Abstract
Gallbladder carcinoma (GBC) is a rare tumor with a dismal survival rate overall. Hence, there is an urgent need for exploring more specific and sensitive biomarkers for the diagnosis and treatment of GBC. At first, amplified total RNAs from two paired GBC tumors and adjacent non-tumorous tissues (ANTTs) were subjected to RNA sequencing. 161 genes were identified differentially expressed between tumors and ANTTs. Functional enrichment analysis indicated that the up-regulated genes in tumor were primarily associated with signaling molecules and enzyme modulators, and mainly involved in cell cycles and pathways in cancer. Twelve differentially expressed genes (DEGs) were further confirmed in another independent cohort of 35 GBC patients. Expression levels of BIRC5, TK1, TNNT1 and MMP9 were found to be positively related to postoperative relapse. There was also a significant correlation between BIRC5 expression and tumor-node-metastasis (TNM) stage. Besides, we observed a positive correlation between serum CA19–9 concentration and the expression levels of TNNT1, MMP9 and CLIC3. Survival analysis revealed that GBC patients with high TK1 and MMP9 expression levels had worse prognosis. These identified DEGs might not only be promising biomarkers for GBC diagnosis and prognosis, but also expedite the discovery of novel therapeutic strategies.
Keywords: biomarker, oncology, oncogene, tumor suppressor, tumorgenesis
INTRODUCTION
Gallbladder carcinoma (GBC), as the fifth most common gastrointestinal cancer, is a relatively uncommon but challenging cancer. In these years its incidence is increasing in China [1]. The risk factors for GBC include gender, aging, obesity, chronic cholecystitis and cholelithiasis [1–3]. Besides, various single nucleotide polymorphisms (SNPs) have also been determined to be associated with the susceptibility to GBC [4, 5]. GBC is the most common cause of mortality among biliary tract cancers (BCTs). Because patients with GBC often diagnosed at an advanced stage for lack of effective early diagnostic method, prognosis of GBC is generally dismal. The median survival time for individuals with GBC is no more than one year [2]. In spite of multiple studies on novel molecules for diagnosis and prediction of clinical outcomes in GBC [6], the progress in clinical applications has so far been limited. Thus, more precise markers with better sensitivity and specificity for GBC are needed to benefit the patients and expedite the discovery of novel therapeutic strategies.
RNA sequencing (RNA-Seq) technology, as a typical kind of next-generation RNA sequencing, was widely used recently in the studies of oncology, taking advantage of its superior sensitivity, high efficiency and high throughput. Many promising findings have been reported on the underlying molecular etiology or therapeutic targets in diverse malignancies, including prostate cancer [7], liver cancer [8], lung cancer [9], etc. However, there are few reports on the application of RNA-Seq in GBC study.
Here we used RNA-Seq technology to sequence the whole transcriptomes of paired tumor tissues and adjacent non-timorous tissues (ANTTs) from two GBC patients. The differentially expressed genes (DEGs) were further confirmed in another cohort of 35 GBC patients. The association of several DEGs with clinical characteristics and postoperative outcoming were also revealed.
RESULTS
General information of RNA-Seq and DEGs analysis
RNA-Seq was performed on two pairs of matched GBC tumors and ANTTs, and the characteristics of the GBC patients were summarized in Table 2. There was no significant difference between numbers of genes detected in each pair of tumor and ANTT. An average of 16764 genes were detected in sequenced tissue sample, by requiring that the FPKM value was greater than 0.7. The correlation analysis showed that the average global profiles of gene expressions between tumor and ANTT samples are highly correlated (correlation coefficient r = 0.93; Figure 1a). Furthermore, the Q30 value of every library was more than 80%. The evaluations of sequencing randomness and saturation also proved the RNA-Seq system is good in quality.
Table 2. The characteristics summary of the RNA-Seq data.
| Library | total reads | reads length(bp) | Total mapped | mapped% | mapped pairs | mapped paired% | Mapped genes* |
|---|---|---|---|---|---|---|---|
| T1 | 23,708,937 | 101 | 33,274,003 | 70.17% | 15,623,535 | 65.90% | 15,594 |
| N1 | 16,433,264 | 101 | 18,224,025 | 55.45% | 8,108,072 | 49.34% | 16,245 |
| T2 | 46,059,548 | 101 | 38,593,916 | 83.79% | 18,494,595 | 80.31% | 17,549 |
| N2 | 40,830,844 | 101 | 34,288,810 | 83.98% | 16,370,875 | 80.19% | 17,584 |
FPKM > 0.7 was defined as a mapped gene.
Figure 1. Schematic view of DEGs in GBC detected by RNA-Seq.
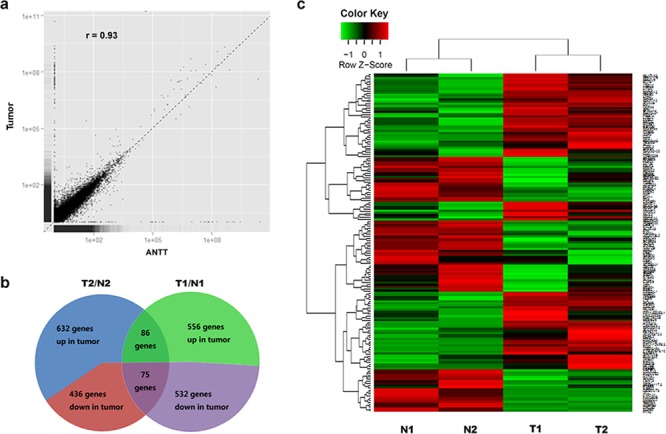
a. Expression analysis of GBC tumor and ANTT samples. The Pearson correlation coefficient is shown. b. Composition of DEGs in two paired samples used for RNA-Seq. c. Heat map was generated from normalized FPKM of 161 consistent DEGs across two paired samples. Expression level for each transcript is represented by a color range from green (low) to red (high).
