Abstract
Berchowitz et al. establish that transient amyloid-like forms of Rim4, a yeast RNA-binding protein with a predicted prion domain, translationally repress cyclin CLB3 in meiosis I, thereby ensuring homologous chromosome segregation. These findings suggest that prion domains might enable formation of tightly regulated amyloid-like effectors in diverse functional settings.
Meiosis yields haploid gametes from diploid progenitors via two consecutive rounds of chromosome segregation, meiosis I and II, following a single S phase. During meiosis I, homologous chromosomes segregate, whereas in meiosis II sister chromatids separate. Proper meiotic progression requires translational regulation of specific cyclins, such as the B-type cyclin, CLB3 in budding yeast, which is repressed during meiosis I but translated during meiosis II (Carlile and Amon, 2008). Aberrant expression of CLB3 during meiosis I prevents the essential segregation of homologous chromosomes and triggers premature sister chromatid separation (Carlile and Amon, 2008). In this issue of Cell, an elegant study by Berchowitz et al. (2015) now establishes that Rim4, a meiosis-specific RNA-binding protein (RBP) with a predicted prion domain (Figure 1A) (Alberti et al., 2009), forms functional amyloid-like conformers that directly repress CLB3 translation during meiosis I (Berchowitz et al., 2015).
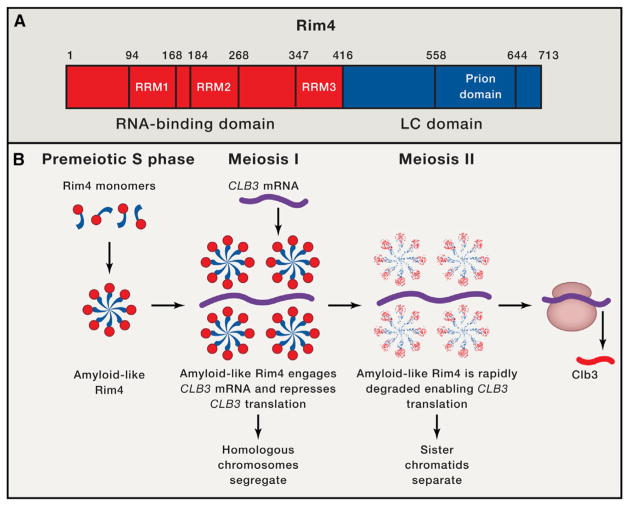
Figure 1. Amyloid-like Forms of Rim4 Repress CLB3 Translation During Meiosis I.
(A) Domain architecture of Rim4. Rim4 is divided into an N-terminal RNA-binding domain (red) harboring three RNA-recognition motifs (RRM1-3) and a C-terminal low-complexity (LC) domain (blue) harboring a predicted prion domain.
(B) Rim4 expression is induced in premeiotic S phase, and the LC domain of Rim4 (blue) enables assembly into amyloid-like species. Upon entry into meiosis I, amyloid-like Rim4 represses the translation of cyclin CLB3 by binding the 5′UTR of the CLB3 mRNA (purple). Repression of CLB3 translation enables segregation of homologous chromosomes. Upon transition to meiosis II, Ime2 kinase activity increases and elicits rapid degradation of amyloid-like Rim4, which relieves inhibition of CLB3 translation thereby facilitating sister chromatid separation.
Rim4 is a 713-residue RBP with three N-terminal RNA-recognitions motifs (RRMs) and a C-terminal low-complexity (LC) domain predicted to be predominantly disordered, which harbors a putative prion domain (Figure 1A) (Alberti et al., 2009). Prion domains have an unusual, low-complexity amino acid composition comprised of predominantly uncharged polar amino acids (particularly glutamine, asparagine, and tyrosine) and glycine (Alberti et al., 2009; King et al., 2012). In canonical yeast prion proteins, the prion domain can switch from an intrinsically disordered conformation to an infectious cross-β conformation (Alberti et al., 2009; King et al., 2012). In the present study, the authors showed that in isolation, Rim4 spontaneously assembled into β sheet-rich fibrils, and that Rim4 fibrillization was RNA-independent and required the C-terminal LC domain (Berchowitz et al., 2015). Moreover, in vivo, the predicted prion domain plus C-terminal determinants in the LC domain of Rim4 mediated formation of stable, SDS-resistant aggregates during premeiotic S phase, which sequestered specific mRNAs and repressed cyclin CLB3 translation exclusively during meiosis I (Figure 1B) (Berchowitz et al., 2015). Indeed, Rim4 variants bearing C-terminal deletions in the LC domain were unable to form amyloid-like species or elicit translational repression in this context (Berchowitz et al., 2015). Remarkably, at the onset of meiosis II, increased Ime2 kinase activity triggers rapid degradation of amyloid-like Rim4 to enable CLB3 translation and sister chromatid segregation (Figure 1B) (Berchowitz et al., 2015). Collectively, these data suggest that the formation and elimination of amyloid-like Rim4 is exquisitely timed and tightly orchestrated to enable a specific translational repression modality critical for meiosis I.
Is this mode of regulation specific to yeast? The answer is probably no. In fact, humans possess four meiosis-specific RBPs with predicted prion domains, DAZ1-4 (King et al., 2012). Moreover, a related RBP, DAZL (DAZ-like), is crucial for gametogenesis in mice. In the current work, the authors observed that DAZL forms SDS-resistant aggregates in cells undergoing meiosis but not in non-meiotic tissue (Berchowitz et al., 2015). Thus, similar to Rim4 in yeast, an amyloid-like form of DAZL, and perhaps also DAZ1-4, might play a role in translational regulation during mammalian gametogenesis.
