Abstract
Bacterial type II toxin-antitoxin modules are protein–protein complexes whose functions are finely tuned by rapidly changing environmental conditions. E. coli toxin YafQ is suppressed under steady state growth conditions by virtue of its interaction with its cognate antitoxin, DinJ. During stress, DinJ is proteolytically degraded and free YafQ halts translation by degrading ribosome-bound mRNA to slow growth until the stress has passed. Although structures of the ribosome with toxins RelE and YoeB have been solved, it is unclear what residues among ribosome-dependent toxins are essential for mediating both recognition of the ribosome and the mRNA substrate given their low sequence identities. Here we show that YafQ coordinates binding to the 70S ribosome via three surface-exposed patches of basic residues that we propose directly interact with 16S rRNA. We demonstrate that YafQ residues H50, H63, D67 and H87 participate in acid-base catalysis during mRNA hydrolysis and further show that H50 and H63 functionally complement as general bases to initiate the phosphodiester cleavage reaction. Moreover YafQ residue F91 likely plays an important role in mRNA positioning. In summary, our findings demonstrate the plasticity of ribosome-dependent toxin active site residues and further our understanding of which toxin residues are important for function.
INTRODUCTION
Bacteria rapidly adapt their transcriptome and proteome to facilitate the halting of cellular replicative processes to promote survival in changing environments such as limited nutrients. Toxin-antitoxin genes are central to this adaptation through their regulation of growth during stress (1). These stress modules are ubiquitous throughout bacteria and their prominent role in facilitating the transition into the bacterial persistence state is now well established (2–11). Type II toxin-antitoxin gene pairs encode two small proteins (∼10–15 kDa) that form a transcriptional repressor complex during nutrient-rich growth. Diminishing nutrients and other stresses trigger a number of downstream events that cause degradation of the antitoxin by cellular proteases Lon, ClpXP and ClpAP, resulting in de-repression at the toxin-antitoxin locus and freeing of the toxin protein (12–16). Toxin activation causes an arrest in replication, transcription or translation due to degradation of essential protein or RNA molecules that reduce the available pools of building blocks for sufficient growth. The dual roles of toxin-antitoxin modules therefore allow for rapid transcriptional and translation regulation to alter expression patterns that are dependent upon the environment encountered.
Type II toxin proteins repress translation by degradation of free mRNA, ribosome-bound mRNA, rRNA, tRNA or even modification of translation factors (17–27). Toxins RelE, YafQ, YoeB and HigB cleave only ribosome-bound mRNA and display loose codon specificity (17,18,22,26,28). These ribosome-dependent, RelE/YoeB family members adopt microbial RNase folds containing a compact, antiparallel β-sheet flanked by 2–4 α-helices and a distinctive concave active site where mRNA is likely engaged (29–33). Despite the overall topological similarity of the toxins, there is high variability in the residues that comprise the active site (34) (Figure 1A), leading to the suggestion that mRNA cleavage may be distinct for each toxin or that the ribosome may play an active role in hydrolysis (17,35). However it is currently thought that although the ribosome positions the mRNA substrate in an orientation that is optimal for catalysis, the ribosome does not actively participate in hydrolysis (36).
Figure 1.

YafQ sequence conservation and modeling of its interactions with the 70S ribosome. (A) Sequence alignments of E. coli YafQ, E. coli RelE, E. coli YoeB and P. vulgaris HigB share low sequence identities (11–20%). There are no 70S structures available with YafQ and HigB bound (upper sequences). RelE and YoeB residues (bottom sequence) that interact with 16S rRNA helices (shaded blue) and proposed catalytic residues (shaded pink) are from RelE- and YoeB-bound 70S structures (PDB codes 4V7J and 4V8X). The proposed HigB catalytic residue H92 (26) is shaded pink. YafQ residues predicted to interact with 16S rRNA (shaded light blue) or proposed to be catalytic (shaded light pink) are tested in this study. (B) The crystal structure of YafQ (PDB code 4Q2U; blue) docked onto 70S by structurally aligning with the 70S-RelE complex (middle; PDB code 4V7J). YafQ residues tested in this study are depicted as sticks. The three mRNA nucleotides in the A site are numbered +4, +5 +6 according to the AUG start codon (+1, +2, +3) in the P site. A structure of the 70S-RelE active site (middle; PDB code 4V7J) and the 70S-YoeB active site (right; PDB code 4V8X) are shown for comparison. RelE residues R81* and R45* were mutated to alanine in 70S structural studies. (C) A 120° rotation of (B) emphasizing the predicted and known interactions of YafQ (left), RelE (middle) and YoeB (right) with 16S rRNA helices h18, h31 and h34. A YoeB dimer interacts with the ribosomal A site (depicted as purple and white monomers) which alters how YoeB interacts with 16S rRNA h31 as compared to RelE (red dotted box).
Structures of RelE and YoeB bound to the A site of the 70S provided significant insights into how toxins recognize their mRNA targets (36,37). From the 70S-RelE structures, it was proposed that R81 and Y87 function as acid-base pairs, R61 stabilizes the transition state and K52 and K54 provide transition state charge stabilization (36) (Figure 1B). Single turnover kinetic experiments further demonstrated that R81 and K52 facilitate acid-base catalysis and R61 and K54 stabilize the transition state (38). No detailed mechanistic studies of YoeB exist but functional and structural studies suggest that H83 and E46 promote mRNA degradation as the acid-base pairs while R65 likely stabilizes the transition state (32,37). Despite these advances, it is difficult to extend our understanding to other proposed ribosome-dependent toxins such as YafQ and HigB because they lack similar active site residues and instead, contain histidines and acidic residues such as aspartic acid and glutamic acid (Figure 1A). Therefore it is unclear if a unified mechanism to describe how toxins degrade mRNA on translating ribosomes can be developed.
The crystal structure of the DinJ-YafQ toxin-antitoxin complex confirmed that the YafQ toxin is a RelE/YoeB family member; YafQ adopts a four-stranded, β-sheet with three solvent-exposed α-helices and a distinctive concave cleft located between its β-sheet and α3 (39,40). In the context of the DinJ-YafQ complex, three histidines (H50, H63 and H87) chelate a single sulfate ion, suggesting these residues interact with the negatively charged nucleic acid phosphate backbone (39,40). Modeling of YafQ onto other ribosome-dependent toxins RelE and YoeB bound to the 70S ribosome (36,39) provides hints as to which YafQ residues may be important for catalysis and ribosome recognition (Figure 1B and C). In this study we demonstrate that YafQ residues H50, H63, D67, H87 and F91 play significant roles in mRNA catalysis and that three patches of basic residues on the surface of YafQ facilitate ribosome recognition. Comparison with other RelE/YoeB toxin family members and microbial endonucleases reveals conserved toxin α-helices are the likely determinants for recognition of 16S rRNA providing a structural basis for the selectivity of these toxins for the ribosomal A site.
