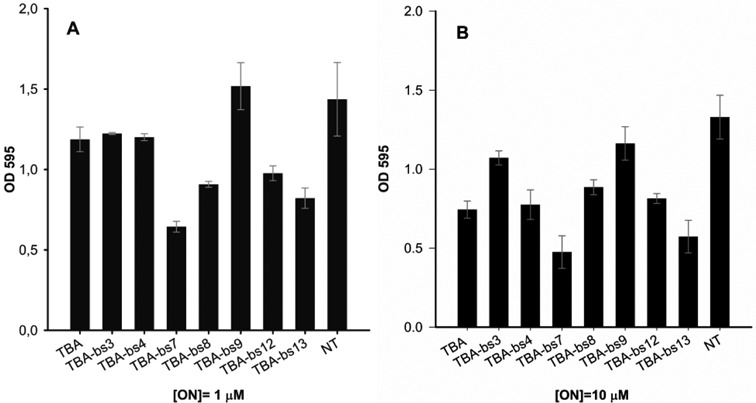
Antiproliferative activity on He-La cervical carcinoma. Cells were treated with two different doses of ON, 1 (A) and 10 (B) μM, annealed in K+ buffer (see experimental section). The bar NT (Not Treated) reports the cell viability in absence of ON. Cell viability was assayed seven days after addition of ON using the MTT assay. A pool of three different sets of experiments (each repeated in triplicate) was performed, and each value expressed as mean ± Standard Deviation.
Figure 11.
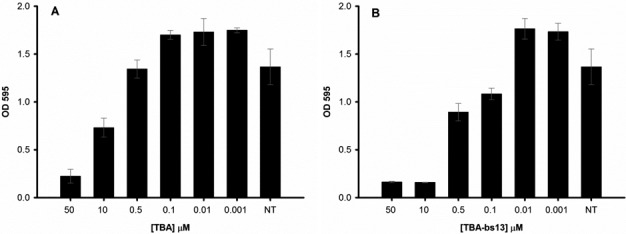
Concentration-dependent response in MTT assay for TBA (A) and TBA-bs13 (B). Cell viability was assayed seven days after addition of ON using the MTT assay. A pool of three different sets of experiments (each repeated in triplicate) was performed, and each value expressed as mean ± Standard Deviation.
DISCUSSION
The findings that specific modifications of TT loops of TBA negatively interfere with its thrombin affinity without affecting the G-quadruplex stability (64), and that the antiproliferative activity is a general property of GT-rich ON strands (11) make ready the perspectives of modified TBAs as selective antiproliferative ONs provided with the notable advantage to fold into well defined G-quadruplex structures.
Aimed by this idea, we selected, among the commercially available acyclic diols, the unusual dibenzyl-linker 1 (Figure 1) as the modifying agent for the obtainment of a series of new TBA-based sequences (see Table 1) in which 1 (indicated as X in Table 1) replaced one at the time the loop nucleotides of TBA. Indeed, the linker 1 provided for: (i) two hydroxyls, to make the required phosphoramidite building block 3; (ii) an aromatic portion, to mimic the aptitude of some loop residues in the TBA G-quadruplex to stack on the adjacent G-quartet (37–43); (iii) a moderate bulky portion, to cause steric crash between the TBA G4 and thrombin (64). Finally, as the requirements were fulfilled into symmetrical molecules, it was also ensured that the insertion of the linker did not give rise to diastereomeric nucleic acid sequences (54).
CD and 1H NMR studies performed on the new synthesized TBA-bs suggested that the resulting G4 structures strongly resembled that of the TBA (Figure 2, Supplementary Figures S1 and S4). Indeed, all CD profiles showed the stronger positive band around 295 nm and a weak negative band around 270 nm, that are generally accepted as distinctive for antiparallel G4s structures (74–76). According to the CD data, the G4 diagnostic imino signals were observed in the 11.5–12.5 ppm region of the 1H NMR spectra of the new derivatives (for some examples see Figure 3), as for the TBA, with the exception of TBA-bs8. Furthermore, in line with previous structural data which arranged the nucleobases 3, 12 and 7 outside the G-quadruplex core (37–43), the apparent CD melting temperatures of the G4 TBA-bs3, TBA-bs12 and TBA-bs7 were similar to that of TBA (Supplementary Figure S2 and Table 2). Differently, substantial destabilization of the G4 structure occurred when the linker 1 occupied the position 8 or 9 of the TGT loop (ΔTm = -4.3 and -15.5°C, respectively, compared to that of TBA in K+ buffer; Supplementary Figure S2 and Table 2). Given that T8 and G9 stack on the G1-G15-G10-G6 quartet, thus stabilizing the TBA G-quadruplex core (37–43), the obtained Tm values suggested that neither of the two benzene rings of the linker 1 efficiently mimic the role of the replaced nucleobase in the G4 structure formed by TBA-bs8 and TBA-bs9. Interestingly, stable antiparallel G4s were obtained from the folding of TBA-bs4 and TBA-bs13 (Tm = 51.9 and 53.3°C, respectively, in K+ buffer; 37.2 and 38.9°C in PBS; Table 2), thus suggesting that the aromatic moieties at position 4 or 13 could provide for the stacking interactions with the nucleobase on the adjacent quartet (G2-G5-G11-G14).