The expression levels of 632 and 556 genes were observed up-regulated in tumor tissues (T1/T2) compared with paired ANTTs (N1/N2) respectively. Meanwhile the expression levels of 436 and 532 genes were down-regulated (Figure 1b). Among all those genes, there were 86 up-regulated and 75 down-regulated consistently in both GBC pairs. Then these 161 DEGs were regarded as candidates for further study, whose expression levels were shown as heat map in Figure 1c.
Functional enrichment analysis of DEGs
Functional enrichment analysis was used to investigate DEGs in GBC tumorigenesis. From GO results, 56 terms composed of 13 of cellular compound (CC), 12 of molecular function (MF), and 31 of biology process (BP) were significantly over-represented for all the DEGs (adjusted P values < 0.01) (Supplementary Table S2). For protein class by PANTHER website, the up-regulated genes in tumor tissues were mainly associated with “signaling molecule and enzyme modulator”, while the genes with decreased expression in tumor tissues were mainly associated with “receptor and extracellular matrix protein” (Figure 2). For KEGG pathway analysis, significantly enriched pathways of the up-regulated genes are “cell cycle” and “pathways in cancer”.
Figure 2. PANTHER protein class categories of total DEGs, increased and decreased DEGs in pie chart.

The chart legends showed each detailed category.
Validation of RNA-Seq data by qRT-PCR
To verify the RNA-Seq data, the original two RNA samples used for RNA-Seq were tested again by qRT-PCR on a panel of fifteen selected DEGs (Figure 3a). Log2 fold change of genes of qRT-PCR was compared with that of RNA-Seq. Finally, these DEGs were demonstrated concordant expression change in these two gene expression analysis platforms, with the correlation coefficient of validation cohort and RNA-Seq cohort was 0.958 (Figure 3b). Hence, our RNA-Seq method could reliably measure gene expression differences.
Figure 3. Validation of DEGs in other 35 paired tissue samples.
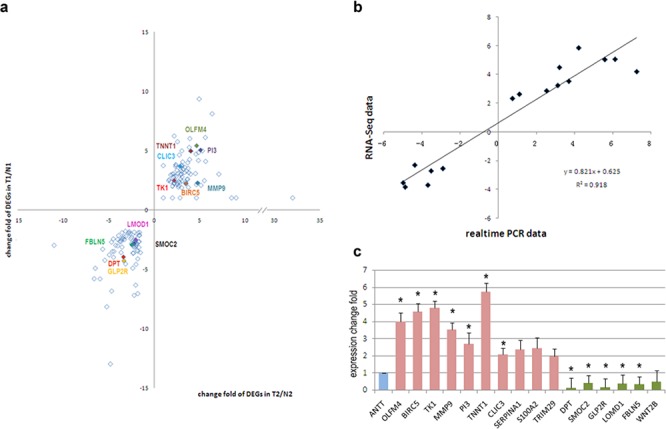
a. Validated DEGs by realtime PCR (colored) in all 161 DEGs. b. Correlation of expression of DEGs in RNA-Seq and validation cohort. c. Expression levels of DEGs in validation cohort, including up-regulated in tumor tissues (red) and down-regulated in tumor tissues (green). Expression level in ANTTs was used as a reference (blue). *, significant differences (P < 0.05).
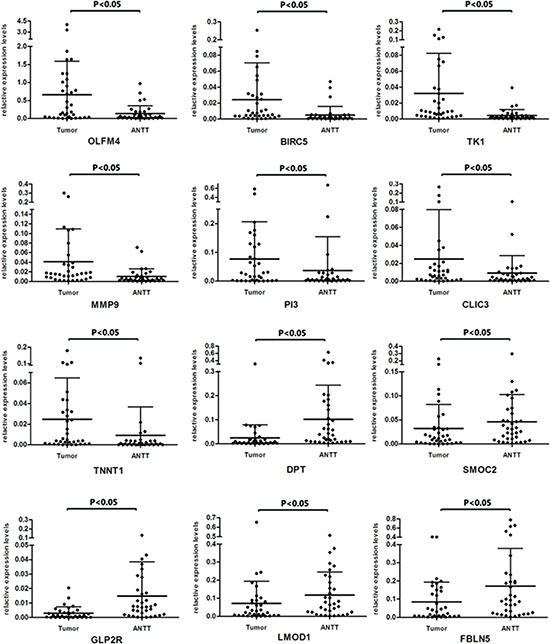
On the other hand, confirmation of RNA-Seq data was performed by qRT-PCR using samples from another independent cohort of 35 patients diagnosed as GBC. Among the selected 15 DEGs, 12 genes showed consistent expression differences in validation cohort as shown by RNA-Seq data (Figure 3c). Among the identified genes, OLFM4, BIRC5, TK1, PI3, TNNT1, CLIC3 and MMP9 levels were distinctly increased in GBC tissues. Meanwhile expression of gene DPT, SMOC2, GLP2R, FBLN5 and LMOD1 were down-regulated in tumor tissues. Figure 4 represented the expression levels of each validated DEG in paired GBC tissues and ANTTs respectively.
Figure 4. Expression levels of twelve DEGs in paired GBC tissues and ANTTs respectively.
Correlation analysis between DEGs and clinic pathological characteristics
Stratification analysis of DEGs up-regulated in tumor tissues was performed according to the GBC patients' preoperative laboratory and pathologic parameters. As shown in Table 3, the expression levels of OLFM4 and TK1 were both significantly positively related to patients' age. The expression levels of BIRC5, TK1, TNNT1 and MMP9 were found to be positively related to tumor recurrence after cystic resection. Besides, we observed a positive correlation with serum CA19–9 value and the expression levels of TNNT1, MMP9 and CLIC3. There was also a positive correlation between another serum tumor marker CEA and PI3 expression levels. Furthermore, BIRC5 expression level was found to be correlated with tumor-node-metastasis (TNM) grade in GBC patients. The patients with higher TNM grades possessed more BIRC5 expression in tumors. However, the presence of cholelithiasis had no correlation with expression levels of these DEGs.