MATERIALS AND METHODS
Sequence alignments and molecular modeling
Ribosome-dependent toxins were aligned with E. coli YafQ (PDB code 4Q2U) (39) based upon their similar tertiary structures represented by having an overall high Z-scores using the program Dali (41): E. coli RelE (PDB code 4FXI, Z-score = 9.5, RMSD of 2.3 Å, 11% sequence identity) (30), E. coli YoeB (PDB code 2A6S, Z-score = 12.0, RMSD of 2.0 Å, 20% sequence identity) (32) and P. vulgaris HigB (PDB code 4MCX, Z-score = 9.3, RMSD of 2.3 Å, 15% sequence identity) (33). YafQ was modeled onto E. coli RelE bound to the Thermus thermophilus 70S structure (PDB code 4V7J) (36) in DaliLite using the pairwise structural alignment feature (42). All figures were generated using PyMOL (43).
Site-directed mutagenesis
The plasmid pBAD33-YafQ was a kind gift from the Woychik laboratory (Rutgers University). The design of plasmid pET-DinJ-(His6)YafQ was described previously (39). Both of these plasmids served as templates for site-directed mutagenesis with indicated primers using the QuikChange Lightning Kit (Agilent) (Supplementary Table S1).
Growth assays
The E. coli BW25113 strain was used in the toxicity assays (44). Growth assays were performed in M9 medium supplemented with 0.05% (w/v) casamino acids, 10 mM magnesium sulfate and 25 μg ml−1 chloramphenicol at 37°C as previously described (18). Overnight cultures containing 0.21% (v/v) glucose to suppress leaky expression were diluted 1:100 and grown in M9 medium containing 0.21% (v/v) glycerol until an OD600 of 0.2. Each culture was then split with one receiving 0.2% arabinose and the other no arabinose. Time points were taken every hour after induction for 5 h. All growth curves were performed at least three independent times.
Western blot analysis
Expression levels of DinJ-YafQ from the growth assays were detected by Western blot analysis using polyclonal antibodies against the complex (kind gift from the Woychik laboratory) (18). Anti-rabbit IgG (Sigma) and ECL-prime kit (GE Healthcare) were used for chemiluminescent detection. YafQ mutants (10.8 kDa, untagged) were compared to both components of the DinJ-(His6)YafQ complex (9.4 kDa and 12.1 kDa, respectively).
[35S]-labeling and purification of YafQ and DinJ
The E. coli BL21 Gold (DE3) pLysS strain was used for purification of [35S]-labeled YafQ or [35S]-DinJ. Overnight cultures were grown in M9 minimal medium supplemented with 0.05% (w/v) casamino acids, 0.2% (v/v) glycerol, 10 mM magnesium sulfate, 10 μg ml−1 thiamine and 200 μg ml−1 ampicillin at 37°C. The cultures were diluted to 1:100 and grown to an OD600 of 0.5. [35S]-L-Methionine (final concentration of 2 μCi/ml) was added to the cells and grown for 15 min to suppress methionine biosynthesis. Cultures were induced with 0.4 mM IPTG (final concentration), grown for 2 h and cells were lysed by sonication in 20 mM Tris-HCl pH 7.5, 250 mM KCl, 0.1% (v/v) Triton X-100, 5 mM β-mercaptoethanol (β-Me), 0.1 mM phenylmethanesulfonylfluoride and 0.1 mM benzamidine. Cell debris was removed by centrifugation at 35,000 x g. Recombinant protein was purified by affinity chromatography on a HisSpin TrapTM column (GE Healthcare) pre-equilibrated with a binding buffer containing 40 mM Tris-HCl pH 7.5, 250 mM KCl, 5 mM imidazole, 5 mM MgCl2, 10% (v/v) glycerol and 5 mM β-Me. To disrupt the DinJ-(His6)YafQ complex and elute DinJ, binding buffer in addition to 6 M guanidine-HCl was applied. Denatured YafQ was eluted by the addition of 300 mM imidazole and 6 M guanidine-HCl to the binding buffer. Both DinJ and YafQ were shock refolded by rapid dilution, concentrated and buffer exchanged into a storage buffer of 40 mM Tris-HCl pH 7.5, 250 mM KCl, 5 mM MgCl2 and 5 mM β-Me, flash frozen in liquid nitrogen and stored at −80°C. Protein purity was determined to be of >95% by SDS-PAGE in addition to imaging using a Typhoon Trio phosphorimager (GE Healthcare).
Purification of wild-type YafQ, DinJ and DinJ-YafQ
The same strain and buffers used for [35S]-labeling of YafQ and DinJ were used when purifying the DinJ-(His)6YafQ complex, YafQ and DinJ. Overnight cultures were grown in LB medium supplemented with 200 μg ml−1 ampicillin at 37°C. Cultures were diluted 1:100, grown until an OD600 of 0.5–0.7, protein expression was induced with 0.4 mM IPTG (final concentration) and cultures were incubated for 3 h. Cells were lysed by sonication, centrifuged at 35,000 x g to remove cell debris, and the supernatant containing DinJ-(His)6YafQ was affinity purified on a HisTrap FF CrudeTM Ni2+ Sepharose column (GE Healthcare). The DinJ-(His6)YafQ complex was eluted by the addition of 300 mM imidazole in the binding buffer. For DinJ or YafQ purification, the complex was disrupted by applying the denaturing binding buffer containing 6 M guanidine-HCl to elute DinJ, while denatured (His6)YafQ was column refolded by slowly reducing the concentration of guanidine-HCl over 8 h. Eluted DinJ was shock refolded as described above. Finally, (His6)YafQ was eluted by addition of 300 mM imidazole and further purified on a Sephadex 75 16/60 gel filtration column (GE Healthcare) into the storage buffer (40 mM Tris-HCl pH 7.5, 250 mM KCl, 5 mM MgCl2 and 5mM β-Me). The purity of all proteins was >95% as determined by SDS-PAGE. Typical yields were 25 mg of the DinJ-YafQ complex from one liter of culture with a final yield of 6–7 mg of YafQ after the denaturation/renaturation protocol. Proteins were flash frozen in liquid nitrogen and stored at −80°C.
