Summary
Although many distinct mutations in a variety of genes are known to cause Amyotrophic Lateral Sclerosis (ALS), it remains poorly understood how they selectively impact motor neuron biology and whether they converge on common pathways to cause neuronal degeneration. Here, we have combined reprogramming and stem cell differentiation approaches with genome engineering and RNA sequencing to define the transcriptional and functional changes that are induced in human motor neurons by mutant SOD1. Mutant SOD1 protein induced a transcriptional signature indicative of increased oxidative stress, reduced mitochondrial function, altered sub-cellular transport as well as activation of the ER stress and unfolded protein response pathways. Functional studies demonstrated that these pathways were perturbed in a manner dependent on the SOD1 mutation. Finally, interrogation of stem cell-derived motor neurons produced from ALS patients harboring a repeat expansion in C9orf72 indicates at least a subset of these changes are more broadly conserved in ALS.
Introduction
ALS is a fatal neurological condition characterized by death of motor neurons (MNs) (Hardiman et al., 2011). Classical linkage studies and DNA sequencing approaches have demonstrated that ALS can be caused by a variety of mutations in more than two dozen genes acting on diverse cellular functions (Sreedharan and Brown, 2013). Mutations in SUPEROXIDE DISMUTASE 1 (SOD1) were originally identified through their autosomal dominant inheritance pattern (Rosen et al., 1993). More recently, genome wide association studies, DNA sequencing efforts and linkage analysis have all contributed to the identification of a hexanucleotide repeat expansion at C9orf72 as a cause of ALS in an ample fraction of both familial and sporadic cases (DeJesus-Hernandez et al., 2011; Renton et al., 2011; Robberecht and Philips, 2013).
The discovery of SOD1 mutations led to widely studied transgenic rodent models of ALS (Gurney et al., 1994; Howland et al., 2002). While indisputably valuable, these animals as well as many cell-based models (Oh et al., 2008; Wada et al., 2012), overexpress heterologous human SOD1 at super-physiological levels. Therefore, it is generally accepted that findings from these systems carry the caveat that they could be artifacts of protein overexpression (Gladman et al., 2012). Moreover, there is little information on how SOD1 impacts human MNs, leaving open to what extent results from the SOD1 rodent models are relevant to understanding ALS in the human nervous system.
In addition, identification of patient mutations in other genes, such as TDP43 (Sreedharan et al., 2008) and C9orf72 (DeJesus-Hernandez et al., 2011; Renton et al., 2011) have not yet translated into the creation of animal models that are as widely accepted as SOD1 transgenic mice (Sreedharan and Brown, 2013). As a result, understanding whether mutant SOD1-induced changes in MNs are relevant to other genetic types of ALS, has been slow to develop. Identification of shared mechanisms of MN disease could inform the selection of pathways for therapeutic intervention with the greatest relevance to a broader patient population.
We and others have proposed that induced pluripotent stem cells (iPSCs) from ALS patients and their differentiation into spinal MNs (Dimos et al., 2008) could complement existing animal models, allowing hypotheses to be tested in human MNs with the patients' unique genetic constellation (Bilican et al., 2012; Donnelly et al., 2013; Egawa et al., 2012; Sareen et al., 2013).
Here, we have combined reprogramming and stem cell differentiation approaches with genome engineering and RNA sequencing technologies to identify the transcriptional and functional changes induced by the SOD1A4V mutation in human MNs. In addition to supporting hypotheses concerning the actions of mutant SOD1 protein developed using transgenic mouse models, such as the disruption of mitochondrial function and transport, our studies identified novel mechanisms that may contribute to MN dysfunction. Notably, we found that mutant SOD1 disrupts a delicate balance between ER stress and neuronal excitability that is inherent to MNs. Finally, studies using iPSCs derived from patients harboring C9orf72 repeat expansions indicate that at least a subset of the changes induced by mutant SOD1 in human MNs are relevant to both forms of ALS.
Results
Generation of iPSCs and Functional Motor Neurons from SOD1+/A4V ALS Patients
We derived skin fibroblasts from two female ALS patients (study participants 39 and RB9) carrying the same dominantly acting SOD1A4V mutation (SOD1+/A4V). We then generated iPSCs via retroviral transduction of OCT4, SOX2 and KLF4 and validated their integrity through a battery of standard pluripotency assays (Table S1). We then obtained differentiated spinal MNs through modest modifications to a previously reported protocol (Figure 1A, Figure S1A) (Boulting et al., 2011), which resulted in highly neuralized cultures (TUJ1+>97%), with significant percentage of ISL+ and HB9+ postmitotic MNs (Figures S1-2). Our MN cultures were electrophysiologically active (Figure S1D-G) and could functionally integrate into the developing chick spinal cord (Figure S2D-E).
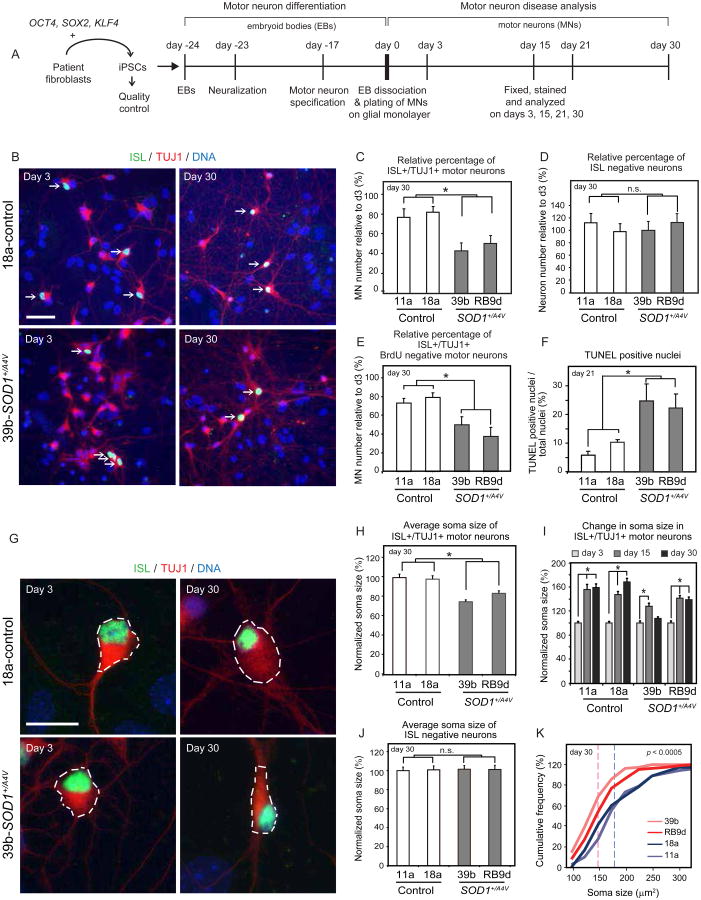
Figure 1. iPSC-Derived Motor Neurons From SOD1+/A4V ALS Patients Exhibit Survival and Morphometric Differences Relative to Healthy Controls.
(A) Experimental outline: patient fibroblasts were reprogrammed to generate iPSCs, which were differentiated into MNs and assessed for ALS-related phenotypes. (B) Neuronal cultures on glial monolayers 3 and 30 days post-differentiation from control and SOD1+/A4V iPSCs. MNs co-stained for ISL and TUJ1 are indicated by white arrows (scale bar=50μm). (C) Quantifications of ISL positive MNs (n=3, m>8000, +/-SEM, P<0.05), and (D) ISL negative neurons (n=3, m>25000, +/-SEM, P<0.05), after 30 days in culture. (E) Quantifications of MN numbers that are BrdU negative after 30 days in culture (n=1, m>3600, +/-SD, P<0.05). Differential motor neurogenesis does not explain the lower numbers in SOD1+/A4V cases. (F) Quantifications of TUNEL positive nuclei of neuronal cultures without glia after 21 days in culture, (n=2, m>13000, +/-SD, P<0.05). (G) Representative images of measured soma size (white-dotted circumference) of control and SOD1+/A4V MNs, (scale bar=20μm). (H) Quantifications of ISL positive MN soma size, values normalized to controls (n=3, m=340, +/-SEM, P<0.01). (I) MN soma size after 3, 15 and 30 days in culture normalized to day 3 for each cell line. Although MNs increase in size over time in all 4 cell lines, they do less so in SOD1+/A4V cases. (J) Quantifications of ISL negative neuron soma size, values normalized to controls (n=3, m=446, +/-SEM, P: n.s.). (K) Cumulative frequency graphs of MN soma size after 30 days in culture. Dotted lines indicate averages for control and disease. n.s: not significant; n=experiment, m=cell number.
Increased Apoptosis and Altered Morphometry in SOD1+/A4V Motor Neurons
To ask whether SOD1+/A4V MNs manifest a phenotype under standard culture conditions we compared them with MNs produced in parallel from two control iPSC lines (11a, 18a), which were similar in their neuronal differentiation capacity, reprogramming method and donor age (Table S1). Differentiated preparations were plated on primary glial monolayers and the total number of ISL/TUJ1+ MNs was assessed after 3, 15 and 30 days in culture (Figure 1A-B). We found that in comparison to the number of MNs present in cultures made from each line at day 3, there was a trend towards a reduced number of SOD1+/A4V MNs at 15 days (Figure S3A), which was further reduced and became statistically significant at day 30 (Figure 1C). Interestingly, we found no significant difference in the number of ISL-negative, TUJ1+, presumptive non-MNs between cases and controls at day 30 (Figure 1D).
