Abstract
Polyunsaturated fatty acids (PUFAs) have a major impact on human health. Recent genome-wide association studies (GWAS) have identified several genetic loci that are associated with plasma levels of n-3 and n-6 PUFAs in primarily subjects of European ancestry. However, the relevance of these findings has not been evaluated extensively in other ethnic groups. The primary aim of this study was to evaluate for genetic loci associated with n-3 and n-6 PUFAs and to validate the role of recently identified index loci using data from a Singaporean Chinese population. Using a GWAS approach, we evaluated associations with plasma concentrations of three n-3 PUFAs [alphalinolenic acid (ALA), eicosapentaenoic acid and docosahexaenoic acid], four n-6 PUFAs [linoleic acid (LA), gammalinolenic acid, dihomogammalinolenic acid (DGLA) and arachidonic acid], and estimates of delta-5 desaturase and delta-6 desaturase activities among the participants (N = 1361) of the Singaporean Chinese Health Study. Our results reveal robust genome-wide associations (p value <5 × 10−8) with ALA, all four n-6 PUFAs, and delta-6 desaturase activity at the FADS1/FADS2 locus. We further replicated the associations between common index variants at the NTAN1/PDXDC1 locus and n-6 PUFAs LA and DGLA, and between the JMJD1C locus and n-6 PUFA LA (p value between 0.0490 and 9.88 × 10−4). These associations were independent of dietary intake of PUFAs. In aggregate, we show that genetic loci that influence plasma concentrations of n-3 and n-6 PUFAs are shared across different ethnic groups.
Electronic supplementary material
The online version of this article (doi:10.1007/s12263-015-0502-2) contains supplementary material, which is available to authorized users.
Keywords: Genome-wide association study, Polyunsaturated fatty acids, n-3 Fatty acids, n-6 Fatty acids
Introduction
Dietary changes aimed to reduce the risk of cardiovascular diseases traditionally involve limiting saturated fatty acids intakes, replacing them in part by polyunsaturated fatty acids (PUFAs). These PUFAs are characterized by multiple desaturations in their aliphatic tail and are classified as n-3 or n-6, depending on the position of the first double bond proximate to the methyl end. PUFAs in plasma and cell membranes originate from the diet or are synthesized from the two essential PUFAs, linoleic acid [LA; 18:2n-6] and alphalinolenic acid [ALA; 18:3n-3], through a series of desaturation and elongation processes (Youdim et al. 2000).
The association of dietary and membrane long-chain n-3 PUFAs with a lower risk of cardiovascular disease (Dewailly et al. 2001; Albert et al. 2002; Yokoyama et al. 2007; Wang et al. 2006) as well as non-cardiovascular diseases (Cherubini et al. 2007; Lorente-Cebrián et al. 2013) has been documented in various studies. For example, a diet rich in n-3 PUFAs, especially marine derived n-3 PUFAs—eicosapentaenoic acid (EPA; 20:5n3) and docosahexaenoic acid (DHA; c22:6n3), associated with high seafood intake is thought to be responsible for the lower incidence of heart disease in Inuit populations (Dewailly et al. 2001). An important and yet unanswered question pertains to the importance of n-6 PUFAs on cardiovascular risk. Some studies have suggested that these may convey beneficial health effects (Wu et al. 2014); however, such findings are still controversial (Harris and Shearer 2014). For example, certain n-6 PUFAs, such as LA, can be converted in humans to arachidonic acid [AA; 20:4n-6], and the latter serves as the precursor of important molecules involved in inflammation (e.g., eicosanoids) that is thought to play a role in the atherosclerotic process (Wu et al. 2014).
Data from family and twin studies suggest high heritability of circulating levels of these groups of PUFAs (Shah et al. 2009). Candidate gene studies focusing on desaturase genes cluster, fatty acid desaturase 1 and 2 (FADS1 and FADS2), have demonstrated significant associations with multiple n-3 and n-6 PUFAs (Mathias et al. 2010; Xie and Innis 2008). GWAS studies of n-3 and n-6 plasma levels conducted in an Italian population-based dataset (Tanaka et al. 2009) have found significant associations between rs174537 near FADS1 and levels of eicosadienoic acid (EDA; 20:2n6) and EPA. Another SNP mapped to chromosome 6 in the region encoding an elongase of very long fatty acids 2 (ELOVL2) was also found to be associated with EPA.
Recently, two large-scale meta-analyses of GWAS from the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE, 14–15) were conducted to identify common genetic variants associated with plasma n-3 and n-6 PUFA phenotypes. In the first study, findings showed that common variation in n-3 metabolic pathway genes [FADS1, FADS2 and ELOVL2] and in glucokinase regulator (GCKR) influence plasma phospholipid levels of n-3 PUFAs in these populations of European ancestry (Lemaitre et al. 2011). In the second study the CHARGE consortium confirmed previous findings of the FADS cluster with n-6 PUFA plasma levels and observed five additional loci that influence n-6 PUFA plasma concentrations (Guan et al. 2014).
These recent GWAS, however, have been carried out primarily in populations of European ancestry (especially the recent discoveries for n-6 PUFA associations) and may have missed loci unique to other populations. Furthermore, the significance of these recent findings has not been evaluated extensively in other ethnicities (e.g., East-Asian populations). Therefore, in this study, we performed a GWAS for n-3 and n-6 PUFA plasma levels and sought to evaluate the role of recently identified common index variants in a Singaporean Chinese population.
Materials and methods
Study population
This study utilized data from a case–control GWAS study for myocardial infarction (MI, N = 1540) that was nested within the Singapore Chinese Health Study (SCHS), a prospective cohort study of 63,257 Singaporean Chinese men and women aged 45–74 years living in Singapore between 1993 and 1998 (Hankin et al. 2001; Koh et al. 2011). The cohort study recruited only participants belonging to one of the two major Chinese dialect groups in Singapore, the Hokkiens or the Cantonese, who originated from two contiguous prefectures in southern China.
In-person interviews and phlebotomy were conducted before the onset of disease. Baseline assessment was conducted through a face-to-face structured interview during recruitment. Information was recorded by a trained interviewer using a structured questionnaire that included demographics, medical history, cigarette smoking, alcohol consumption, physical activity and detailed menstrual and reproductive history (women only). Food frequency questionnaires were used to estimate usual diet among the SCHS participants and daily proportions of macronutrients intake including n-3 and n-6 PUFA intakes were estimated (Hankin et al. 2001). Between April 1994 and December 1999, we collected blood and single-void urine specimens from a random 3 % sample of study enrollees. Starting in January 2000, the bio-specimen collection was extended to all surviving cohort members. By April 2005 when all subjects had been contacted, bio-specimens were collected from 32,543 participants, representing an approximately 60 % consent rate. Various components (plasma, red blood cell, serum and white blood cell) of blood were separated and have been stored continuously at −80 °C. Age was defined at the time of blood collection, and sex and ethnicity was recorded as indicated in the participant’s National Registration Identity Card (NRIC).