Binding assays
The binding assays were performed as previously reported (45) with a few minor changes. E. coli 70S were purified from strain MRE600 as previously described (46) but after the initial sucrose pelleting step, the lysate was applied to a hydrophobic interaction column (Toyopearl Butyl-650S) to further purify from RNases. E. coli 70S ribosomes (2.5 μM final concentration) were programmed with a two molar excess of mRNA (IDT) at 37°C for 6 min in 50 mM KCl, 10 mM NH4Cl, 10 mM Mg(OAc)2 and 5 mM HEPES, pH 7.5 (47). The mRNA sequence 5′-GGCAAGGAGGUAAAA AUG A1A2A3 GAAA-3′ contains an A-site AAA lysine codon (Shine-Dalgarno is underlined, P-site codon is in italics and subscripts denote the A-site codon nucleotides) or an uncleavable mRNA with 2′-OCH3 groups at the three A-site nucleotides to prevent cleavage. A four-fold molar excess of P-site E. coli tRNAfMet (Chemical Block) was incubated at 37°C for 30 min. This programmed 70S complex was serially diluted (0.125 -2 μM [70S] final) and either [35S]-YafQ or [35S]-DinJ (0.25 μM final concentration) were added and incubated for 10 min at 37°C. The 70S complex was separated from unbound [35S]-YafQ or [35S]-DinJ by filtration over a MicroconTM YM-100 column (EMD Millipore). The samples were centrifuged to dryness at 5000 x g for 10 min, the 70S complex was recovered by the addition of 25 μl of buffer to the membrane for 10 min and the amount of radioactivity was determined by liquid scintillation counting. The fraction of YafQ/DinJ bound to the 70S was determined by the amount of radioactivity in the retentate (cpm) divided by the total radioactivity (cpm). The previously published protocol measured the amount of unbound [35S]-IF1 in the filtrate (45), however we observed nonspecific binding of free [35S]-YafQ to the membrane therefore the amount of 70S-[35S]-YafQ bound in the retentate was instead measured. Equilibrium binding of [35S]-YafQ to 70S were fit to a quadratic equation taking into account ligand depletion (45,48) using the GraphPad Software (49).
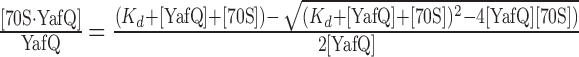 |
K d is the equilibrium dissociation constant and [70S·Yaf Q]/[YafQ] is the fraction of YafQ bound to the ribosome. The fraction of 70S bound [35S]-YafQ was plotted against the [70S] as determined by three independent experiments. The use of the ligand depletion binding allows for a statistical analysis of the goodness of fit of the data (R2).
mRNA cleavage assays
E. coli 70S ribosomes (1.2 μM) were programmed with 5′- [32P] mRNA (0.6 μM, IDT; same mRNA as used in the binding studies) at 37°C for 6 min. A two fold molar excess of 70S to mRNA was used to ensure all mRNA was bound to the ribosome. E. coli tRNAfMet was next added (3 μM) to fill the P site at 37°C for 30 min and then further incubated for 10 min at 25°C. The final concentration of the 70S-[32P]mRNA-tRNAfMet complex was 0.6 μM. Similar to previous qualitative 70S-RelE cleavage assays (36), one molar equivalent of YafQ (0.6 μM) was next incubated at 25°C, aliquots were removed at different time points, quenched with an equal volume of formamide dye (80% formamide, 0.2 mg ml−1 bromophenol blue, 0.2 mg ml−1 xylene cyanol) and incubated at 65°C for 2 min. Samples were loaded onto an 18% polyacrylamide/8 M urea gel and the amount of mRNA cleaved was quantified with ImageQuant TL (GE Healthcare). To determine whether a 3′ phosphate remained after YafQ cleavage, samples were heated to 65°C for 10 min to precipitate ribosomal RNA and proteins, cooled for 10 min, and then T4 polynucleotide kinase (T4 PNK; New England Biolabs) was incubated for 6 h at 37°C before analysis on the denaturing 8M urea PAGE gel. All of the mRNA cleavage assays were performed in three independent experiments.
RESULTS
H50, H63, D67, H87 and F91 are important for YafQ toxicity
YafQ residues that may be potentially important for catalysis were identified from modeling studies of the X-ray crystal structure of YafQ onto previously solved 70S-RelE structures (36) (Figure 1B) and upon mutation to alanine, tested their effect on YafQ function using in vivo growth assays. Overexpression of YafQ inhibits cell growth and a mutation in a residue required for YafQ activity should restore growth (18). YafQ H50A, H63A, D67A, H87A and F91A variants all result in the restoration of growth upon overexpression indicating that each residue is important for YafQ function (Figure 2). YafQ mutants D61A and Q53A did not affect growth, suggesting these residues are not individually required for function. Western blot analysis of each growth assay using antibodies specific for the DinJ-YafQ complex demonstrate that restoration of growth is not from insoluble YafQ but from an inactive YafQ (Supplementary Figure S1).
Figure 2.

Identification of YafQ residues important for function. (A and B) Potential YafQ residues important for function were mutated to alanine and upon overexpression, their effect on E. coli growth was monitored. Overexpression of wild-type YafQ (WT) results in inhibition of cell growth (red line) while restoration of cell growth indicates YafQ inactivity suggesting an essential role for the residue. An uniduced WT YafQ is shown for comparison. Error bars display standard deviations from at least three replicates.
Extended basic patches on the YafQ surface are also important for YafQ toxicity
RelE α1 and α2 interact with 16S rRNA helices 31 and 18 (h31 and h18), respectively and do not contact the 50S or ribosomal proteins (Figure 1C) (36). YoeB also interacts with 16S rRNA h31 and h18 but the details of this interaction are slightly different because YoeB binds to the ribosome as a dimer unlike other ribosome-dependent toxins which are monomeric on and off the ribosome (Figure 1C) (30,32,33,37,39,40). An additional interaction between the ribosome and YoeB is with loop 3 residues K42 and K44 while in RelE this loop is shorter and does not contact the ribosome (Figure 1C). Although these interactions have been seen in 70S-toxin bound structures, whether these interactions are important for function has never been established. From our modeling experiments of YafQ in the ribosomal A site, we predict that α1 and α2 are adjacent to 16S rRNA while loop 3 would need to rearrange to prevent clashing with mRNA (Figure 1B and C). To determine if YafQ residues in α1 and α2 are important for function, we mutated three patches of positively charged residues (K14A/K17A, K21A/R22A/K24A and K28A/K30A) and loop 3 residues Y47 and K48 to alanine, and tested the growth of bacteria expressing mutant toxins. If restoration of growth occurs, this potentially suggests we have ablated YafQ binding to the ribosome. Our results show that all three patches of lysine and arginine mutations and K48A result in a similar partial restoration of cell growth (Figure 3A and B). These data indicate that while each single residue may not individually be essential for YafQ recognition of the ribosome, the residues may be collectively required for recognition of the A-site 16S rRNA surface. The Y47 residue appears to play a significant functional role as bacterial growth is fully restored upon mutation to an alanine (Figure 3B).
Figure 3.

Ribosome-dependent toxins use two α-helices to directly interact with 16S rRNA. (A and B) Potential YafQ residues important for interacting with 16S rRNA were mutated to alanine and their effect on E. coli growth was monitored. Overexpression of wild-type YafQ (WT) results in inhibition of cell growth (red line) while restoration of growth indicates a possible disruption of YafQ's interaction with the ribosome. An uniduced WT YafQ is shown for comparison. Error bars display standard deviations from at least three replicates.
