Summary
Biofilms - communities of bacteria encased in a polymer-rich matrix- confer bacteria with the ability to persist in pathologic host contexts, such as the cystic fibrosis (CF) airways. How bacteria assemble polymers into biofilms is largely unknown. We find that the extracellular matrix produced by Pseudomonas aeruginosa self-assembles into a liquid crystal through entropic interactions between polymers and filamentous Pf bacteriophages, which are long, negatively charged filaments. This liquid crystalline structure enhances biofilm function by increasing adhesion and tolerance to desiccation and antibiotics. Pf bacteriophages are prevalent amongst P. aeruginosa clinical isolates and were detected in CF sputum. The addition of Pf bacteriophage to sputum polymers or serum was sufficient to drive their rapid assembly into viscous liquid crystals. Fd, a related bacteriophage of Escherichia coli, has similar biofilm-building capabilities. Targeting filamentous bacteriophage or the liquid crystalline organization of the biofilm matrix may represent antibacterial strategies.
Introduction
Pseudomonas aeruginosa is a major bacterial pathogen that causes about 10% of nosocomial infections and is responsible for much of the morbidity and mortality associated with cystic fibrosis (CF) airway infections (Hoiby et al., 2010; Jarvis, 2003). In CF, viscous secretions accumulate in the airways (Shak et al., 1990), promoting chronic infection. The capacity of P. aeruginosa to establish chronic infections is dependent, in part, upon its ability to form biofilms - communities of bacteria encased in a polymer-rich matrix. Bacteria within biofilms display increased tolerance to antibiotics and desiccation, allowing them to persist in host tissues and on medical device surfaces (Costerton et al., 1999).
As P. aeruginosa biofilms develop, genes belonging to filamentous Pf1-like bacteriophage (Pf phage) are amongst the most highly transcribed (Rice et al., 2009; Webb et al., 2003; Whiteley et al., 2001). Pf phage belong to the genus Inovirus which are long, negatively charged filaments about 2 μm in length and 6-7 nm in diameter. Their production by P. aeruginosa is stimulated in viscous environments (Yeung et al., 2009) and under anaerobic growth conditions (Platt et al., 2008), such as would be encountered within infected regions of CF lungs. Indeed, many CF clinical isolates carry Pf prophage (Finnan et al., 2004; Kirov et al., 2007; Manos et al., 2008; Mathee et al., 2008), including epidemic strains (Fothergill et al., 2012; Winstanley et al., 2009). Laboratory strains of P. aeruginosa do as well, including PAO1 and PA14 (Mooij et al., 2007), which contain Pf4 and Pf5 prophage, respectively. Pf phage can also infect P. aeruginosa without integrating into the genome, as Pf1 does, whose host is P. aeruginosa strain K (PAK).
Pf phage are involved in the progression of the P. aeruginosa biofilm life cycle by inducing cell death and the subsequent release of bacterial DNA (Rice et al., 2009), a major component of the biofilm matrix (Whitchurch et al., 2002). However, the production of large amounts of Pf phage by P. aeruginosa biofilms - up to 1011 plaque forming units (PFU)/ml - does not result in biofilm eradication (McElroy et al., 2014). Moreover, Pf phage production can be regulated by bacterial factors (Castang and Dove, 2012). We hypothesized that Pf phage could serve a more symbiotic role within P. aeruginosa biofilms.
Here, we report such a symbiotic role for filamentous phage in bacterial biofilm formation. Production of Pf phage by P. aeruginosa biofilms resulted in the spontaneous assembly of a highly ordered liquid crystalline matrix that enhanced biofilm function and, thereby, bacterial fitness.
Results
Mixtures of Pf phage and host and microbial polymers self-assemble into well-organized structures
While performing unrelated experiments that involved supplementing cultures of P. aeruginosa with hyaluronan (HA), a host glycosaminoglycan present in CF airways and abundant in inflamed tissues (Gill et al., 2010; Schulz et al., 2010), we observed the formation of morphologically complex biofilms composed of interlaced structures (Figure 1A and B). The addition of hyaluronidase (HA’ase) dissolved these structures (Figure S1A and B), indicating that HA was essential for their formation. When filtered biofilm supernatants were mixed with HA, DNA, or alginate (a microbial polymer important in CF), similar interwoven structures assembled (Figure 1C; Figure S1C-E), suggesting that P. aeruginosa biofilms produce an extracellular factor that interacts with polymers to assemble well-organized structures.
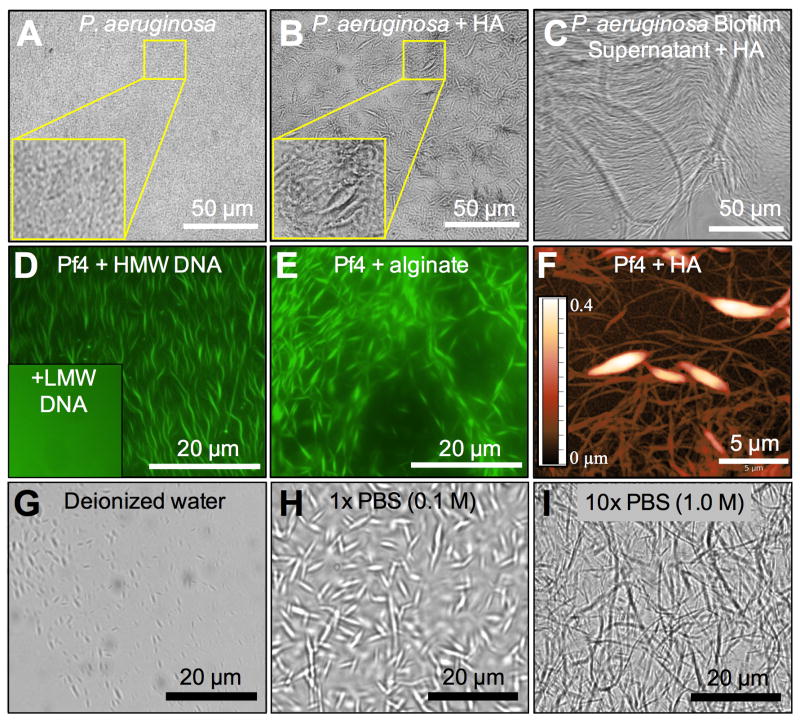
Figure 1. The Filamentous Phage Pf4 Interacts With Host and Microbial Polymers To Spontaneously Assemble Higher Order Structures.
(A) P. aeruginosa forms a flat confluent biofilm in vitro.
(B) P. aeruginosa supplemented with 5 mg/ml HA forms morphologically complex biofilms in vitro.
(C) The addition of P. aeruginosa biofilm supernatant to HA (5 mg/ml) results in the spontaneous formation of higher order structures.
(D) Purified, fluorescently labeled Pf4 (green, 8.8 × 109 PFU/ml) mixed with 5 mg/ml DNA 2 kbp in size (HMW) forms large, interwoven structures while DNA <0.3 kbp in size (LMW) does not (inset).
(E) Purified, fluorescently labeled Pf4 (green, 8.8 × 109 PFU/ml) mixed with 5 mg/ml alginate forms large, interwoven structures.
(F) Visualization of structures formed from Pf4 and HA by AFM semi-contact topography. The color scale indicates height.
(G-I) Pf4 (8.8 × 109 PFU/ml) and HA (5 mg/ml) were suspended in: DI water, 1× PBS, or 10× PBS.
See also Figure S1 and S2.
Proteomic analysis demonstrated that CoaB, the major coat protein of the filamentous bacteriophage Pf4, was abundant in P. aeruginosa supernatants. Thus, we assessed if Pf phage were the factor responsible for the assembly of these structures. Addition of HA to a biofilm formed by a strain of P. aeruginosa lacking the Pf phage integrase (ΔPA0728), a gene essential for Pf4 production (Castang and Dove, 2012) (Figure S5C and D), did not promote structure formation (Figure S1F and G). Conversely, purified Pf4 mixed with HA rapidly (~1-5 minutes) formed similar structures (Movie S1). These structures could be destroyed with HA’ase treatment (Figure S1H and I). Of note, similar structures were observed whether Pf4 phage were purified using PEG precipitation or a cesium chloride gradient method (Figure S1J). Similar structures were also seen upon the mixing of Pf4 with DNA, alginate, HA, or a broad, chemically diverse range of microbial and host polymers, including human serum (Figure 1D-F; Figure S1K-Q). Additionally, the filamentous phage fd, which infects Escherichia coli, formed similar structures when mixed with HA or DNA (Figure S1R and S). These observations show that filamentous phage derived from different bacterial species interact with diverse polymers to rapidly assemble higher order structures.