To determine whether the selective decline in SOD1+/A4V MN numbers was due to differential proliferation, we monitored progenitor activity via long-term BrdU incorporation (Figure S4). After chronic BrdU administration to cultures from day 0 to day 30, we found that only 2-3% of MNs were labeled (BrdU+, ISL+, TUJ1+) and that this modest rate of labeling was similar in both genotypes (Figure S4D-F). To confirm the effective labeling of progenitors, we administered BrdU from day -17 of directed differentiation until day 0, finding that the vast majority of MNs (>95%) were BrdU+ (Figure S4A-C). These results suggest that the vast majority of MNs in our cultures were postmitotic prior to “day 0” and that the reduced SOD1+/A4V MN numbers were not due to a decrease in progenitor activity. Importantly, when we specifically quantified only the MNs born before day 0, we again found that significantly more SOD1+/A4V MNs were lost over the 30 days in culture (Figure 1E). In addition, at day 21, cultures from the two SOD1+/A4V cases exhibited an increase in TUNEL+ cells in compared to the two controls (Figure 1F), indicating an increased apoptotic rate. When we cultured control and patient derived MNs on primary glia from the SOD1G93A mouse model, we found that both genotypes of MNs survived significantly less than on control glia (Figure S3B). However, survival of SOD1+/A4V MNs was even lower than controls, suggesting a strong cell-autonomous contribution to the effects we observed.
The higher rate of apoptosis in SOD1+/A4V MNs was accompanied by altered morphological characteristics similar to those seen in patients and in the SOD1G93A mouse model (Kiernan and Hudson, 1991; McIlwain, 1991). In particular, 30-day old SOD1+/A4V MNs but not ISL-negative neurons, showed a significant reduction in relative soma size and fewer, as well as shorter processes compared to controls (Figure 1G-K and Figure S3C-F). MNs larger than 150μm2 made up less than 40% of the total MN population in SOD1+/A4V cases, compared to ∼65% for controls (Figure 1K).
Gene Targeting and Correction of the SOD1A4V Mutation
To determine whether the phenotypes we observed were dependent on the SOD1A4V allele, we used a two-step, zinc-finger nuclease (ZFN)-mediated gene targeting strategy to correct the mutation in iPSC line 39b (Figure 2A, methods). Gene-targeting and correction of SOD1A4V were confirmed through sequencing of a single PCR product amplified with primers outside the 5′ and 3′ homology arms, as well as through the elimination of a PshA1 RFLP in the SOD1 cDNA, which is specific to the mutant allele (Figure 2B-C and Figure S5A). Quantitative PCR demonstrated that the corrected 39b-SOD1+/+ line did not harbor multiple insertions of the targeting vector, and that SOD1 transcript levels were unchanged between the parental 39b-SOD1+/A4V and corrected cell lines (Figure 2D-E). We next examined SOD1 protein levels and found that while the intermediate 39b-SOD1+/- line, which expresses only one allele, exhibited approximately half of the SOD1 levels of the parental 39b-SOD1+/A4V cell line, correcting the mutation restored SOD1 protein levels (Figure 2F). As expected, based on the lower protein stability of the SOD1A4V variant (Cardoso et al., 2002), correction of the SOD1 mutant allele resulted in a modest increase in SOD1 levels relative to the parental cells.
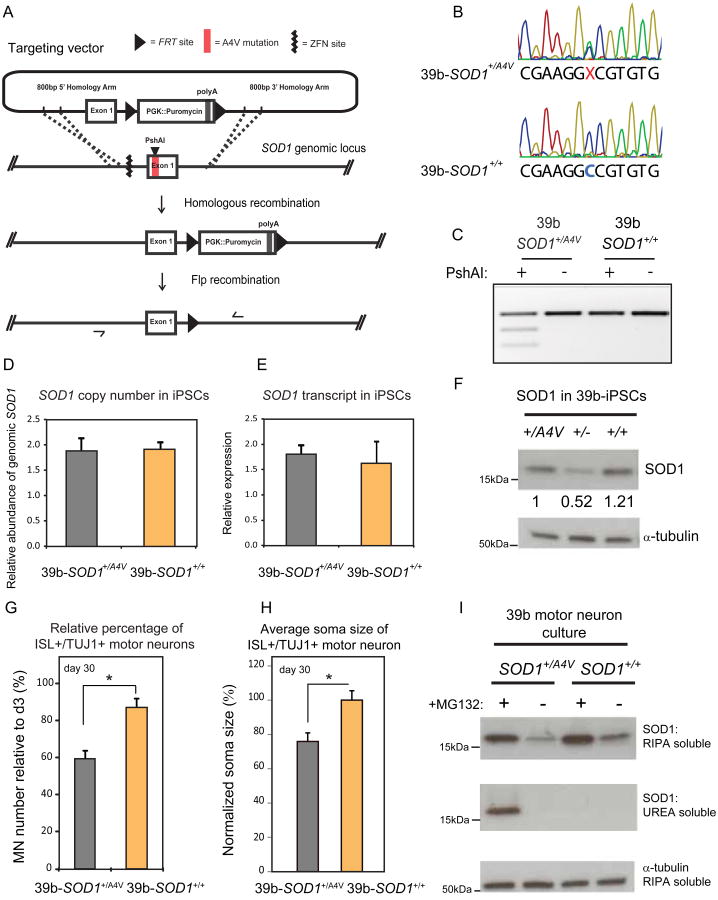
Figure 2. Genetic Correction of the SOD1A4V Mutant Allele Rescues Motor Neuron Survival and Soma Size Deficits.
(A) Gene targeting strategy used to generate isogenic 39b-SOD1+/+ iPSC line. Nucleases targeting the SOD1 locus created a double strand break upstream of Exon1; homologous recombination of the genomic locus with a targeting plasmid with control sequence of Exon1 coupled to PGK∷Puromycin replaced the SOD1A4V mutant allele; after antibiotic selection, the resistance cassette was removed by Flp-mediated recombination, leaving only an FRT footprint.; ZFN: Zinc Finger Nuclease, FRT: Flippase Recognition Target. (B) Sequencing chromatograms of Exon1 of SOD1 in iPSC line 39b before and after targeting, demonstrate the correction of the A4V mutation. (C) PshAI restriction digestion of amplified SOD1 cDNA before and after gene targeting. The mutation creates a PshAI restriction site that is absent in the corrected line. (D) qPCR of genomic SOD1 shows that there are no extra copies of the gene in the corrected line and (E) qRT-PCR of cDNA shows that the expression levels of SOD1 are the same in the corrected line (n=3, +/-SEM). (F) SOD1 protein levels assessed by western blot (WB) analysis in parental (39b-SOD1+/A4V), targeted hemizygous knockout (39b-SOD1+/-) and corrected (39b-SOD1+/+) iPSC clones. (G) Isogenic 39b-SOD1+/+ MNs exhibit increased cell survival (n=6, m>13000, +/-SEM, P<0.05) and (H) soma size (n=3, m=280, +/-SEM, P<0.01). (I) WB analysis of SOD1 protein in detergent- soluble (RIPA) and detergent-insoluble (UREA) fractions in day 12 neuronal cultures. Insoluble SOD1 is not detected under normal conditions. After proteasome inhibition by MG132 treatment, insoluble SOD1 selectively accumulates only in SOD1+/A4V MNs and not in the corrected line.
Given that undesired genetic variations could potentially arise as a result of ZFN off-target activity or clonal expansion, we carefully evaluated the genomic sequence and integrity of the parental and edited cell lines. We carried out whole genome sequencing for the 39b-SOD1+/A4V (33.4× coverage) and 39b-SOD1+/+(31.1× coverage) cell lines, and determined that they closely matched each other throughout the genome for sequencing read depth (Figure S5B), excluding the possibility that large-scale copy-number variations (CNVs) had arisen during passaging, genome editing, or clonal expansion. This lack of large-scale CNVs was validated using a lower resolution Nanostring nCounter Karyotyping Panel (Figure S5C). Moreover, the top 12,000 genomic loci with sequence similarity to the binding site of the ZFN pair we used, did not deviate between the two lines, indicating highly specific nuclease activity.
We also compared the fine-scale (SNP and indel) sequence calls between the two cell lines across their genomes. Overall, we found the corrected line to be free of such events. However, these analyses were sufficiently sensitive to identify a likely mitotic recombination event on the q arm of chr12 (from 108Mb to the end of the chromosome). Deeper analysis of this region demonstrated that the event had not induced novel or rare protein-coding variants. Neither had it induced coding variants associated with any known disease state, suggesting this event was likely to be phenotypically neutral. Analysis of the coding sequences of 26 genes implicated in ALS (Sreedharan and Brown, 2013), identified a single protein-altering difference, which corresponded to the genome edit of the A4V variant in SOD1.
Following the correction of the mutation, we used the 39b-SOD1+/A4V and 39b-SOD1+/A4V lines to ask whether the SOD1A4V-encoding variant was necessary for the MN survival and soma size phenotypes observed in the earlier patient-control comparisons. Importantly, correction of the mutation resulted in a significant rescue of both MN survival and soma size (Figure 2G-H).
