Significance
Type II polyketide natural products are powerful antimicrobial agents that are biosynthesized within bacteria by enzyme-encoding clusters of genes. We present a method to elucidate the evolution of these gene clusters as a whole, illuminating how natural selection has led to the chemical diversity of type II polyketides. Our approach can be applied to understand how other natural product gene clusters evolve. This understanding may aid efforts to access novel natural products and to design rational enzyme assemblies that produce chemicals of desired structures and activities.
Keywords: evolution, polyketide, natural products, gene cluster
Abstract
Natural product biosynthetic pathways generate molecules of enormous structural complexity and exquisitely tuned biological activities. Studies of natural products have led to the discovery of many pharmaceutical agents, particularly antibiotics. Attempts to harness the catalytic prowess of biosynthetic enzyme systems, for both compound discovery and engineering, have been limited by a poor understanding of the evolution of the underlying gene clusters. We developed an approach to study the evolution of biosynthetic genes on a cluster-wide scale, integrating pairwise gene coevolution information with large-scale phylogenetic analysis. We used this method to infer the evolution of type II polyketide gene clusters, tracing the path of evolution from the single ancestor to those gene clusters surviving today. We identified 10 key gene types in these clusters, most of which were swapped in from existing cellular processes and subsequently specialized. The ancestral type II polyketide gene cluster likely comprised a core set of five genes, a roster that expanded and contracted throughout evolution. A key C24 ancestor diversified into major classes of longer and shorter chain length systems, from which a C20 ancestor gave rise to the majority of characterized type II polyketide antibiotics. Our findings reveal that (i) type II polyketide structure is predictable from its gene roster, (ii) only certain gene combinations are compatible, and (iii) gene swaps were likely a key to evolution of chemical diversity. The lessons learned about how natural selection drives polyketide chemical innovation can be applied to the rational design and guided discovery of chemicals with desired structures and properties.
Microorganisms produce structurally diverse secondary metabolites, many of which have been successfully repurposed by mankind as pharmaceutical agents. These molecules are manufactured by multienzyme assemblies, many of which are encoded by biosynthetic gene clusters. Elucidating the history of how gene clusters evolved to produce a powerhouse of structurally diverse and biologically active molecules could reveal how synthases can be engineered to produce new therapeutic agents. Phylogenetic analyses have revealed evolutionary histories of individual biosynthetic genes, but the mechanisms of evolution of entire gene clusters are not well understood (1–4).
Here, we present an approach to study gene cluster evolution on a cluster-wide scale, and we apply it to type II polyketide gene clusters. In their native bacterial hosts, type II polyketides are thought to confer a selective advantage by serving important roles in chemical defense, signaling, and virulence (5). This class is rich in pharmacologically relevant compounds, including potent antibiotics (e.g., tetracycline) and anticancer agents (e.g., doxorubicin) (5, 6). The historical success of type II polyketides in the clinic, coupled with the need for new antibiotics, has spurred great interest in identifying and engineering new compounds in this class (7). Type II polyketide gene clusters encode discrete and dissociable polyketide synthase (PKS) enzyme assemblies. The core proteins of type II PKS gene clusters are a ketosynthase (KS)-alpha subunit and a KS-beta subunit, also known as a chain length factor (CLF), which collaborate with the acyl carrier protein (ACP) to construct a nascent polyketide chain. Reactive beta-keto chains are converted into structurally diverse molecules by the action of tailoring enzymes, including cyclases and reductases, giving rise to the final branching, oxidation state, and cyclization pattern of the polyaromatic product. The remarkable chemical diversity observed in this class of molecules is thought to originate from variations in chain length and tailoring reactions. Previous phylogenetic studies have revealed the role of the CLF in controlling the chain length of type II polyketides (8–15), but the evolution of the KS-CLF within the context of the entire protein assembly is not well understood.
Our analyses trace the evolution of type II PKS gene clusters, from the initial divergence of an ancestral KS into the homologous KS-CLF pair, and the gain of several key classes of accessory enzymes. We identified 544 putative type II PKSs in public genome databases, ∼15% of which encode a product that has been structurally characterized. Our studies revealed that the ancient pairing of the KS and CLF coincided with the gain of two accessory genes responsible for ring cyclization, an evolutionary shift that likely resulted in the introduction of the characteristic polyaromatic structure of type II polyketides. Subsequent gene swaps of accessory enzymes were highly coordinated with mutations to the KS-CLF, thereby enabling PKSs to diversify the chain length, oxidation state, and overall shape of their molecular products. These findings provide an unprecedented glimpse into the mechanisms by which evolution has led to the chemical diversity of natural products. The application of these methods to other gene collectives could unveil additional modes of chemical diversity generation in nature.
Results and Discussion
Evolution of the Core KS and CLF Proteins.