Ionic strength as well as polymer size and concentration govern structure formation
We next asked what environmental factors influenced the formation of these phage-mediated structures. We found that structure formation was dependent upon ionic strength; they did not efficiently assemble in deionized water and were more condensed and extended in monovalent buffers of increasing tonicity (Figure 1G-I). Raman spectroscopy revealed that HA tightly interacts with Pf4, transforming its secondary structure by suppressing the vibrational freedom of amide groups on Pf4 coat proteins (Figure S2A). This suppression was lost when monovalent salts were washed out, indicating HA was not as tightly associated with Pf4 at low ionic strength (Figure S2B). We also found that the tactoidal structures formed in monovalent solutions dissociated when diluted with water (Figure S2C), indicating the assembly of these structures is reversible.
The formation of these structures was also dependent upon polymer size and concentration. High molecular weight (HMW, ~2 kbp) DNA of similar length to those found in CF sputum (Brandt et al., 1995) form structures with Pf4 (Figure 1D) while low molecular weight (LMW, <0.3 kbp) DNA did not (Figure 1D inset). Similar results were seen with HA, where digested HA fragments did not support structure formation (Figure S1H and I).
Mixtures of Pf phage and polymers interact entropically to form liquid crystals
Taken together, these observations are consistent with the spontaneous self-assembly of filamentous phage and microbial or host polymers into higher order structures by depletion attraction. Depletion attraction is a cohesive force that operates between crowded, like-charged elements in environments where sufficient ionic strength exists to screen their repulsive forces, thus allowing tightly packed structures to assemble (Asakura and Oosawa, 1958). When filamentous particles, like phage, are confined to such structures, they spontaneously align and form liquid crystals with tactoidal morphology (Dogic and Fraden, 2006; Onsager, 1949). Polymer size and concentration are proportional to the range and magnitude of depletion attraction, respectively (Poon, 2002), with larger and/or more concentrated polymers favoring the assembly of liquid crystals from lower concentrations of filamentous phage (Yang et al., 2012). The disassembly of these structures upon dilution is also characteristic of depletion attraction (Tuinier et al., 2003).
One prediction of this model is that the higher-order structures we observed in mixtures of filamentous phage and polymers in monovalent solutions are liquid crystals. To test this, we assessed their optical properties. Liquid crystals are birefringent; they split light into two beams with perpendicular polarization, a consequence of their highly ordered state. Therefore, birefringence is a direct measurement of the molecular alignment of the sample.
To quantify birefringence, we used an optical imaging system developed for birefringent media called Rotopol (Glazer et al., 1996). This device quantitates the phase difference of the polarized light beams emerging from the liquid crystal (i.e., the optical anisotropy) as |sin(δ)| where δ=2πΔnL/λ; Δn=birefringence, L=optical path, and λ=wavelength (550 nm). A computer driven rotatable polarizer probes the light intensities of incoming circularly polarized light as it is changed by the birefringence of the sample.
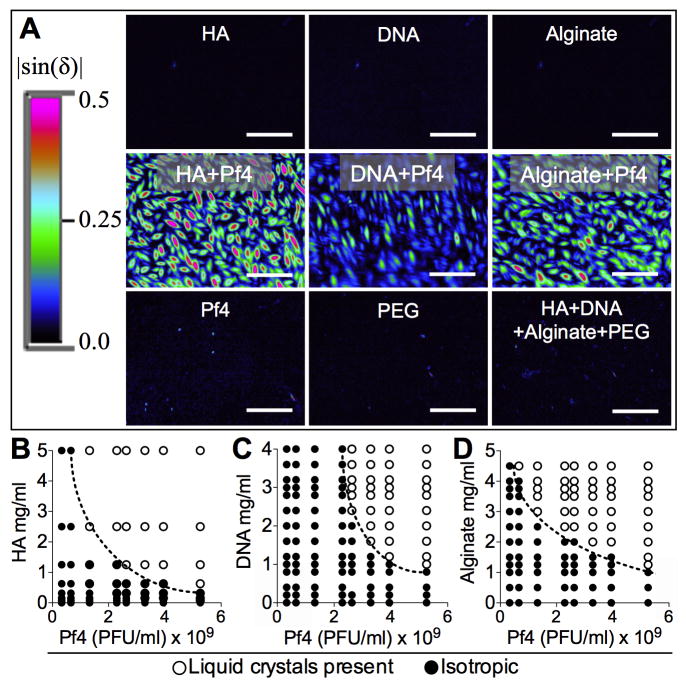
Using this analytical tool, we found that, in monovalent buffer, neither Pf4 nor polymer solutions (alginate, DNA, HA, and PEG, used for phage purification) alone were birefringent. A mixture of Pf4 and these polymers, however, resulted in the assembly of intensely birefringent structures (Figure 2A) indicating that Pf phage are necessary for the observed birefringence. Increasing the concentration of either polymer or Pf4 promotes the assembly of these structures, as indicated in the phase diagrams in Figure 2B-D. This is consistent with organization via depletion attraction; as polymer concentration increases, the system becomes increasingly crowded, allowing lower concentrations of phage to assemble into liquid crystals (Poon, 2002; Yang et al., 2012).
Figure 2. Mixtures of Pf4 and Polymers Assemble Birefringent Liquid Crystals.
(A) The birefringence of the indicated polymer (all 10 mg/ml) were quantified (|sin(δ)|) alone or in the presence of Pf4 (mixed 1:1 with 1×1010 PFU/ml). Scale bars, 10 μm.
(B-D) Phase diagrams showing the relationship between polymer concentration, Pf4 concentration, and liquid crystal assembly. Note the negative slope of the phase boundary (dotted lines), indicative of depletion attraction. See text for details.
See also Figure S3.
We also considered an alternative model where, rather than liquid crystals, Pf phage and polymers crosslink into a birefringent gel. However, if this were the case, we would expect a positive relationship between phage concentration, polymer concentration, and structure formation, as more polymer would be required to crosslink increasing amounts of phage. Instead, we find a negative relationship between these variables and liquid crystal formation (dashed lines in Figure 2B-D), consistent with depletion attraction. To test the impact of crosslinking on birefringence, we added calcium (Ca2+), which can crosslink polyanions like Pf phage (Janmey et al., 2014), to mixtures of Pf4 and HA. We found that the tactoidal morphology was disrupted by Ca2+ and replaced by large aggregates (Figure S2D and E). These aggregates remained intact after dilution with water (Figure S2F), consistent with irreversible crosslinking (Tuinier et al., 2003). Crosslinking also decreased the birefringence of these structures (Figure S3) indicating that the ordered, liquid crystalline structure was disrupted. These data indicate that divalent cations can bind and crosslink Pf phage and polymers, but depletion attraction was likely responsible for the birefringent structures we observed. However, crosslinking of polymers and Pf phage may be relevant to biofilm formation in particular contexts, such as settings with high concentrations of multivalent cations. While the physics of the interactions between filamentous phage and polymers are well established (Dogic and Fraden, 2001), their relevance to microbiology, biofilms, and disease is unknown.
Mixtures of Pf phage and polymers abundant in CF sputum self-assemble into liquid crystals and increase sample viscosity
To understand how Pf phage numbers relate to liquid crystal formation, we first needed accurate methods to quantify phage. Phage are typically enumerated by plaque forming units (PFUs). However, PFU enumeration might be expected to underestimate phage numbers, either because multiple phage could produce a single PFU, or because non-infectious phage particles cannot be detected by PFU enumeration. For this reason, we also measured total phage content using spectroscopy (Tomar et al., 2007). This revealed that PFU enumeration did indeed underestimate total Pf phage content (Figure S4A), suggesting a large amount of non-infectious phage particles were present in our samples. Therefore, results involving Pf4 quantitation are presented in both PFU/ml (to facilitate comparison to the existing microbiology literature) and adjusted total phage content in mg/ml (calculated from the standard curve in Figure S4A).