Table 3. Correlation analysis between DEGs up-expressed in GBC with clinical characteristics.
| Gene | Age | Gender | Cholelithiasis | TNM grade | relapse | Tbil | TBA | ALT | AST | GGT | ALP | CEA | CA199 | CA242 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| OLFM4 | Spearman Correlation | –0.435** | 0.118 | 0.122 | 0.033 | –0.011 | 0.106 | 0.125 | 0.039 | 0.015 | 0.027 | 0.038 | 0.035 | 0.165 | –0.134 |
| Sig. (2-tailed) | 0.007 | 0.488 | 0.474 | 0.845 | 0.966 | 0.534 | 0.495 | 0.819 | 0.931 | 0.875 | 0.823 | 0.837 | 0.329 | 0.506 | |
| BIRC5 | Spearman Correlation | –0.260 | 0.118 | –0.086 | 0.388* | 0.494* | –0.051 | –0.007 | 0.028 | 0.183 | –0.037 | 0.034 | 0.096 | 0.160 | 0.076 |
| Sig. (2-tailed) | 0.120 | 0.488 | 0.612 | 0.018 | 0.037 | 0.762 | 0.968 | 0.868 | 0.279 | 0.829 | 0.843 | 0.572 | 0.346 | 0.707 | |
| TK1 | Spearman Correlation | –0.401* | 0.194 | –0.015 | 0.260 | 0.824** | –0.165 | –0.226 | –0.081 | 0.009 | –0.151 | –0.059 | 0.060 | 0.187 | 0.184 |
| Sig. (2-tailed) | 0.014 | 0.250 | 0.929 | 0.120 | < 0.001 | 0.329 | 0.213 | 0.636 | 0.958 | 0.374 | 0.730 | 0.724 | 0.267 | 0.358 | |
| MMP9 | Spearman Correlation | 0.170 | –0.062 | –0.086 | 0.142 | 0.653** | 0.186 | 0.153 | 0.230 | 0.198 | 0.168 | 0.213 | 0.020 | 0.430** | 0.349 |
| Sig. (2-tailed) | 0.321 | 0.718 | 0.620 | 0.408 | 0.004 | 0.276 | 0.410 | 0.176 | 0.246 | 0.328 | 0.213 | 0.906 | 0.009 | 0.075 | |
| TNNT1 | Spearman Correlation | –0.191 | 0.036 | –0.046 | 0.072 | 0.626** | –0.052 | –0.143 | –0.125 | –0.102 | –0.245 | –0.070 | –0.025 | 0.494** | 0.172 |
| Sig. (2-tailed) | 0.258 | 0.834 | 0.789 | 0.672 | 0.005 | 0.761 | 0.434 | 0.459 | 0.550 | 0.144 | 0.680 | 0.884 | 0.002 | 0.391 | |
| PI3 | Spearman Correlation | –0.254 | 0.312 | –0.010 | 0.277 | –0.011 | –0.033 | 0.158 | –0.029 | 0.026 | –0.004 | 0.118 | 0.340* | 0.167 | –0.244 |
| Sig. (2-tailed) | 0.129 | 0.060 | 0.953 | 0.097 | 0.966 | 0.846 | 0.388 | 0.863 | 0.879 | 0.980 | 0.487 | 0.039 | 0.322 | 0.219 | |
| CLIC3 | Spearman Correlation | –0.241 | 0.110 | 0.121 | 0.255 | 0.477 | 0.208 | –0.090 | 0.002 | 0.018 | –0.002 | 0.131 | 0.102 | 0.505** | 0.292 |
| Sig. (2-tailed) | 0.158 | 0.522 | 0.484 | 0.133 | 0.053 | 0.224 | 0.630 | 0.992 | 0.919 | 0.989 | 0.448 | 0.555 | 0.002 | 0.148 | |
P < 0.05;
P < 0.01.
Survival analysis
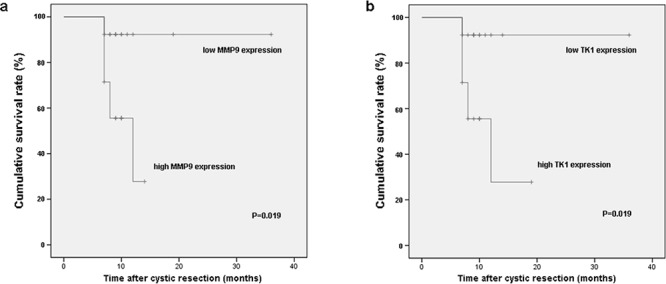
The median follow-up duration in all involved 27 GBC patients was 10 months (range 7–36 months). Kaplan–Meier analysis revealed that survival rate of GBC patients with high MMP9 expression was lower than that with low MMP9 expression (P = 0.019) (Figure 5a). The expression level of TK1 gene was found to bear similar effect on survival rate in present cohort. GBC patients with high TK1 expression had worse prognosis (P = 0.019) (Figure 5b). However, there were no statistically significant differences between subgroups with different expression levels of the other DEGs.
Figure 5. Survival rates of GBC patients who underwent cystic resection were compared between the high and low expression of MMP9 a. and TK1 b. The median expression level was used as the cutoff.
DISCUSSION
GBC is a highly malignant and fatal disease with a mean 5-year survival rate of 5% [10]. Its dismal prognosis attributes to its late presentation, early lymph node metastases, adjacent organ invasion, and poor response to chemotherapy. To improve the prognosis of GBC, it is important to screen out the risk factors or precursor diseases relevant to gallbladder carcinogenesis, and establish appropriate molecular biomarkers, favoring early diagnosis and progress monitoring.