The results from PT and fibrinogen assays (Figures 8 and 9) allowed us to assess the involvement of the linker 1 in the formation of TBA-thrombin complex, while those from the MTT assay (Figure 10) the influence of 1 on the TBA cytotoxicity. By modifying the TGT loop, it resulted that TBA-bs8 and TBA-bs9, that folded into the least stable G4 structures, still affected, although with reduced efficiency, the coagulation time measured in both the fibrinogen and PT assays (Figures 8 and 9; Supplementary Tables S5 and S6), and showed the lowest antiproliferative effects in the MTT test on He-la carcinoma cell line (Figure 10 and Supplementary Figure S5). On the other hand, both the anticoagulant activity and the cytotoxic effect of TBA-bs7 resulted increased. These results could reflect a tighter correlation between the structural stability of G4s and the antiproliferative activity (11,21), than that observed with the antithrombin efficiency (54,57,59–61,64). Indeed, among these three ONs, the best-structured TBA variant (TBA-bs7) was also the strongest growth-inhibitor agent and viceversa (TBA-bs9). However, none of these ONs showed a selective action with respect to the two functional effects. Consequently, their G4 structures were not further investigated. The most relevant outcomes emerged from TBA variants modified at one residue of the TT loops. In fact, although the corresponding G4s had Tm values very similar to each others (ΔT < 3.3°C; Table 2), their biological properties were significantly different. In the MTT assay (Figure 10 and Supplementary Figure S5), TBA-bs4 and TBA-bs13, which resulted totally unable to act as anticoagulant agents (Figures 8 and 9; Table 2, Supplementary Figures S5 and S6), showed significant cytotoxic effects on He-La cells, and TBA-bs13 was a better inhibitor of cell growth than TBA. Contrarily, TBA-bs3 and TBA-bs12 afforded to a good correlation between structure stability, antithrombin and antiproliferative activities (Figures 8–10 and Table 2). The biological properties of the last two sequences were almost comparable to that of TBA. Nevertheless, it was also observed that the increased antithrombin activity of TBA-bs12 with respect to TBA, measured in fibrinogen assay (Figure 8 and Supplementary Table S5), was not observed in the PT test (Figure 9, Table 2 and Supplementary Figure S6). Such incoherence between the fibrinogen and PT data was previously observed by us (60,61) and others (59,78) for other TBA variants, and supports the hypothesis that the clotting inhibitory activity of some modified TBAs could not wholly depend on the ability of the aptamers to compete with fibrinogen for thrombin ABE I. In fact, the presence in the plasma medium of other thrombin effectors and/or precursors (53), and the thrombin allostery (79), which regulates the complex thrombin action during homeostasis, could also play an important role in determining the anticoagulant effect of such type of TBA derivatives. However, the overall results evidenced that, despite the TBA G-quadruplex well sustained the replacement of one T with the linker 1 at any position of the TT loops, the TBA-thrombin complex did not tolerate such modification at T4 or T13. Furthermore, the results were consistent with the TBA tertiary structure, which leads residues T3 and T12, as well as T4 and T13, to occupy equivalent positions with respect to the G quartets (37–43). Nevertheless, TBA-bs12 and TBA-bs13 are slightly more potent inhibitors of cell growth than TBA-bs3 and TBA-bs4, respectively (Figure 10 and Supplementary Figure S5). At the best of our knowledge, this is the first evidence that loop structure is a further parameter capable to affect the antiproliferative efficiency of the TBA G-quadruplex in addition to G4 stability.
To better understand the requirements to discriminate between the antiproliferative and antithrombin effects, we undertook an extensive 2D NMR study of the G4s formed by TBA-bs12 and TBA-bs13 in PBS buffer. The inspection of the NMR-restrained molecular mechanics average G4 structure of TBA-bs12 (Figure 5) confirmed that the two benzene rings of 1 are projected outside the G4 core. In contrast, in the corresponding average structure obtained for TBA-bs13, the benzene ring coloured in red in Figure 7 stacked exactly above the 6-membered ring of G11, whereas the blue ring turned away the G2-G5-G11-G14 tetrad pointing at the T3-T4 loop. These findings agree in full with the anticoagulant activities showed by TBA-bs12 and TBA-bs13. In fact, a lot of structure-activity relationships and the analysis of several X-ray TBA-Thrombin complexes (38,39,42,43), showed that the TBA G-quadruplex interacts with thrombin mainly through the TT loops region. Particularly, essential polar interactions between the nucleobases T4, T13 and G5 of the tertiary structure and the side chains of Arg 77, Arg 75 and Tyr 76 located in the ABE I of the thrombin take place, whereas T3 and T12 are essentially involved into hydrophobic contacts (43). Given that the dibenzyl linker was positioned outside the TT loops, the modification at T12 did not provoke substantial alteration in the TT loops region of the resulting G4, and almost all the above mentioned contacts between TBA and thrombin could occur. As opposite to this, the G-quadruplex bioactive conformation could be significantly affected by the replacement of T13 with the dibenzyl linker 1, seeing that the two benzene rings occupied the region between the TT loops. Consequently, despite the overall topology of the G4 formed by TBA-bs13 strongly resembled that of TBA, the formation of a stable TBA-thrombin complex was completely prevented.