Cases of myocardial infarction (MI) patients (N = 744) were identified through the Singapore Registry of Births and Deaths and the Hospital Discharge Database, respectively, and who had donated blood for research before suffering MI or death. MI cases were confirmed by review of medical records by a cardiologist and through criteria recommended by the Multi-Ethnic Study of Atherosclerosis (available at: http://www.mesa-nhlbi.org/manuals.aspx) or through the Singapore Myocardial Infarction Registry which uses similar procedures. Controls (N = 796) were participants who were alive and free of coronary heart disease at the time of the diagnosis or death of the index cases and were matched for sex, dialect group, year of birth (±2 years), year of recruitment (±1 year) and date of blood collection (±6 months). Informed consent was obtained from all subjects and approval by the Institutional Review Board of the National University of Singapore was given for this study.
Main outcome variables
Plasma subtypes of n-3 and n-6 PUFAs were measured using GC/MS/MS analysis implemented on the Agilent 7890 GC system (Shanghai, China) equipped with a 7001B QQQ triple quadrupole mass detector (PA, USA). Three plasma n-3 PUFAs [ALA, EPA and DHA] and four n-6 PUFAs [LA, GLA (Gammalinolenic acid; c18:3n6), DGLA (Dihomogammalinolenic acid; c20:3n6) and AA] were available for analysis (Table 1). Delta-5 desaturase (D5D) was estimated as the AA:DGLA ratio and delta-6 desaturase (D6D) as the DGLA:LA ratio (Warensjö et al. 2009) (Table 1).
Table 1.
Demographic, dietary PUFA intake (presented as means and SDs) and fatty acid levels (presented as means of % fatty acid subtype from total plasma fatty acids) in the SCHS study
| SCHS cases | SCHS controls | p value | ||
|---|---|---|---|---|
| N | 717 | 644 | ||
| % Males | 64.5 | 64.0 | ||
| Age | 66.6 (7.80) | 66.2 (7.7) | ||
| Daily n-3 PUFA intake (g/day) | 15.3 (7.6) | 15.1 (7.4) | ||
| Daily n-3 PUFA intake as proportion of total daily energy intake (%) | 8.4 (2.2) | 8.3 (2.0) | ||
| Daily n-6 PUFA intake (g/day) | 8.9 (4.8) | 9.1 (4.9) | ||
| Daily n-6 PUFA intake as proportion of total daily energy intake (%) | 4.9 (1.8) | 5.1 (1.8) | ||
| Plasma n-3 PUFA subtypes | ||||
| c183n3 (%) | Alphalinolenic acid (ALA) | 0.334 | 0.345 | 0.415 |
| c205n3 (%) | Eicosapentaenoic acid (EPA) | 0.443 | 0.522 | <0.001 |
| c226n3 (%) | Docosahexaenoic acid (DHA) | 2.263 | 2.459 | 0.009 |
| Plasma n-6 PUFA subtypes | ||||
| c182n6c (%) | Linoleic acid (LA) | 36.137 | 36.440 | 0.232 |
| c183n6 (%) | Gammalinolenic acid (GLA) | 0.232 | 0.223 | 0.237 |
| c203n6 (%) | Dihomogammalinolenic acid (DGLA) | 0.932 | 0.918 | 0.460 |
| c204n6 (%) | Arachidonic acid (AA) | 7.561 | 7.658 | 0.307 |
Genotyping and imputation
Genome-wide genotyping was performed for SCHS samples using the Illumina HumanOmni ZhongHua-8 Bead Chip. Quality control (QC) procedures of samples and SNPs are detailed in Supplementary Table 1 and 2. Samples with call-rate <98.0 % (N = 8), extremes in heterozygosity (N = 11) and known duplicates (positive controls, N = 42) were excluded from analyses. Correlation between positive controls were >98.0 %. Identity-by-state measures were performed by pair-wise comparison of samples to detect cryptic relatedness (Supplementary Table 1) and one sample from each relationship was excluded from further analysis (N = 16). Principle component analysis (PCA) (Price et al. 2006) together with HapMap reference populations (International HapMap Consortium 2003) was performed to identify possible outliers from reported ethnicity and 18 samples were excluded. 717 MI cases and 644 controls were available for subsequent statistical analysis (Supplementary Table 1).
Details of quality-control measures for SNPs are provided in Supplementary Table 2. Briefly, sex-linked and mitochondrial SNPs were removed, together with gross HWE outliers (p value <1 × 10−5). SNPs that were monomorphic or with a MAF <1.0 % and SNPs with low call-rates (<95.0 %) were excluded. We imputed for additional autosomal SNPs with IMPUTE v2 (Marchini et al. 2007) using a posterior probability of at least 0.90 and a call-rate of at least 95.0 % for all imputed SNPs. Genotype calls were based on HapMap East-Asian samples (CHB and JPT) and imputed SNPs with info score <0.5 were excluded from analyses. 1,972,057 SNPs in the MI cases and 1,980,188 SNPs in the MI controls were available for statistical analyses after QC procedures (Supplementary Table 2).
Statistical analysis
GWAS association analysis for PUFA subtypes
As performed in previous studies, raw PUFA concentrations were converted to proportions of total fatty acids (Lemaitre et al. 2011; Guan et al. 2014). All data were normalized by natural log transformation and truncated to 4 SD from respective means. SNP-based trend tests for PUFA associations as well as D5D and D6D activities were carried out separately in the MI cases and controls. Associations between genetic variants (SNPs) and plasma n-3 or n-6 PUFA concentrations were analyzed in an additive model and adjusted for age, sex, population stratification (the first 3 principal components) and proportion of daily n-3 or n-6 PUFA intake from total calories (derived from food frequency questionnaires) under investigation. These analyses were performed using the genome association toolset, SNPTEST (version 2) (Marchini et al. 2007). Genomic inflation factor (λ) of association results from each analysis was used to evaluate levels of inflation of study results. Individual study results from MI cases and controls were subsequently combined using the inverse variance-weighted meta-analysis, assuming a fixed effects model. Heterogeneity of effects was examined using Cochran’s Q, and a Qp value cut-off <0.05 was used to determine SNPs with between-study heterogeneity (Marchini et al. 2007). All meta-analysis procedures were performed using the meta package in STATA (version 11.2). At loci where genome-wide signals were identified, we further evaluated for additional independent SNPs by repeating all analyses and conditioning SNP associations on the genotypes of the index SNP at that region. For lead SNPs with genome-wide association identified in our study, we estimated the proportion of the respective PUFA variance explained by these SNPs by evaluating adjusted R2 values of regression models before and after inclusion of the lead SNP into the model. Finally, at genome-wide loci, we also evaluated changes in effects after further adjustments for concentrations of the precursor fatty acids.