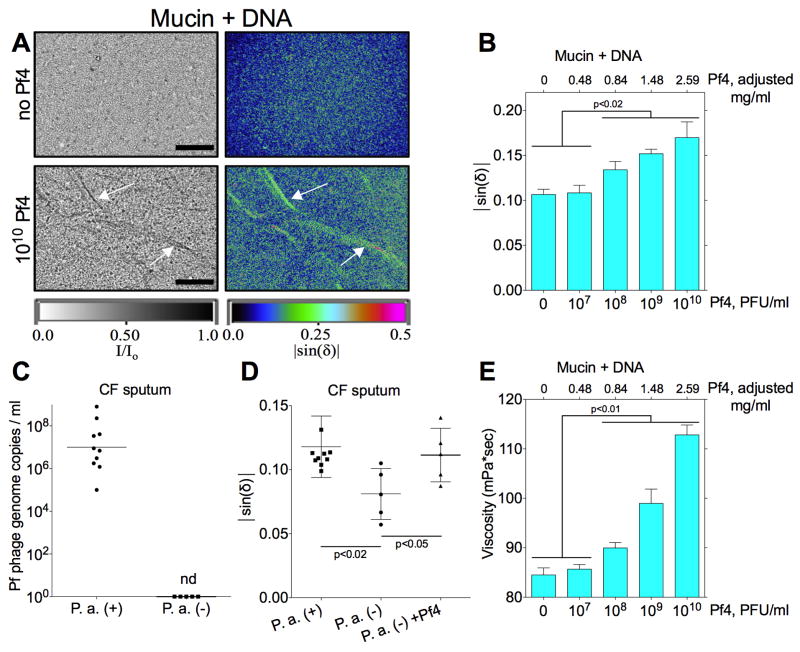
To assess if Pf phage promote liquid crystal formation under physiologically relevant conditions, we added Pf4 to disease relevant concentrations of mucin (8% solids) mixed with DNA (HMW, 4 mg/ml) (Brandt et al., 1995; Matsui et al., 2006). Supplementation of this host polymer mixture with Pf4 resulted in a dose-dependent increase in birefringence at phage concentrations ≥108 PFU/ml, with filamentous structures forming at the highest Pf4 concentrations (Figure 3A and B). Of note, this was lower than the concentration of phage required to form liquid crystals in 4 mg/ml of DNA alone (109 PFU/ml, Figure 2B), suggesting that the presence of multiple polymers amplified the depletion force, allowing lower concentrations of phage to assemble into liquid crystals.
Figure 3. Pf4 Assembles Liquid Crystalline Structures in the Presence of Disease Relevant Polymers and Increases Viscosity.
(A) Representative images of mixtures of mucin (8% solids) and DNA (HMW, 4 mg/ml) in the presence or absence of Pf4. Transmitted light is displayed as I/Io where I = intensity of light emerging from the sample and Io = intensity of incident light. Birefringence is displayed as |sin(δ)|. Arrows indicate filament assembly. Scale bars, 200 μm.
(B) Birefringence was quantified as |sin(δ)| in mixtures of mucin and DNA described in (A) supplemented with increasing amounts of Pf4. Results are mean ± SD of 3 experiments.
(C) Pf phage were quantified by qPCR in sputum collected from CF patients infected by P. aeruginosa (P. a. (+), n = 10) or patients not infected by P. aeruginosa (P. a. (-), n = 5). Results are plotted as the geometric mean, nd; not detected.
(D) The birefringence (|sin(δ)|) of sputum samples described in (C) was quantified. The addition of 108 PFU/ml Pf4 to P. a. (-) sputum augmented birefringence. Results are mean ± SD of 3 experiments.
(E) Changes in the viscosity (mPa*sec) of mucin + DNA samples described in (A) were measured after supplementation with Pf4. Results are mean ± SD of 4 experiments.
See also Figure S4.
To determine if Pf phage might play a similar role in clinical samples, we quantified Pf phage in CF sputum. Because plaques could be generated by other bacteriophage in sputum, we developed a specific quantitative PCR (qPCR) method to detect intact Pf phage in CF sputum, as detailed in the Methods section. Quantitation of purified Pf4 by qPCR produced similar results to enumeration by PFU (Figure S4B). All CF patients colonized with P. aeruginosa were positive for PF phage (Figure 3C). Conversely Pf phage were not detected in sputum collected from patients not infected with P. aeruginosa.
We then asked if Pf phage conferred liquid crystalline properties to CF sputum. Sputum from Pf-positive patients was more birefringent than sputum from Pf-negative patients (Figure 3D). Supplementation of Pf-negative sputum with Pf4 made these samples more birefringent. It is notable that sputum samples containing Pf phage displayed increased birefringence despite the fact that the average Pf phage content in these samples (~107 Pf phage/ml, Figure 3C) was lower than what was required in vitro to assemble liquid crystals in DNA (109 Pf phage/ml, Figure 2C) or mixtures of mucin and DNA (108 Pf phage/ml, Figure 3B). We speculate that in addition to DNA and mucin, multiple polymers present in CF sputum (e.g. F-actin, alginate, and HA) might provide a sufficiently crowded environment at high enough ionic strength to allow the assembly of liquid crystals from lower concentrations of Pf phage. Moreover, there may be phage variants within CF sputum not detected by our qPCR assay, as the mutation rate for Pf phage is quite high (McElroy et al., 2014).
We also investigated the functional significance of liquid crystal assembly by Pf phage in sputum polymers. Given that liquid crystals are inherently viscous (Meyer et al., 1985) and impaired clearance of viscous airway secretions contributes to airway obstruction in CF (Shak et al., 1990), we hypothesized that the addition of Pf4 would increase the viscosity of mucin + DNA mixtures. To test this, viscosity was measured with a capillary viscometer. Indeed, the viscosity of mucin + DNA mixtures increased with the addition of Pf4 (Figure 3E), demonstrating that filamentous phage enhance the viscosity of disease relevant polymers. While crosslinking would also increase viscosity, the associated increase in birefringence implicates liquid crystal formation in the increased viscosity of these disease relevant polymer mixtures.
Filamentous bacteriophage organize the biofilm matrix into a liquid crystal
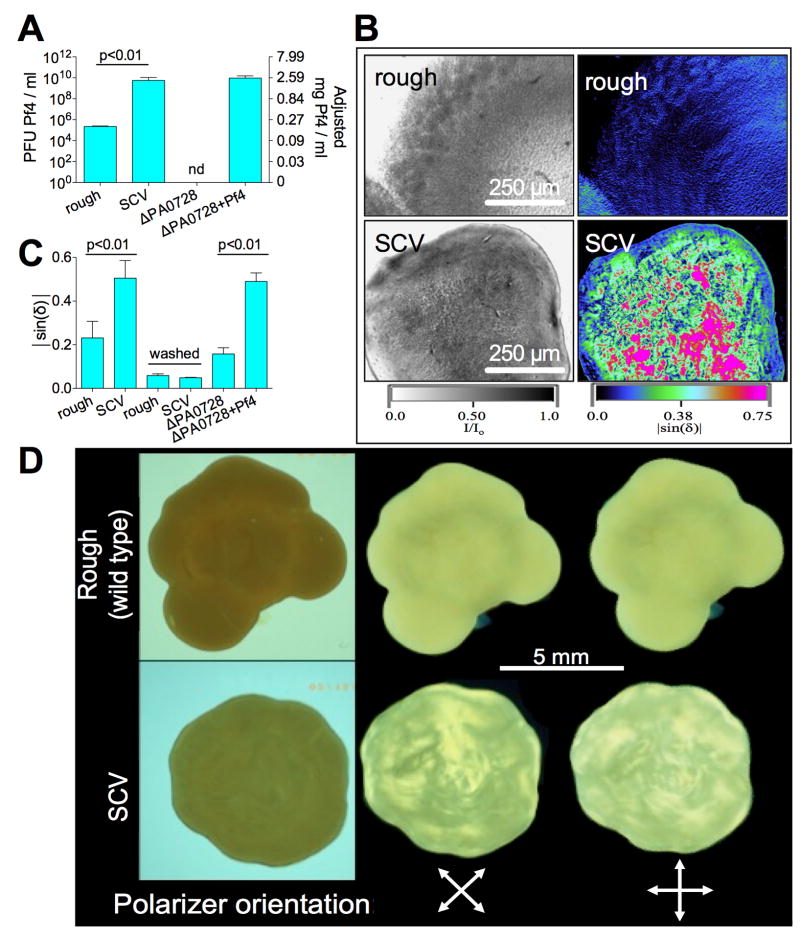
In light of these findings, we hypothesized that Pf phage might promote similar liquid crystalline organization of the P. aeruginosa biofilm matrix. To test this, we analyzed colony biofilms formed from P. aeruginosa PAO1 small colony variants (SCVs). SCVs arise spontaneously within P. aeruginosa biofilm communities and produce abundant amounts of Pf4 (Webb et al., 2004). SCVs display enhanced adherence and antibiotic tolerance, properties that are often implicated in intractable CF airway infections (Haussler, 2004). Compared to the larger wild type colony biofilms with rough edges (Figure S5A), SCVs produced ~5 × 104 fold more endogenous Pf4 (Figure 4A). When subcultured, SCVs stopped producing Pf4 and reverted back to the rough morphology (Figure S5B), consistent with a role for Pf4 in producing the SCV phenotype (Webb et al., 2004). SCVs were intensely birefringent while rough colonies were minimally so (Figure 4B and C). The extended birefringence of SCV biofilms could be visualized by rotating the biofilm between crossed polarizing lenses (Figure 4D; Movie S2 and S3). These data are consistent with a role for Pf phage in the organization of bacterial colonies into liquid crystals.