Finally, the cytotoxic effect of TBA-bs13 on the He-La cell line was studied in comparison with that of TBA by means of MTT dose response experiments at concentrations in the range of 0.001–50 μM (Figure 11; Supplementary Tables S7 and S8). Inhibition of cell growth by TBA-bs13 started at 0.1 μM and reached the maximum effect at 10 μM, whereas TBA produced the same effects at concentration in the range of 1–50 μM. Consequently, TBA-bs13 was significantly more potent than TBA in the inhibition of He-La cell line growth.
Interestingly, it has been ascertained that some G4s bind NOA1 (51), a large GTP binding protein involved in important mitochondrial functions such as the synthesis of specific proteins, cellular respiration and apoptosis (52,80–83). In particular, both parallel and antiparallel DNA G4s are able to bind NOA1, but only the antiparallel ones also cause an increasing of about 4–5 folds of the NOA1 GTP-ase activity (51). The GTP dependent allosteric regulation has also been established for other GTP binding proteins involved in cancer development, survival and resistance (84–86). In light of the ability of TBA (antiparallel G-quadruplex) to accumulate into mitochondria (47), it will be of interest to verify the existence of such connection between the antiproliferative activities of new TBAs on He-La cells and the NOA1 functionality into mitochondria and/or in other cellular districts.
CONCLUSION
In conclusion, we showed that artful modifications in the TBA could provide cytotoxic GROs which are able to fold into the well known TBA G-quadruplex topology but devoided of the TBA anticoagulant action. In particular, the replacement of the residue 4 or 13 with the dibenzyl linker 1 afforded two TBA variants, specifically TBA-bs4 and TBA-bs13, which resulted unable to inhibit thrombin but showed significant antiproliferative activity against the He-La cell line. TBA-bs13 showed a 5-fold increased cell growth inhibition relative to the TBA as determined by the MTT dose response assay. The data concerning the other modified TBAs showed that none of them had a selective action relative to either the biological properties of TBA. It is noteworthy that TBA-bs9, which folded into the least stable G4, showed the lowest antiproliferative activity at all the explored concentrations. A good correlation between the structure stability and the antiproliferative action on He-La cells was also observed for the other two TBA-bs modified at one residue of the TGT loop, TBA-bs7 and TBA-bs8. Among the here reported new analogues, TBA-bs7 showed to be the best inhibitor of He-La cell line growth. Nevertheless, the presented data also demonstrated that more complex phenomena than the G-quadruplex stability control the difference in the antiproliferative effects of the TBA-bs obtained by replacing one of the residues in the TT loops of the TBA with the dibenzyl linker 1.
Although further deepened studies on these new TBA variants are certainly necessary, we herein described a potential route to obtain helpful G-quadruplex tools for the understanding of the molecular factors that contribute to the cytotoxicity of quadruplex-forming GROs (27,28,30,31).
Supplementary Material
Acknowledgments
The authors are grateful to Prof Caterina Fattorusso for the helpful discussion about GTP binding proteins and their involvement in cancer development and resistance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Department of Pharmacy of the University of Naples ‘Federico II’; the Italian Association for Cancer Research [IG10341 to D.C.]; PNR-CNR Aging Program [2012–2015]. Funding for open access charge: Department of Pharmacy of the University of Naples “Federico II”.
Conflict of interest statement. None declared.