In silico replication analysis of previously reported candidate SNPs
All recently discovered index plasma n-3 PUFA loci (7 SNPs in or near 3 loci, FADS1, ELOVL2 and GCKR) and index plasma n-6 PUFA loci [5 SNPs in or near 4 loci, FADS1, jumonji domain containing 1C (JMJD1C), N-terminal asparagine amidase (NTAN1) and palmitoyl-protein thioesterase 2 (PPT2)] from recent GWAS studies (Lemaitre et al. 2011; Guan et al. 2014) were selected for in silico replication among the SCHS GWAS data. QUANTO power calculator (Gauderman and Morrison 2006) was used to estimate the power to detect similar associations of index SNPs in the SCHS dataset using reported effect estimates (Lemaitre et al. 2011; Guan et al. 2014) and observed MAF from the SCHS datasets.
To determine significance, consistency of effect direction with previously reported GWAS data was ensured in the SCHS datasets and as these loci have been well-validated [in 8866 subjects of European ancestry (Lemaitre et al. 2011; Guan et al. 2014)], a p value threshold <0.05 was used (Dorajoo et al. 2013; Dorajoo et al. 2012). At loci that failed to replicate, we evaluated associations of SNPs within the recombination block containing the reported index SNP (between 2 recombination peaks of at least 20 cM/Mb) (Dorajoo et al. 2012). These analyses were performed using LocusZoom ver1.1 (Pruim et al. 2010).
Results
Robust genome-wide signals at FADS1/2 locus in chromosome 11 detected for ALA, LA, GLA, DGLA and AA associations and delta-6 desaturase activity in the Singapore Chinese
Supplementary Fig. 1–3 shows QQ plots for all meta-analysis association results of 7 n-3 and n-6 PUFAs and 2 desaturase activities assessed in the study. Genomic inflation in these meta-analysis results was low (λ between 1.004 and 1.019 for n-3 PUFA associations, between 1.004 and 1.006 for n-6 PUFA associations and between 1.002 and 1.005 for desaturase activities) indicating that our QC procedures had effectively excluded aberrant samples and SNPs. Supplementary Tables 3–11 provide association results for SNPs with meta-analysis p value associations <1.0 × 10−4 for the 9 traits evaluated in SCHS.
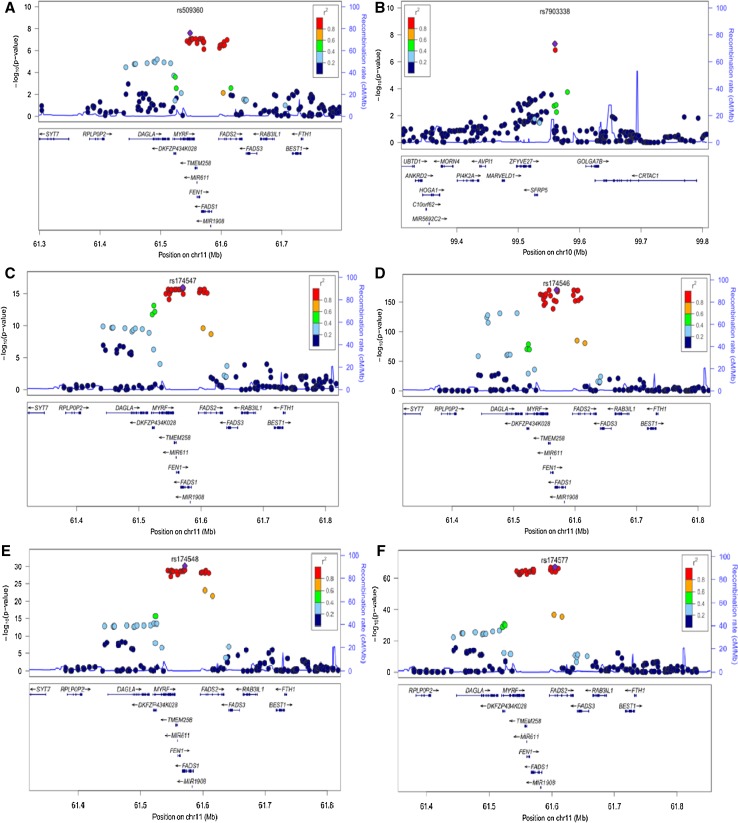
SNPs at the chr11q12.2 locus, in or near the FADS1/FADS2 gene, were detected at genome-wide levels for ALA, LA, GLA, DGLA, AA and D6D activity associations in our Singaporean Chinese dataset (Fig. 1; Table 2). These signals explained between 2.0 % (for ALA) and 36.5 % (for GLA) of the phenotypic variation in PUFA traits assessed. All genome-wide SCHS lead SNPs (SNPs with strongest associations) at the chr11q12.2 locus were either the same index SNP reported in previous European studies for these traits (rs174547) (Lemaitre et al. 2011; Guan et al. 2014) or were in strong LD (r2 between 0.973 and 1.000) with this European index SNP (Table 2). Conditioning analyses on the genotype of the index SNP (rs174547) did not reveal additional independent signals at the chr11q12.2 locus in our study (Supplementary Fig. 4). Although strong genome-wide associations were observed at this locus for D6D, no associations were detected for D5D in the SCHS (Supplementary Fig. 5). Further adjustment of results with preceding plasma PUFA concentrations also did not affect associations of lead SNPs except at rs174548 for DGLA association, where the effect estimate was observed to be in the opposite direction (as compared to the unadjusted study results) (Table 2).
Fig. 1.
Regional plots of SNPs within 500 kb region of lead genome-wide SNP (SNP with strongest association level) for a ALA association in chromosome 11, b ALA association in chromosome 10, c LA association in chromosome 11, d GLA association in chromosome 11, e DGLA association in chromosome 11 and f AA association in chromosome 11. LD (r 2) data of SNPs based on ASN panels of 1000 Genome database. Plots plotted using LocusZoom (http://csg.sph.umich.edu/locuszoom/)
Table 2.