Rim4 is not the first example of amyloid-like species regulating mRNA translation. However, its repression of CLB3 translation stands in stark contrast to the translational activation of specific transcripts sparked by amyloid-like forms of a neuronal cytoplasmic polyadenylation element binding protein from Aplysia californica (ApCPEB) (Shorter and Lindquist, 2005; Si et al., 2010). ApCPEB also harbors a prion domain, which drives assembly of amyloid-like forms of ApCPEB in response to specific neurotransmitter cues that stimulate a translational program that enables synaptic robustness and long-term facilitation (Shorter and Lindquist, 2005; Si et al., 2010). Like Rim4, the amyloid-like form of ApCPEB engages specific RNAs more avidly (Berchowitz et al., 2015; Shorter and Lindquist, 2005). This shared ability of amyloid-like forms of Rim4 and ApCPEB to modulate mRNA translation suggests that this type of regulation may be more widespread than presently appreciated. Remarkably, ~1% of human proteins harbor a predicted prion domain, including at least ~50 RBPs (Kim et al., 2013; King et al., 2012; Li et al., 2013). The pervasiveness of these domains is cause to ask—is the formation of amyloid or amyloid-like states a widely employed mechanism of cellular regulation underlying translational control and perhaps a spectrum of other molecular processes?
It is important to note that amyloid or amyloid-like conformers are not the only structures encoded by predicted prion domains. Indeed, prior to populating self-templating amyloid-like states, these intrinsically disordered regions can undergo concentration-dependent liquid demixing phase transitions. These transitions enable partitioning of various dynamic compartments with liquid- or gel-like properties, including P bodies, stress granules, and paraspeckles (Hennig et al., 2015; Li et al., 2013; Patel et al., 2015). Despite the clear utility of both preamyloid and amyloid-like conformations encoded by predicted prion domains, this type of domain is a double-edged sword. Indeed, RBPs with predicted prion domains, including FUS, TDP-43, hnRNPA1, and hnRNPA2, have emerged as causative agents in several fatal neurodegenerative disorders such as amyotrophic lateral sclerosis and frontotemporal dementia (Kim et al., 2013; King et al., 2012; Li et al., 2013). Here, the predicted prion domains enable RBP misfolding into deleterious oligomeric and fibrillar structures that likely underpin disease, and disease-linked mutations can accelerate these misfolding events (Kim et al., 2013; King et al., 2012; Li et al., 2013; Patel et al., 2015). In particular, an aberrant phase transition from liquid- or gel-like states to more intractable solid phases mediated by the prion domain is likely a key pathogenic event (Li et al., 2013; Patel et al., 2015).
A critical goal is to accurately understand what primary sequence elements and auxiliary factors determine whether predicted prion domains assemble into functional, beneficial structures or deleterious, misfolded conformers that are pathogenic. Such understanding will empower the design of therapeutics to prevent or reverse detrimental RBP mis-folding trajectories. In this context, a mechanistic understanding of the developmentally regulated formation and elimination of amyloid-like Rim4 is likely to be extremely valuable. For example, the mechanism by which amyloid-like Rim4 is rapidly degraded during meiosis II remains uncharted. It is not clear how increased Ime2 kinase activity elicits this degradation process or whether it requires protein disaggregases (e.g., Hsp110 or Hsp104), proteasomes, or autophagy. Delineation of this temporally restricted clearance pathway could inform effective strategies to eliminate pathogenic amyloid conformers.
References
- Alberti S, Halfmann R, King O, Kapila A, Lindquist S. Cell. 2009;137:146–158. doi: 10.1016/j.cell.2009.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berchowitz LE, Walker MR, Kabachinski G, Carlile TM, Gilbert WV, Schwartz TU, Amon A. Cell. 2015;163:406–418. doi: 10.1016/j.cell.2015.08.060. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlile TM, Amon A. Cell. 2008;133:280–291. doi: 10.1016/j.cell.2008.02.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennig S, Kong G, Mannen T, Sadowska A, Kobelke S, Blythe A, Knott GJ, Iyer KS, Ho D, Newcombe EA, et al. J Cell Biol. 2015;210:529–539. doi: 10.1083/jcb.201504117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Kim NC, Wang YD, Scarborough EA, Moore J, Diaz Z, MacLea KS, Freibaum B, Li S, Molliex A, et al. Nature. 2013;495:467–473. doi: 10.1038/nature11922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King OD, Gitler AD, Shorter J. Brain Res. 2012;1462:61–80. doi: 10.1016/j.brainres.2012.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li YR, King OD, Shorter J, Gitler AD. J Cell Biol. 2013;201:361–372. doi: 10.1083/jcb.201302044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- Shorter J, Lindquist S. Nat Rev Genet. 2005;6:435–450. doi: 10.1038/nrg1616. [DOI] [PubMed] [Google Scholar]
- Si K, Choi YB, White-Grindley E, Majumdar A, Kandel ER. Cell. 2010;140:421–435. doi: 10.1016/j.cell.2010.01.008. [DOI] [PubMed] [Google Scholar]