REFERENCES
- 1.Kedzierski S., Khoshnejad M., Caltagirone G.T. Synthetic Antibodies: The Emerging Field of Aptamers. Bioprocessing J. 2012/2013;11:46–49. [Google Scholar]
- 2.Keefe A.D., Pai S., Ellington A. Aptamers as therapeutics. Nature Rev. 2010;9:537–550. doi: 10.1038/nrd3141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shum K.-T., Zhou J., Rossi J.J. Aptamer-Based Therapeutics: New Approaches to Combat Human Viral Diseases. Pharmaceuticals. 2013;6:1507–1542. doi: 10.3390/ph6121507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan W., Donovan M.J., Jiang J. Aptamers from Cell-Based Selection for Bioanalytical Applications. Chem. Rev. 2013;113:2842–2862. doi: 10.1021/cr300468w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han K., Liang Z., Zhou N. Design Strategies for Aptamer-Based Biosensors. Sensors. 2010;10:4541–4557. doi: 10.3390/s100504541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cho E.J., Lee J.-W., Ellington A.D. Applications of Aptamers as Sensors. Annu. Rev. Anal. Chem. 2009;2:241–264. doi: 10.1146/annurev.anchem.1.031207.112851. [DOI] [PubMed] [Google Scholar]
- 7.Bochman M.L., Paeschke K., Zakian V.A. DNA secondary structures: stability and function of G-quadruplex structures. Nature Rev. 2012;13:770–780. doi: 10.1038/nrg3296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collie G.W., Parkinson G.N. The application of DNA and RNA G-quadruplexes to therapeutic medicines. Chem. Soc. Rev. 2011;40:5867–5892. doi: 10.1039/c1cs15067g. [DOI] [PubMed] [Google Scholar]
- 9.Lech C.J., Heddi B., Phan A.T. Guanine base stacking in G-quadruplex nucleic acids. Nucleic Acids Res. 2013;41:2034–2046. doi: 10.1093/nar/gks1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang D., Okamoto K. Structural insights into G-quadruplexes: towards new anticancer drugs. Future Med. Chem. 2010;2:619–646. doi: 10.4155/fmc.09.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choi E.W., Nayak L.V., Bates P.J. Cancer-selective antiproliferative activity is a general property of some G-rich oligodeoxynucleotides. Nucleic Acids Res. 2010;38:1623–1635. doi: 10.1093/nar/gkp1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bates P.J., Laber D.A., Miller D.M., Thomas S.D., Trent J.O. Discovery and Development of the G-rich Oligonucleotide AS1411 as a Novel Treatment for Cancer. Exp. Mol. Pathol. 2009;86:151–164. doi: 10.1016/j.yexmp.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pei X., Zhang J., Liu J. Clinical applications of nucleic acid aptamers in Cancer. Mol. Clin. Oncol. 2014;2:341–348. doi: 10.3892/mco.2014.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Girvan A.C., Teng Y., Casson L.K., Thomas S.D., Juliger S., Ball M.W., Klein J.B., Pierce W.M. Jr, Barve S.S., Bates P.J. AGRO100 inhibits activation of nuclear factor-KB (NF-kB) by forming a complex with NF-kB essential modulator (NEMO) and nucleolin. Mol. Cancer Ther. 2006;5:1790–1799. doi: 10.1158/1535-7163.MCT-05-0361. [DOI] [PubMed] [Google Scholar]
- 15.Ireson C.R., Kelland R.L. Discovery and development of anticancer aptamers. Mol. Cancer Ther. 2006;5:2957–2962. doi: 10.1158/1535-7163.MCT-06-0172. [DOI] [PubMed] [Google Scholar]
- 16.Bates P.J., Kahlon J.B., Thomas S.D., Trent J.O., Miller D.M. Antiproliferative Activity of G-rich Oligonucleotides Correlates with Protein Binding. J. Biol. Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 17.Dempsey L.A., Sun H., Hanakahi L.A., Maizels N. G4 DNA Binding by LR1 and Its Subunits, Nucleolin and hnRNP D. A Role for G-G pairing in Immunoglobulin Switch Recombination. J. Biol. Chem. 1999;274:1066–1071. doi: 10.1074/jbc.274.2.1066. [DOI] [PubMed] [Google Scholar]
- 18.Hanakahi L.A., Sun H., Maizels N. High Affinity Interactions of Nucleolin with G-G-paired rDNA. J. Biol. Chem. 1999;274:15908–15912. doi: 10.1074/jbc.274.22.15908. [DOI] [PubMed] [Google Scholar]
- 19.Jing N., Li Y., Xu X., Sha W., Li P., Feng L., Tweardy D.J. Targeting Stat3 with G-Quartet Oligodeoxynucleotides in Human Cancer Cells. DNA and Cell Biol. 2003;22:685–696. doi: 10.1089/104454903770946665. [DOI] [PubMed] [Google Scholar]
- 20.Jing N., Li Y., Xiong W., Sha W., Jing L., Tweardy D.J. G-Quartet Oligonucleotides: A New Class of Signal Transducer and Activator of Transcription 3 Inhibitors That Suppresses Growth of Prostate and Breast Tumors through Induction of Apoptosis. Cancer Res. 2004;64:6603–6609. doi: 10.1158/0008-5472.CAN-03-4041. [DOI] [PubMed] [Google Scholar]