Association results of lead SNPs for various plasma polyunsaturated fatty acids and delta-6 desaturase activity (D6D) detected at genome-wide levels (p value <5 × 10−8) in SCHS
| Outcome variable | Lead SNP | Test allele | TAF | Chr | Position | β | SE | p value | Qp value | Variance explained by lead SNP (%) | r 2 with index FADS1 SNP (rs174757) |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ALA | rs509360 | G | 0.673 | 11 | 61548559 | 0.1087 | 0.0195 | 2.51 × 10−8 | 0.9499 | 2.0 | 0.972 |
| LA | rs174547 | T | 0.338 | 11 | 61570783 | −0.0480 | 0.0056 | 1.12 × 10−16 | 0.6123 | 5.3 | NA |
| GLA | rs174546 | T | 0.662 | 11 | 61569830 | −0.5227 | 0.0187 | 5.18 × 10−171 | 0.5720 | 36.5 | 1.000 |
| GLA adjusted for LA | −0.5312 | 0.0183 | 1.72 × 10−175 | 36.5 | |||||||
| DGLA | rs174548 | G | 0.649 | 11 | 61571348 | −0.1789 | 0.0152 | 6.91 × 10−31 | 0.6884 | 8.4 | 0.973 |
| DGLA adjusted for GLA | 0.1282 | 0.0129 | 5.42 × 10−22 | 7.9 | |||||||
| AA | rs174577 | C | 0.335 | 11 | 61604814 | 0.1542 | 0.0088 | 4.22 × 10−68 | 0.2448 | 18.0 | 0.994 |
| AA adjusted for DGLA | 0.2090 | 0.0118 | 1.58 × 10−68 | 18.6 | |||||||
| D6D | rs174548 | G | 0.649 | 11 | 61571348 | −0.2384 | 0.0181 | 2.26 × 10−38 | 0.7245 | 9.1 | 0.973 |
Association results are from meta-analysis of MI cases and control datasets (N = 1361). The index FADS1 SNP (rs174757) was identified in previous GWAS studies for the same FA subtypes in Lemaitre et al. (2011) and Guan et al. (2014)
Lead SNP: strongest SNP association in SCHS meta-analysis; TAF: test allele frequency; Qp value: Cochran’s Q heterogeneity measure
We also identified a novel SNP at chr10q24.2 for the association with ALA at marginal genome-wide significance levels in SCHS (rs7903338, meta-analysis p value = 4.33 × 10−8, Supplementary Table 3), but this did not replicate in 8866 European subjects from the Cohorts for Heart and Aging Research in Genomic Epidemiology Consortium (CHARGE), (β = −0.0002, p value = 0.825, C allele frequency = 0.743, http://faculty.washington.edu/rozenl/files/CHARGE_N3_ALA.txt).
Replication of index PUFA subtypes SNPs in the Singaporean Chinese
With the SCHS PUFA subtype data, we were able to replicate known index SNP associations at 3 out of 4 n-3 PUFAs (ALA, EPA and DHA) and 4 out of 5 n-6 PUFAs (LA, GLA, DGLA and AA). Tables 3 and 4 details association levels of these recently identified index n-3 and n-6 PUFA SNPs in the SCHS meta-analysis data.
Table 3.
The associations between reported n-3 and n-6 FA subtype index SNPs (Lemaitre et al. 2011; Guan et al. 2014) and n-3 fatty acids (ALA, EPA and DHA) in the meta-analysis data of SCHS
| Index SNPs from reported GWAS studies for omega-3 and omega-6 fatty acid subtypes (Lemaitre et al. 2011; Guan et al. 2014) | SCHS meta-analysis (N = 1361) | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Power (%) | Comment | Test allele | TAF | ALA | EPA | DHA | |||||||||||
| Trait | SNP | Chr | Position | Reported Gene | β | SE | p value | β | SE | p value | β | SE | p value | ||||
| ALA | rs174547 | 11 | 61570783 | FADS1 | 50.42 | Typed | T | 0.3382 | −0.100 | 0.019 | 1.88 × 10 −7* | 0.059 | 0.019 | 0.0017 | 0.007 | 0.022 | 0.7646 |
| EPA | rs3798713 | 6 | 11116608 | ELOVL2 | 40.39 | Imp | G | 0.278 | 0.020 | 0.019 | 0.3086 | 0.004 | 0.019 | 0.8182 | 0.004 | 0.022 | 0.8456 |
| rs174538 | 11 | 61560081 | FADS1 | 99.99 | Typed | G | 0.3419 | −0.099 | 0.019 | 2.15 × 10 −7 | 0.061 | 0.019 | 0.0011* | 0.012 | 0.021 | 0.5866 | |
| rs174535 | 11 | 61551356 | FADS1 | 99.99 | Typed | T | 0.3324 | −0.103 | 0.019 | 9.21 × 10 −8 | 0.060 | 0.019 | 0.0016* | 0.007 | 0.022 | 0.7625 | |
| DPA | rs780094 | 2 | 27741237 | GCKR | NA | Typed | T | 0.4535 | 0.027 | 0.017 | 0.1239 | 0.028 | 0.017 | 0.0983 | 0.062 | 0.020 | 0.0019 |
| rs3734398 | 6 | 10982973 | ELOVL2 | NA | Typed | T | 0.0669 | −0.006 | 0.035 | 0.8640 | −0.027 | 0.034 | 0.4315 | −0.001 | 0.040 | 0.9738 | |
| rs174547 | 11 | 61570783 | FADS1 | NA | Imp | T | 0.3382 | −0.100 | 0.019 | 1.88 × 10 −7 | 0.059 | 0.019 | 0.0017 | 0.007 | 0.022 | 0.7646 | |
| DHA | rs2236212 | 6 | 11103001 | ELOVL2 | 52.23 | Imp | G | 0.282 | −0.004 | 0.018 | 0.8396 | 0.008 | 0.018 | 0.6618 | 0.002 | 0.022 | 0.9392 |
| LA | rs10740118 | 10 | 64771213 | JMJD1C | 15.43 | Imp | G | 0.7048 | 0.005 | 0.019 | 0.7824 | 0.016 | 0.019 | 0.405 | 0.048 | 0.022 | 0.0321 |
| rs174547 | 11 | 61570783 | FADS1 | 99.99 | Typed | T | 0.3382 | −0.100 | 0.019 | 1.88 × 10 −7 | 0.059 | 0.019 | 0.0017 | 0.007 | 0.022 | 0.7646 | |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 32.36 | Typed | G | 0.6054 | −0.008 | 0.018 | 0.6452 | −0.005 | 0.017 | 0.7866 | −0.016 | 0.020 | 0.4231 | |
| GLA | rs174547 | 11 | 61570783 | FADS1 | 58.38 | Typed | T | 0.3382 | −0.100 | 0.019 | 1.88 × 10 −7 | 0.059 | 0.019 | 0.0017 | 0.007 | 0.022 | 0.7646 |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 12.05 | Typed | G | 0.6054 | −0.008 | 0.018 | 0.6452 | −0.005 | 0.017 | 0.7866 | −0.016 | 0.020 | 0.4231 | |
| DGLA | rs174547 | 11 | 61570783 | FADS1 | 99.99 | Typed | T | 0.3382 | −0.100 | 0.019 | 1.88 × 10 −7 | 0.059 | 0.019 | 0.0017 | 0.007 | 0.022 | 0.7646 |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 99.99 | Typed | G | 0.6054 | −0.008 | 0.018 | 0.6452 | −0.005 | 0.017 | 0.7866 | −0.016 | 0.020 | 0.4231 | |
| AA | rs174547 | 11 | 61570783 | FADS1 | 99.99 | Typed | T | 0.3382 | −0.100 | 0.019 | 1.88 × 10 −7 | 0.059 | 0.019 | 0.0017 | 0.007 | 0.022 | 0.7646 |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 60.62 | Typed | G | 0.6054 | −0.008 | 0.018 | 0.6452 | −0.005 | 0.017 | 0.7866 | −0.016 | 0.020 | 0.4231 | |
| Adrenic acid | rs174547 | 11 | 61570783 | FADS1 | NA | Typed | T | 0.3382 | −0.100 | 0.019 | 1.88 × 10 −7 | 0.059 | 0.019 | 0.0017 | 0.007 | 0.022 | 0.7646 |
| rs3134950 | 6 | 32235455 | PPT2 | NA | Imp | C | 0.4111 | −0.036 | 0.018 | 0.0493 | −0.013 | 0.018 | 0.4801 | 0.007 | 0.021 | 0.7354 | |
Significant (p value <0.05) associations indicated in bold
Power: indicates power in the SCHS study to detect reported associations at these SNPs given reported effect estimates (Yokoyama et al. 2007; Wang et al. 2006) and MAF of SCHS dataset; NA: power not calculated for previously reported traits that were not measured in SCHS; TAF: test allele frequency
* Indicates significant association for the same n-3 trait in the SCHS data
Table 4.