Solubility of Mutant SOD1 in Motor Neuron Cultures
SOD1 protein variants that cause ALS are prone to unfolding, misfolding and ultimately aggregation (Pasinelli and Brown, 2006). The pair of isogenic iPSC lines we developed allowed us to address the state of SOD1 in human MNs expressing physiological levels of this protein. Using immunoblotting assays on detergent-soluble and UREA fractions obtained from SOD1+/+ and SOD1+/A4V MN cultures, we found no evidence of insoluble SOD1 protein under basal culture conditions (Figure 2I). Treatment with the proteasome inhibitor MG132, which increased soluble SOD1 levels 2-4 fold, resulted in the accumulation of detergent- insoluble SOD1 only in SOD1+/A4V neurons but not in isogenic controls (Figure 2I). In agreement with the biochemical analysis, aggregation of SOD1 was observed by immunocytochemistry only after treatment with MG132 and selectively in SOD1+/A4V MNs (Figure S6 C-E). Although we cannot rule out the possibility that undetectable levels of aggregated SOD1 protein were the cause of the degeneration of SOD1+/A4V MNs (Figure 1-2), our findings are consistent with recent claims that soluble mutant SOD1 can have substantial phenotypic effects (Brotherton et al., 2012).
RNA sequencing of Purified SOD1+/A4V and Isogenic Control Motor Neurons
In order to gain insight into the molecular pathways affected by the SOD1A4V mutation in human MNs, we performed RNA sequencing. As in earlier experiments, we differentiated 39b-SOD1+/A4V and isogenic control iPSCs and plated them on a glial monolayer (Figure 3A). We next transduced these cultures with an Hb9∷RFP lentiviral reporter (Marchetto et al., 2008) and used FACS to purify MNs after 15 days of additional culture. We reasoned that at on day 15 we might identify the transcriptional changes that predisposed mature MNs to apoptosis, as at this stage MNs were physiologically active (Figure S1 D-G), but there was only a trend towards reduced MN survival in SOD1+/A4V cultures (Figure S3A).
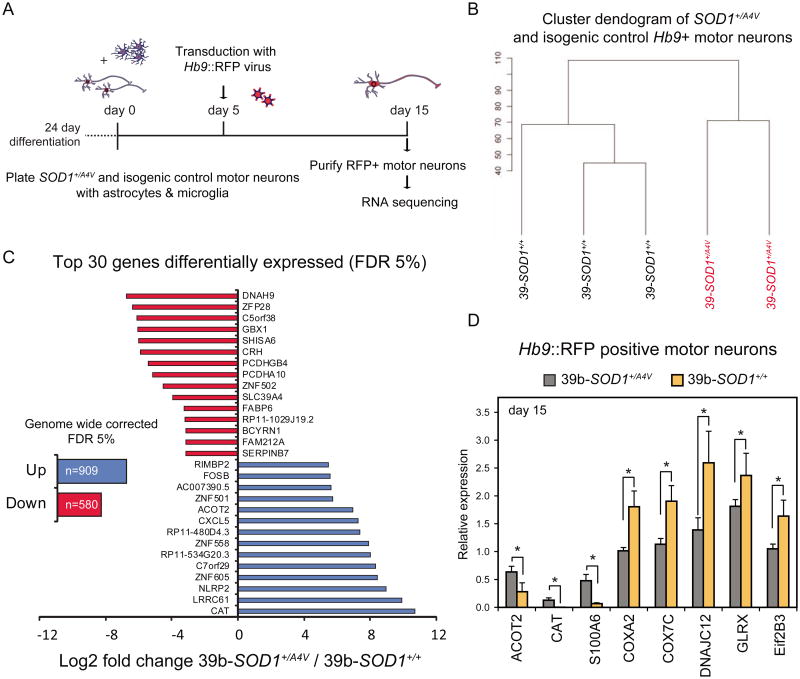
Figure 3. Transcriptional Analysis of 39b-SOD1+/A4V and Isogenic Control Motor Neurons by RNA-Seq Reveals Genotypic Regulatory Changes.
(A) Experimental outline: SOD1+/A4V and isogenic control MN cultures were co-cultured with primary glial cells, transduced with an Hb9∷RFP lentivirus and FACS-purified on day 15 for RNA isolation and sequencing. (B) Dendogram demonstrating clear genotypic clustering based on transcriptional changes measured by RNA sequencing. (C) Total number of genes and top 30 genes (based on fold change) misregulated in SOD1+/A4V MNs with an FDR 5%. (D) qRT-PCR validation of misregulated genes in independent samples (n=3, +/-SEM).
Unsupervised hierarchical clustering segregated samples based on their SOD1 genotype, suggesting that effects of the mutant allele were driving measurable transcriptional differences between SOD1+/A4V and SOD1+/+ MNs (Figure 3B). As a measure of the transcripts most affected by the SOD1A4V mutation we determined the identity of the 30 transcripts most increased and decreased in abundance at a false discovery rate (FDR) of 5% (Figure 3C, Table S2). Analysis of a representative subset of these genes by qRT-PCR in independent experiments validated our findings (Figure 3D). To understand whether the changes found by RNA-seq were specific to MNs and not present in cell types less affected in ALS we determined the expression of a subset of differentially expressed transcripts in the parental 39b and 39b corrected iPSCs (Figure S6A). Of 22 genes analyzed, only 19% were found to be differentially expressed in both iPSCs and MNs. Moreover, RNA-seq analysis on fibroblast cultures isolated from 5 healthy control individuals and the two ALS patients harboring the SOD1A4V mutation, failed to segregate transcriptomes based on genotype after unsupervised hierarchical clustering (Figure S6B). Importantly, a number of genes we identified to be misregulated in SOD1+/A4V MNs have not previously been implicated as being modulated by mutant SOD1 (Table S2).
Ontology of Transcripts Modulated in SOD1+/A4V Motor Neurons
In order to probe the RNA-seq data for biological meaning we utilized two bioinformatic tools that query for enriched gene ontology terms (Table S3). We first performed gene-annotation enrichment analysis with DAVID (Huang da et al., 2009), using all the genes that were significantly altered (909 upregulated and 580 downregulated) in SOD1+/A4V MNs at a FDR of 5% (Table S3A-B). A total of 27 and 65 gene terms were enriched when increased and reduced transcripts were considered respectively. Transcripts implicated in ‘cytoskeleton’ organization, where amongst the most significantly induced in SOD1+/A4V MNs, consistent with the morphological alterations that we observed in these cells relative to isogenic controls (Figure 2H). Transcripts involved in ‘transcriptional regulation’ and ‘motor proteins’, were also induced as a result of the SOD1 mutation. Amongst the significantly decreased transcripts in SOD1+/A4V MNs, there was a very strong enrichment for genes implicated in mitochondrial function and structure. In particular, 60% of all downregulated ontology terms were related to mitochondria, while genes implicated in ‘protein translation’ were also repressed (Table S3B).
As an alternative approach for querying our RNA-seq data, we performed Gene Set Enrichment Analysis (GSEA) (Mootha et al., 2003; Subramanian et al., 2005). GSEA identified 16 gene sets that were significantly induced in SOD1+/A4V MNs (NES<1.5; Table S3C). Amongst these gene sets, were the motor proteins ‘kinesins’. GSEA also identified 100 gene sets to be significantly repressed in SOD1+/A4V MNs (NES<1.5), and notably, gene sets associated with mitochondrial function and protein translation were again amongst the most significantly suppressed (Table S3D).
SOD1+/A4V Motor Neurons Exhibit Disturbances in Mitochondrial Morphology and Motility
To determine whether the transcriptional changes in mitochondrial genes that we identified by RNA-seq in SOD1+/A4V MNs (Table S2-3) were indicative of actual disturbances to mitochondria, we initially performed electron microscopy (EM) studies (Figure 4B). Whereas mitochondrial morphology was normal in MNs derived from a control cell line (18a), mitochondria in SOD1+/A4V MNs (39b and RB9d) were commonly deranged and more vacuolar in appearance. These differences were mostly apparent in neuronal processes. We concluded that distortion in mitochondrial morphology was mediated by expression of the SOD1A4V mutant allele as correction of the mutation eliminated this phenotype (Figure 4B). To further validate mitochondrial damage in SOD1+/A4V MNs, we used immunoblotting assays to quantify the levels of two mitochondrial proteins, SDHA, which is encoded in the nucleus, and MT-COX1, encoded by mitochondrial DNA. We found that correction of the SOD1A4V allele in 39b-SOD1+/+ MNs increased the protein levels of both SDHA and MT-COX1, relative to the parental 39b-SOD1+/A4V MNs (Figure 4C).
Figure 4. SOD1+/A4V Motor Neurons Exhibit Mitochondrial Defects That are Rescued by Genetic Correction of the SOD1A4V Mutation.