H50 and H87 are not important for 70S binding
Our in vivo growth assays, in addition to previous studies (34), indicate that YafQ residues Y47, H50, H63, D67, H87 and F91 are functionally important (Figure 2). To delineate whether these, and other residues that cluster around the mRNA according to our modeling studies (Figure 1B), are important for recognition of the ribosomal A site, we performed binding assays where we incubated [35S]-YafQ with increasing concentrations of 70S ribosomes programmed with mRNA containing an A-site lysine codon (5′-AAA-3′; all codons are denoted in the 5′ to 3′ direction) and a tRNAfMet in the P site. The mRNA also contained a 2′-OCH3 ribose modification at all three A-site nucleotide positions which prevents toxin-mediated cleavage (36). In the 70S-RelE and 70S-YoeB structures, as there are no interactions with the 2′-OH groups at the +4 and +6 positions of the mRNA, we rationalize that these groups play little to no role in recognition of the A-site mRNA and therefore should have little effect when modified with a 2′-OCH3 (36,37). Our results indicate that wild-type YafQ binds to the A site of the ribosome with an apparent dissociation constant (Kd) of 358 nM, binding within the same order of magnitude as other A-site binding translation factors (IF1, 600 nM; IF3, 440 nM) and tRNAs (20–500 nM) (Figure 4A; Table 1)(45,50). YafQ shows weak or no binding to a 70S programmed with mRNA containing a non-preferred UUC codon or an empty 70S (e.g. without mRNA or P-site tRNA), respectively (Figure 4A). Likewise as expected, cognate [35S]-DinJ antitoxin does not show detectable binding to a programmed 70S. We next tested YafQ binding to a 70S programmed with an AAA codon (i.e. cleavable lysine codon lacking the 2′-OCH3 modification). In this case, the apparent dissociation constant is reduced 10-fold to 4.5 μM, consistent with YafQ departing the programmed 70S after mRNA degradation (Figure 4A). For all subsequent binding assays, we used an uncleavable lysine codon for consistency.
Figure 4.

YafQ α1 and α2 helices are important for recognition of the A site of the 70S ribosome. (A) Increasing amounts of E. coli 70 were incubated with [35S]-YafQ and the amount of YafQ bound to the A site were plotted against fraction bound. Control experiments include: A-site AAA lysine mRNA, uncleavable mRNA (AAAOMe456 where ‘OMe456’ denotes a 2′-OCH3 group at the A-site nucleotides +4, +5 and +6), non-preferred UUC phenylalanine codon, cognate antitoxin DinJ, and experiments lacking mRNA bound to the 70S. Binding assays in (B), (C), and (D) were performed with an uncleavable mRNA (AAAOMe45 lysine codon). Data were fit to a one site binding with ligand depletion model using the GraphPad Prism program. Error bars display standard deviations from at least three replicates.
Table 1. Summary of the effects YafQ mutations have on bacterial growth, in vitro binding to a programmed 70S and in vitro mRNA degradation.
| E. coli YafQ | mRNA codona | Growth rateb | 70S bindingc | mRNA cleavaged | Possible role for YafQ residues | |
|---|---|---|---|---|---|---|
| Kd (μM)c | R2 | |||||
| Wild-type | AAA | NA | 4.5 ± 0.78 | 0.9402 | +++ | |
| AAAOMe | NA | 0.36 ± 0.07 | 0.9489 | NA | ||
| UUC | NA | >15e | 0.8979 | NA | ||
| Q53A | - | 0.50 ± 0.13 | 0.9497 | NA | No role | |
| Y47A | +++ | 0.49 ± 0.08 | 0.9692 | +++ | Unknown | |
| D61A | - | 0.61 ± 0.14 | 0.9157 | ++ | Minimal role in positioning | |
| F91A | +++ | 3.37 ± 0.48 | 0.9709 | ++ | mRNA positioning | |
| H63A | +++ | 3.94 ± 0.92 | 0.8362 | ++ | Catalysis/mRNA positioning | |
| D67A | +++ | 0.81 ± 0.10 | 0.9616 | ++ | Indirect catalysis | |
| H50A | +++ | 0.99 ± 0.27 | 0.9053 | + | Catalysis | |
| H87A | +++ | 0.62 ± 0.20 | 0.9229 | - | Catalysis | |
| Triple mutant | NA | NA | NA | - | Catalysis | |
| K48A | ++ | 14.7 ± 3.1 | 0.9456 | NA | 16S rRNA recognition | |
| K14A/K17A | ++ | >15e | 0.8607 | NA | 16S rRNA recognition | |
| K21A/R22A/K24A | ++ | >15e | 0.9636 | NA | 16S rRNA recognition | |
| K28A/K30A | ++ | 1.44 ± 0.28 | 0.9545 | NA | 16S rRNA recognition | |
NA, not attempted; Triple mutant is YafQ H50A, D61A and H63A.
aA-site codons are shown for the 70S binding studies with wild-type YafQ. AAAOMe represents a codon containing a 2′-OCH3 modification at all three A-site nucleotides.
bGrowth rates in minimal M9 medium at 37°C with wild-type YafQ overexpression resulting in no growth (-) while resumption of growth is indicated by ++ (partial reversal) or +++ (complete reversal of growth inhibition).
cKd values are the mean ± SEM from at least three independent experiments with their corresponding goodness of fit (R2). For the studies with the different YafQ variants, all were performed with an uncleavable AAAOMe A-site codon.
dExtent to which YafQ and YafQ variants cleave mRNA are indicated by +++ (robust degradation), ++ (partial degradation), + (minimal degradation) and – (no degradation).
eLittle to no binding to 70S in our assay.
YafQ proteins with mutations at residues determined to be important for activity from functional assays were tested for their ability to bind a programmed 70S (Figure 2). YafQ carrying H50A and H87A mutations had only a ∼2–2.5-fold effect on binding (990 nM and 620 nM, respectively). In contrast, the H63A mutant displayed a reduction in affinity by 10-fold to 4 μM (Figure 4B). YafQ residue D67 is proposed to be functionally important (Figure 2B) yet did not exhibit any reduction in binding upon alanine substitution, indicating its main role is likely catalytic (Figure 4C). Conversely, the main role of proposed YafQ active site residue F91A appears to be in recognition of either the mRNA or the ribosome as mutation to alanine results in a 10-fold reduction in binding (Kd = 3.4 μM) (Figure 4C). YafQ binding to 70S is affected slightly when Q53 is mutated to alanine (502 nM) suggesting this residue does not play any role in productive binding (Figure 4C). Lastly although the D61A variant exhibits wild-type YafQ toxicity in growth assays suggesting an important functional role (Figure 2D), our binding results show that the D61A variant has wild-type affinity (610 nM) providing support that D61 is not critical for recognition of the ribosomal complex (Figure 4D).