Next generation sequencing (NGS) of the complete RNA transcriptome offers a novel approach for systematically characterizing the underlying molecular etiology of malignancies. Here by RNA-seq, we gained a whole transcriptomic profiling of GBC. To our knowledge, this is the first description of the global transcriptome of human gallbladder tumor tissue using RNA-seq approach. We observed hundreds of genes demonstrating 2-fold or greater increase or decrease in expression compared to matched ANTTs, respectively. Several candidate DEGs were screened out and further confirmed in another cohort of 35 GBC patients. Among these genes, some have already been investigated for their physiological function in gallbladder carcinogenesis, even tumorigenesis.
BIRC5, baculoviral IAP repeat containing 5, is a member of the inhibitor of apoptosis (IAP) gene family [11]. This gene encodes protein Survivin, which is famous for its crucial roles in apoptosis and cell cycle regulation [12]. Survivin over-expressed in a wide variety of tumors, accompanied by worse prognosis, shorter survival and increased tumor recurrence [13, 14]. Furthermore, Survivin is involved in the promotion of angiogenesis, which is necessary for tumor progression and expansion, by inhibiting apoptosis of endothelial cells. Recently, it has already been considered as a new diagnostic and prognostic marker of GBC [15]. As for TK1, thymidine kinase 1, its diagnostic and prognostic role of TK1 has recently been widely investigated. TK1 might be helpful for screening and monitoring of human malignancies [16, 17]. The increased expression of TK1 in tumor is consistent with the former reports. Furthermore, an age-related difference was also observed in our study, which is in accord with Li's finding that more high serum TK1 values were noted in the elderly patient population than low TK1 levels [18]. Another familiar DEG in oncology is MMP9, matrix metallopeptidase 9, which promotes the progression of diverse cancers by improving tumor growth, migration, invasion, metastasis and angiogenesis [19, 20]. In tumor biology, it has been increasingly appreciated that MMP9 is associated with poor disease prognosis, such as glioma [21] and gastric cancer [22]. OLFM4, olfactomedin 4, has been reported to be over-expressed in various cancers including gastric cancer but also colon, breast, and lung cancers [23], and has been proposed as a potential serum biomarker of gastric cancer [24]. Functionally, OLFM4 has been shown to bind to GRIM19 (a cell-death regulatory protein), cadherins and lectins, and could suppress apoptosis and promote tumor growth and invasion [23, 25]. However, a recent finding showed that OLFM4 has a proapoptotic effect in myeloid leukemia cells [26]. Here, we found OLFM4 as well as TK1 was significantly positively related to patients' age. In previous report, positive expression of OLFM4 was already found to be correlated with age in patients with gastric cancer [27], which is consistent with our result. Gene PI3 is either called ESI (Elastase Specific Inhibitor) or SKALP (skin-derived antileukoprotease) or Trappin-2 or Elafin, encoding an elastase-specific inhibitor. PI3 is reported to participate in inflammatory responses e.g. by increasing neutrophil chemotaxis in situ while preventing the excessive protease damage generated by inflammation. This dual mechanism of action suggests that it acts at both ends of the inflammatory process [28]. As for its biological function in tumorigenesis, transcripts of PI3 are extremely abundant in glioblastoma patient samples and significantly correlated with dismal survival [29]. In ovarian and breast carcinoma, PI3 expression was also found to be transcriptionally up-regulated through activation of the NFκB and MEK/ERK signaling pathway, and correlate with unfavourable overall survival [30]. However, controversial report has already emerged that PI3 could suppress tumorigenesis through inhibition of elastase and thus could serve as a prognostic indicator in breast cancer [31]. Chloride intracellular channel 3 (CLIC3) is a member of the p64 family and facilitates chloride ion movement and regulates cellular processes associated with the movement of chloride in placental and fetal membrane cells [32]. In addition, this protein may participate in cellular growth control, based on its association with ERK7, a member of the MAP kinase family [33]. CLIC3 expression predicts lymph node metastasis and poor prognosis in operable cases of pancreatic ductal adenocarcinoma (PDAC) [34]. The finding indicated that CLIC3 might be a potential molecule to promote cancer progression.
Compared with above increased genes detected in GBC, the role of gene TNNT1 in carcinogenesis has been little unraveled. TNNT1, troponin T type 1, encodes a protein that is a subunit of troponin, which is a regulatory complex located on the thin filament of the sarcomere [35]. This complex regulates striated muscle contraction in response to fluctuations in intracellular calcium concentration. Serum TNNT1 levels had a positive association with increased risk for hypertrophic cardiomyopathy [36] and also with different conditions related to the severity of the disease [37]. However, most studies on TNNT1 are about its function in cardiovascular diseases [38, 39], whether it is involved in tumorigenesis is rarely known.
In this study, we also found correlations between these up-regulated DEGs and clinical characteristics by stratification analysis. In spite of some verified DEGs have already been studied intensively, most of their function and clinical application have not been previously reported in GBC. The correlation between TNNT1 expression and tumor progression (TNM grade) or serum tumor marker CA19–9 might catch more attention on its potential application in diagnosis for GBC. The association between BIRC5, TK1, TNNT1 and MMP9 expression and postoperative relapse, together with the repression of TK1 and MMP9 expression on survival rate suggested their clinical prognostic value for GBC patients. However, only 27 GBC patients had intact follow-up information. The overall survival may have been influenced by the limitation of patients' number. Further follow-up studies with a large amount of GBC patients are needed to make up this shortcoming and confirm the effect of these DEGs expression on prognosis.
Although we put more attention on the increased DEGs, the verified decreased DEGs are also very noteworthy. The most significant decreased gene in cancer samples is DPT, encoding protein Dermatopontin, an extracellular matrix protein with possible functions in cell-matrix interactions assembly [40], cell adhesion [41], wound healing [42], and positive modification of the growth inhibition activity of TGF-β1 [43]. DPT was significantly down-regulated in HCC tissues compared with non-tumor [44]. Down-regulation of DPT is also observed in oral squamous cell carcinoma (OSCC)-derived cells, which suggested that it might inhibit tumor invasion and metastasis [45]. DPT together with other decreased genes, which have been less investigated, might be potential tumor suppressor and need to be further studied.