- 21.Qi H., Lin C.-P., Fu X., Wood L.M., Liu A.A., Tsai Y.-C., Chen Y., Barbieri C.M., Pilch D.S., Liu L.F. G-Quadruplexes Induce Apoptosis in Tumor Cells. Cancer Res. 2006;66:11808–11816. doi: 10.1158/0008-5472.CAN-06-1225. [DOI] [PubMed] [Google Scholar]
- 22.Longe H.O., Romesser P.B., Rankin A.M., Faller D.V., Eller M.S., Gilchrest B.A., Denis G.V. Telomere homolog oligonucleotides induce apoptosis in malignant but not in normal lymphoid cells: Mechanism and therapeutic potential. Int. J. Cancer. 2009;2:473–482. doi: 10.1002/ijc.23946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rankin A.M., Sarkar S., Faller D.V. Mechanism of T-oligo-induced cell cycle arrest in Mia-Paca pancreatic cancer cells. J. Cell Physiol. 2012;227:2586–2594. doi: 10.1002/jcp.22997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rankin A.M., Forman L., Sarkar S., Faller D.V. Enhanced citotoxicity from Deoxyguanosine-enriched T-oligo in prostate cancer cells. Nucleic Acids Ther. 2013;23:311–321. doi: 10.1089/nat.2013.0420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sedoris K.C., Thomas S.D., Clarkson C.R., Muench D., Islam A., Singh R., Miller D.M. Genomic c-Myc Quadruplex DNA Selectively Kills Leukemia. Mol. Cancer Ther. 2012;11:66–76. doi: 10.1158/1535-7163.MCT-11-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cogoi S., Quadrifoglio F., Xodo L.E. G-rich Oligonucleotides inhibits the Binding of a Nuclear Protein to the Ki-ras Promoter and Strongly Reduces Cell Growth in Human Carcinoma Pancreatic Cells. Biochem.-Us. 2004;43:2512–2523. doi: 10.1021/bi035754f. [DOI] [PubMed] [Google Scholar]
- 27.Reyes-Reyes E.M., Teng Y., Bates P.J. A New Paradigm for Aptamer Therapeutic AS1411 Action: Uptake by Macropinocytosis and Its Stimulation by a Nucleolin-Dependent Mechanism. Cancer Res. 2010;70:8617–8629. doi: 10.1158/0008-5472.CAN-10-0920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhang N., Bing T., Liu X., Qi C., Shen L., Wang L., Shangguan D. Cytotoxicity of Guanine-Based Degradation Products Contributes to the Antiproliferative Activity of Guanine-rich Oligonucleotides. Chem. Sci. 2015;6:3831–3838. doi: 10.1039/c4sc03949a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosenberg J.E., Bambury R.M., Van Allen E.M., Drabkin H.A., Lara P.N. Jr, Harzstark A.L., Wagle N.R., Figlin A., Smith G.W., Garraway L.A., et al. A phase II trial of AS1411 (a novel nucleolin-targeted DNA aptamer) in metastatic renal cell carcinoma. Invest. New Drugs. 2014;32:178–187. doi: 10.1007/s10637-013-0045-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Reyes-Reyes E.M., Salipurb F.R., Shamsc M., Forsthoefelc M.K., Bates P.J. Mechanistic studies of anticancer aptamer AS1411 reveal a novel role for nucleolin in regulating Rac1 activation. Mol. Oncol. 2015;9:1392–1405. doi: 10.1016/j.molonc.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Islam M.A., Thomas S.D., Murty V.V., Sedoris K.J., Miller D.M. c-Myc Quadruplex-forming Sequence Pu-27 Induces Extensive Damage in Both Telomeric and Nontelomeric Regions of DNA. J. Biol. Chem. 2014;289:8521–8531. doi: 10.1074/jbc.M113.505073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burge S., Parkinson G.N., Hazel P., Todd A.K., Neidle S. DNA quadruplex: sequence topology and structure. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Le H.T., Dean W.L., Buscaglia R., Chaires J.B., Trent J.O. An Investigation of G-Quadruplex Structural Polymorphism in the Human Telomere Using a Combined Approach of Hydrodynamic Bead Modeling and Molecular Dynamics Simulation. J. Phys. Chem. B. 2014;118:5390–5405. doi: 10.1021/jp502213y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dai J., Carver M., Punchihewa C., Jones R.A., Yang D. Cytotoxic G-rich oligodeoxynucleotides: putative protein targets and required sequence motif. Nucleic Acids Res. 2007;35:4927–4940. doi: 10.1093/nar/gkm465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dailey M.M., Miller M.C., Bates P.J., Lane A.N., Trent J.O. Resolution and characterization of the structural polymorphism of a single quadruplex-forming sequence. Nucleic Acids Res. 2010;38:4877–4888. doi: 10.1093/nar/gkq166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bock L.C., Griffin L.C., Latham J.A., Vermaas E.H., Toole J.J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992;355:564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- 37.Macaya R.F., Schultze P., Smith F.W., Roe J.A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex in solution. Proc. Natl. Acad. Sci. U.S.A. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Padmanabhan K., Tulinsky A. An ambiguous structure of a DNA 15-mer thrombin complex. Acta. Crystallogr. D Biol. Crystallogr. 1996;D52:272–282. doi: 10.1107/S0907444995013977. [DOI] [PubMed] [Google Scholar]