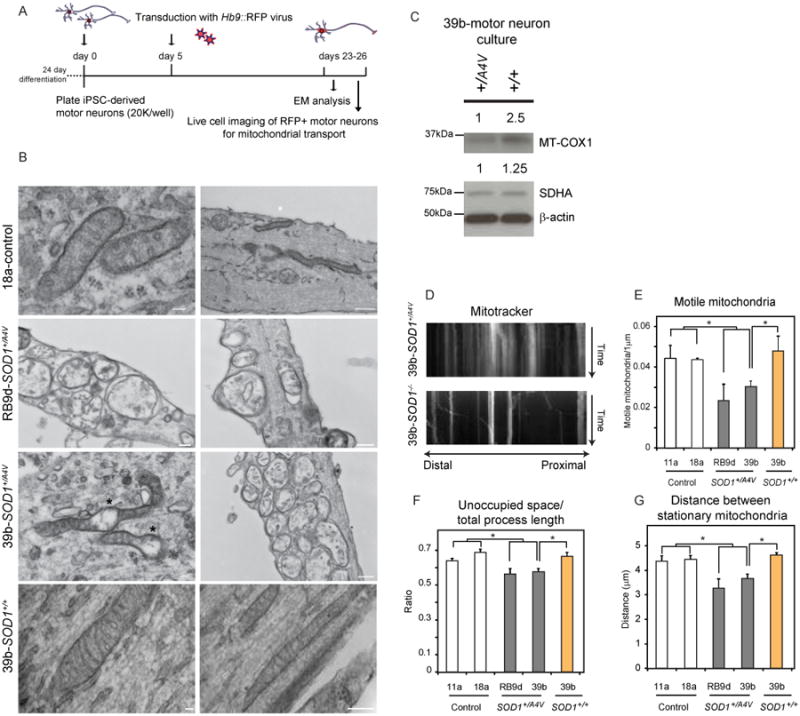
(A) Experimental outline: SOD1+/A4V and isogenic control MN cultures were plated at low densities, transduced with an Hb9∷RFP lentivirus on day 5 and analyzed on days 23-26. (B) Electron microscopy analysis demonstrating that in contrast to control neurons (top panel), SOD1+/A4V neurons (middle panels) often exhibited swelling, morphological disorganization and clustering in neurites. Mitochondria in genetically corrected neurons (bottom panel) exhibited normal morphological characteristics. Representative images out of 3 independent experiments are shown. (C) WB analysis of mitochondrial proteins. (D) Representative kymographs of MitoTracker-stained mitochondria in MNs after live cell imaging. Distal and proximal orientation relative to cell body is indicated. SOD1+/A4V MNs exhibit mitochondria transportation deficiencies relative to controls with (E) fewer motile mitochondria, (F) less space unoccupied by mitochondria relative to axon length and (G) shorter distances between stationary mitochondria (n=4, +/-SEM, P<0.05).
We next sought to measure the movements of mitochondria within the axons of our in vitro-derived MNs (Figure 4D-G). MN cultures differentiated from control and SOD1+/A4V iPSCs were co-labeled with Hb9∷RFP and MitoTracker-Green, which selectively stained mitochondria. We then carried out live cell imaging to register mitochondrial movement along MN processes over the course of 5 minutes, and generated time/distance kymographs for further analysis (Figure 4D). We found that the A4V mutation resulted in a significant decrease in the number of motile mitochondria (Figure 4E). This was coupled to an increase in mitochondrial density in processes, as measured by the shorter distance between stationary mitochondria and the significantly smaller amount of space unoccupied by these organelles (Figure 4F, G).
SOD1+/A4V Motor Neurons Exhibit Signatures of an Unfolded Protein Response (UPR) and ER Stress Induction
RNA-seq analysis showed that 93% of all transcripts involved in protein translation were reduced in SOD1+/A4V MNs (Figure 5A). Translational inhibition is a well-established hallmark of ER stress and the UPR, which is activated upon accumulation of misfolded proteins (Trusina et al., 2008). While the UPR is initially cytoprotective, its persistent activation can lead to apoptosis. In one of the branches of the pathway, pEIF2α leads to a global attenuation of translation and to selective translation of ATF4, while in another branch, IRE1 cleaves the mRNA of XBP1, creating its active spliced form (sXBP1) (Figure 5B) (Ron and Walter, 2007). The transcription factors, sXBP1 and ATF4 modulate expression of downstream effectors including chaperone proteins. Importantly, RNA-seq analysis identified the heat-shock proteins DNAJC12 and HSBP1, the prefoldin subunits PFDN2 and PFDN5, as well as the chaperonin subunits CCT4 and CCT7 as being differentially expressed in SOD1+/A4V MNs (Table S2). In addition, SOD1+/A4V MNs exhibited increased levels of pEIF2α (Figure 5C), as well as significantly elevated sXBP1 transcript (Figure 5D) relative to isogenic controls, consistent with an active UPR.
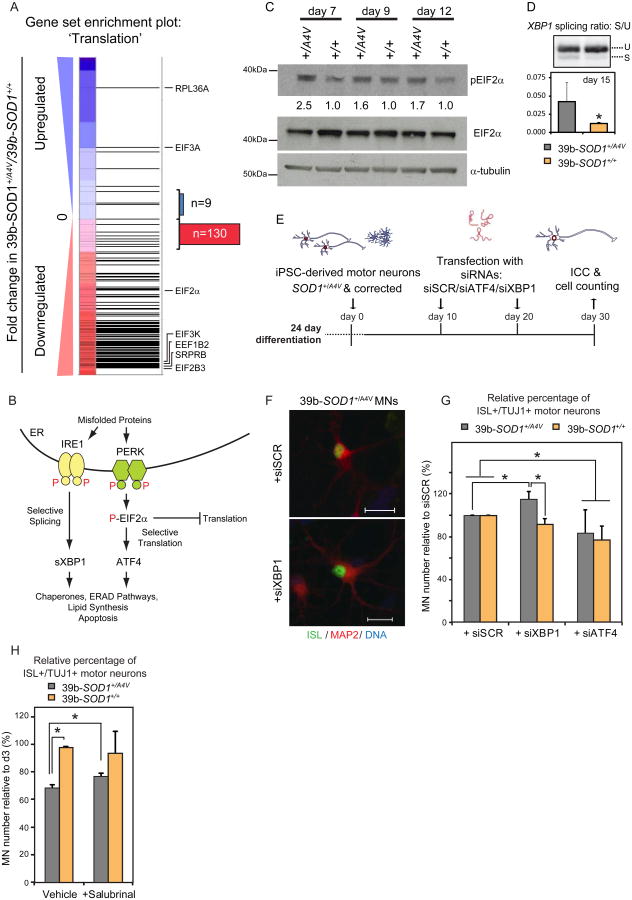
Figure 5. SOD1+/A4V Motor Neurons Exhibit Signatures of an Unfolded Protein Response and are Selectively Vulnerable to ER Stress Induction.
(A) Gene set enrichment analysis of transcriptional changes in SOD1+/A4V MNs shows strong downregulation (normalized enrichment score: -3.31) for genes involved in translational capacity. Horizontal black bars represent individual genes with representative examples indicated. Blue or red represents genes which were up or downregulated respectively in SOD1+/A4V relative to isogenic control MNs. Out of 139 genes detected that are annotated as involved in regulation of translation, 130 were downregulated, consistent with activation of the UPR pathway. (B) Diagram illustrating the canonical unfolded protein response. (C) WB analysis demonstrates increased levels of phosphorylated EIF2α, a marker for activation of the UPR pathway, in SOD1+/A4V MN cultures. Percentage relative to control samples for each time point is shown. (D) SOD1+/A4V MNs exhibit increased levels of XBP1 splicing, a marker of ER stress. RNA was isolated from purified Hb9∷RFP MNs after 15 days in culture (n=3, +/-SEM, P<0.05). U: unspliced, S: spliced. (E) Experimental strategy used to assess the contribution of XBP1 and ATF4 in the survival of SOD1+/A4V MNs. (F) Representative images of untreated and treated MN cultures are shown. (G) SOD1+/A4V MN numbers selectively increase after XBP1 knockdown, while ATF4 knockdown significantly decreases numbers in both control and SOD1+/A4V cases (n=3, m>9800 and m>8500, P<0.05), +/-SEM, P<0.05). (H) Salubrinal has a modest but positive effect on survival of SOD 1+/A4V MNs (n=2, m>10000, +/-SEM, P<0.05).
To test if ER stress directly contributed to mutant SOD1-mediated toxicity in our culture system, we genetically manipulated the two UPR branches using siRNA knockdown of XBP1 and ATF4 and assessed the effect on MN survival (Figure 5E, Figure S7B). Knockdown of XBP1 resulted in a modest but significant increase in the number of SOD1+/A4V MNs after 30 days in culture (Figure 5F-G). In contrast, there was a trend for reduced MN numbers in the isogenic controls, suggesting that XBP1 might provide a protective function in this context. Knockdown of ATF4 depressed survival of both SOD1+/A4V and SOD1+/+ MNs, implying that this protein plays a protective role in both contexts (Figure 5F). Given that reducing ATF4 levels was detrimental to the survival of MNs we asked whether a further induction of pEIF2α would confer MN protection. Salubrinal is a selective inhibitor of phosphatases, which dephosphorylate pEIF2α (Boyce et al., 2005), and has previously been shown to extend the survival of the SOD1G93A mouse model (Saxena et al., 2009). To identify the optimal concentration for these experiments, we performed dose response treatments on FACS purified MNs and analyzed the expression of known UPR genes by qRT-PCR, leading to the selection of a 15μM concentration (Figure S6G). Treatment of cultures from d15-30 led to a modest but significant increase in the numbers of SOD1+/A4V MNs, whereas controls were not affected (Figure 5H).