Lately, Li reported their work about whole-exome and targeted gene sequencing of gallbladder carcinoma [46]. They identified several genes with a significant frequency of non-silent mutations including TP53, KRAS and ERBB3, and the most extensively mutated pathway ErbB signaling. Here we also checked expression levels of above genes in our RNA-Seq data. However, besides CDKN2A, which is extremely over-expressed in tumor tissues, other genes mentioned by Li did not show significant difference on transcription level. This discrepancy indicates the different significance between researches on transcriptome and exome. RNA-Seq offers another view of transcriptomics to make up the shortcoming of whole-exome sequencing on DNA level.
In conclusion, we screened out twelve DEGs from GBC tissues by RNA-seq and confirmed them in 35 GBC patients. The up-regulated DEGs in this study may be promising molecular candidates for gallbladder cancer diagnosis or prognosis. All these DEGs in GBC revealed a pool of novel molecules which could be available for furthermore investigation of mechanism for GBC development and progression, and even for targeted therapy.
MATERIALS AND METHODS
Patients' information
A total of 37 GBC patients were recruited for RNA-Seq and validation in this study, including the specimens of one male GBC patient (T1/N1) and one female GBC patient (T2/N2) used for RNA-Seq. Tumor tissues and paired ANTTs were collected at the time of operation. Pathological diagnosis was made by two independent and expert pathologists. All of the tumor tissues were confirmed as primary adenosquamous carcinoma of gallbladder. Demographic, preoperative laboratory and pathologic data of all patients were collected from electronic medical records (EMR) and retrospectively reviewed. The whole cohort consists of 16 men and 21 women, with a mean age of 59. Among these GBC patients there were fourteen patients with gallstones. According to the seventh edition of the Union for International Cancer Control (UICC)and American Joint Committee on Cancer (AJCC) on Tumor Node Metastasis (TNM) classification, twenty-three and thirteen of these patients presented at tumor-node-metastasis (TNM) stage III and IV respectively. The demographic and laboratory parameters of all subjects included in the present study were listed in Table 1. Tumor recurrence and survival data were collected during the follow-up study. Informed consents of all patients were obtained in advance, and the data were analyzed anonymously. This study was approved by the Committee on Ethics of Biomedicine Research, Eastern Hepatobiliary Surgery Hospital, Second Military Medical University. All experiments were conducted in accordance with the official recommendations of the Chinese Community Guidelines.
Table 1. Demographic and laboratory parameters of the subjects included in the study.
| Samples | Validation Cohort | RNA-seq cohort | |
|---|---|---|---|
| (n = 35) | T1/N1 | T2/N2 | |
| Demographic parameters | |||
| Gender (M/F) | 15/20 | M | F |
| Age (Y) ( ± SD) | 59.66 ± 7.69 | 80 | 56 |
| TNM stages | III | IV | |
| II | 2 | ||
| III | 23 | ||
| IV | 10 | ||
| Cholecystolithiasis | 17 | positive | positive |
| Laboratory parameters [median (range), Positive percentage] | |||
| T-Bil (μmol/L) | 12.4 (3.9–441.1), 10/35 | 6 | 227.6 |
| D-Bil (μmol/L) | 5.25 (1.4–341.1), 13/34 | 2.4 | 184.5 |
| ALP (U/L) | 86 (38–539), 13/35 | 99 | 1096 |
| AFP (μg/L) | 2.3 (0.7–187.9), 2/35 | 2.5 | 1.2 |
| CEA (μg/L) | 3.9 (0.6–91.3), 5/35 | 57.5 | 277.4 |
| CA19-9 (U/ml) | 157.3 (1.1– > 1000), 24/35 | >1000 | 404.1 |
| CA242 (IU/ml) | 71 (0.8–200), 13/22 | 150 | 104.7 |
TNM stages, Tumor Node Metastasis stages; T-Bil, serum total bilirubin level; D-Bil, serum direct bilirubin level; ALP, serum alkaline phosphatase; AFP, serum alpha fetoprotein level; CEA, serum carcinoembryonic antigen level.
RNA library preparation and sequencing
Total RNAs of GBC tumor tissues and paired ANTTs from two GBC patients were isolated using Trizol reagent (Life Technologies, USA). Four libraries were constructed using Illumina standard kit according to the manufacturer's protocol. All sequencing was performed on Illumina Hiseq2000.
Read mapping and screening of DEGs
Tophat v2.0.0 with default parameters in pair-end mode was used to map reads to reference genomes (Ensembl Human Genome v73). Pair-end reads with one-end drop in one chromosome and the other end drop in different chromosome had been discarded. As for reads could be mapped to multiple positions, we uniformly divided each multi-mapped read to all of positions it mapped to. In other words, a read mapping to 10 positions would count as 0.1 of a read at each position. Then, the expression levels for each gene normalized by fragments per kilo base of transcript per million mapped reads (FPKM) were calculated based on filtered mapping results. FPKM > 0.7 was defined as a mapped gene.
The numerical measure of mapped reads was used to evaluate the expression level of certain gene or alternative splicing isoforms using TopHat and Cufflinks. Expression difference of each gene between tumor tissues and ANTTs was analyzed based on Cufflinks. The significant DEGs were selected with threshold P-value less than 0.05 and fold-change more than 2 between each paired tumor and ANTT libraries. Meanwhile, their expression change tendencies were consistent in both two GBC patients for RNA-Seq. As the total number of reads varied between libraries, the fold-change was calculated after normalization.
Functional enrichment analysis of DEGs
Functional enrichment analysis was used to investigate DEGs in GBC tumorigenesis. Gene ontology (GO) (http://www.geneontology.org/) is a standard classification system of gene function and gene products. Besides GO analysis, PANTHER website (http://go.pantherdb.org/) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis (http://www.kegg.jp/) were also used to reveal the physiological function of DEGs in gallbladder carcinogenesis.