The associations between reported n-3 and n-6 FA subtype index SNPs (Lemaitre et al. 2011; Guan et al. 2014) and n-6 fatty acids (LA, GLA, DGLA and AA) in the meta-analysis data from SCHS
| Index SNPs from reported GWAS studies for omega-3 and omega-6 fatty acid subtypes (Lemaitre et al. 2011; Guan et al. 2014) | SCHS meta-analysis (N = 1361) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Power | Comment | Test allele | TAF | LA | ||||||
| Trait | SNP | Chr | Position | Reported gene | β | p value | ||||
| LA | rs10740118 | 10 | 64771213 | JMJD1C | 15.43 | Imp | G | 0.705 | −0.009 | 0.1128 |
| rs174547 | 11 | 61570783 | FADS1 | 99.99 | Typed | T | 0.338 | −0.048 | 1.12 × 10 −16* | |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 32.36 | Typed | G | 0.605 | 0.009 | 0.0498* | |
| GLA | rs174547 | 11 | 61570783 | FADS1 | 58.38 | Typed | T | 0.338 | −0.048 | 1.12 × 10 −16 |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 12.05 | Typed | G | 0.605 | 0.009 | 0.0598 | |
| DGLA | rs174547 | 11 | 61570783 | FADS1 | 99.99 | Typed | T | 0.338 | −0.048 | 1.12 × 10 −16 |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 99.99 | Typed | G | 0.605 | 0.009 | 0.0598 | |
| AA | rs174547 | 11 | 61570783 | FADS1 | 99.99 | Typed | T | 0.338 | −0.048 | 1.12 × 10 −16 |
| rs16966952 | 16 | 15135943 | NTAN1/PDXDC1 | 60.62 | Typed | G | 0.605 | 0.009 | 0.0598 | |
| Adrenic acid | rs174547 | 11 | 61570783 | FADS1 | NA | Typed | T | 0.338 | −0.048 | 1.12 × 10 −16 |
| rs3134950 | 6 | 32235455 | PPT2 | NA | Imp | C | 0.411 | −0.008 | 0.1109 | |
| ALA | rs174547 | 11 | 61570783 | FADS1 | 50.42 | Typed | T | 0.338 | −0.048 | 1.12 × 10 −16 |
| EPA | rs3798713 | 6 | 11116608 | ELOVL2 | 40.39 | Imp | G | 0.278 | 0.005 | 0.3276 |
| rs174538 | 11 | 61560081 | FADS1 | 99.99 | Typed | G | 0.342 | −0.045 | 2.22 × 10 −16 | |
| rs174535 | 11 | 61551356 | FADS1 | 99.99 | Typed | T | 0.332 | −0.047 | 3.99 × 10 −16 | |
| DPA | rs780094 | 2 | 27741237 | GCKR | NA | Typed | T | 0.454 | −0.014 | 0.005 |
| rs3734398 | 6 | 10982973 | ELOVL2 | NA | Typed | T | 0.067 | −0.011 | 0.2521 | |
| rs174547 | 11 | 61570783 | FADS1 | NA | Imp | T | 0.338 | −0.048 | 1.12 × 10 −16 | |
| DHA | rs2236212 | 6 | 11103001 | ELOVL2 | 52.23 | Imp | G | 0.282 | −0.002 | 0.6754 |
| Index SNPs from reported GWAS studies for omega-3 and omega-6 fatty acid subtypes (Lemaitre et al. 2011; Guan et al. 2014) | SCHS meta-analysis (N = 1361) | |||||||
|---|---|---|---|---|---|---|---|---|
| GLA | DGLA | DGLAa | AA | |||||
| Trait | β | p value | β | p value | β | p value | β | p value |
| LA | −0.003 | 0.8861 | −0.022 | 0.1284 | −0.019 | 0.4126 | −0.003 | 0.7314 |
| 0.523 | 1.68 × 10 −170 | 0.177 | 1.06 × 10 −29 | −0.139 | 1.46 × 10 −24 | 0.152 | 3.88 × 10 −66 | |
| 0.008 | 0.7179 | 0.034 | 0.0104 | 0.030 | 9.88 × 10 −4 | −0.004 | 0.6162 | |
| GLA | 0.523 | 1.68 × 10 −170* | 0.177 | 1.06 × 10 −29 | −0.139 | 1.46 × 10 −24 | 0.152 | 3.88 × 10 −66 |
| 0.008 | 0.7179 | 0.034 | 0.0104 | 0.030 | 9.88 × 10 −4 | −0.004 | 0.6162 | |
| DGLA | 0.523 | 1.68 × 10 −170 | 0.177 | 1.06 × 10−29 | −0.139 | 1.46 × 10 −24* | 0.152 | 3.88 × 10 −66 |
| 0.008 | 0.7179 | 0.034 | 0.0104* | 0.030 | 9.88 × 10 −4* | −0.004 | 0.6162 | |
| AA | 0.523 | 1.68 × 10 −170 | 0.177 | 1.06 × 10 −29 | −0.139 | 1.46 × 10 −24 | 0.152 | 3.88 × 10 −66* |
| 0.008 | 0.7179 | 0.034 | 0.0104 | 0.030 | 9.88 × 10 −4 | −0.004 | 0.6162 | |
| Adrenic acid | 0.523 | 1.68 × 10 −170 | 0.177 | 1.06 × 10 −29 | −0.139 | 1.46 × 10 −24 | 0.152 | 3.88 × 10 −66 |
| 0.013 | 0.5676 | 0.007 | 0.6049 | 0.007 | 0.5962 | 0.012 | 0.1767 | |
| ALA | 0.523 | 1.68 × 10 −170 | 0.177 | 1.06 × 10 −29 | −0.139 | 1.46 × 10 −24 | 0.152 | 3.88 × 10 −66 |
| EPA | 0.008 | 0.7389 | 0.006 | 0.665 | 0.006 | 0.9063 | −0.008 | 0.3949 |
| 0.508 | 1.74 × 10 −170 | 0.170 | 2.65 × 10 −29 | −0.132 | 2.51 × 10 −23 | 0.148 | 1.90 × 10 −63 | |
| 0.512 | 2.18 × 10 −170 | 0.176 | 2.79 × 10 −29 | −0.135 | 2.02 × 10 −23 | 0.151 | 1.99 × 10 −64 | |
| DPA | 0.014 | 0.5094 | −0.016 | 0.2143 | −0.023 | 0.011 | −0.012 | 0.1811 |
| −0.026 | 0.5481 | −0.013 | 0.6276 | −0.001 | 0.9573 | 0.002 | 0.9037 | |
| 0.523 | 1.68 × 10 −170 | 0.177 | 1.06 × 10 −29 | −0.139 | 1.46 × 10 −24 | 0.152 | 3.88 × 10 −66 | |
| DHA | 0.001 | 0.9618 | −0.002 | 0.8946 | −0.002 | 0.9925 | −0.005 | 0.6063 |
Significant (p value <0.05) associations indicated in bold
Power indicates power in the SCHS study to detect reported associations at these SNPs given reported effect estimates (Yokoyama et al. 2007; Wang et al. 2006) and MAF of SCHS dataset; NA: power not calculated for previously reported traits that were not measured in SCHS; and TAF: test allele frequency
* Indicates significant association for same n-6 trait in the SCHS data
aDGLA trait adjusted for preceding FA level (GLA) in SCHS as this adjustment showed differences in effect estimates
Index SNPs at the FADS1/FADS2 gene region on chromosome 11 were validated for all PUFA subtype traits available in SCHS (p value between 0.0011 and 1.68 × 10−170, Tables 3 and 4). For n-3 subtypes these FADS1 index SNPs showed opposite effect directions for ALA as compared with EPA (Table 3). For n-6 fatty acids, effect directions of LA and DGLA associations were opposite to that of GLA and AA associations (Table 4). Besides these SNPs, rs16966952 at the NTAN1/PDXDC1 locus was also validated for increased LA and DGLA concentrations (p value between 0.0490 and 9.88 × 10−4, Table 4) in the SCHS. At index SNPs where we did not observe any direct replication for the specific traits they were reported to be associated with, we examined associations for SNPs within the recombination block containing the reported index SNPs (Supplementary Fig. 6). Only at the JMJD1C locus for LA associations, we identified a regional SNP with nominal associations (rs7896783, p value = 0.0098, Supplementary Fig. 6C) in strong LD with the reported index SNP (rs10740118, r2 = 0.82).