Figure 4. Pf4 Organizes the P. aeruginosa Biofilm Matrix Into a Liquid Crystalline Structure.
(A) Pf4 production by colony biofilms formed from the indicated strains were enumerated as PFU/ml. Adjusted total Pf phage content are also plotted on the right axis, see text and Figure S4A. Results are mean ± SD of 3 experiments.
(B) Representative images of rough and SCV colony biofilms showing transmitted light (displayed as I/Io) and birefringence (|sin(δ)|).
(C) Birefringence (|sin(δ)|) of the indicated colony biofilms was quantified after normalizing for sample thickness. Birefringence was again measured after washing of the bacteria to remove the extracellular matrix. Results are mean ± SD of 4 experiments.
(D) Representative images for SVC and “rough” colony biofilms (placed between glass plates) visualized through crossed polarizing lenses. Birefringence is visualized as bright areas where light passes through both polarizing lenses. The birefringence patterns change when the sample is rotated with respect to the polarizing lenses, revealing extended areas of birefringence.
See also Figure S6.
Many biological structures, due to their ordered nature, are birefringent. For example, lipid membranes, cell walls, and polymers can be birefringent, so long as they display sufficient order (Mashaghi et al., 2008; Quatrano and Stevens, 1976; Si et al., 2013). This could explain the basal levels of birefringence in biofilms producing little or no phage (Figure 4C). Removal of the matrix by washing of the bacterial cells resulted in a substantial decrease in birefringence (Figure 4C), illustrating that the liquid crystalline nature of the biofilm was manifested primarily from the extracellular matrix and not the bacterial cells.
Given that growth in a biofilm can produce genetic diversity (Boles and Singh, 2008), the SCV strain used in our studies could contain mutations that promote other mechanisms of liquid crystal formation in the biofilm matrix. As a control, birefringence was measured in colony biofilms formed by P. aeruginosa ΔPA0728, which cannot produce Pf4 (Castang and Dove, 2012). Birefringence in this strain was low, similar to that of wild type “rough” P. aeruginosa, indicating that the low amounts of Pf4 produced by rough wild type P. aeruginosa are insufficient to promote liquid crystal assembly. Supplementation of ΔPA0728 with Pf4 produced colony biofilms with a smooth morphology similar to SCVs and increased birefringence (Figure 4C). Similar results were obtained with P. aeruginosa strain K (PAK) and its Pf phage, Pf1 (Figure S6A). We also observed increased birefringence in flowcell biofilms that correlated with Pf4 production (Figure S6B-D), indicating that Pf-dependent matrix organization can occur under diverse growth conditions. Conversely, when grown on agar supplemented with Ca2+, the birefringence of SCV colony biofilms was reduced (Figure S6E) showing that divalent cations disrupt the liquid crystalline order of the biofilm matrix, likely through crosslinking interactions.
Taken together, these data are consistent with the model that, in the absence of sufficient amounts of multivalent cations, Pf phage assemble the P. aeruginosa biofilm matrix into a highly ordered liquid crystalline structure.
The liquid crystalline biofilm matrix confers enhanced adhesiveness
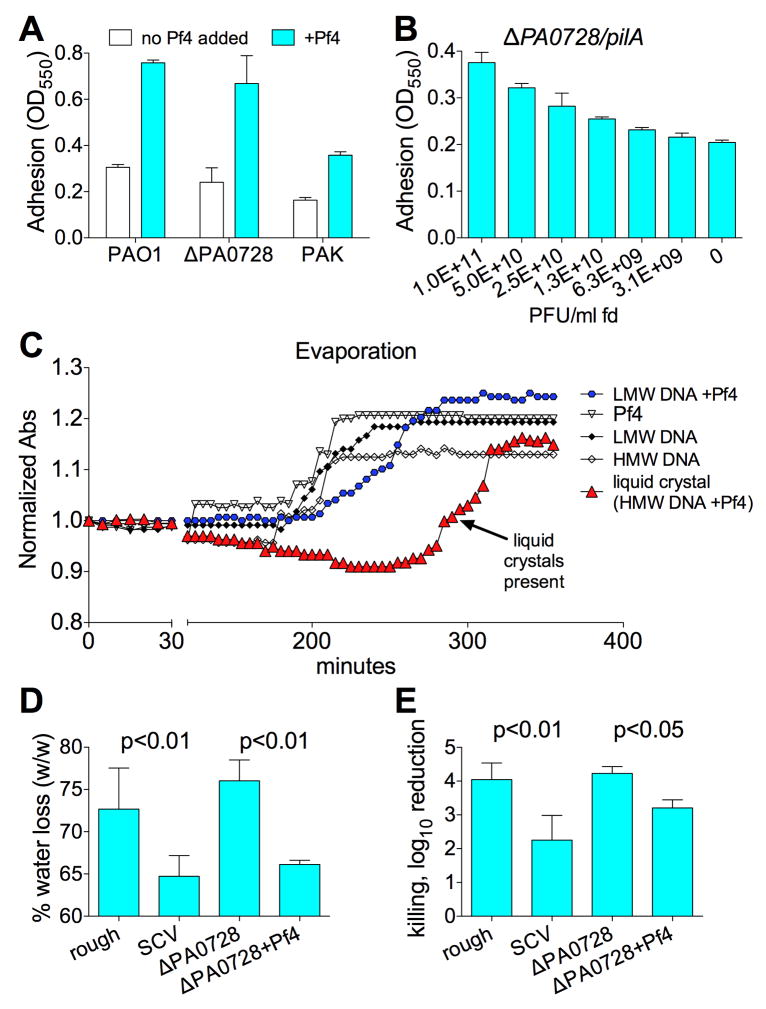
We then asked how Pf4 and the liquid crystalline matrix contribute to biofilm functionality. One canonical feature of biofilms is their ability to adhere to surfaces. We measured the adhesiveness of biofilms with a liquid crystalline matrix and found that the addition of Pf4 promoted biofilm adhesion in PAO1 and ΔPA0728 (Figure 5A). Adhesion is also increased when Pf4 is supplied to P. aeruginosa strain K (PAK) (Figure 5A), which cannot be infected by Pf4 (Figure S5E). To rule out infection-mediated effects on adhesion, we repeated these studies using a strain of P. aeruginosa (ΔPA0728/pilA) that can neither produce nor be infected by Pf4 (Figure S5E-F). This strain lacks PA0728 and type IV pili (pilA), the receptor that Pf4 use to infect P. aeruginosa (Castang and Dove, 2012). To further exclude any possibility of infection, fd phage from E. coli were used instead of Pf phage. Using this system, we again find that phage addition promotes adhesion, as measured by an increase in adherent biomass (Figure 5B). Together with previous work implicating Pf phage in P. aeruginosa adhesion (Webb et al., 2004), these data indicate that filamentous phage make structural contributions to biofilms that increase adhesion.
Figure 5. P. aeruginosa Biofilms With Liquid Crystalline Matrices Display Increased Adhesion and Tolerance to Desiccation.
(A) Biofilm adhesion after 24 hours of growth as measured by the crystal violet adhesion assay. Results are mean ± SD of 3 experiments.
(B) Adhesion of biofilms formed by strain ΔPA0728/pilA in the setting of increasing concentrations of fd phage.
(C) Evaporation of isotropic and liquid crystalline phases of Pf4 and DNA was monitored by absorbance (Abs, 600 nm). Absorbance was normalized to initial readings for each sample. As samples dried, the absorbance increased and stabilized once dried to completeness. In this way, the retention of water was monitored over time. Results are mean of 3 experiments; error bars are omitted for clarity.
(D) Colony biofilms formed from the indicated strains were desiccated for 18-h at 37°C in an ambient incubator. The percent water loss was measured as the wet weight pre desiccation / wet weight post desiccation. Results are mean ± SD of 6 experiments for rough and SCV biofilms and 3 experiments for ΔPA0728 +/- Pf4 biofilms.
(E) Killing by desiccation is represented as the log10 reduction of viable cells recovered from control biofilms compared to desiccated biofilms. Results are mean ± SD of 3 experiments.