DEGs verification and stratification analysis
The differential expression genes (DEGs) were initially validated by quantitative real-time PCR (qRT-PCR) on the same set of RNA sequencing. Furthermore, total RNAs were extracted from tumor tissues and paired ANTTs of an independent cohort of 35 GBC patients using an RNeasy Mini Kit (Qiagen, the Netherlands). The concentration and quality of RNAs were measured using EpochTM (BioTek, USA). The cDNAs were synthesized using ReverTra Ace-α-RT-PCR kit (TOYOBO, Japan) according to the manufacturer's instructions. The qRT-PCR of DEGs was performed using the SYBR Green Real-time PCR Master Mix kit (TOYOBO, Japan) and was analyzed on ABI system 7300 (LifeTechnologies, USA). All assays were carried out independently in triplicate. The gene ACTB was used as the reference. Relative gene expression values expressed as fold change were subsequently determined using the Delta-Delta Ct method. The primers for qRT-PCR were listed in Supplementary Table S1.
Furthermore, the GBC patients were retrospectively stratified based on their clinical characteristics, including preoperative laboratory and pathologic parameters such as CA19–9, TNM stages and the presence of cholelithiasis. The presence of relapse was also included. Then, the expression levels of these validated DEGs were compared between subgroups to detect their relationship with clinical characteristics.
Statistical analysis
All data were analyzed using SPSS 18.0 software. Data were presented as mean ± standard deviation (mean ± S.D.) or median and range as appropriate. The t test was used to compare data with normal distribution between two independent groups, while Mann-Whitney U test was used for non-normal data. Comparison between two-paired groups was carried out using paired t test or Wilcoxon signed ranks test when it is necessary. The correlation of the gene expression between GBC and ANTT samples was analyzed using Spearman's correlation, as well as the correlation between DEG expression levels and clinical characteristics. Survival curves were calculated using the Kaplan–Meier method and compared using the log-rank test. The chi-square test was used for categorical data. P < 0.05 was defined as indicating a statistical significance.
SUPPLEMENTARY TABLES
Acknowledgments
This work was supported by the China National Key Projects for Infectious Disease (grant number 2012ZX10002–016); and the Shanghai Natural Science Foundation (grant number 15ZR1413700). We are deeply grateful to Dr. Weiqiang Zheng (Changhai Hospital) for his excellent technical support in pathological diagnosis.
Footnotes
CONFLICTS OF INTEREST
The authors declared to have no conflicts of interest.
REFERENCES
- 1.Hsing AW, Gao YT, Han TQ, Rashid A, Sakoda LC, Wang BS, Shen MC, Zhang BH, Niwa S, Chen J, Fraumeni JF., Jr Gallstones and the risk of biliary tract cancer: a population-based study in China. Br J Cancer. 2007;97:1577–1582. doi: 10.1038/sj.bjc.6604047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boutros C, Gary M, Baldwin K, Somasundar P. Gallbladder cancer: past, present and an uncertain future. Surg Oncol. 2012;21:e183–191. doi: 10.1016/j.suronc.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Stinton LM, Shaffer EA. Epidemiology of gallbladder disease: cholelithiasis and cancer. Gut Liver. 2012;6:172–187. doi: 10.5009/gnl.2012.6.2.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mishra K, Behari A, Kapoor VK, Khan MS, Prakash S, Agrawal S. Vascular endothelial growth factor single-nucleotide polymorphism in gall bladder cancer. J Gastroenterol Hepatol. 2013;28:1678–1685. doi: 10.1111/jgh.12343. [DOI] [PubMed] [Google Scholar]
- 5.Rai R, Sharma KL, Misra S, Kumar A, Mittal B. PSCA gene variants (rs2294008 and rs2978974) confer increased susceptibility of gallbladder carcinoma in females. Gene. 2013;530:172–177. doi: 10.1016/j.gene.2013.08.058. [DOI] [PubMed] [Google Scholar]
- 6.Srivastava K, Srivastava A, Mittal B. Potential biomarkers in gallbladder cancer: present status and future directions. Biomarkers. 2013;18:1–9. doi: 10.3109/1354750X.2012.717105. [DOI] [PubMed] [Google Scholar]
- 7.Pflueger D, Terry S, Sboner A, Habegger L, Esgueva R, Lin PC, Svensson MA, Kitabayashi N, Moss BJ, MacDonald TY, Cao X, Barrette T, Tewari AK, et al. Discovery of non-ETS gene fusions in human prostate cancer using next-generation RNA sequencing. Genome Res. 2011;21:56–67. doi: 10.1101/gr.110684.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ho DW, Yang ZF, Yi K, Lam CT, Ng MN, Yu WC, Lau J, Wan T, Wang X, Yan Z, Liu H, Zhang Y, Fan ST. Gene expression profiling of liver cancer stem cells by RNA-sequencing. PLoS One. 2012;7:e37159. doi: 10.1371/journal.pone.0037159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beane J, Vick J, Schembri F, Anderlind C, Gower A, Campbell J, Luo L, Zhang XH, Xiao J, Alekseyev YO, Wang S, Levy S, Massion PP, et al. Characterizing the impact of smoking and lung cancer on the airway transcriptome using RNA-Seq. Cancer Prev Res (Phila) 2011;4:803–817. doi: 10.1158/1940-6207.CAPR-11-0212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shukla HS. Gallbladder cancer. J Surg Oncol. 2006;93:604–606. doi: 10.1002/jso.20524. [DOI] [PubMed] [Google Scholar]