Mutation of three patches of positively charged residues on the surface of YafQ that we predict might interact with the negatively charged phosphate backbone of 16S rRNA each resulted in a partial restoration of growth (Figure 3A). Two groups of YafQ mutations (K14A/K17A and K21A/R22A/K24A) did not bind to the 70S at all, while the K28A/K30A mutation exhibited a greater than 4-fold reduction in affinity (Kd = 1.4 μM) (Figure 4D). Single YafQ point mutations of Y47A and K48A that we predict to interact with 16S rRNA based upon our modeling studies (Figure 1C) show a complete and a partial restoration of growth, respectively (Figure 3B). Therefore the restoration of growth by the K48A mutation was most likely due to its reduced ability to bind to the 70S (Kd reduced by 40-fold to 14.7 μM). This is in contrast to the Y47A mutant which displayed a wild-type YafQ binding affinity (Figure 3B). These seemingly contradictory results for residue Y47 could be explained by the possibility that the Y47A variant undergoes rapid turnover in the cell allowing for a phenotype that resembles an inactive YafQ (i.e. normal growth). This interpretation is consistent with the low levels of YafQ Y47A protein we observe by Western blot analysis (Supplementary Figure S1).
Proposed mechanism of YafQ-mediated mRNA cleavage
To determine how YafQ cleaves mRNA, we performed in vitro assays where we monitor the 5′-[32P]-mRNA cleavage products after addition of YafQ over time. We first incubated 70S with mRNA containing a strong Shine-Dalgarno sequence optimally placed from the P-site AUG start codon and next added tRNAfMet to the P site to ensure correct positioning of the mRNA (data not shown). Our results show that YafQ, the DinJ-YafQ complex, or antitoxin DinJ alone, are unable to cleave free mRNA (Figure 5A). As expected, YafQ cleaves an mRNA containing an A-site lysine codon (AAA) almost to completion when bound to the 70S supporting its predicted role as a ribosome-dependent endonuclease (Figure 5A) (18).
Figure 5.

YafQ residues important for degrading the AAA lysine codon in the ribosomal A site. The cleavage of programmed 5′-[32P]-mRNA (25 mer; filled arrowhead) containing an AAA lysine codon in the 70S A site was monitored by the appearance of a degradation product (∼19 mer; open arrowhead). (A) mRNA cleavage experiments demonstrate the requirement for the ribosome by YafQ. Antitoxin DinJ and YafQ without the 70S are unable to cleave an AAA lysine-containing mRNA transcript alone. 19 mer, 20 mer and 25 mer standards are shown at the left of each gel. (B) mRNA containing an AAA codon in the A site was incubated with YafQ and then further incubated with T4 PNK. The removal of the 3′ phosphate by T4 PNK shows the mRNA run as a 20 mer (gray arrowhead). (C) YafQ residues potentially important for activity were mutated to alanine and assayed for their ability to cleave mRNA on a programmed 70S ribosome and monitored over time. If YafQ is absent from the reaction, no mRNA is cleaved (lanes 1–5; 0s, 60s, 600s, 1800s and 3600s). Wild-type YafQ (lanes 6–13) and YafQ Y47A variant (lanes 14–21) were monitored over the following eight time points: 10s, 30s, 60s, 120s, 300, 600s, 1800s and 3600s. YafQ H50A and YafQ D61A reactions were monitored over seven time points: 30s, 60s, 120s, 300, 600s, 1800s and 3600s. (D) Further degradation assays were performed with YafQ H63A (lanes 1–7), YafQ D67A (lanes 8–14), YafQ H87A (lanes 15–21), YafQ F91A (22–28) and a YafQ triple mutant of H50A, D61A and H63A denoted as ‘tm’ (lanes 29–35) over seven time points similar to YafQ H50A and D61A. The percent mRNA degraded by each YafQ variant at 2 min (E; Y axis) and 30 min (F) were plotted with error bars showing the standard deviation from at least three replicates.
Ribonucleases mediate RNA hydrolysis by facilitating nucleophilic attack on an adjacent scissile phosphate resulting in products that either contain a phosphate at the 3′ or 5′ end (51). RelE and YoeB structurally resemble endonucleases such as RNase T1, RNase Sa2 and RNase A which all leave a 3′ phosphate after cleavage, in contrast to RNase H, which produces a 5′ phosphate (52,53). Cleavage by RelE results in a 2′-3′ cyclic phosphate end while YoeB has been proposed to leave a 3′ phosphate (36,37). To determine whether YafQ cleavage results in a 3′-phosphate product, we performed the same in vitro cleavage assay as described previously, treated the reaction with T4 PNK to remove a 3′ phosphate and then ran the reactions on a denaturing polyacrylamide gel. Our results show that without T4 PNK treatment, the reaction product ran faster than after incubation with T4 PNK (Figure 5B). As RNAs containing a 2′-3′ cyclic phosphate or a 3′ phosphate typically migrate one to two nucleotides faster on denaturing gels than 3′ dephosphorylated RNA (54,55), these results are consistent with the presence of a terminal 3′ phosphate after mRNA cleavage by YafQ. Additionally, these results provide supporting evidence that YafQ performs phosphodiester backbone cleavage via a similar mechanism as RNases T1, Sa2 and A.
H50 and H87 are important for mRNA cleavage
To qualitatively test which YafQ residues participate in mRNA hydrolysis, we performed mRNA in vitro cleavage assays with a number of YafQ variants. Our results demonstrate the that YafQ variant Y47A does not display any changes in mRNA degradation as compared to wild-type, indicating this residue plays little to no role in catalysis (Figure 5C & E). In contrast, when YafQ residues D61, H63, D67 and F91 are mutated to alanine, the ability of YafQ to cleave mRNA is reduced (Figure 5C–F). Furthermore, YafQ residue H50 appears to play a significant role in mRNA cleavage as upon mutation to alanine, the amount of mRNA cleaved is reduced substantially, and in the case of H87A variant, no cleavage occurs at all. Lastly, a YafQ triple mutant of H50A, D61A and H63A (‘tm’) completely ablates YafQ activity much like the H87A variant.
DISCUSSION
E. coli YafQ is a member of the RelE/YoeB toxin family, characterized by a conserved microbial RNase fold. This family of endoribonucleases share low sequence identities ranging from 11–20% and high amino acid variability especially within their active sites. One possible reason for this variability is the requirement to recognize diverse three-nucleotide codons in the vicinity of the ribosomal A site. Based upon our initial modeling studies of YafQ bound to the 70S ribosome using the previously solved structures of the YafQ, 70S-RelE and 70S-YoeB, we predict that three histidines (H50, H63 and H87) and two aspartate residues (D61 and D67) of YafQ are adjacent to the scissile phosphate of the mRNA and participate in mRNA cleavage. Our growth and cleavage assays here support a critical role for YafQ residue H87 in mRNA cleavage while residues H50, H63 and D67 play important, though not individually indispensable roles. We propose that H87 likely functions as the general acid by stabilizing the 5′ leaving group based upon its proximal location to the scissile phosphate at the second nucleotide of the A site. Similar to RNase A, possible interactions between D67 and H87 may be to generate the correct tautomer of H87, thus allowing this residue to function as a proton donor (56). Based upon their spatial locations and their functional importance from our studies, either H50 or H63 acts as a general base to abstract the 2′ hydroxyl proton that activates the 2′-oxygen for SN2 in-line attack at the scissile phosphate. Since it is somewhat unclear from our data whether H50 or H63 is the definitive catalytic base, we made a YafQ triple mutant of H50A, D61A and H63A (called ‘tm’), which all singly affect mRNA degradation to a much lesser extent than H87A. This triple mutant ablates YafQ activity. Since residue H50 appears to have a larger effect on catalysis than H63 and is closer to the 2'-OH, we interpret H50 as the primary general base. Upon H50A mutation, the catalytic deficiency may be compensated by the adjacent H63. In the H50A, D61A and H63A triple mutant, there is effectively no residue that can function as a base, and in this situation, YafQ activity is ablated. The functional redundancy of these histidines suggests flexibility in the requirements for YafQ-mediated mRNA catalysis. There does seem to be some precedent for ribosome-dependent active site flexibility as seen in the case of YoeB (32). Substitution of proposed general base E46 of YoeB still retains low activity (32) implying that other neighboring YoeB residues or even perhaps 16S rRNA (37) may be able to functionally compensate (Figure 6). This active site plasticity can be viewed as either an adaption to codon specificity for each toxin or a redundant feature of toxin enzymes that aim to promote mRNA degradation at any cost as the bacterial survival is at stake.