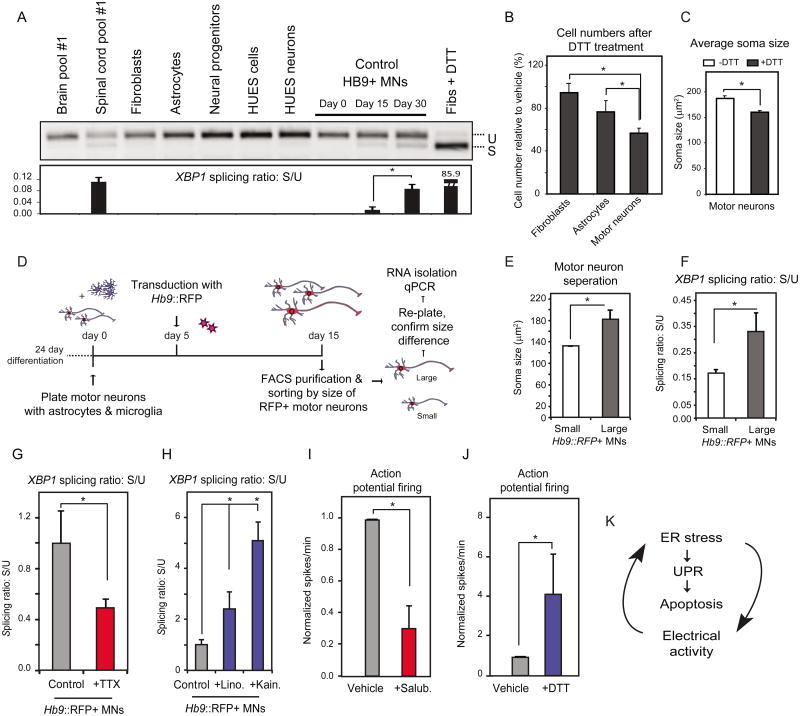
ER Stress is Inherent in Human Motor Neurons and Spinal Cord
Although the SOD1A4V mutation increased the levels of pEIF2α and of spliced XBP1, we noted that control MNs also expressed markers of an UPR, albeit at a lower level. This observation led us to hypothesize that MNs could be more sensitive to unfolded proteins and ER stress because the presence of these liabilities is a constitutive aspect of this neuronal sub-type's inherent biology. To test this, we compared the levels of XBP1 splicing in purified, control HUES3 Hb9∷GFP+ MNs (Di Giorgio et al., 2008) with levels found in a range of other cell types including astrocytes, embryonic stem cells (ESCs), neural progenitors, fibroblasts, Hb9∷GFP-negative neurons and ESC-derived anterior neurons. Of all the cell types examined, only the control MNs displayed detectable levels of sXBP1 that increased as MNs matured in culture (Figure 6A, S7A). When we compared the levels of sXBP1 in RNA isolated from human brain and spinal cord (n=2; each replicate was pooled RNA from 22, non-overlapping, healthy controls), we strikingly saw little or no evidence in the brain, while in the spinal cord there was a marked and significant accumulation of the spliced transcript (Figure 6A, S7A).
Figure 6. Human Motor Neurons Exhibit Increased Levels of Basal ER Stress That is Dependent on Their Physiological Activity.
(A) Purified Hb9∷GFP control MNs show higher levels of spliced XBP1 relative to other human cell types. Human spinal cord RNA also shows higher levels relative to brain RNA. U: unspliced, S: spliced. (B) Control MNs are more vulnerable to acute ER stress induction (DTT, 2mM) relative to fibroblasts or astrocytes, while (C) DTT treatment leads to a reduction in MN soma size (n=2, +/-SD, P<0.05). (D) Experimental strategy used to isolate MNs based on cell size. Control MN cultures were infected with the Hb9∷RFP virus on day 5. On day 15, RFP+ MNs were FACS-purified and sorted by size to separate small and large cells. (E) A subset of cells were re-plated to confirm soma size by measuring MAP2+ cell bodies (F). Basal levels of XBP1 splicing were assessed in RNA isolated from the remaining purified MNs showing that larger MNs had higher spliced XBP1 than smaller ones (n=3, +/-SEM, P<0.05). Treatment of MN cultures with (G) TTX reduces while (H) linopiridine and kainate increase XBP1 splicing respectively. Treatment with (I) salubrinal reduces, while (J) DTT increases action potential firing (n=3, +/-SEM, P<0.05). (K) ER stress, the UPR and electrical activity of MNs are connected.
We wondered whether this ongoing activation of the UPR pathway was associated with an increased sensitivity to ER-stress inducing agents such as DTT. Control MNs, indeed exhibited greater susceptibility to DTT administration than astrocytes and fibroblasts (Figure 6B). When we analyzed the area of the soma of MNs after treatment we found that the average soma size decreased substantially (Figure 6C), which given the acute nature of the treatment seemed most consistent with death of the largest MNs. These observations prompted us to investigate whether MN size correlated with basal ER stress levels. To address this, we purified Hb9∷RFP+ MNs and separated large and small populations (Figure 6D). We validated this approach by re-plating a subset of the purified MNs and measuring their soma size (Figure 6E). We then isolated RNA and examined the levels of sXBP1 as an indicator of ER stress. Larger MNs showed significantly higher levels of sXBP1 than smaller ones (Figure 6F) suggesting that an increased constitutive ER stress may contribute to their increased vulnerability in our cell-culture model.
Inherent ER Stress in Human Motor Neurons is Dependent on Their Electrical Activity
Wainger and colleagues have found that the SOD1+/A4V human MNs that we report here are hyper-excitable in comparison to controls (Wainger et al., 2014). Given that the XBP1 splicing levels increased as MNs matured in culture and became excitable (Figure 6A), we reasoned that a relationship might exist between the inherent ER stress we found in MNs and their electrophysiological activity. Treatment of MN cultures with sufficient tetrodotoxin (TTX) to effectively block action potentials (Figure S6C-D) significantly reduced spliced XBP1 (Figure 6G). Reciprocally, treatment of cultures with the glutamatergic agonist kainate, which depolarizes MNs, led to a significant increase in spliced XBP1 (Figure 6H). In addition, linopiridine, a compound that blocks Kv7 voltage gated potassium channels (Brown and Passmore, 2009), and increased MN activity (Figure S7D), also increased XBP1 splicing (Figure 6H).
We next addressed the inverse question and assessed whether manipulating ER stress would affect the electrical activity of MNs. Treatment with salubrinal, resulted in a relative reduction in the number of spikes per minute (Figure 6I). Conversely, an acute treatment of MN cultures with DTT, which robustly induced sXBP1 (Figure S6F), resulted in an increase in the number of spikes per minute (Figure 6J). These two data sets suggest that ER stress, the UPR and the physiological activity of human MNs are interconnected and that alterations in one of these pathways can affect the other (Figure 6K).
A Subset of Transcriptional Changes in SOD1+/A4V are Shared in C9orf72 MNs
A central question in the ALS field is whether mutations in the diverse causative genes converge on shared molecular pathways. To begin to address this question we focused on the most prevalent genetic ALS type and selected two familial patients (19, RB8) that carried GGGGCC repeat expansions in the C9orf72 locus. We generated iPSCs, confirmed the presence of the expansion in both the parental fibroblasts as well as in multiple passages of the resulting iPSC lines and demonstrated the ability of these lines to differentiate into ISL/HB9+ MNs (Figure 7A-C, Table S1). To determine whether these MNs exhibited transcriptional disturbances similar to the ones that we identified in SOD1+/A4V MNs, we simultaneously differentiated them with six iPSC lines originating from five healthy individuals (Boulting et al., 2011) and FACS-purified Hb9∷RFP+ MNs in multiple biological replicates.
Figure 7. Transcriptional Changes Detected in SOD1+/A4V Motor Neurons are Partially Conserved in Motor Neurons With C9orf72 Expansions.
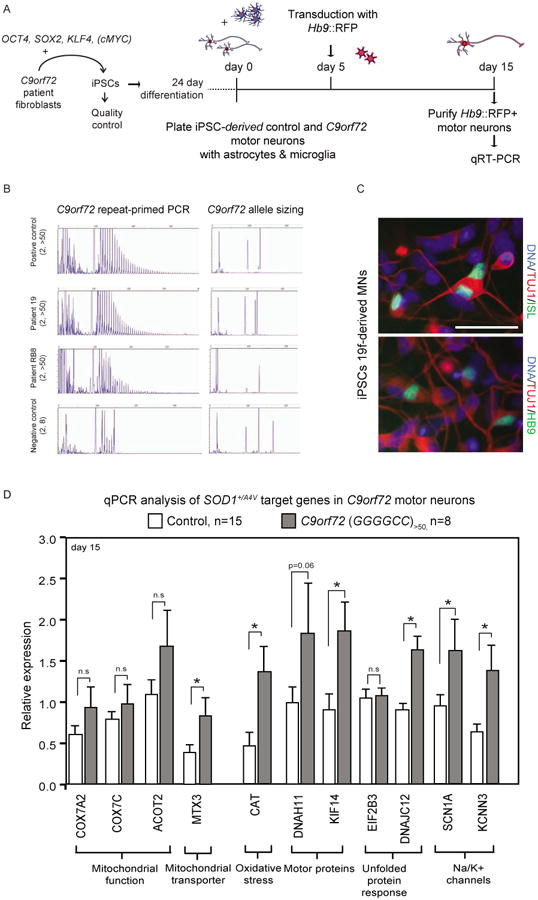
(A) Experimental outline: ALS patient fibroblasts harboring C9orf72 repeat expansions were reprogrammed to generate iPSCs, which were differentiated into MNs and plated in co-culture with glial cells. On day 5 cultures were infected with an Hb9∷RFP lentivirus and on day 15, RFP-labeled MNs were FACS-purified for RNA isolation. (B) Genotyping of samples by repeat-primed PCR and allele sizing for C9orf72.Patient samples RB8 and 19 exhibited more than 50 GGGGCC repeats. (C) Representative images of MN cultures differentiated from C9orf72 ALS patients (scale bar=50μm). (D) A subset of transcripts found to be differentially expressed in SOD1+/A4V MNs also exhibited significant transcriptional changes in MNs derived from C9orf72 (n=2 lines, n=8 replicates, +/-SEM, P<0.05) compared to a large number of control samples (n=6 lines, n=15 replicates, +/-SEM, P<0.05).