- 39.Padmanabhan K., Padmanabhan K.P., Ferrara J.D., Sadler J.E., Tulinsky A. The structure of alpha thrombin inhibited by a 15-mer single-stranded DNA aptamer. J. Biol. Chem. 1993;268:17651–17654. doi: 10.2210/pdb1hut/pdb. [DOI] [PubMed] [Google Scholar]
- 40.Wang K.Y., Krawczyk S.H., Bischofberger N., Swaminathan S., Bolton P.H. The tertiary structure of a DNA aptamer which binds to and inhibits thrombin determines activity. Biochem.-Us. 1993;32:11285–11292. doi: 10.1021/bi00093a004. [DOI] [PubMed] [Google Scholar]
- 41.Wang K., Mccurdy S., Shea R., Swaminathan S., Bolton P.A. Dna Aptamer Which Binds to and Inhibits Thrombin Exhibits a New Structural Motif for Dna. Biochem.-Us. 1993;32:1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- 42.Kelly J.A., Feigon J., Yeates T.O. Reconciliation of the X-ray and NMR structures of the thrombin binding aptamer d(GTTGGTGTGGTTGG) J. Mol. Biol. 1996;256:417–422. doi: 10.1006/jmbi.1996.0097. [DOI] [PubMed] [Google Scholar]
- 43.Russo Krauss I., Merlino A., Randazzo A., Novellino E., Mazzarella L., Sica F. High-resolution structures of two complexes between thrombin and thrombin-binding aptamer shed light on the role of cations in the aptamer inhibitory activity. Nucleic Acids Res. 2012;40:8119–8128. doi: 10.1093/nar/gks512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schultze P., Macaya R.F., Feigon J. Three-Dimensional Solution Structure of the Thrombin-Binding DNA Aptamer D(GGTTGGTGTGGTTGG) J. Mol. Biol. 1994;235:1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- 45.Đapiĉ V., Abdomeroviĉ V., Marrington R., Peberdy J., Rodger A., Trent J.O., Bates P.J. Biophysical and biological properties of quadruplex oligodeoxyribonucleotides. Nucleic Acids Res. 2003;31:2097–2107. doi: 10.1093/nar/gkg316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tasset D.M., Kubik M.F., Steiner W. Oligonucleotide Inhibitors of Human Thrombin that Bind Distinct Epitopes. J. Mol. Biol. 1997;272:688–698. doi: 10.1006/jmbi.1997.1275. [DOI] [PubMed] [Google Scholar]
- 47.Tseng T.Y., Wang Z.-F., Chien C.-H., Chang T.-C. In-cell optical imaging of exogenous G-quadruplex DNA by fluorogenic ligands. Nucleic Acids Res. 2013;41:10605–10618. doi: 10.1093/nar/gkt814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wanrooija P.H., Uhlera J.P., Simonssona T., Falkenberga M., Gustafssona C.M. G-quadruplex structures in RNA stimulate mitochondrial transcription termination and primer formation. Proc. Natl. Acad. Sci. 2010;107:16072–16077. doi: 10.1073/pnas.1006026107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zheng K., Wu R., He Y., Xiao S., Zhang J., Liu J., Hao Y., Tan Z. A competitive formation of DNA:RNA hybrid G-quadruplex is responsible to the mitochondrial transcription termination at the DNA replication priming site. Nucleic Acids Res. 2014;42:10832–10844. doi: 10.1093/nar/gku764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bharti S.K., Sommers J.A., Zhou J., Kaplan D.L., Spelbrink J.N., Mergny J.-L., Brosh R.M. DNA Sequences Proximal to Human Mitochondrial DNA Deletion Breakpoints Prevalent in Human Disease Form G-quadruplexes, a Class of DNA Structures Inefficiently Unwound by the Mitochondrial Replicative Twinkle Helicase. J. Biol. Chem. 2014;289:29975–29993. doi: 10.1074/jbc.M114.567073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Furoukh N., Goffart S., Szibor M., Wanrooij S., Braun T. Binding to G-quadruplex RNA activates the mitochondrial GTPase NOA1. Bioch. Bioph. Acta. 2013;1833:2933–2942. doi: 10.1016/j.bbamcr.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 52.Tang T., Zheng B., Chen S.-h., Murphy A.N., Kudlicka K., Zhou H., Farquhar M.G. hNOA1 Interacts with Complex I and DAP3 and Regulates Mitochondrial Respiration and Apoptosis. J. Biol. Chem. 2009;284:5414–5424. doi: 10.1074/jbc.M807797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kretz C.A., Stafford A.R., Fredenburgh J.C., Weitz J.I. HD1, a Thrombin-directed Aptamer, Binds Exosite 1 on Prothrombin with High Affinity and Inhibits Its Activation by Prothrombinase. J. Biol. Chem. 2006;281:37477–37485. doi: 10.1074/jbc.M607359200. [DOI] [PubMed] [Google Scholar]