Among the set of known index SNPs, we also identified nominal associations for additional PUFA subtypes available in the SCHS. These include increased levels of DHA (T allele, p value = 0.0019) and decreased levels of LA (T allele, p value = 0.0050) at the GCKR index SNP rs780094 (Tables 3, 4), increased levels of DHA (G allele, p value = 0.0321) at the JMJD1C index SNP rs10740118 (Table 3) and decreased levels of ALA (C allele, p value = 0.0493) at the PPT index SNP rs3134950 (Table 3).
Discussion
PUFAs have a major impact on human health (Youdim et al. 2000; Dewailly et al. 2001; Albert et al. 2002; Yokoyama et al. 2007; Wang et al. 2006; Cherubini et al. 2007; Lorente-Cebrián et al. 2013; Wu et al. 2014). The present study details associations of common genetic variants with specific n-3 and n-6 plasma PUFA concentrations in the Singaporean Chinese population. It is known that PUFA plasma concentrations can originate from dietary sources and that dietary choices vary greatly among different ethnic populations (Ogce et al. 2008). For example in East-Asians, where rice is a staple, diet is usually higher in starch and carbohydrates as compared to European populations (Drewnowski and Popkin 1997). In our study we corroborated and further characterized the role of numerous common genetic loci, first identified in European populations among the Singaporean Chinese and suggest that these identified associations were independent of dietary n-3 and n-6 PUFA intakes.
We confirmed the importance of common variants in/near the central desaturase genes, FADS1 and FADS2 on chromosome 11, which showed robust associations with n-3 PUFA (ALA) and n-6 PUFAs (LA, GLA, DGLA and AA) in the Singaporean Chinese. The associations were likely to stem from a single causal locus and conditional probability analyses did not reveal independent associations. It was only for the DGLA association that we observed differences in effect estimates when adjustment for precursor PUFA subtype concentrations was made (GLA concentration). The especially strong associations with GLA at this locus, explaining approximately 36 % of this phenotypic variance, and the known positive correlations between GLA and DGLA concentrations may explain these varied results and it is likely that adjustment with plasma GLA concentrations refined the DGLA genetic association signal observed at the FADS1/FADS2 locus in our study.
Increased delta-6 desaturase activity is associated with an increased plasma GLA:LA ratio. Consistently, the T allele of the rs174547 index SNP in the region of the FADS1 gene was associated with higher GLA and lower LA concentrations among the Singaporean Chinese (Hankin et al. 2001) and showed robust genome-wide associations with increased indirect estimate of delta-6 activity. This indicates the prominent role these desaturation enzymes play in the conversion of the LA essential fatty acid to GLA. The same allele also showed robust associations with higher AA and lower DGLA concentrations in our dataset, as previously reported (Guan et al. 2014), suggesting a higher delta-5 desaturase activity. However, we do not find evidence that the FADS1/2 locus is associated with delta-5 desaturase activity. This lack of association between delta-5 desaturase activity, and the FADS1/2 locus is also consistent with a previous study that analyzed subjects of European ancestry (Bokor et al. 2010). Nevertheless, it is noteworthy that a rare variant (rs968567) in weak LD with the common variants evaluated in our study (r2 = 0.4 between rs968567 and rs174547 in HapMap CEU population; rs968567 not available for HapMap East-Asian reference population) at the FADS2 promoter site may still be associated with delta-5 desaturase (Bokor et al. 2010).
Besides the validations for n-3 and n-6 PUFA at the FADS1/2 locus, we further replicated SNPs at the NTAN1/PDXDC1 on chromosome 13 (for LA and DGLA associations) and the JMJD1C locus on chromosome 10 (for LA associations) for n-6 PUFAs. Importantly, these recent findings for plasma n-6 PUFA subtypes associations have been only evaluated in subjects of European ancestry (Guan et al. 2014). The PDXDC1 locus has been identified in a separate genome-wide study for human serum metabolite concentrations (LA/PUFA ratio) and may be involved in eicosanoid biosynthesis (Kettunen et al. 2012). JMJDC1 encodes a probable histone demethylase and has recently been shown to be involved in hematopoiesis (Soranzo et al. 2009) as well as triglyceride levels (Teslovich et al. 2010). In aggregate, our data suggest that diverse ethnic groups share some degree of genetic predisposition to both n-3 and n-6 PUFA metabolism.
For a few loci, however, we were unable to detect significant associations, namely ELVOL2 SNP associations for EPA and DHA n-3 PUFA concentrations and the NTAN1/PDXDC1 SNP associations for GLA and AA n-6 PUFA concentrations. One possibility is that there is significant genetic heterogeneity at these loci between East-Asians and Europeans. However, index SNPs are unlikely to be causal and differences in allele frequencies and LD between populations may have masked associations. Moreover, due to modest effects of these variants, the sample size of our study may not have been adequately powered to detect all associations. The loci that failed to replicate tended to be those for which we had lower power in our study. Further large-scale studies in non-European populations would be warranted to fully elucidate the role of all common variants involved in n-3 and n-6 PUFA metabolism and to evaluate for possible ethnic specific associations.