Using qRT-PCR we interrogated a subset of transcripts representative of pathways or cellular functions, which we had found to be altered by the SOD1A4V mutation (Figure 7D). Interestingly, in C9orf72 mutant MNs, we did not detect a significant change in the transcript levels of genes implicated in electron transport in mitochondria, but we did detect a significant change in levels of the mitochondrial transporter MTX3. We also found a significant induction of catalase (CAT), indicative of oxidative stress. To determine whether intracellular transport might be impacted, we examined the two most highly upregulated motor proteins in SOD1+/A4V MNs and found that expression of the kinesin KIF14 was significantly induced in C9orf72 MNs. Finally, we analyzed the levels of 4 additional transcripts (DNAJC12, EIF2B3, SCN1A, KCNN3) to determine whether there was evidence for an unfolded protein response and/or shared changes in the expression of channels that might be associated with changes in physiological activity. Of these transcripts, 3 of 4, including both cation channels and the protein chaperone DNAJC12 were transcribed at significantly different levels between control and C9orf72 MNs, supporting the notion that at least a subset of changes are shared between MNs of these two types.
Recently, Donnelly et. al., reported transcriptional changes occurring in human iPSC-differentiated neurons from patients carrying either a C9orf72 repeat expansion or an SOD1+/D90A mutant allele (Donnelly et al., 2013). We compared the transcriptional profile of our SOD1+/A4V mutant iPSC-derived MNs to these data sets by intersecting the respective gene lists. Of the 1,489 differentially expressed genes discovered in our SOD1+/A4V MNs, 357 were found to be misregulated in SOD1+/D90A neurons and 79 in the C9orf72 expansion lines. This is consistent with the interpretation that C9orf72 repeat expansions and SOD1 mutations result in distinct but partially overlapping changes in transcript abundance.
Discussion
Transgenic rodent models of ALS have been indispensable for developing hypotheses concerning how mutant SOD1 proteins induce MN degeneration. However, studies in these animals have yet to yield an effective treatment (Gladman et al., 2012). We reasoned that the gulf between successful animal studies and more positive clinical outcomes might be bridged with model systems that enable hypotheses originating from animals to be validated and extended in the context of human MNs bearing patient mutations. Utilizing iPSCs, genome editing and directed differentiation, we established a well-controlled cell culture system and interrogated the differential properties of patient-derived and healthy control MNs. Advances in gene targeting tools, which we have employed here, allowed our studies of the SOD1A4V variant to be elevated beyond correlative distinctions between ALS cases and controls to the demonstration of causal connections between this mutation and transcriptional as well as functional phenotypes.
Notably, our studies demonstrate that the SOD1A4V missense mutation was necessary to cause a pro-apoptotic phenotype in cultured human MNs, restricting their long-term survival. By employing RNA sequencing, we defined the transcriptional differences between human SOD1+/A4V and isogenic control MNs. Curating these data supported the view that patient-specific ALS iPSC-derived MNs display hallmarks of disease found in both patients and in animal models. We identified defects in mitochondrial transport and morphology, oxidative and ER-related stress and an activated UPR, all of which were dependent on the presence of the SOD1A4V mutation. Importantly, we also identified novel candidate genes, previously unstudied in the context of ALS, which were significantly affected by this disease-causing mutation. These newly identified genes will require further investigation as they may represent potential therapeutic targets. Our functional validation of the pathways in which these genes act suggest they will serve as an invaluable resource for many future studies of ALS.
We also found that other cell and neural-types were relatively unaffected by the SOD1 mutation. As is observed in ALS patients, our molecular and pharmacological studies suggest that human MNs were more susceptible to mutant SOD1. We propose that this susceptibility may originate from a pre-existing burden of ER stress that we found to be constitutively present in healthy, physiologically active MNs, but absent from a variety of other cell-types. Interestingly, this inherent ER stress positively correlated with MN size, drawing parallels to the fact that the largest α-MNs are the most vulnerable to degeneration in ALS patients (Kiernan and Hudson, 1991). It has previously been proposed that combinations of stressors may converge and reinforce each other leading to dysfunction and eventual degeneration of vulnerable neurons (Saxena and Caroni, 2011). We have found that the UPR, ER stress and electrical activity of MNs appear interconnected. Therefore distinct categories of compounds may be of substantial therapeutic benefit to ALS patients; those that support folding of proteins generally, those that specifically aid MNs in handling ER stress, and finally those that alter MN physiological activity. Our studies with salubrinal as well as those of Wainger and colleagues (Wainger et al., 2014) with retigabine, support this view. It is noteworthy that neither treatment with salubrinal, nor knockdown of XBP1 alone resulted in a complete rescue of the survival deficit, implying that perhaps ER stress is only one of many components that contribute to MN death in our system.
While the function of the C9ORF72 protein remains unknown, the mechanism by which the hexanucleotide repeat expansion predisposes individuals to ALS has been suggested to range from haploinsufficiency to toxic gain-of-function properties of the mutant RNA or protein (Ash et al., 2013; DeJesus-Hernandez et al., 2011; Mori et al., 2013; Renton et al., 2011). Our studies indicate a partial conservation of transcriptional changes between SOD1+/A4V and C9orf72 cases. Of particular note are transcripts reflecting a heightened oxidative stress response, reduced mitochondrial function, as well as changes in cation channels and motor proteins. Our discoveries of transcriptional and functional aberrations particularly in relation to mitochondria and ER stress in these patient-specific MNs could potentially relate to the typically late clinical onset of ALS, as these pathways are known to be involved in ageing. Taken together with the manuscript by Wainger et al. (Wainger et al., 2014), our work validates the utility of iPSCs and genome engineering strategies for probing relationships between the genetic variants responsible for ALS in the MNs that selectively degenerate in this disease.
Experimental Procedures
Cell culture
Stem cells were maintained on Matrigel (BD Biosciences) with mTeSR1 media (Stem Cell Technologies) and passaged by dispase (Gibco, 1mg/mL). All cell cultures were maintained at 37°C, 5% CO2.
Derivation of human fibroblasts and iPSC generation
Fibroblasts were generated from 3mm forearm dermal biopsies following informed consent as described previously (Dimos et al., 2008). Generation of iPSCs was done as reported previously by retroviral transduction of KLF4, SOX2, OCT4 and (cMYC) (Boulting et al., 2011).
Motor neuron differentiation
MNs differentiation was carried out as previously described (Boulting et al., 2011) with a few modifications (see also Figure S1). Briefly, iPSC colonies were dissociated to single cells with accutase and plated in suspension in low-adherence flasks, at a 400K/ml density with 10μM ROCK inhibitor (Sigma) in mTeSR1 media for 24hrs. Embryoid bodies (EBs) were formed and media was gradually diluted (50% on day 3 and 100% on day 4) to KOSR (DMEM/F12, 15% KOSR) between days 1-4 and to a neural induction medium (NIM: DMEM/F12 with L-glutamine, NEAA, Heparin (2μg/ml), N2 supplement (Gibco) for days 5-24. Treatment with small molecules and recombinant proteins was as follows: on d1-d6, 10μM SB431542 (Sigma) + 1μM Dorsmorphin (Stemgent); on d5-d24 10ng/mL BDNF (R&D), 0.4μg/ml ascorbic acid (AA, Sigma), 1μM Retinoic Acid (RA, Sigma) and 1μM Smoothened Agonist 1.3 (SAG 1.3, Calbiochem). At day 24 EBs were dissociated to single cells with Papain/DNase (Worthington Bio) and plated onto lysine/laminin-coated surfaces (BD Biosciences) for relevant experiments.
Motor neuron survival assay
20K differentiated MN cultures were plated on 8-well chamber slides (BD biosciences) containing a confluent monolayer of primary cortical mouse glia. Primary glial preparations from P0-P2 mouse pups were obtained as described previously (Di Giorgio et al., 2008). Co-cultures were maintained in Neurobasal media (NB, Invitrogen), supplemented with B27 and N2 supplement (Gibco), 10ng/mL of each of BDNF, GDNF, CNTF (R&D) and 0.4μg/ml ascorbic acid (Sigma) and fed every 2-3 days. Slides were fixed at various time points, cultures were stained and cell numbers assessed. Whole-well images were quantified in a manner blinded to the genotype and condition of the experiment. Neuronal numbers on day 3 were set as 100% and numbers on subsequent time points were expressed as a percentage of day 3. To evaluate cell death, neuronal cultures were plated without glia on coverslips and live cells were assayed using the In Situ Cell Death Kit (Roche Diagnostics) according to manufacturer's instructions.