Figure 6.

Proposed mechanism for phosphodiester cleavage of ribosome-bound mRNA by YafQ. The second A-site nucleotide or A+5 stacks with Phe91 (double arrows) orienting the mRNA optimally for in-line attack at the scissile phosphate. His50 or His63 functions as a general base to abstract the 2′-proton of the 2′-OH of mRNA residue A+5 and His87 stabilizes the 5′-leaving group as a general acid. Asp67 interacts with His87 to shift its pKa to facilitate leaving group stabilization. The specific role of Asp61 is unclear although it does appear important for overall function.
Structural similarities between the RelE/YoeB toxin family and general endoribonucleases such as RNases T1 and Sa2 have been noted previously (36,37). These enzymes are also compact nucleases with concave active sites similar to the ribosome-dependent toxins yet instead recognize free single stranded RNA. An important question is therefore, why do toxin endonucleases require the ribosome for RNase activity while general endoribonucleases do not? Both actives sites of RNase T1 and Sa2 contain aromatic residues (H40/H92/F100, and F39/H86/Y87, respectively) that cluster around the RNA for optimal positioning for catalysis (57,58) (Figure 7A). RelE lacks such active site aromatic residues, and their absence was proposed as a possible explanation for the requirement of the ribosome for RelE activity (36). Moreover, 16S rRNA nucleotide C1054 was proposed to functionally compensate for aromatic residues to aid in mRNA substrate positioning. However our present study demonstrates that YafQ aromatic active site residues (H50, H87 and F91) are important for mRNA cleavage similar to RNase T1 and Sa2. Structural alignment of YafQ with RNase T1 and Sa2 reveals YafQ residues H50, H87 and F91 are located at equivalent positions as the corresponding RNase T1 and Sa2 active site residues (Figure 7A). Thus the lack of active site aromatic residues in toxin endonucleases is not why the ribosome is critical for toxin activity. Our new insights into which YafQ residues are important for catalysis and binding indicate that juxtaposed toxin α-helices, with the α1 motif absent in both RNase T1 and Sa2, confer specificity for the ribosome (Figure 7B). In the place of α1, RNase Sa2 contains two β strands which upon structural alignment are located further from 16S rRNA than α1 and, moreover, lack basic residues to interact with an RNA backbone (Figure 7C). RNase T1 completely lacks this region altogether. While these data clearly implicate toxin α-helices as critical for ribosome-dependent activity, we also found that wild-type YafQ is unable to bind to an empty ribosome (i.e. without mRNA), further demonstrating toxin-16S rRNA interactions are not sufficient alone for productive ribosome binding. Taken together, these results point to an mRNA cleavage mechanism by ribosome-dependent toxins that rely on a conserved structural toxin scaffold appended with divergent active site residues that direct mRNA sequence specificity. These requirements allow for recognition of different mRNA sequences but concurrently, the conserved, surface α-helices mediate recognition of the negatively charged, ribosomal A site.
Figure 7.

Structural similarities between ribosome-dependent toxins, RNase T1 and RNase Sa2. (A) Structural alignment of YafQ, RNase T1 and RNase Sa2 (PDB codes 4Q2U, 5HOH, 3D4A) reveals the aromatic residues cluster in similar positions likely around the RNA. (B) YafQ and RelE both adopt a structurally conserved, microbial RNase fold (PDB codes 4Q2U and 4FXI) with interactions between α1 and α2 helices with the backbone of 16S rRNA in the A site (only α1 interactions are shown for simplicity). Orientation is similar to Figure 1C. (C) Despite a common structural scaffold, microbial RNase T1 and RNase Sa2 (PDB codes 5HOH and 3D4A) lacks α1 possibly explaining how they are unable to recognize the 16S rRNA in the A site.
Supplementary Material
Acknowledgments
We thank Kurt Fredrick with advice for the [35S]-binding experiments and Graeme Conn and Dunham lab members Marc Schureck and Eric Hoffer for critical reading of the manuscript.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation CAREER award [MCB 0953714 to C.M.D.]. C.M.D. is a Pew Scholar in the Biomedical Sciences. Funding for open access charge: National Science Foundation.
Conflict of interest statement. None declared.