One advantage of our study was the availability of a number of n-3 and n-6 PUFAs for the evaluation of possible pleiotropic effects at these established genetic loci. Besides the FADS1/FADS2 locus that was strongly associated with all n-3 and n-6 PUFA concentrations, perhaps of note is the GCKR locus (T allele of rs780094) that has been implicated with DPA concentration (Lemaitre et al. 2011). The T allele of this SNP has been associated with decreased blood glucose and insulin levels as well as with increased blood lipids (Teslovich et al. 2010; Dupuis et al. 2010; Wallace et al. 2008; Coram et al. 2013). We further identified nominal associations with both increased n-3 DHA and decreased n-6 LA concentrations among the Singaporean Chinese. Given the strong associations between DHA and LA subtypes on blood lipid levels, the GCKR locus may potentially play a central role modulating both glucose metabolism and lipogenesis pathways that impact upon plasma n-3 and n-6 PUFA concentrations in varied ways. These finding should be verified in additional large-scale studies, and future functional studies should aim to tease out the true pleiotropic role of GCKR in these metabolic pathways.
Our study is not without limitations and an important one would be the modest sample size of our study population. Thus we were unlikely to be well-powered to identify additional novel associations (especially associations with modest effect sizes) for the traits we analyzed in this Singaporean Chinese population. For ALA association, however, we identified a novel locus at chr10q24.2 at marginal genome-wide levels. This signal, however, did not replicate among 8866 European subjects from the CHARGE consortium and given that the SNP was also common in the European population (MAF = 25.7 %), this finding is likely to be a false positive one due to random statistical fluctuation and highlights the importance to follow-up novel leads with additional independent replications. Nevertheless, at the specific genetic loci for n-3 and n-6 PUFA identified recently, we were primarily adequately powered to detect similar associations identified first in Europeans, among our Singaporean Chinese dataset and this limitation does not impact on the main finding of our study that a proportion of genetic predisposition influencing plasma n-3 and n-6 PUFA concentrations are shared across ethnic groups, at least among European and Chinese ancestry populations.
In conclusion, we firmly corroborated the role of FADS1/FADS2 common variants as determinants of plasma n-3 and n-6 PUFA concentrations and show that this locus is an important determinant of delta-6 desaturase activity in the Singaporean Chinese population. Several additional associations at the NTAN1/PDXDC1 and the JMJD1C loci were also replicated for n-6 PUFA levels for the first time in a non-European ancestry population, suggesting that similar common genetic variants may play a role as primary determinants of plasma PUFA concentrations among Chinese and European ancestry populations.
Electronic supplementary material
Acknowledgments
We thank Siew-Hong Low of the National University of Singapore for supervising the fieldwork and Renwei Wang for the maintenance of the database in the Singapore Chinese Health Study. We also thank the Ministry of Health in Singapore for assistance with the identification of fatal and non-fatal myocardial infarction cases via database linkages. Finally, we acknowledge the founding Principal Investigator of the Singapore Chinese Health Study, Mimi C. Yu. The Singapore Chinese Health Study was supported by the National Institutes of Health, USA (RO1 CA144034 and UM1 CA182876), the nested case–control study of myocardial infarction by the Singapore National Medical Research Council (NMRC 1270/2010) and genotyping by the HUJ-CREATE Programme of the National Research Foundation, Singapore (Project Number 370062002).
Compliance with ethical standards
Conflict of interest
All authors have no conflicts of interest.
Contributor Information
Chew-Kiat Heng, Email: paehck@nus.edu.sg.
Yechiel Friedlander, Phone: +972-2-6777805, Email: yechielf@ekmd.huji.ac.il.
References
- Albert CM, Campos H, Stampfer MJ, Ridker PM, Manson JE, Willett WC, Ma J. Blood levels of long-chain n-3 fatty acids and the risk of sudden death. N Engl J Med. 2002;346(15):1113–1118. doi: 10.1056/NEJMoa012918. [DOI] [PubMed] [Google Scholar]
- Bokor S, Dumont J, Spinneker A, Gonzalez-Gross M, Nova E, Widhalm K, Moschonis G, Stehle P, Amouyel P, De Henauw S, et al. Single nucleotide polymorphisms in the FADS gene cluster are associated with delta-5 and delta-6 desaturase activities estimated by serum fatty acid ratios. J Lipid Res. 2010;51(8):2325–2333. doi: 10.1194/jlr.M006205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherubini A, Andres-Lacueva C, Martin A, Lauretani F, Iorio AD, Bartali B, Corsi A, Bandinelli S, Mattson MP, Ferrucci L. Low plasma N-3 fatty acids and dementia in older persons: the InCHIANTI study. J Gerontol A Biol Sci Med Sci. 2007;62(10):1120–1126. doi: 10.1093/gerona/62.10.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coram MA, Duan Q, Hoffmann TJ, Thornton T, Knowles JW, Johnson NA, Ochs-Balcom HM, Donlon TA, Martin LW, Eaton CB, et al. Genome-wide characterization of shared and distinct genetic components that influence blood lipid levels in ethnically diverse human populations. Am J Hum Genet. 2013;92(6):904–916. doi: 10.1016/j.ajhg.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewailly E, Blanchet C, Lemieux S, Sauve L, Gingras S, Ayotte P, Holub BJ. n-3 Fatty acids and cardiovascular disease risk factors among the Inuit of Nunavik. Am J Clin Nutr. 2001;74:464–473. doi: 10.1093/ajcn/74.4.464. [DOI] [PubMed] [Google Scholar]
- Dorajoo R, Blakemore AI, Sim X, Ong RT, Ng DP, Seielstad M, Wong TY, Saw SM, Froguel P, Liu J, et al. Replication of 13 obesity loci among Singaporean Chinese, Malay and Asian-Indian populations. Int J Obes (Lond) 2012;36(1):159–163. doi: 10.1038/ijo.2011.86. [DOI] [PubMed] [Google Scholar]
- Dorajoo R, Li R, Ikram MK, Liu J, Froguel P, Lee J, Sim X, Ong RT, Tay WT, Peng C, et al. Are C-reactive protein associated genetic variants associated with serum levels and retinal markers of microvascular pathology in Asian populations from Singapore? PLoS One. 2013;8(7):e67650. doi: 10.1371/journal.pone.0067650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drewnowski A, Popkin BM. The nutrition transition: new trends in the global diet. Nutr Rev. 1997;55(2):31–43. doi: 10.1111/j.1753-4887.1997.tb01593.x. [DOI] [PubMed] [Google Scholar]
- Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauderman WJ, Morrison JM (2006) QUANTO 1.1: a computer program for power and sample size calculations for genetic-epidemiology studies. http://hydra.usc.edu/gxe/