Electrophysiology recordings
MNs were plated at 20K cells/cm2 on coverslips, in the presence of primary mouse glia and allowed to mature for 2-4 weeks. MNs were identified by RFP fluorescence, after transduction with the Hb9∷RFP lentivirus (Marchetto et al., 2008). Whole-cell voltage-clamp or current-clamp recordings were made using a Multiclamp 700B (Molecular Devices). Data were digitized with a Digidata 1440A A/D interface and recorded using pCLAMP 10_software (Molecular Devices). For MEA recordings, equal numbers of MN cultures were plated on lysine/laminin coated M768-GLx 12-well plates (Axion BioSystems) at typical densities of 40-80K/well and recorded after approximately 14 days using an Axion Maestro device and analyzed using Axion Integrated Studio software.
RNA preparation, qRT-PCR and RNA-seq
RFP+ MNs for RNA assays were purified by FACS 8-10 days after transduction with Hb9∷RFP lentiviral reporter, following a total of 15 days of co-culture with mouse glia. Total RNA was isolated using Trizol LS (Invitrogen) according to manufacturer's instructions. A total of 300-1000ng was used to synthesize cDNA by reverse transcription according to the iSCRIPT kit (Bio-Rad). qRT-PCR was then performed using SYBR green (Bio-Rad) and the iCycler system (Bio-Rad). Quantitative levels for all genes were normalized to the average levels of 3 housekeeping genes (GAPDH/β-Actin/YWHAZ) and expressed relative to the relevant control samples or the lowest expressing sample in the experiment. For RNA-Seq, libraries were generated from ∼250ng total RNA using the illumina TruSeq RNA kit v2, according to the manufacturer's directions. Libraries were sequenced at the Harvard Bauer Core Sequencing facility on a HiSeq 2000. All FASTQ files were analyzed using FastQC software (v 0.10.1) to confirm that Phred scores were acceptable at all read positions (median Phred score>25 and lower quartile>20). The FASTQ files were aligned to the GRCh37/hg19 reference genome using Tophat (v 2.0.7). Data analysis was performed by DSeq, GEO submission number for RNA-seq is GSE54409.
Mitochondrial transport assays and EM analysis
Hb9∷RFP+ MNs were stained with 50nM MitoTracker® Green FM (Invitrogen) and transferred to a custom observation chamber mounted on the stage of a Nikon Eclipse Ti microscope equipped with an automated stage and In Vivo Scientific incubator. Mitochondrial movements were recorded for 5 minutes with 4-second time-lapse intervals using NIS-Elements (Nikon) using a 63× lens. Kymographs were generated from each video using NIS-Elements Analyzing Software (Nikon). Mitochondria were considered motile if they traveled faster than 0.017 μm/second. For Electron Microscopy analysis, ∼60nm thick sections of MN cultures were fixed with 2.5% glutaraldehyde-2% paraformaldehyde in 0.1M sodium cacodylate buffer (pH 7.4) and maintained at 4°C O/N. Following post-fixing, cells were then embedded in plastic and ∼60nm thick sections were cut, stained with lead citrate and analyzed in a JEOL 1200EX Transmission Electron Microscope. At least 3 independent differentiation experiments were analyzed in each case and pictures were taken by a technician blinded for sample IDs.
XBP1 splicing assay
300ng of RNA was used to generate cDNA. PCR products were analyzed after electrophoresis on a 2% low-melting agarose gel. The ratio of spliced/unspliced bands was quantified using Image J software.
Gene targeting
Zinc finger nucleases (ZFNs) targeting the SOD1 locus were constructed using a modified version of the OPEN method (Maeder et al., 2008) and their nuclease activity was validated in HEK293 cells. For genetic correction of 39b-SOD1+/A4V iPSC line, 2.5 million cells were nucleofected (Amaxa™) with 1 μg of ZFN plasmid and 5μg of targeting plasmid. 48hrs after, puromycin selection was applied for 1 week. Surviving colonies were expanded and PCR was used to confirm proper targeting. To remove the puromycin cassette from the intermediate SOD1+/- cells, 2.5 million cells were nucleofected with 5μg of a mammalian expression plasmid containing the FLp recombinase. Sequencing of the genomic DNA was used to confirm removal of the puromycin cassette. Copy number qPCR was performed as described previously (D'Haene et al., 2010) to rule out random integration events.
Genome sequencing and analysis
DNA samples were obtained from the parental 39b-SOD1+/A4V cell line and the gene corrected clone using phenol chloroform extraction. The sequencing libraries were made with 50ng of genomic DNA using the Illumina Nexterra DNA kit. Deep (30×) WGS was performed using the Illumina HiSeq 2500 Platform (500 bp library, 101 bp reads). All subsequent alignments and analysis were performed with hg19 as a reference. To investigate whether there were changes in copy number we used Genome STRiP (Handsaker et al., NG, 2011). To look for regions of copy number change, we evaluated the ratio of normalized read depth in the derived cell line compared to the parental cell line in each window. To find rare coding SNPs in ALS genes, we annotated coding variants called by Haplotype caller with SNPeff (Abecasis et al., 2012). SNPs classified as missense, silent, or nonsense were retained. We then integrated allele frequencies for the European population from the thousand genomes project (Cingolani et al., 2012). Variants were selected that overlapped target genes for ALS. To find variants that differed between cell lines, we compared the genotypes of both lines in a stringent manner similar to the methodologies used to discover de novo mutations. To examine the off-target effects of the designed nuclease, variants within the top 12,000 potential off-target nuclease cut sites were selected from this filtered set of confident variants.
Immunocytochemistry
Cell cultures were fixed in 4% PFA for 15 minutes at 4°C, permeabilized with 0.2% Triton-X in PBS for 45 minutes and blocked with 10% donkey serum in PBS-T (Triton 0.1%). Cells were then incubated in primary antibody overnight and in secondary antibodies for 1 hour in 2% donkey serum in PBS-T after several washes in between. See supplemental methods for a full list of antibodies.
Western Blot assays
For analysis of Phospho-eIF2α protein, cells were lysed in RIPA buffer with protease and phosphatase inhibitors (Roche). 20μg of protein were separated by SDS-PAGE and transferred to nitrocellulose membranes. Anti-Phospho-eIF2α (#3597, Cell Signaling Technology), anti-α–Tubulin (abcam, ab4074) and anti-eIF2α (Cell Signaling Technology, #9722) antibodies were used. For SOD1 protein, detergent-soluble (RIPA buffer) and detergent-insoluble (UREA buffer) fractions were obtained and 5μg of protein samples were separated by SDS-PAGE, transferred to PDVF membranes and probed with anti-SOD1 (Agrisera #AS09 540) and anti-α–Tubulin (Sigma Aldrich # T6199). For mitochondrial biogenesis analysis, 6μg of protein samples were analyzed using the MitoBiogenesis™ Western Blot Cocktail (ab123545).
Statistical analysis
Statistical significance was assessed by a standard Student's T test (1 tail & 2 tail); P<0.05 was considered significant. Two-tailed, unpaired tests were used except to confirm specific hypotheses, in which case one-tailed, unpaired tests were used.
Supplementary Material
Highlights.
iPSC-derived motor neurons harboring SOD1 mutations exhibit cell survival deficits
Genetic correction rescues ALS-related phenotypes
RNA-seq reveals novel expression changes, mitochondrial and ER stress disturbances
MNs exhibit inherent ER stress linked to electrical activity
Acknowledgments
We thank H. Mitsumoto and D. McKenna-Yasek for performing skin biopsies. K. Koszka for maintaining mice; all members of the Eggan Lab, C. Henderson and MW. Amoroso for helpful comments on the manuscript. RHB acknowledges generous support from the ALS Therapy Alliance, Project ALS, P2ALS, the Angel Fund, the Pierre L. de Bourgknecht ALS Research Foundation, the Al-Athel ALS Research Foundation, the ALS Family Charitable Foundation and the NIH/NINDS (1R01NS050557 and NINDS ARRA Award RC2-NS070-342). This work was funded by Target ALS, Project A.L.S., P2ALS, NINDS GO grant (5RC2NS069395-02), NINDS R24 (1U24NS078736-01), HHMI and NIH Director's Pioneer Award DP1 (OD006862). E.K. is a Charles A. King Trust Postdoctoral fellow; K.E. is an HHMI early career scientist.