REFERENCES
- 1.Pandey D.P., Gerdes K. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 2005;33:966–976. doi: 10.1093/nar/gki201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu Y., Kwan B.W., Osbourne D.O., Benedik M.J., Wood T.K. Toxin YafQ Increases Persister Cell Formation by Reducing Indole Signaling. Environ. Microbiol. 2014;17:1275–1285. doi: 10.1111/1462-2920.12567. [DOI] [PubMed] [Google Scholar]
- 3.Helaine S., Cheverton A.M., Watson K.G., Faure L.M., Matthews S.A., Holden D.W. Internalization of Salmonella by macrophages induces formation of nonreplicating persisters. Science. 2014;343:204–208. doi: 10.1126/science.1244705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maisonneuve E., Castro-Camargo M., Gerdes K. (p)ppGpp controls bacterial persistence by stochastic induction of toxin-antitoxin activity. Cell. 2013;154:1140–1150. doi: 10.1016/j.cell.2013.07.048. [DOI] [PubMed] [Google Scholar]
- 5.Maisonneuve E., Shakespeare L.J., Jorgensen M.G., Gerdes K. Bacterial persistence by RNA endonucleases. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13206–13211. doi: 10.1073/pnas.1100186108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 6.Dorr T., Vulic M., Lewis K. Ciprofloxacin causes persister formation by inducing the TisB toxin in Escherichia coli. PLoS Biol. 2010;8:e1000317. doi: 10.1371/journal.pbio.1000317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pedersen K., Christensen S.K., Gerdes K. Rapid induction and reversal of a bacteriostatic condition by controlled expression of toxins and antitoxins. Mol. Microbiol. 2002;45:501–510. doi: 10.1046/j.1365-2958.2002.03027.x. [DOI] [PubMed] [Google Scholar]
- 8.Harrison J.J., Wade W.D., Akierman S., Vacchi-Suzzi C., Stremick C.A., Turner R.J., Ceri H. The chromosomal toxin gene yafQ is a determinant of multidrug tolerance for Escherichia coli growing in a biofilm. Antimicrob. Agents Chemother. 2009;53:2253–2258. doi: 10.1128/AAC.00043-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keren I., Shah D., Spoering A., Kaldalu N., Lewis K. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 2004;186:8172–8180. doi: 10.1128/JB.186.24.8172-8180.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim Y., Wood T.K. Toxins Hha and CspD and small RNA regulator Hfq are involved in persister cell formation through MqsR in Escherichia coli. Biochem. Biophys. Res. Commun. 2010;391:209–213. doi: 10.1016/j.bbrc.2009.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vazquez-Laslop N., Lee H., Neyfakh A.A. Increased persistence in Escherichia coli caused by controlled expression of toxins or other unrelated proteins. J. Bacteriol. 2006;188:3494–3497. doi: 10.1128/JB.188.10.3494-3497.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gerdes K., Christensen S.K., Lobner-Olesen A. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 2005;3:371–382. doi: 10.1038/nrmicro1147. [DOI] [PubMed] [Google Scholar]
- 13.Christensen S.K., Maenhaut-Michel G., Mine N., Gottesman S., Gerdes K., Van Melderen L. Overproduction of the Lon protease triggers inhibition of translation in Escherichia coli: involvement of the yefM-yoeB toxin-antitoxin system. Mol. Microbiol. 2004;51:1705–1717. doi: 10.1046/j.1365-2958.2003.03941.x. [DOI] [PubMed] [Google Scholar]
- 14.Diago-Navarro E., Hernandez-Arriaga A.M., Kubik S., Konieczny I., Diaz-Orejas R. Cleavage of the antitoxin of the parD toxin-antitoxin system is determined by the ClpAP protease and is modulated by the relative ratio of the toxin and the antitoxin. Plasmid. 2013;70:78–85. doi: 10.1016/j.plasmid.2013.01.010. [DOI] [PubMed] [Google Scholar]
- 15.Lehnherr H., Yarmolinsky M.B. Addiction protein Phd of plasmid prophage P1 is a substrate of the ClpXP serine protease of Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 1995;92:3274–3277. doi: 10.1073/pnas.92.8.3274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Van Melderen L., Bernard P., Couturier M. Lon-dependent proteolysis of CcdA is the key control for activation of CcdB in plasmid-free segregant bacteria. Mol. Microbiol. 1994;11:1151–1157. doi: 10.1111/j.1365-2958.1994.tb00391.x. [DOI] [PubMed] [Google Scholar]
- 17.Pedersen K., Zavialov A.V., Pavlov M.Y., Elf J., Gerdes K., Ehrenberg M. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell. 2003;112:131–140. doi: 10.1016/s0092-8674(02)01248-5. [DOI] [PubMed] [Google Scholar]
- 18.Prysak M.H., Mozdzierz C.J., Cook A.M., Zhu L., Zhang Y., Inouye M., Woychik N.A. Bacterial toxin YafQ is an endoribonuclease that associates with the ribosome and blocks translation elongation through sequence-specific and frame-dependent mRNA cleavage. Mol. Microbiol. 2009;71:1071–1087. doi: 10.1111/j.1365-2958.2008.06572.x. [DOI] [PubMed] [Google Scholar]
- 19.Winther K.S., Gerdes K. Enteric virulence associated protein VapC inhibits translation by cleavage of initiator tRNA. Proc. Natl. Acad. Sci. U.S.A. 2011;108:7403–7407. doi: 10.1073/pnas.1019587108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Christensen S.K., Gerdes K. Delayed-relaxed response explained by hyperactivation of RelE. Mol. Microbiol. 2004;53:587–597. doi: 10.1111/j.1365-2958.2004.04127.x. [DOI] [PubMed] [Google Scholar]
- 21.Christensen-Dalsgaard M., Jorgensen M.G., Gerdes K. Three new RelE-homologous mRNA interferases of Escherichia coli differentially induced by environmental stresses. Mol. Microbiol. 2010;75:333–348. doi: 10.1111/j.1365-2958.2009.06969.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y., Yamaguchi Y., Inouye M. Characterization of YafO, an Escherichia coli toxin. J. Biol. Chem. 2009;284:25522–25531. doi: 10.1074/jbc.M109.036624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cruz J.W., Rothenbacher F.P., Maehigashi T., Lane W.S., Dunham C.M., Woychik N.A. Doc toxin is a kinase that inactivates elongation factor Tu. J. Biol. Chem. 2014;289:7788–7798. doi: 10.1074/jbc.M113.544429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schifano J.M., Edifor R., Sharp J.D., Ouyang M., Konkimalla A., Husson R.N., Woychik N.A. Mycobacterial toxin MazF-mt6 inhibits translation through cleavage of 23S rRNA at the ribosomal A site. Proc. Natl. Acad. Sci. U.S.A. 2013;110:8501–8506. doi: 10.1073/pnas.1222031110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schifano J.M., Vvedenskaya I.O., Knoblauch J.G., Ouyang M., Nickels B.E., Woychik N.A. An RNA-seq method for defining endoribonuclease cleavage specificity identifies dual rRNA substrates for toxin MazF-mt3. Nat. Commun. 2014;5:3538–3548. doi: 10.1038/ncomms4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hurley J.M., Woychik N.A. Bacterial toxin HigB associates with ribosomes and mediates translation-dependent mRNA cleavage at A-rich sites. J. Biol. Chem. 2009;284:18605–18613. doi: 10.1074/jbc.M109.008763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Castro-Roa D., Garcia-Pino A., De Gieter S., van Nuland N.A., Loris R., Zenkin N. The Fic protein Doc uses an inverted substrate to phosphorylate and inactivate EF-Tu. Nat. Chem. Biol. 2013;9:811–817. doi: 10.1038/nchembio.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hurley J.M., Cruz J.W., Ouyang M., Woychik N.A. Bacterial toxin RelE mediates frequent codon-independent mRNA cleavage from the 5′ end of coding regions in vivo. J. Biol. Chem. 2011;286:14770–14778. doi: 10.1074/jbc.M110.108969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Takagi H., Kakuta Y., Okada T., Yao M., Tanaka I., Kimura M. Crystal structure of archaeal toxin-antitoxin RelE-RelB complex with implications for toxin activity and antitoxin effects. Nat. Struct. Mol. Biol. 2005;12:327–331. doi: 10.1038/nsmb911. [DOI] [PubMed] [Google Scholar]