- 11.Ambrosini G, Adida C, Altieri DC. A novel anti-apoptosis gene, survivin, expressed in cancer and lymphoma. Nat Med. 1997;3:917–921. doi: 10.1038/nm0897-917. [DOI] [PubMed] [Google Scholar]
- 12.Church DN, Talbot DC. Survivin in solid tumors: rationale for development of inhibitors. Curr Oncol Rep. 2012;14:120–128. doi: 10.1007/s11912-012-0215-2. [DOI] [PubMed] [Google Scholar]
- 13.Fukuda S, Pelus LM. Survivin, a cancer target with an emerging role in normal adult tissues. Mol Cancer Ther. 2006;5:1087–1098. doi: 10.1158/1535-7163.MCT-05-0375. [DOI] [PubMed] [Google Scholar]
- 14.Altieri DC. Survivin and IAP proteins in cell-death mechanisms. Biochem J. 2010;430:199–205. doi: 10.1042/BJ20100814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nigam J, Chandra A, Kazmi HR, Parmar D, Singh D, Gupta V M N. Expression of survivin mRNA in gallbladder cancer: a diagnostic and prognostic marker? Tumour Biol. 2014;35:9241–9246. doi: 10.1007/s13277-014-2200-x. [DOI] [PubMed] [Google Scholar]
- 16.He E, Xu XH, Guan H, Chen Y, Chen ZH, Pan ZL, Tang LL, Hu GZ, Li Y, Zhang M, Zhou J, Eriksson S, Fornander T, et al. Thymidine kinase 1 is a potential marker for prognosis and monitoring the response to treatment of patients with breast, lung, and esophageal cancer and non-Hodgkin's lymphoma. Nucleosides Nucleotides Nucleic Acids. 2010;29:352–358. doi: 10.1080/15257771003738535. [DOI] [PubMed] [Google Scholar]
- 17.Li Z, Wang Y, He J, Ma J, Zhao L, Chen H, Li N, Zhou J, He E, Skog S. Serological thymidine kinase 1 is a prognostic factor in oesophageal, cardial and lung carcinomas. Eur J Cancer Prev. 2010;19:313–318. doi: 10.1097/CEJ.0b013e32833ad320. [DOI] [PubMed] [Google Scholar]
- 18.Li Z, Wang Y, Ma J, He J, Zhou J, He E, Skog S. Transient increase in serum thymidine kinase 1 within one week after surgery of patients with carcinoma. Anticancer Res. 2010;30:1295–1299. [PubMed] [Google Scholar]
- 19.Egeblad M, Werb Z. New functions for the matrix metalloproteinases in cancer progression. Nat Rev Cancer. 2002;2:161–174. doi: 10.1038/nrc745. [DOI] [PubMed] [Google Scholar]
- 20.Vandooren J, Van den Steen PE, Opdenakker G. Biochemistry and molecular biology of gelatinase B or matrix metalloproteinase-9 (MMP-9): the next decade. Crit Rev Biochem Mol Biol. 2013;48:222–272. doi: 10.3109/10409238.2013.770819. [DOI] [PubMed] [Google Scholar]
- 21.Hu F, Ku MC, Markovic D, a Dzaye OD, Lehnardt S, Synowitz M, Wolf SA, Kettenmann H. Glioma-associated microglial MMP9 expression is upregulated by TLR2 signaling and sensitive to minocycline. Int J Cancer. 2014;135:2569–2578. doi: 10.1002/ijc.28908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao XH, Yang XQ, Wang BC, Liu SP, Wang FB. Overexpression of twist and matrix metalloproteinase-9 with metastasis and prognosis in gastric cancer. Asian Pac J Cancer Prev. 2013;14:5055–5060. doi: 10.7314/apjcp.2013.14.9.5055. [DOI] [PubMed] [Google Scholar]
- 23.Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Oue N, Sentani K, Noguchi T, Ohara S, Sakamoto N, Hayashi T, Anami K, Motoshita J, Ito M, Tanaka S, Yoshida K, Yasui W. Serum olfactomedin 4 (GW112, hGC-1) in combination with Reg IV is a highly sensitive biomarker for gastric cancer patients. Int J Cancer. 2009;125:2383–2392. doi: 10.1002/ijc.24624. [DOI] [PubMed] [Google Scholar]
- 25.Zhang X HQ, Yang Z, Li Y, Li CY. GW112, a novel antiapoptotic protein that promotes tumor growth. Cancer Res. 2004;64:2474–2481. doi: 10.1158/0008-5472.can-03-3443. [DOI] [PubMed] [Google Scholar]
- 26.Liu WL, Lee HW, Liu YQ, Wang RH, Rodgers GP. Olfactomedin 4 is a novel target gene of retinoic acids and 5-aza-2-deoxycytidine involved in human myeloid leukemia cell growth, differentiation, and apoptosis. Blood. 2010;116:4938–4947. doi: 10.1182/blood-2009-10-246439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo ZY, Zhang Q, Zhao ZS, Li B, Chen JF, Wang YY. OLFM4 is associated with lymph node metastasis and poor prognosis in patients with gastric cancer. J Cancer Res Clin. 2011;137:1713–1720. doi: 10.1007/s00432-011-1042-9. [DOI] [PubMed] [Google Scholar]
- 28.Verrier T, Solhonne B, Sallenave JM, Garcia-Verdugo I. The WAP protein Trappin-2/Elafin: a handyman in the regulation of inflammatory and immune responses. Int J Biochem Cell Biol. 2012;44:1377–1380. doi: 10.1016/j.biocel.2012.05.007. [DOI] [PubMed] [Google Scholar]
- 29.Saidi A, Javerzat S, Bellahcene A, De Vos J, Bello L, Castronovo V, Deprez M, Loiseau H, Bikfalvi A, Hagedorn M. Experimental anti-angiogenesis causes upregulation of genes associated with poor survival in glioblastoma. Int J Cancer. 2008;122:2187–2198. doi: 10.1002/ijc.23313. [DOI] [PubMed] [Google Scholar]