See also Figure S5.
The liquid crystalline biofilm matrix provides protection from desiccation
The biofilm matrix protects bacteria against desiccation, promoting their survival (Ophir and Gutnick, 1994). Given that liquid crystals are inherently viscous (Meyer et al., 1985) and viscous solutions generally display reduced rates of evaporation, we hypothesized that the liquid crystalline biofilm matrix might increase water retention and, in turn, augment desiccation tolerance.
To study water retention in mixtures of polymers and phage in the isotropic (unordered) or liquid crystalline phases, we measured evaporation rates using a plate reader. Samples were placed into a 96-well plate and incubated uncovered in a plate reader at 37°C. Liquid crystals assembled from Pf4 and DNA evaporated at a slower rate compared to Pf4 or DNA alone (Figure 5C), indicating that the liquid crystalline phase retains water more efficiently than polymers in the isotropic phase.
To investigate the impact of liquid crystal assembly on evaporation independent of polymer concentration, the HMW-DNA in liquid crystalline mixtures was replaced with LMW-DNA. The resulting mixtures contained the same concentrations of Pf4 and DNA, but since LMW-DNA did not promote liquid crystal assembly at the concentrations tested (Figure 1D inset), the mixtures remained isotropic. These isotropic mixtures evaporated faster than liquid crystalline mixtures containing the same concentrations of DNA and Pf4, indicating that liquid crystal assembly, rather than high polymer concentration, promotes water retention (Figure 5C). Consistent with this conclusion, when biofilms with liquid crystalline matrices were desiccated, they retained more water (wet weight) and displayed increased desiccation tolerance compared to biofilms lacking liquid crystalline order (Figure 5D and E). Together, these results suggest that the assembly of the liquid crystalline biofilm matrix enhances water retention, increasing tolerance to desiccation.
The liquid crystalline biofilm matrix promotes tolerance to aminoglycoside antibiotics
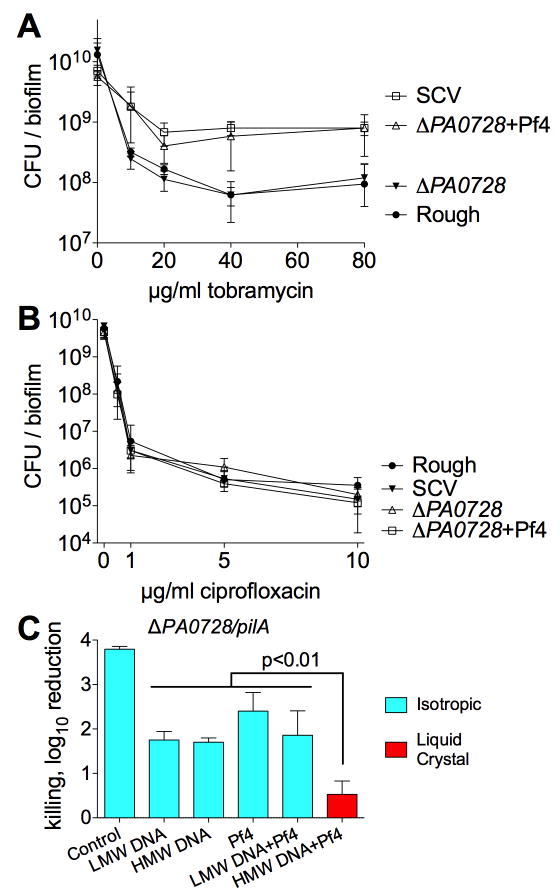
Antibiotic tolerance is a hallmark of bacterial biofilms (Costerton et al., 1999). Hence, we investigated if the liquid crystalline organization of biofilm matrix might further increase antibiotic tolerance in P. aeruginosa biofilms. Indeed, relative to biofilms not organized into a liquid crystal, bacteria within liquid crystalline biofilms were more tolerant to aminoglycoside antibiotics, but were equally sensitive to the fluoroquinolone ciprofloxacin (Figure 6A and B; Figure S7A).
Figure 6. The Liquid Crystalline Biofilm Matrix Protects Against Tobramycin but not Ciprofloxacin.
(A and B) CFUs recovered from colony biofilms after treatment (90 minutes) with increasing concentrations of (A) tobramycin or (B) ciprofloxacin are plotted. Results are mean ± SD of 3 experiments.
(C) To investigate whether or not Pf phage mediate tolerance to tobramycin through an extracellular mechanism, planktonic ΔPA0728/pilA, which cannot produce nor be infected by Pf4, was suspended in isotropic or liquid crystalline phases of Pf4 (1010 PFU/ml) and DNA (2.5 mg/ml). Killing is represented as the log10 reduction of viable cells recovered from cultures treated with tobramycin (10 μg/ml, 90 min) compared to untreated controls. Results are mean ± SD of 3 experiments.
See also Figures S5 and S7.
To address the possibility that tolerance to aminoglycosides was due to physiological changes induced by Pf4 or undefined genetic mutations in the SCV strain, we again used the Pf phage-null strain ΔPA0728/pilA. Planktonic ΔPA0728/pilA were added to isotropic or liquid crystalline mixtures of Pf4 and DNA containing tobramycin. Mixtures containing liquid crystals offered P. aeruginosa the most protection (Figure 6C) even though equivalent amounts of DNA and phage from the same stocks were present in both isotropic and liquid crystal-containing samples. Strains containing deletions of pilA, PA0728, or the double mutant ΔPA0728/pilA were equally sensitive to tobramycin, showing that these genes do not influence antibiotic tolerance (Figure S7B). Further, when Pf4 was replaced with the filamentous phage fd, similar trends were observed (Figure S7C and D). Together, these results indicate that liquid crystal formation by filamentous phage promote tolerance to aminoglycosides such as tobramycin.
Liquid crystalline phases of Pf phage sequester aminoglycoside antibiotics
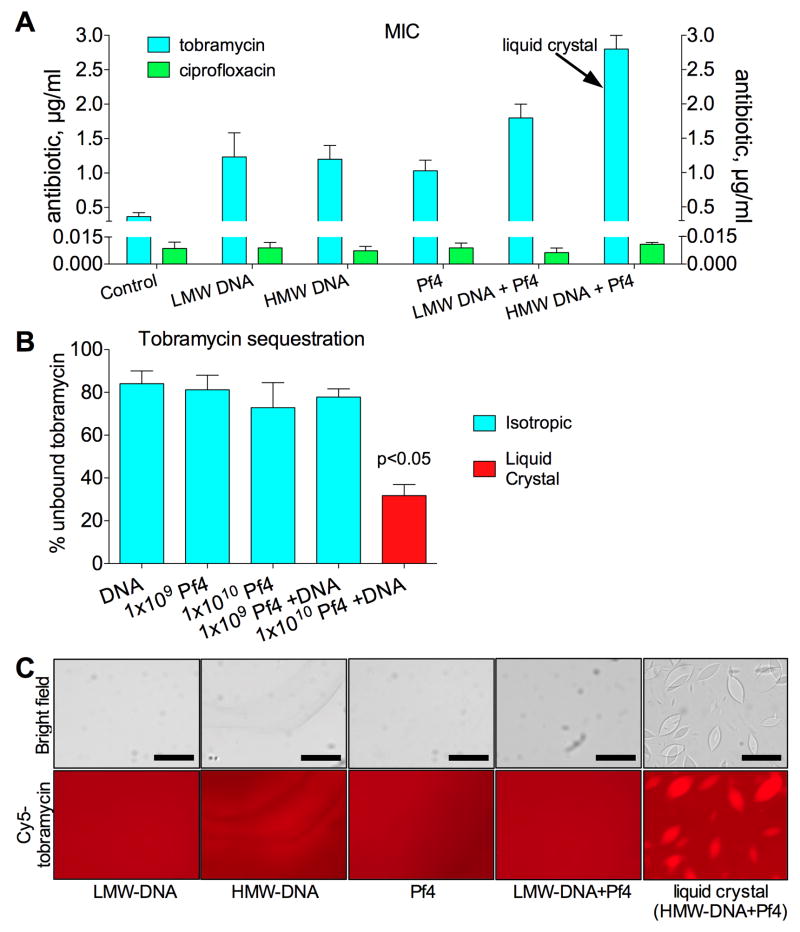
Aminoglycosides are cationic and, hence, are bound by polyanions like DNA in the biofilm matrix (Tseng et al., 2013) and at sites of infection (Hunt et al., 1995), reducing their efficacy. In contrast, ciprofloxacin does not interact electrostatically with polyanions and readily penetrates P. aeruginosa biofilms (Tseng et al., 2013). Given that biofilms with liquid crystalline matrices were more tolerant to aminoglycosides, we hypothesized that liquid crystal assembly might promote binding and sequestration of aminoglycosides in ways that prevent bacterial killing.