- 30.Boggild A., Sofos N., Andersen K.R., Feddersen A., Easter A.D., Passmore L.A., Brodersen D.E. The crystal structure of the intact E. coli RelBE toxin-antitoxin complex provides the structural basis for conditional cooperativity. Structure. 2012;20:1641–1648. doi: 10.1016/j.str.2012.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li G.Y., Zhang Y., Inouye M., Ikura M. Inhibitory mechanism of Escherichia coli RelE-RelB toxin-antitoxin module involves a helix displacement near an mRNA interferase active site. J. Biol. Chem. 2009;284:14628–14636. doi: 10.1074/jbc.M809656200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kamada K., Hanaoka F. Conformational change in the catalytic site of the ribonuclease YoeB toxin by YefM antitoxin. Mol. Cell. 2005;19:497–509. doi: 10.1016/j.molcel.2005.07.004. [DOI] [PubMed] [Google Scholar]
- 33.Schureck M.A., Maehigashi T., Miles S.J., Marquez J., Cho S.E., Erdman R., Dunham C.M. Structure of the Proteus vulgaris HigB-(HigA)2-HigB toxin-antitoxin complex. J. Biol. Chem. 2014;289:1060–1070. doi: 10.1074/jbc.M113.512095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Armalyte J., Jurenaite M., Beinoraviciute G., Teiserskas J., Suziedeliene E. Characterization of Escherichia coli dinJ-yafQ toxin-antitoxin system using insights from mutagenesis data. J. Bacteriol. 2012;194:1523–1532. doi: 10.1128/JB.06104-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hayes C.S., Sauer R.T. Toxin-antitoxin pairs in bacteria: killers or stress regulators. Cell. 2003;112:2–4. doi: 10.1016/s0092-8674(02)01282-5. [DOI] [PubMed] [Google Scholar]
- 36.Neubauer C., Gao Y.G., Andersen K.R., Dunham C.M., Kelley A.C., Hentschel J., Gerdes K., Ramakrishnan V., Brodersen D.E. The structural basis for mRNA recognition and cleavage by the ribosome-dependent endonuclease RelE. Cell. 2009;139:1084–1095. doi: 10.1016/j.cell.2009.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng S., Chen Y., Kamada K., Wang H., Tang K., Wang M., Gao Y.G. YoeB-ribosome structure: a canonical RNase that requires the ribosome for its specific activity. Nucleic Acids Res. 2013;41:9549–9556. doi: 10.1093/nar/gkt742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Griffin M.A., Davis J.H., Strobel S.A. Bacterial toxin RelE: a highly efficient ribonuclease with exquisite substrate specificity using atypical catalytic residues. Biochemistry. 2013;52:8633–8642. doi: 10.1021/bi401325c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ruangprasert A., Maehigashi T., Miles S.J., Giridharan N., Liu J.X., Dunham C.M. Mechanisms of toxin inhibition and transcriptional repression by Escherichia coli DinJ-YafQ. J. Biol. Chem. 2014;289:20559–20569. doi: 10.1074/jbc.M114.573006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang Y., Gao Z., Wang F., Zhang Y., Dong Y., Liu Q. Structural and functional characterization of escherichia coli toxin-antitoxin complex DinJ-YafQ. J. Biol. Chem. 2014;289:21191–21202. doi: 10.1074/jbc.M114.559773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holm L., Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hasegawa H., Holm L. Advances and pitfalls of protein structural alignment. Curr. Opin. Struct. Biol. 2009;19:341–348. doi: 10.1016/j.sbi.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 43.The PyMOL Molecular Graphics System. 2010 Version 1.7.4 Schrödinger, LLC. [Google Scholar]
- 44.Datsenko K.A., Wanner B.L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qin D., Fredrick K. Control of translation initiation involves a factor-induced rearrangement of helix 44 of 16S ribosomal RNA. Mol. Microbiol. 2009;71:1239–1249. doi: 10.1111/j.1365-2958.2009.06598.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Powers T., Noller H.F. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Selmer M., Dunham C.M., Murphy F.V.t., Weixlbaumer A., Petry S., Kelley A.C., Weir J.R., Ramakrishnan V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–1942. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 48.Yan K., Hunt E., Berge J., May E., Copeland R.A., Gontarek R.R. Fluorescence polarization method to characterize macrolide-ribosome interactions. Antimicrob. Agents Chemother. 2005;49:3367–3372. doi: 10.1128/AAC.49.8.3367-3372.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swillens S. Interpretation of binding curves obtained with high receptor concentrations: practical aid for computer analysis. Mol. Pharmacol. 1995;47:1197–1203. [PubMed] [Google Scholar]
- 50.Tapprich W.E., Goss D.J., Dahlberg A.E. Mutation at position 791 in Escherichia coli 16S ribosomal RNA affects processes involved in the initiation of protein synthesis. Proc. Natl. Acad. Sci. U.S.A. 1989;86:4927–4931. doi: 10.1073/pnas.86.13.4927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang W. Nucleases: diversity of structure, function and mechanism. Q. Rev. Biophys. 2011;44:1–93. doi: 10.1017/S0033583510000181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Loverix S., Steyaert J. Deciphering the mechanism of RNase T1. Methods Enzymol. 2001;341:305–323. doi: 10.1016/s0076-6879(01)41160-8. [DOI] [PubMed] [Google Scholar]
- 53.Sevcik J., Zegers I., Wyns L., Dauter Z., Wilson K.S. Complex of ribonuclease Sa with a cyclic nucleotide and a proposed model for the reaction intermediate. Eur. J. Biochem. 1993;216:301–305. doi: 10.1111/j.1432-1033.1993.tb18145.x. [DOI] [PubMed] [Google Scholar]
- 54.Cruz-Reyes J., Piller K.J., Rusche L.N., Mukherjee M., Sollner-Webb B. Unexpected electrophoretic migration of RNA with different 3′ termini causes a RNA sizing ambiguity that can be resolved using nuclease P1-generated sequencing ladders. Biochemistry. 1998;37:6059–6064. doi: 10.1021/bi972868g. [DOI] [PubMed] [Google Scholar]
- 55.Schurer H., Lang K., Schuster J., Morl M. A universal method to produce in vitro transcripts with homogeneous 3′ ends. Nucleic Acids Res. 2002;30:e56. doi: 10.1093/nar/gnf055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schultz L.W., Quirk D.J., Raines R.T. His…Asp catalytic dyad of ribonuclease A: structure and function of the wild-type, D121N, and D121A enzymes. Biochemistry. 1998;37:8886–8898. doi: 10.1021/bi972766q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bauerova-Hlinkova V., Dvorsky R., Perecko D., Povazanec F., Sevcik J. Structure of RNase Sa2 complexes with mononucleotides–new aspects of catalytic reaction and substrate recognition. FEBS J. 2009;276:4156–4168. doi: 10.1111/j.1742-4658.2009.07125.x. [DOI] [PubMed] [Google Scholar]
- 58.Langhorst U., Loris R., Denisov V.P., Doumen J., Roose P., Maes D., Halle B., Steyaert J. Dissection of the structural and functional role of a conserved hydration site in RNase T1. Protein Sci. 1999;8:722–730. doi: 10.1110/ps.8.4.722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