- 30.Labidi-Galy SI, Clauss A, Ng V, Duraisamy S, Elias KM, Piao HY, Bilal E, Davidowitz RA, Lu Y, Badalian-Very G, Gyorffy B, Kang UB, Ficarro S, et al. Elafin drives poor outcome in high-grade serous ovarian cancers and basal-like breast tumors. Oncogene. 2015;34:373–383. doi: 10.1038/onc.2013.562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hunt KK, Wingate H, Yokota T, Liu Y, Mills GB, Zhang F, Fang B, Su CH, Zhang M, Yi M, Keyomarsi K. Elafin, an inhibitor of elastase, is a prognostic indicator in breast cancer. Breast Cancer Res. 2013;15:R3. doi: 10.1186/bcr3374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Money TT, King RG, Wong MH, Stevenson JL, Kalionis B, Erwich JJ, Huisman MA, Timmer A, Hiden U, Desoye G, Gude NM. Expression and cellular localisation of chloride intracellular channel 3 in human placenta and fetal membranes. Placenta. 2007;28:429–436. doi: 10.1016/j.placenta.2006.08.002. [DOI] [PubMed] [Google Scholar]
- 33.Qian Z, Okuhara D, Abe MK, Rosner MR. Molecular cloning and characterization of a mitogen-activated protein kinase-associated intracellular chloride channel. J Biol Chem. 1999;274:1621–1627. doi: 10.1074/jbc.274.3.1621. [DOI] [PubMed] [Google Scholar]
- 34.Dozynkiewicz MA, Jamieson NB, Macpherson I, Grindlay J, van den Berghe PV, von Thun A, Morton JP, Gourley C, Timpson P, Nixon C, McKay CJ, Carter R, Strachan D, et al. Rab25 and CLIC3 collaborate to promote integrin recycling from late endosomes/lysosomes and drive cancer progression. Dev Cell. 2012;22:131–145. doi: 10.1016/j.devcel.2011.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kee AJ, Hardeman EC. Tropomyosins in skeletal muscle diseases. Adv Exp Med Biol. 2008;644:143–157. doi: 10.1007/978-0-387-85766-4_12. [DOI] [PubMed] [Google Scholar]
- 36.Shand JA, Menown IB, McEneaney DJ. A timely diagnosis of myocardial infarction. Biomark Med. 2010;4:385–393. doi: 10.2217/bmm.10.16. [DOI] [PubMed] [Google Scholar]
- 37.Moreno V, Hernandez-Romero D, Vilchez JA, Garcia-Honrubia A, Cambronero F, Casas T, Gonzalez J, Martinez P, Climent V, de la Morena G, Valdes M, Marin F. Serum levels of high-sensitivity troponin T: a novel marker for cardiac remodeling in hypertrophic cardiomyopathy. J Card Fail. 2010;16:950–956. doi: 10.1016/j.cardfail.2010.07.245. [DOI] [PubMed] [Google Scholar]
- 38.Kratnov AE. Markers of inflammation and intracellular metabolism of neutrophils in patients with acute coronary syndrome and normal troponin T levels. Bull Exp Biol Med. 2011;151:467–469. doi: 10.1007/s10517-011-1358-0. [DOI] [PubMed] [Google Scholar]
- 39.Pinto JR, Gomes AV, Jones MA, Liang J, Nguyen S, Miller T, Parvatiyar MS, Potter JD. The functional properties of human slow skeletal troponin T isoforms in cardiac muscle regulation. J Biol Chem. 2012;287:37362–37370. doi: 10.1074/jbc.M112.364927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takeda U, Utani A, Wu J, Adachi E, Koseki H, Taniguchi M, Matsumoto T, Ohashi T, Sato M, Shinkai H. Targeted disruption of dermatopontin causes abnormal collagen fibrillogenesis. J Invest Dermatol. 2002;119:678–683. doi: 10.1046/j.1523-1747.2002.01863.x. [DOI] [PubMed] [Google Scholar]
- 41.Kato A, Okamoto O, Ishikawa K, Sumiyoshi H, Matsuo N, Yoshioka H, Nomizu M, Shimada T, Fujiwara S. Dermatopontin interacts with fibronectin, promotes fibronectin fibril formation, and enhances cell adhesion. J Biol Chem. 2011;286:14861–14869. doi: 10.1074/jbc.M110.179762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takemoto S, Murakami T, Kusachi S, Iwabu A, Hirohata S, Nakamura K, Sezaki S, Havashi J, Suezawa C, Ninomiya Y, Tsuji T. Increased expression of dermatopontin mRNA in the infarct zone of experimentally induced myocardial infarction in rats: comparison with decorin and type I collagen mRNAs. Basic Res Cardiol. 2002;97:461–468. doi: 10.1007/s00395-002-0371-x. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto O, Fujiwara S, Abe M, Sato Y. Dermatopontin interacts with transforming growth factor beta and enhances its biological activity. Biochem J. 1999;337:537–541. [PMC free article] [PubMed] [Google Scholar]
- 44.Li X, Feng P, Ou J, Luo Z, Dai P, Wei D, Zhang C. Dermatopontin is expressed in human liver and is downregulated in hepatocellular carcinoma. Biochemistry (Mosc) 2009;74:979–985. doi: 10.1134/s0006297909090053. [DOI] [PubMed] [Google Scholar]
- 45.Yamatoji M, Kasamatsu A, Kouzu Y, Koike H, Sakamoto Y, Ogawara K, Shiiba M, Tanzawa H, Uzawa K. Dermatopontin: a potential predictor for metastasis of human oral cancer. Int J Cancer. 2012;130:2903–2911. doi: 10.1002/ijc.26328. [DOI] [PubMed] [Google Scholar]
- 46.Li M, Zhang Z, Li X, Ye J, Wu X, Tan Z, Liu C, Shen B, Wang XA, Wu W, Zhou D, Zhang D, Wang T, et al. Whole-exome and targeted gene sequencing of gallbladder carcinoma identifies recurrent mutations in the ErbB pathway. Nat Genet. 2014;46:872–876. doi: 10.1038/ng.3030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