- Guan W, Steffen BT, Lemaitre RN, Wu JH, Tanaka T, Manichaikul A, Foy M, Rich SS, Wang L, Nettleton JA, et al. Genome-wide association study of plasma N6 polyunsaturated fatty acids within the cohorts for heart and aging research in genomic epidemiology consortium. Circ Cardiovasc Genet. 2014;7(3):321–331. doi: 10.1161/CIRCGENETICS.113.000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankin JH, Stram DO, Arakawa K, Park S, Low SH, Lee HP, Yu MC. Singapore Chinese Health Study: development, validation, and calibration of the quantitative food frequency questionnaire. Nutr Cancer. 2001;39(2):187–195. doi: 10.1207/S15327914nc392_5. [DOI] [PubMed] [Google Scholar]
- Harris WS, Shearer GC. Omega-6 fatty acids and cardiovascular disease: friend, not foe? Circulation. 2014;130(18):1562–1564. doi: 10.1161/CIRCULATIONAHA.114.012534. [DOI] [PubMed] [Google Scholar]
- International HapMap Consortium The international HapMap project. Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- Kettunen J, Tukiainen T, Sarin AP, Ortega-Alonso A, Tikkanen E, Lyytikäinen LP, Kangas AJ, Soininen P, Würtz P, Silander K, et al. Genome-wide association study identifies multiple loci influencing human serum metabolite levels. Nat Genet. 2012;44(3):269–276. doi: 10.1038/ng.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh WP, Yuan JM, Wang R, Lee YP, Lee BL, Yu MC, Ong CN. Plasma carotenoids and risk of acute myocardial infarction in the Singapore Chinese Health Study. Nutr Metab Cardiovasc Dis. 2011;21:685–690. doi: 10.1016/j.numecd.2009.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre RN, Tanaka T, Tang W, Manichaikul A, Foy M, Kabagambe EK, Nettleton JA, King IB, Weng LC, Bhattacharya S, et al. Genetic loci associated with plasma phospholipid n-3 fatty acids: a meta-analysis of genome-wide association studies from the CHARGE Consortium. PLoS Genet. 2011;7(7):e1002193. doi: 10.1371/journal.pgen.1002193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorente-Cebrián S, Costa AG, Navas-Carretero S, Zabala M, Martínez JA, Moreno-Aliaga MJ. Role of omega-3 fatty acids in obesity, metabolic syndrome, and cardiovascular diseases: a review of the evidence. J Physiol Biochem. 2013;69(3):633–651. doi: 10.1007/s13105-013-0265-4. [DOI] [PubMed] [Google Scholar]
- Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies via imputation of genotypes. Nat Genet. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- Mathias RA, Vergara C, Gao L, Rafaels N, Hand T, Campbell M, Bickel C, Ivester P, Sergeant S, Barnes KC, et al. FADS genetic variants and omega-6 polyunsaturated fatty acid metabolism in a homogeneous island population. J Lipid Res. 2010;51(9):2766–2774. doi: 10.1194/jlr.M008359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogce F, Ceber E, Ekti R, Oran NT. Comparison of mediterranean, Western and Japanese diets and some recommendations. Asian Pac J Cancer Prev. 2008;9(2):351–356. [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38(8):904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- Pruim RJ, Welch RP, Sanna S, Teslovich TM, Chines PS, Gliedt TP, Boehnke M, Abecasis GR, Willer CJ. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26(18):2336–2337. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah SH, Hauser ER, Bain JR, Muehlbauer MJ, Haynes C, Stevens RD, Wenner BR, Dowdy ZE, Granger CB, Ginsburg GS, et al. High heritability of metabolomic profiles in families burdened with premature cardiovascular disease. Mol Syst Biol. 2009;5:258. doi: 10.1038/msb.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soranzo N, Spector TD, Mangino M, Kühnel B, Rendon A, Teumer A, Willenborg C, Wright B, Chen L, Li M, et al. A genome-wide meta-analysis identifies 22 loci associated with eight hematological parameters in the HaemGen consortium. Nat Genet. 2009;41(11):1182–1190. doi: 10.1038/ng.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Shen J, Abecasis GR, Kisialiou A, Ordovas JM, Guralnik JM, Singleton A, Bandinelli S, Cherubini A, Arnett D, et al. Genome-wide association study of plasma polyunsaturated fatty acids in the InCHIANTI Study. PLoS Genet. 2009;5(1):e1000338. doi: 10.1371/journal.pgen.1000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teslovich TM, Musunuru K, Smith AV, Edmondson AC, Stylianou IM, Koseki M, Pirruccello JP, Ripatti S, Chasman DI, Willer CJ, et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature. 2010;466(7307):707–713. doi: 10.1038/nature09270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace C, Newhouse SJ, Braund P, Zhang F, Tobin M, Falchi M, Ahmadi K, Dobson RJ, Marçano AC, et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am J Hum Genet. 2008;82(1):139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C, Harris WS, Chung M, Lichtenstein AH, Balk EM, Kupelnick B, Jordan HS, Lau J. n-3 Fatty acids from fish or fish-oil supplements, but not alpha-linolenic acid, benefit cardiovascular disease outcomes in primary- and secondary-prevention studies: a systematic review. Am J Clin Nutr. 2006;84(1):5–17. doi: 10.1093/ajcn/84.1.5. [DOI] [PubMed] [Google Scholar]
- Warensjö E, Rosell M, Hellenius ML, Vessby B, De Faire U, Risérus U. Associations between estimated fatty acid desaturase activities in serum lipids and adipose tissue in humans: links to obesity and insulin resistance. Lipids Health Dis. 2009;8:37. doi: 10.1186/1476-511X-8-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JH, Lemaitre RN, King IB, Song X, Psaty BM, Siscovick DS, Mozaffarian D. Circulating omega-6 polyunsaturated fatty acids and total and cause-specific mortality: the Cardiovascular Health Study. Circulation. 2014;130(15):1245–1253. doi: 10.1161/CIRCULATIONAHA.114.011590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Innis SM. Genetic variants of the FADS1 FADS2 gene cluster are associated with altered (n-6) and (n-3) essential fatty acids in plasma and erythrocyte phospholipids in women during pregnancy and in breast milk during lactation. J Nutr. 2008;138(11):2222–2228. doi: 10.3945/jn.108.096156. [DOI] [PubMed] [Google Scholar]
- Yokoyama M, Origasa H, Matsuzaki M, Matsuzawa Y, Saito Y, Ishikawa Y, Oikawa S, Sasaki J, Hishida H, Itakura H, et al. Effects of eicosapentaenoic acid on major coronary events in hypercholesterolaemic patients (JELIS): a randomised open-label, blinded endpoint analysis. Lancet. 2007;369(9567):1090–1098. doi: 10.1016/S0140-6736(07)60527-3. [DOI] [PubMed] [Google Scholar]
- Youdim KA, Martin A, Joseph JA. Essential fatty acids and the brain: possible health implications. Int J Dev Neurosci. 2000;18(4–5):383–399. doi: 10.1016/S0736-5748(00)00013-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.