To test this, increasing concentrations of tobramycin or ciprofloxacin were added to isotropic or liquid crystalline mixtures of Pf4 and DNA. After a 4-h incubation to allow binding of antibiotics, E. coli was added and the minimum inhibitory concentration (MIC) was determined (Figure 7A). E. coli was used because it cannot be infected by Pf4 and to highlight the generality of the mechanism. Pf4 and DNA did not offer bacteria protection against ciprofloxacin, even in mixtures containing liquid crystals. In contrast, Pf4 and DNA did provide bacteria protection against tobramycin, consistent with the binding of tobramycin to polyanions.
Figure 7. The Liquid Crystalline Biofilm Matrix Increases Antibiotic Tolerance to Antibiotics by Enhancing Aminoglycoside Binding.
(A) Antibiotic killing in the presence of DNA and Pf4 was investigated. Increasing concentrations of tobramycin or ciprofloxacin were added to isotropic or liquid crystal containing mixtures of Pf4 (1010 PFU/ml) and DNA (2.5 mg/ml). The arrow indicates the only sample with liquid crystals. E. coli was then added and samples were incubated overnight. The MIC for each antibiotic was plotted. Results are mean ± SD of 3 experiments.
(B) Tobramycin (200 μg/ml) was mixed with the indicated amounts of Pf4 and DNA (2.5 mg/ml) and placed into a dialysis cassette. The diffusion of unbound tobramycin across the membrane was monitored by HPLC-MS. Results were normalized to controls containing no Pf4 or DNA. Results are mean ± SD of duplicate experiments.
(C) Binding of fluorescent tobramycin (Cy5-tobramycin, 40 μg/ml) to isotropic and liquid crystalline phases of Pf4 (1010 PFU/ml) and HMW-DNA (2.5 mg/ml) was visualized by fluorescent microscopy. Scale bars, 20 μm.
See also Figures S5 and S7.
To differentiate the contributions of polymer concentration and liquid crystal assembly towards antibiotic killing, we compared mixtures of Pf4 and HMW-DNA (which form liquid crystals) against mixtures of LMW-DNA and Pf4 (which do not), as described for the desiccation experiments above. Mixtures containing liquid crystals inhibited the activity of tobramycin more so than isotropic mixtures of Pf4 and DNA (Figure 7A). Differential binding of tobramycin to HMW-DNA and LMW-DNA cannot explain these results (Figure 7A), suggesting that liquid crystal assembly further enhances tobramycin binding. Further, when mixtures of Pf4 and DNA were separated from buffer by a membrane, samples containing liquid crystals sequestered more tobramycin compared to isotropic samples (Figure 7B). To investigate sequestration further, we used a fluorescent Cy5-conjugated form of tobramycin to assess the binding of tobramycin to liquid crystals. Fluorescent imaging revealed that tobramycin was sequestered within liquid crystals (Figure 7C). The binding of microgram quantities of tobramycin did not affect the morphology of the liquid crystal, suggesting that the underlying molecular order was not severely disrupted. These results are consistent with the idea that liquid crystal assembly enhances the binding of aminoglycoside antibiotics, thereby mitigating bacterial killing.
Discussion
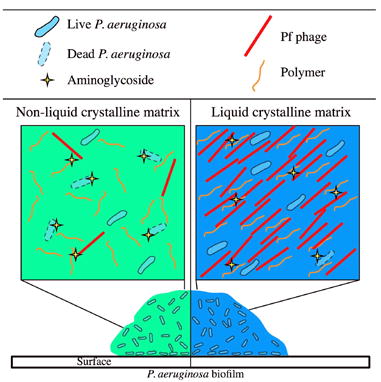
The production of filamentous Pf phage by P. aeruginosa biofilms is associated with the organization of the matrix into a liquid crystalline structure that promotes the fundamental, pathogenic features of biofilms, including adhesion and tolerance to desiccation and antibiotics.
These findings are particularly relevant to the pathophysiology of CF, where the high viscosity and polymer content of airway secretions are thought to contribute potently to disease symptoms. Here, we show that Pf phage enhance the viscosity of polymers abundant in CF airway secretions. Although an increase in birefringence was correlated with an increase in viscosity, crosslinking interactions could also be operating between Pf phage and components of these complex polymer mixtures, contributing to viscosity.
Pf phage may also promote the transmissibility of P. aeruginosa. The transmission of P. aeruginosa from one CF patient to another can occur through aerosols or contaminated surfaces, and desiccation tolerance is thought to be critical to transmission (Panagea et al., 2005). Our results suggest that the liquid crystal organization of the matrix slows the evaporation of water, allowing biofilms to better tolerate desiccation. This property alone could have implications on the transmissibility of P. aeruginosa between CF patients, particularly in epidemic strains that contain Pf prophage, such as the Liverpool epidemic strain (Winstanley et al., 2009).
The liquid crystalline biofilm matrix also provides P. aeruginosa protection against aminoglycoside antibiotics. Given that tobramycin accumulates within liquid crystals composed of Pf phage, the increased tolerance of P. aeruginosa biofilms to these antibiotics may be mediated by enhanced binding to the liquid crystalline matrix. Binding is perhaps facilitated by the highly ordered nature of these liquid crystals; they are densely packed with filamentous phage, so much so that they change the polarization of light. Thus, the negative charge density within these liquid crystals would be high, attracting cationic antibiotics. In addition to aminoglycosides, liquid crystals composed of Pf phage might be expected to efficiently bind other cationic antimicrobials important in airway defense. For example, cationic antimicrobial peptides bind to Pf phage through ionic interactions (Wnorowska et al., 2015). Together, our data on adhesion, sputum polymer viscosity, and antibiotic tolerance suggest that Pf phage contribute to the persistence of P. aeruginosa biofilm infections and may help explain how Pf phage influence P. aeruginosa virulence in vivo (Rice et al., 2009).
In addition to influencing persistence phenotypes, Pf phage are important to the progression of the biofilm lifecycle by inducing cell lysis (at least in a subset of the bacterial population), resulting in the accumulation of extracellular DNA in the biofilm matrix (Rice et al., 2009). The concurrent increase in both DNA and phage concentrations would favor the organization of the biofilm matrix into a liquid crystalline structure. In addition to P. aeruginosa, filamentous phage of the genus Inovirus are produced by many Gram-negative bacterial species, including pathogens such as E. coli, Stenotrophomonas maltophilia, Vibrio cholerae, and others. Given that the E. coli phage fd likewise assembled liquid crystals, we postulate that filamentous phage may contribute to other chronic infections by affecting biofilm structure and function.
Finally, our data point towards potential strategies to prevent or treat biofilm infections by targeting filamentous phage and the liquid crystal organization of the biofilm matrix. Perturbing the soft matter physics of the biofilm matrix might increase the susceptibility of bacterial biofilms to host defenses or existing therapeutics.
Experimental Procedures
Bacterial strains, media, and culture conditions
Bacterial strains, plasmids, and PCR primers are listed in Table S1. Unless specified otherwise, bacteria were grown at 37°C with shaking in Luria-Bertani (LB) medium. P. aeruginosa strain PAO1 was used for all experiments unless stated otherwise. Biofilms were grown as previously described (Walters et al., 2003) and elaborated further in the Supplemental Experimental Procedures.
Evaporation and Desiccation experiments
Evaporation of the indicated mixtures of Pf4 (1010 PFU/ml) and DNA (2.5 mg/ml) was monitored using a plate reader at 37°C. Samples (60 μl) were placed into random wells of a 96-well plate. The plate was placed into a warm plate reader without a cover and absorbance (600 nm) was monitored until the samples were completely dried. Absorbance readings were normalized to initial readings for each sample.
Colony biofilms (48-h) of the indicated strains were placed onto 1% Noble agar lacking carbon and nutrient sources (moisture control) or onto a dry plastic dish for 18-h at 37°C in an ambient incubator. Percent water loss was calculated by subtracting the final and initial wet weights of desiccated and moisture control biofilms. Viable bacteria were enumerated by CFUs.