- 54.Coppola T., Varra M., Oliviero G., Galeone A., D'Isa G., Mayol L., Morelli E., Bucci M., Vellecco V., Cirino G., et al. Synthesis and biological properties of new TBA analogues containing an acyclic nucleotide. Bioorg. Med. Chem. 2008;16:8244–8253. doi: 10.1016/j.bmc.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 55.Buff M.C.R., Schafer F., Wulffen B., Muller J., Potzsch B., Heckel A., Mayer G. Dependence of aptamer activity on opposed terminal extensions: improvement of light-regulation efficiency. Nucleic Acids Res. 2010;38:2111–2118. doi: 10.1093/nar/gkp1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bonifacio L., Church F.C., Jarstfer M.B. Effect of Locked-Nucleic Acid on a Biologically Active G-Quadruplex. A Structure-Activity Relationship of the Thrombin Aptamer. Int. J. Mol. Sci. 2008;9:422–433. doi: 10.3390/ijms9030422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagatoishi S., Isono N., Tsumoto K., Sugimoto N. Loop residues of thrombin-binding DNA aptamer impact G-quadruplex stability and thrombin binding. Biochimie. 2011;93:1231–1238. doi: 10.1016/j.biochi.2011.03.013. [DOI] [PubMed] [Google Scholar]
- 58.Jensen T.B., Henriksen J.R., Rasmussen B.E., Rasmussen L.M., Andresen T.L., Wengel J., Pasternak A. Thermodynamic and biological evaluation of a thrombin binding aptamer modified with several unlocked nucleic acid (UNA) monomers and a 20-C-piperazino-UNA monomer. Bioorg. Med. Chem. 2011;19:4739–4745. doi: 10.1016/j.bmc.2011.06.087. [DOI] [PubMed] [Google Scholar]
- 59.Pasternak A., Hernandez F.J., Rasmussen L.M., Vester B., Wengel J. Improved thrombin binding aptamer by incorporation of a single unlocked nucleic acid monomer. Nucleic Acids Res. 2011;39:1155–1164. doi: 10.1093/nar/gkq823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Borbone N., Bucci M., Oliviero G., Morelli E., Amato J., D'Atri V., D'Errico S., Vellecco V., Cirino G., Piccialli G., et al. Investigating the role of T7 and T12 residues on biological properties of Thrombin-Binding-Aptamer: enhancement of anticoagulant activity by a single nucleobase modification. J. Med. Chem. 2012;55:10716–10728. doi: 10.1021/jm301414f. [DOI] [PubMed] [Google Scholar]
- 61.Virgilio A., Petraccone L., Scuotto M., Vellecco V., Bucci M., Mayol L., Varra M., Esposito V., Galeone A. Investigating Anticoagulant Activity by Making a Tiny Chemical Modification. Chem. Bio. Chem. 2014;15:2427–2434. doi: 10.1002/cbic.201402355. [DOI] [PubMed] [Google Scholar]
- 62.Esposito V., Scuotto M., Capuozzo A., Santamaria R., Varra M., Mayol L., Virgilio A., Galeone A. A straightforward modification in the thrombin binding aptamer improving the stability, affinity to thrombin and nuclease resistance. Org. Biom. Chem. 2014;12:8840–8843. doi: 10.1039/c4ob01475h. [DOI] [PubMed] [Google Scholar]
- 63.Cai B., Yang X., Sun L., Fan X., Li L., Jin H., Wu Y., Guan Z., Zhang L., Zhanga L., Yang Z. Stability and bioactivity of thrombin binding aptamers modified with D-/L-isothymidine in the loop regions. Org. Biom. Chem. 2014;12:8866–8876. doi: 10.1039/c4ob01525h. [DOI] [PubMed] [Google Scholar]
- 64.Scuotto M., Persico M., Bucci M., Vellecco V., Borbone N., Morelli E., Oliviero G., Novellino E., Piccialli G., Cirino G., et al. Outstanding effects on antithrombin activity of modified TBA diastereomers containing an optically pure acyclic nucleotide analogue. Org. Biom. Chem. 2014;12:5235–5242. doi: 10.1039/c4ob00149d. [DOI] [PubMed] [Google Scholar]
- 65.Virgilio A., Varra M., Scuotto M., Capuozzo A., Irace C., Mayol L., Esposito V., Galeone A. Expanding the potential of G-quadruplex structures: Formation of a heterochiral TBA analogue. Chem. Bio. Chem. 2014;15:652–655. doi: 10.1002/cbic.201300775. [DOI] [PubMed] [Google Scholar]
- 66.Gait M.J. Oligonucleotide Synthesis: A practical approach. NY: Oxford University Press; 1990. [Google Scholar]