Footnotes
Supplemental Information: Supplemental Information for this article includes 7 figures and 3 tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash PE, Bieniek KF, Gendron TF, Caulfield T, Lin WL, Dejesus-Hernandez M, van Blitterswijk MM, Jansen-West K, Paul JW, 3rd, Rademakers R, et al. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilican B, Serio A, Barmada SJ, Nishimura AL, Sullivan GJ, Carrasco M, Phatnani HP, Puddifoot CA, Story D, Fletcher J, et al. Mutant induced pluripotent stem cell lines recapitulate aspects of TDP-43 proteinopathies and reveal cell-specific vulnerability. Proc Natl Acad Sci U S A. 2012;109:5803–5808. doi: 10.1073/pnas.1202922109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulting GL, Kiskinis E, Croft GF, Amoroso MW, Oakley DH, Wainger BJ, Williams DJ, Kahler DJ, Yamaki M, Davidow L, et al. A functionally characterized test set of human induced pluripotent stem cells. Nat Biotechnol. 2011;29:279–286. doi: 10.1038/nbt.1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyce M, Bryant KF, Jousse C, Long K, Harding HP, Scheuner D, Kaufman RJ, Ma D, Coen DM, Ron D, et al. A selective inhibitor of eIF2alpha dephosphorylation protects cells from ER stress. Science. 2005;307:935–939. doi: 10.1126/science.1101902. [DOI] [PubMed] [Google Scholar]
- Brotherton TE, Li Y, Glass JD. Cellular toxicity of mutant SOD1 protein is linked to an easily soluble, non-aggregated form in vitro. Neurobiol Dis. 2012;49C:49–56. doi: 10.1016/j.nbd.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown DA, Passmore GM. Neural KCNQ (Kv7) channels. Br J Pharmacol. 2009;156:1185–1195. doi: 10.1111/j.1476-5381.2009.00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardoso RM, Thayer MM, DiDonato M, Lo TP, Bruns CK, Getzoff ED, Tainer JA. Insights into Lou Gehrig's disease from the structure and instability of the A4V mutant of human Cu,Zn superoxide dismutase. J Mol Biol. 2002;324:247–256. doi: 10.1016/s0022-2836(02)01090-2. [DOI] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Haene B, Vandesompele J, Hellemans J. Accurate and objective copy number profiling using real-time quantitative PCR. Methods. 2010;50:262–270. doi: 10.1016/j.ymeth.2009.12.007. [DOI] [PubMed] [Google Scholar]
- DeJesus-Hernandez M, Mackenzie IR, Boeve BF, Boxer AL, Baker M, Rutherford NJ, Nicholson AM, Finch NA, Flynn H, Adamson J, et al. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Giorgio FP, Boulting GL, Bobrowicz S, Eggan KC. Human embryonic stem cell-derived motor neurons are sensitive to the toxic effect of glial cells carrying an ALS-causing mutation. Cell Stem Cell. 2008;3:637–648. doi: 10.1016/j.stem.2008.09.017. [DOI] [PubMed] [Google Scholar]
- Dimos JT, Rodolfa KT, Niakan KK, Weisenthal LM, Mitsumoto H, Chung W, Croft GF, Saphier G, Leibel R, Goland R, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321:1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
- Donnelly CJ, Zhang PW, Pham JT, Heusler AR, Mistry NA, Vidensky S, Daley EL, Poth EM, Hoover B, Fines DM, et al. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egawa N, Kitaoka S, Tsukita K, Naitoh M, Takahashi K, Yamamoto T, Adachi F, Kondo T, Okita K, Asaka I, et al. Drug screening for ALS using patient-specific induced pluripotent stem cells. Sci Transl Med. 2012;4:145ra104. doi: 10.1126/scitranslmed.3004052. [DOI] [PubMed] [Google Scholar]
- Gladman M, Cudkowicz M, Zinman L. Enhancing clinical trials in neurodegenerative disorders: lessons from amyotrophic lateral sclerosis. Curr Opin Neurol. 2012;25:735–742. doi: 10.1097/WCO.0b013e32835a309d. [DOI] [PubMed] [Google Scholar]
- Gurney ME, Pu H, Chiu AY, Dal Canto MC, Polchow CY, Alexander DD, Caliendo J, Hentati A, Kwon YW, Deng HX, et al. Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science. 1994;264:1772–1775. doi: 10.1126/science.8209258. [DOI] [PubMed] [Google Scholar]
- Hardiman O, van den Berg LH, Kiernan MC. Clinical diagnosis and management of amyotrophic lateral sclerosis. Nat Rev Neurol. 2011;7:639–649. doi: 10.1038/nrneurol.2011.153. [DOI] [PubMed] [Google Scholar]
- Howland DS, Liu J, She Y, Goad B, Maragakis NJ, Kim B, Erickson J, Kulik J, DeVito L, Psaltis G, et al. Focal loss of the glutamate transporter EAAT2 in a transgenic rat model of SOD1 mutant-mediated amyotrophic lateral sclerosis (ALS) Proc Natl Acad Sci U S A. 2002;99:1604–1609. doi: 10.1073/pnas.032539299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Kiernan JA, Hudson AJ. Changes in sizes of cortical and lower motor neurons in amyotrophic lateral sclerosis. Brain. 1991;114(Pt 2):843–853. doi: 10.1093/brain/114.2.843. [DOI] [PubMed] [Google Scholar]
- Maeder ML, Thibodeau-Beganny S, Osiak A, Wright DA, Anthony RM, Eichtinger M, Jiang T, Foley JE, Winfrey RJ, Townsend JA, et al. Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol Cell. 2008;31:294–301. doi: 10.1016/j.molcel.2008.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchetto MC, Muotri AR, Mu Y, Smith AM, Cezar GG, Gage FH. Non-cell-autonomous effect of human SOD1 G37R astrocytes on motor neurons derived from human embryonic stem cells. Cell Stem Cell. 2008;3:649–657. doi: 10.1016/j.stem.2008.10.001. [DOI] [PubMed] [Google Scholar]
- McIlwain DL. Nuclear and cell body size in spinal motor neurons. Adv Neurol. 1991;56:67–74. [PubMed] [Google Scholar]
- Mootha VK, Bunkenborg J, Olsen JV, Hjerrild M, Wisniewski JR, Stahl E, Bolouri MS, Ray HN, Sihag S, Kamal M, et al. Integrated analysis of protein composition, tissue diversity, and gene regulation in mouse mitochondria. Cell. 2003;115:629–640. doi: 10.1016/s0092-8674(03)00926-7. [DOI] [PubMed] [Google Scholar]
- Mori K, Weng SM, Arzberger T, May S, Rentzsch K, Kremmer E, Schmid B, Kretzschmar HA, Cruts M, Van Broeckhoven C, et al. The C9orf72 GGGGCC Repeat Is Translated into Aggregating Dipeptide-Repeat Proteins in FTLD/ALS. Science. 2013 doi: 10.1126/science.1232927. [DOI] [PubMed] [Google Scholar]
- Oh YK, Shin KS, Yuan J, Kang SJ. Superoxide dismutase 1 mutants related to amyotrophic lateral sclerosis induce endoplasmic stress in neuro2a cells. J Neurochem. 2008;104:993–1005. doi: 10.1111/j.1471-4159.2007.05053.x. [DOI] [PubMed] [Google Scholar]
- Pasinelli P, Brown RH. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat Rev Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- Renton AE, Majounie E, Waite A, Simon-Sanchez J, Rollinson S, Gibbs JR, Schymick JC, Laaksovirta H, van Swieten JC, Myllykangas L, et al. A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robberecht W, Philips T. The changing scene of amyotrophic lateral sclerosis. Nat Rev Neurosci. 2013;14:248–264. doi: 10.1038/nrn3430. [DOI] [PubMed] [Google Scholar]
- Ron D, Walter P. Signal integration in the endoplasmic reticulum unfolded protein response. Nat Rev Mol Cell Biol. 2007;8:519–529. doi: 10.1038/nrm2199. [DOI] [PubMed] [Google Scholar]
- Rosen DR, Siddique T, Patterson D, Figlewicz DA, Sapp P, Hentati A, Donaldson D, Goto J, O'Regan JP, Deng HX, et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- Sareen D, O'Rourke JG, Meera P, Muhammad AK, Grant S, Simpkinson M, Bell S, Carmona S, Ornelas L, Sahabian A, et al. Targeting RNA foci in iPSC-derived motor neurons from ALS patients with a C9ORF72 repeat expansion. Sci Transl Med. 2013;5:208ra149. doi: 10.1126/scitranslmed.3007529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saxena S, Cabuy E, Caroni P. A role for motoneuron subtype-selective ER stress in disease manifestations of FALS mice. Nat Neurosci. 2009;12:627–636. doi: 10.1038/nn.2297. [DOI] [PubMed] [Google Scholar]
- Saxena S, Caroni P. Selective neuronal vulnerability in neurodegenerative diseases: from stressor thresholds to degeneration. Neuron. 2011;71:35–48. doi: 10.1016/j.neuron.2011.06.031. [DOI] [PubMed] [Google Scholar]
- Sreedharan J, Blair IP, Tripathi VB, Hu X, Vance C, Rogelj B, Ackerley S, Durnall JC, Williams KL, Buratti E, et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreedharan J, Brown RH., Jr Amyotrophic lateral sclerosis: Problems and prospects. Ann Neurol. 2013 doi: 10.1002/ana.24012. [DOI] [PubMed] [Google Scholar]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trusina A, Papa FR, Tang C. Rationalizing translation attenuation in the network architecture of the unfolded protein response. Proc Natl Acad Sci U S A. 2008;105:20280–20285. doi: 10.1073/pnas.0803476105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wada T, Goparaju SK, Tooi N, Inoue H, Takahashi R, Nakatsuji N, Aiba K. Amyotrophic lateral sclerosis model derived from human embryonic stem cells overexpressing mutant superoxide dismutase 1. Stem Cells Transl Med. 2012;1:396–402. doi: 10.5966/sctm.2011-0061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wainger B, Kiskinis E, Mellin C, Wiskow O, Han S, Sandoe J, Perez N, Williams L, Lee S, Boulting G, et al. Intrinsic Membrane Hyperexcitability of ALS Patient-Derived Motor Neurons. Cell Reports. 2014 doi: 10.1016/j.celrep.2014.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.