Antibiotic tolerance
Colony biofilms (48-h) were transferred to fresh LB plates +/- antibiotics at the indicated concentration and incubated at 37°C for 18-h. Biofilms were then resuspended and viable CFUs were enumerated as described in the Supplemental Experimental Procedures. For experiments investigating the role of extracellular phage and polymers in antibiotic tolerance, planktonic ΔPA0728/pilA or ΔPA0728 (2 × 108 CFUs in 50 μl LB broth) were added to 500 μl of the indicated phage and polymer solutions and incubated for 20 minutes at room temperature. Tobramycin (10 μg/ml) or PBS was added and the cells were incubated at 37°C for 90 minutes. CFUs were then enumerated. MIC and binding experiments are described in the Supplemental Experimental Procedures.
Phage purification, detection, and quantification
Phage purification methods are described in the Supplemental Experimental Procedures. Plaque assays were performed as described previously (Castang and Dove, 2012) with ΔPA0728 as the recipient strain. Bacteriophage fd was quantified using the same methodologies using E. coli strain ATCC 15669 as the recipient strain. The presence of Pf4 in filtered bacterial supernatants was confirmed by the amplification of an 839-bp region corresponding to the circular replicative form (RF) of Pf4 using primers Pf4F and Pf4R (Table S1), as described previously (Webb et al., 2004). Quantification of Pf4 by spectroscopy was performed as described (Tomar et al., 2007).
Pf phage in CF sputum were quantified by qPCR. CF sputum was obtained after written informed consent, for biobanking of the patients’ specimens and subsequent use, approved by the Stanford Institutional Review Board. Sputum was diluted 1:1 (vol:vol) with PBS containing DNase (100 μg/ml) and DTT (1 mM). Sputum was incubated at 37°C for 5 hours with occasional vortexing. Debris (including bacterial cells) was removed by centrifuging the sample at 6,000g for 10 minutes. The supernatants were collected and a 100 μl aliquot was incubated at 95°C for 15 minutes, inactivating the DNase and releasing any protected phage DNA. The sample was then added to 250 μl buffer P1 from a Qiagen miniprep kit. DNA extraction was performed as described by the manufacturer’s protocol. For qPCR, primers specific for a conserved region of the Pf genome (Pf-Conserve-F and Pf-Conserve-R) or the 50S rDNA gene rplU (rplU-F and rplU-R, (Mah et al., 2003)) were used (Table S1). The 50S primers controlled for potential bacterial chromosomal DNA contamination that might inflate detected Pf phage numbers (Figure S4C). Cycling conditions and other details are provided in the Supplemental Experimental Procedures.
Birefringence measurements
Birefringence was measured using Rotopol as described (Glazer et al., 1996). Intact colonies grown on LB agar were visualized for birefringence by excising agar containing bacterial colonies and placing it on a glass slide. Separate samples with normalized thickness were prepared for birefringence quantitation by scraping bacterial colonies off of the agar and placing the biomass onto a glass slide. Parafilm was placed around the biomass to provide a spacer with uniform thickness and a glass coverslip was placed on top. Likewise, the birefringence of polymers and sputum were quantified in this way.
Statistical analysis
Statistical analysis was performed using Prism GraphPad software, mean with standard deviation were calculated and plotted unless specified otherwise. An unpaired t-test was used to calculate p-values.
Supplementary Material
Acknowledgments
Our thanks go to L. K. Jennings, D. Relman, P. Janmey, and J. Nepom for critical reading of the manuscript. We are grateful to J. J. Harrison for assistance in constructing mutants, S. L. Dove for sharing pEX-ΔPA0728, B. S. Tseng for sharing Cy5-tobramycin, and Hongxiang Tang for assistance with HPLC-MS analysis. We also thank P. D. von Haller and J. K. Eng of the University of Washington’s Proteomics Resource.
This work was supported by National Institutes of Health grant R01 HL113294-01A1 to P.L.B, a Cystic Fibrosis Foundation Research Development Program Postdoctoral Fellowship to P.R.S., and in part by UWPR95794 to the University of Washington’s Proteomics Resource, a grant from the Child Health Research Institute, Stanford Transdisciplinary Initiatives Program, and a gift to D.A.S. from Mr. John Flatley. A Gabilan Stanford Graduate Fellowship supported J.M.S.
Footnotes
Author contributions: P.R.S., W.C.P., and P.L.B. wrote the manuscript. P.R.S., P.J, W.C.P., P.K.S., J.R., and P.L.B. designed experiments. P.R.S., J.M.S., L.A.M., A.V.M., D.L., E.K., A.A., K.R.B., S.P.E., M.E.B, and P.L.B. performed experiments. W.K. and D.A.S contributed new reagents/analytical tools.
The authors declare no conflict of interest.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Asakura S, Oosawa F. Interaction between Particles Suspended in Solutions of Macromolecules. J Polym Sci. 1958;33:183–192. [Google Scholar]
- Boles BR, Singh PK. Endogenous oxidative stress produces diversity and adaptability in biofilm communities. Proc Natl Acad Sci U S A. 2008;105:12503–12508. doi: 10.1073/pnas.0801499105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt T, Breitenstein S, von der Hardt H, Tummler B. DNA concentration and length in sputum of patients with cystic fibrosis during inhalation with recombinant human DNase. Thorax. 1995;50:880–882. doi: 10.1136/thx.50.8.880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castang S, Dove SL. Basis for the essentiality of H-NS family members in Pseudomonas aeruginosa. J Bacteriol. 2012;194:5101–5109. doi: 10.1128/JB.00932-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284:1318–1322. doi: 10.1126/science.284.5418.1318. [DOI] [PubMed] [Google Scholar]
- Dogic Z, Fraden S. Development of model colloidal liquid crystals and the kinetics of the isotropic-smectic transition. Philos T R Soc A. 2001;359:997–1014. [Google Scholar]
- Dogic Z, Fraden S. Ordered phases of filamentous viruses. Curr Opin Colloid In. 2006;11:47–55. [Google Scholar]
- Finnan S, Morrissey JP, O’Gara F, Boyd EF. Genome diversity of Pseudomonas aeruginosa isolates from cystic fibrosis patients and the hospital environment. J Clin Microbiol. 2004;42:5783–5792. doi: 10.1128/JCM.42.12.5783-5792.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fothergill JL, Walshaw MJ, Winstanley C. Transmissible strains of Pseudomonas aeruginosa in cystic fibrosis lung infections. Eur Respir J. 2012;40:227–238. doi: 10.1183/09031936.00204411. [DOI] [PubMed] [Google Scholar]
- Gill S, Wight TN, Frevert CW. Proteoglycans: key regulators of pulmonary inflammation and the innate immune response to lung infection. Anat Rec (Hoboken) 2010;293:968–981. doi: 10.1002/ar.21094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glazer AM, Lewis JG, Kaminsky W. An automatic optical imaging system for birefringent media. P Roy Soc Lond a Mat. 1996;452:2751–2765. [Google Scholar]
- Haussler S. Biofilm formation by the small colony variant phenotype of Pseudomonas aeruginosa. Environ Microbiol. 2004;6:546–551. doi: 10.1111/j.1462-2920.2004.00618.x. [DOI] [PubMed] [Google Scholar]
- Hoiby N, Ciofu O, Bjarnsholt T. Pseudomonas aeruginosa biofilms in cystic fibrosis. Future Microbiol. 2010;5:1663–1674. doi: 10.2217/fmb.10.125. [DOI] [PubMed] [Google Scholar]
- Hunt BE, Weber A, Berger A, Ramsey B, Smith AL. Macromolecular mechanisms of sputum inhibition of tobramycin activity. Antimicrob Agents Chemother. 1995;39:34–39. doi: 10.1128/aac.39.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janmey PA, Slochower DR, Wang YH, Wen Q, Cebers A. Polyelectrolyte properties of filamentous biopolymers and their consequences in biological fluids. Soft Matter. 2014;10:1439–1449. doi: 10.1039/c3sm50854d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis WR. Benchmarking for prevention: the Centers for Disease Control and Prevention’s National Nosocomial Infections Surveillance (NNIS) system experience. Infection. 2003;31(Suppl 2):44–48. [PubMed] [Google Scholar]