- 67.Roy S., Caruthers M. Synthesis of DNA/RNA and Their Analogs via Phosphoramidite and H-Phosphonate Chemistries. Molecules. 2013;18:14268–14284. doi: 10.3390/molecules181114268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kibbe W.A. OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res. 2007;35:W43–W46. doi: 10.1093/nar/gkm234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.De Cristofaro R., Di Cera E. Phenomenological analysis of the clotting curve. J. Protein Chem. 1991;10:455–468. doi: 10.1007/BF01025473. [DOI] [PubMed] [Google Scholar]
- 70.Hwang T.L., Shaka A.J. Water Suppression That Works. Excitation Sculpting Using Arbitrary Wave-Forms and Pulsed-Field Gradients. J. Magn. Reson. Ser. A. 1995;112:275–279. [Google Scholar]
- 71.Dalvit C. Efficient Multiple-Solvent Suppression for the Study of the Interactions of Organic Solvents with Biomolecules. J. Biomol. NMR. 1998;11:437–444. [Google Scholar]
- 72.Wüthrich K. NMR of Proteins and Nucleic Acids. NY: John Wiley and Sons; 1986. [Google Scholar]
- 73.Hosur R.V., Govil G., Miles H.T. Application of two-dimensional NMR spectroscopy in the determination of solution conformation of nucleic acids. Magn. Reson. Chem. 1988;26:927–944. [Google Scholar]
- 74.Gray D.M., Wen J.-D., Gray C.W., Repges R., Repges C., Raabe G., Fleischhauer J. Measured and calculated CD spectra of G-quartets stacked with the same or opposite polaritie. Chirality. 2008;20:431–440. doi: 10.1002/chir.20455. [DOI] [PubMed] [Google Scholar]
- 75.Paramasivan S., Rujan I., Bolton P.H. Circular dichroism of quadruplex DNAs: Applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 76.Vorlíčková M., Kejnovská I., Sagi J., Renčiuk D., Bednářová K., Motlová J., Kypr J. Circular dichroism and guanine quadruplexes. Methods. 2012;57:64–75. doi: 10.1016/j.ymeth.2012.03.011. [DOI] [PubMed] [Google Scholar]
- 77.Olsen C.M., Lee H.-T., Marky L.A. Unfolding Thermodynamics of Intramolecular G-Quadruplexes: Base Sequence Contributions of the Loops. J. Phys. Chem. B. 2009;113:2587–2595. doi: 10.1021/jp806853n. [DOI] [PubMed] [Google Scholar]
- 78.Mendelboum Raviv S., Horvath A., Aradi J., Bagoly Z., Fazakas F., Batta Z., Muszbek L., Harsfalvi J. 4-Thio-deoxyuridylate-modified thrombin aptamer and its inhibitory effect on fibrin clot formation, platelet aggregation and thrombus growth on subendothelial matrix. J. Thromb. Haemost. 2008;6:1764–1771. doi: 10.1111/j.1538-7836.2008.03106.x. [DOI] [PubMed] [Google Scholar]
- 79.Lechtenberg B.C., Freund S.M., Huntington J.A. An ensemble view of thrombin allostery. Biol. Chem. 2012;393:889–898. doi: 10.1515/hsz-2012-0178. [DOI] [PubMed] [Google Scholar]
- 80.Kolanczyk M., Pech M., Temojtel T., Yamamoto H., Mikula I., Calvaruso M.-A., van den Brand M., Richter R., Fischer B., Kossler N., et al. NOA1 is an essential GTPase required for mitochondrial protein synthesis. Mol. Biol. Cell. 2011;22:1–11. doi: 10.1091/mbc.E10-07-0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.He J., Cooper H.M., Reyes A., Di Re M., Kazak L., Wood S.R., Mao C.C., Fearnley I.M., Walker J.E., Holt I.J. Human C4orf14 interacts with the mitochondrial nucleoid and is involved in the biogenesis of the small mitochondrial ribosomal subunit. Nucleic Acids Res. 2012;40:6097–6108. doi: 10.1093/nar/gks257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Al-Furoukh N., Kardon J.R., Kruger M., Szibor M., Baker T.A., Braun T. NOA1, a Novel ClpXP Substrate, Takes an Unexpected Nuclear Detour Prior to Mitochondrial Import. PLoS ONE. 2014;9:e103141. doi: 10.1371/journal.pone.0103141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Heidler J., Al-Furoukh N., Kukat C., Salwig I., Ingelmann M.-E., Seibel P., Kruger M., Holtz J., Wittig I., Braun T., et al. Nitric Oxide-associated Protein 1 (NOA1) Is Necessary for Oxygen-dependent Regulation of Mitochondrial Respiratory Complexes. J. Biol. Chem. 2011;286:32086–32093. doi: 10.1074/jbc.M111.221986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat. Rev. Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 85.Vogelstein B., Kinzler K.W. Cancer genes and the pathways they control. Nat. Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 86.Andreoli M., Persico M., Kumar A., Orteca N., Kumar V., Pepe A., Mahalingam S., Alegria A.E., Petrella L., Sevciunaite L., et al. Identification of the First Inhibitor of the GBP1:PIM1 Interaction. Implications for the Development of a New Class of Anticancer Agents against Paclitaxel Resistant Cancer Cells. J. Med. Chem. 2014;57:7916–7932. doi: 10.1021/jm5009902. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