- Kirov SM, Webb JS, O’May C Y, Reid DW, Woo JK, Rice SA, Kjelleberg S. Biofilm differentiation and dispersal in mucoid Pseudomonas aeruginosa isolates from patients with cystic fibrosis. Microbiology. 2007;153:3264–3274. doi: 10.1099/mic.0.2007/009092-0. [DOI] [PubMed] [Google Scholar]
- Mah TF, Pitts B, Pellock B, Walker GC, Stewart PS, O’Toole GA. A genetic basis for Pseudomonas aeruginosa biofilm antibiotic resistance. Nature. 2003;426:306–310. doi: 10.1038/nature02122. [DOI] [PubMed] [Google Scholar]
- Manos J, Arthur J, Rose B, Tingpej P, Fung C, Curtis M, Webb JS, Hu H, Kjelleberg S, Gorrell MD, et al. Transcriptome analyses and biofilm-forming characteristics of a clonal Pseudomonas aeruginosa from the cystic fibrosis lung. J Med Microbiol. 2008;57:1454–1465. doi: 10.1099/jmm.0.2008/005009-0. [DOI] [PubMed] [Google Scholar]
- Mashaghi A, Swann M, Popplewell J, Textor M, Reimhult E. Optical anisotropy of supported lipid structures probed by waveguide spectroscopy and its application to study of supported lipid bilayer formation kinetics. Anal Chem. 2008;80:3666–3676. doi: 10.1021/ac800027s. [DOI] [PubMed] [Google Scholar]
- Mathee K, Narasimhan G, Valdes C, Qiu X, Matewish JM, Koehrsen M, Rokas A, Yandava CN, Engels R, Zeng E, et al. Dynamics of Pseudomonas aeruginosa genome evolution. Proc Natl Acad Sci U S A. 2008;105:3100–3105. doi: 10.1073/pnas.0711982105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsui H, Wagner VE, Hill DB, Schwab UE, Rogers TD, Button B, Taylor RM, 2nd, Superfine R, Rubinstein M, Iglewski BH, et al. A physical linkage between cystic fibrosis airway surface dehydration and Pseudomonas aeruginosa biofilms. Proc Natl Acad Sci U S A. 2006;103:18131–18136. doi: 10.1073/pnas.0606428103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McElroy KE, Hui JG, Woo JK, Luk AW, Webb JS, Kjelleberg S, Rice SA, Thomas T. Strain-specific parallel evolution drives short-term diversification during Pseudomonas aeruginosa biofilm formation. Proc Natl Acad Sci U S A. 2014;111:E1419–1427. doi: 10.1073/pnas.1314340111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RB, Lonberg F, Taratuta V, Fraden S, Lee SD, Hurd AJ. Measurements of the Anisotropic Viscous and Elastic Properties of Lyotropic Polymer Nematics. Faraday Discuss. 1985;79:125–132. [Google Scholar]
- Mooij MJ, Drenkard E, Llamas MA, Vandenbroucke-Grauls CM, Savelkoul PH, Ausubel FM, Bitter W. Characterization of the integrated filamentous phage Pf5 and its involvement in small-colony formation. Microbiology. 2007;153:1790–1798. doi: 10.1099/mic.0.2006/003533-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onsager L. The effects of shape on the interaction of colloidal particles. Ann NY Acad Sci. 1949;51 [Google Scholar]
- Ophir T, Gutnick DL. A role for exopolysaccharides in the protection of microorganisms from desiccation. Appl Environ Microbiol. 1994;60:740–745. doi: 10.1128/aem.60.2.740-745.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagea S, Winstanley C, Walshaw MJ, Ledson MJ, Hart CA. Environmental contamination with an epidemic strain of Pseudomonas aeruginosa in a Liverpool cystic fibrosis centre, and study of its survival on dry surfaces. J Hosp Infect. 2005;59:102–107. doi: 10.1016/j.jhin.2004.09.018. [DOI] [PubMed] [Google Scholar]
- Platt MD, Schurr MJ, Sauer K, Vazquez G, Kukavica-Ibrulj I, Potvin E, Levesque RC, Fedynak A, Brinkman FS, Schurr J, et al. Proteomic, microarray, and signature-tagged mutagenesis analyses of anaerobic Pseudomonas aeruginosa at pH 6.5, likely representing chronic, late-stage cystic fibrosis airway conditions. J Bacteriol. 2008;190:2739–2758. doi: 10.1128/JB.01683-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poon WCK. The physics of a model colloid-polymer mixture. J Phys-Condens Mat. 2002;14:R859–R880. [Google Scholar]
- Quatrano RS, Stevens PT. Cell wall assembly in fucus zygotes: I. Characterization of the polysaccharide components. Plant Physiol. 1976;58:224–231. doi: 10.1104/pp.58.2.224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice SA, Tan CH, Mikkelsen PJ, Kung V, Woo J, Tay M, Hauser A, McDougald D, Webb JS, Kjelleberg S. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. Isme J. 2009;3:271–282. doi: 10.1038/ismej.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz T, Schumacher U, Prante C, Sextro W, Prehm P. Cystic fibrosis transmembrane conductance regulator can export hyaluronan. Pathobiology. 2010;77:200–209. doi: 10.1159/000295859. [DOI] [PubMed] [Google Scholar]
- Shak S, Capon DJ, Hellmiss R, Marsters SA, Baker CL. Recombinant human DNase I reduces the viscosity of cystic fibrosis sputum. Proc Natl Acad Sci U S A. 1990;87:9188–9192. doi: 10.1073/pnas.87.23.9188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si F, Busiek K, Margolin W, Sun SX. Organization of FtsZ filaments in the bacterial division ring measured from polarized fluorescence microscopy. Biophys J. 2013;105:1976–1986. doi: 10.1016/j.bpj.2013.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar S, Green MM, Day LA. DNA-protein interactions as the source of large-length-scale chirality evident in the liquid crystal behavior of filamentous bacteriophages. J Am Chem Soc. 2007;129:3367–3375. doi: 10.1021/ja068498d. [DOI] [PubMed] [Google Scholar]
- Tseng BS, Zhang W, Harrison JJ, Quach TP, Song JL, Penterman J, Singh PK, Chopp DL, Packman AI, Parsek MR. The extracellular matrix protects Pseudomonas aeruginosa biofilms by limiting the penetration of tobramycin. Environ Microbiol. 2013;15:2865–2878. doi: 10.1111/1462-2920.12155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuinier R, Rieger J, de Kruif CG. Depletion-induced phase separation in colloid-polymer mixtures. Adv Colloid Interfac. 2003;103:1–31. doi: 10.1016/S0001-8686(02)00081-7. [DOI] [PubMed] [Google Scholar]
- Walters MC, 3rd, Roe F, Bugnicourt A, Franklin MJ, Stewart PS. Contributions of antibiotic penetration, oxygen limitation, and low metabolic activity to tolerance of Pseudomonas aeruginosa biofilms to ciprofloxacin and tobramycin. Antimicrob Agents Chemother. 2003;47:317–323. doi: 10.1128/AAC.47.1.317-323.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Lau M, Kjelleberg S. Bacteriophage and phenotypic variation in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2004;186:8066–8073. doi: 10.1128/JB.186.23.8066-8073.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb JS, Thompson LS, James S, Charlton T, Tolker-Nielsen T, Koch B, Givskov M, Kjelleberg S. Cell death in Pseudomonas aeruginosa biofilm development. J Bacteriol. 2003;185:4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitchurch CB, Tolker-Nielsen T, Ragas PC, Mattick JS. Extracellular DNA required for bacterial biofilm formation. Science. 2002;295:1487. doi: 10.1126/science.295.5559.1487. [DOI] [PubMed] [Google Scholar]
- Whiteley M, Bangera MG, Bumgarner RE, Parsek MR, Teitzel GM, Lory S, Greenberg EP. Gene expression in Pseudomonas aeruginosa biofilms. Nature. 2001;413:860–864. doi: 10.1038/35101627. [DOI] [PubMed] [Google Scholar]
- Winstanley C, Langille MG, Fothergill JL, Kukavica-Ibrulj I, Paradis-Bleau C, Sanschagrin F, Thomson NR, Winsor GL, Quail MA, Lennard N, et al. Newly introduced genomic prophage islands are critical determinants of in vivo competitiveness in the Liverpool Epidemic Strain of Pseudomonas aeruginosa. Genome Res. 2009;19:12–23. doi: 10.1101/gr.086082.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wnorowska U, Niemirowicz K, Myint M, Diamond SL, Wroblewska M, Savage PB, Janmey PA, Bucki R. Bactericidal activity of cathelicidin LL-37 and select cationic lipids against the hypervirulent P. aeruginosa strain LESB58. Antimicrob Agents Chemother. 2015 doi: 10.1128/AAC.00421-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang YS, Barry E, Dogic Z, Hagan MF. Self-assembly of 2D membranes from mixtures of hard rods and depleting polymers. Soft Matter. 2012;8:707–714. doi: 10.1039/C1SM06201H. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeung AT, Torfs EC, Jamshidi F, Bains M, Wiegand I, Hancock RE, Overhage J. Swarming of Pseudomonas aeruginosa is controlled by a broad spectrum of transcriptional regulators, including MetR. J Bacteriol. 2009;191:5592–5602. doi: 10.1128/JB.00157-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.