The KS, CLF, and ACP proteins form the minimal assembly required to build a nascent polyketide chain (SI Appendix, Fig. S1). Ridley et al. (8) proposed that the core KS and CLF genes of type II PKS gene clusters arose from an ancient KS duplication. Using a large set of recent bacterial genomic sequence data, we performed large-scale phylogenetic analysis of many newly sequenced homologs of the KS and CLF genes (SI Appendix, Fig. S2), resulting in two new insights into the nature and timing of this duplication in the context of bacterial evolution. First, type II PKS KSs appear more similar to primary metabolic FabF KS homologs from the fatty acid (FAS) pathway than to the KS of several secondary metabolic type II PKS relatives, such as aurachin and kedarcidin (1, 3, 4, 16–18). These other secondary metabolic gene clusters harbor tandem KSs that appear superficially similar to the tandem KSs of type II PKS gene clusters (SI Appendix, Fig. S2 A and B). However, our detailed phylogeny reveals that the tandem KS pair of these other secondary metabolic gene clusters arose from a separate duplication event, distinct from the duplication leading to canonical type II PKS KS-CLF genes. Second, despite type II PKS clusters having been identified exclusively in Actinobacteria, and despite their high similarity to FabF homologs (SI Appendix, Fig. S2 A and B), the KS and CLF proteins do not clade with actinobacterial FabF (SI Appendix, Fig. S2C). The divergence of the type II KS from the FabF KSs predates major bacterial speciation events, as visible by the FabF sequences grouping by phylum of origin. This finding suggests that type II KS and CLF genes did not evolve from an ancestral actinobacterial FabF, but rather diverged from the FabF common ancestor well before the actinobacterial phylum had formed.
To study the evolution of all type II PKS gene clusters sequenced to date, we searched public genomic sequence data in the GenBank (February 19, 2015) for KS-CLF gene homologs present in tandem (SI Appendix, Methods and Figs. S3–S5). We defined a redundancy threshold of 87% CLF sequence identity (at which gene clusters tend to encode identical rather than unique small molecules; SI Appendix, Methods). Our search identified 544 nonredundant putative type II PKS gene clusters, exclusively in bacteria (Fig. 1). These gene clusters included 78 (all actinobacterial) whose secondary metabolite products have been structurally characterized. We identified many orphan gene clusters in nonactinobacterial species; these gene clusters are very anciently diverged from actinobacterial homologs (not likely recently horizontally transferred), with sparse coverage of sequence space (Fig. 1 and SI Appendix, Fig. S2). The nonactinobacterial gene clusters are attractive targets for bioprospecting, because their origin in phyla such as Firmicutes and Proteobacteria could allow for expression in tractable nonactinobacterial heterologous hosts.
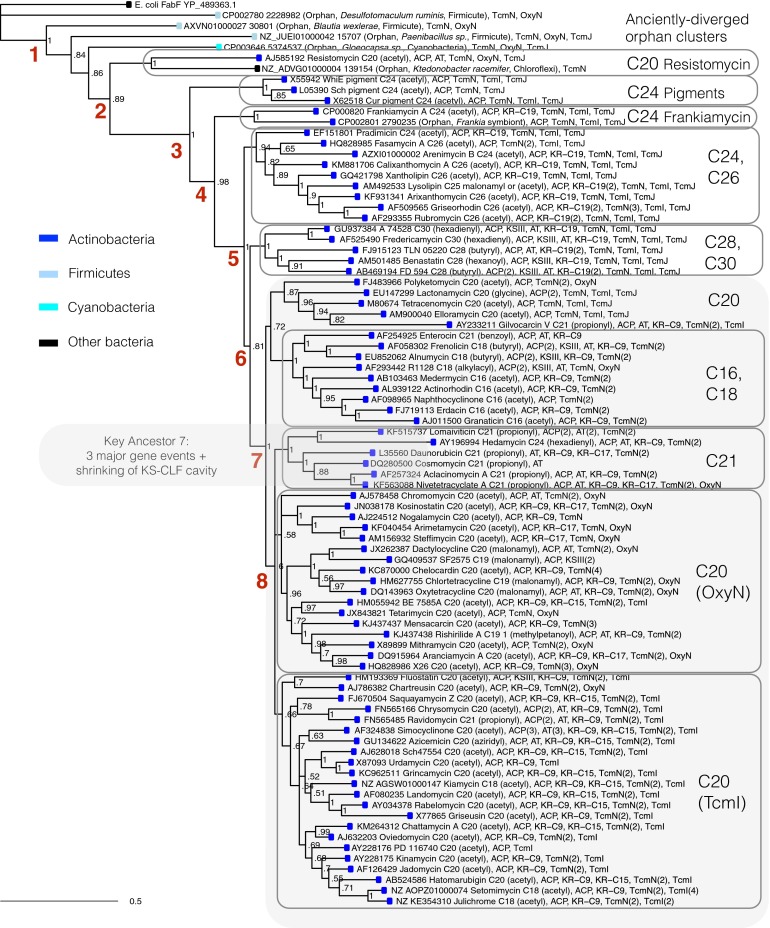
Fig. 1.
Phylogeny of CLF protein sequences from 544 putative type II PKS gene clusters. Red numbers represent key ancestors. Leaf colors represent phylum of origin. Gene clusters clade by polyketide chain length (noted on the right).
Phylogenetic analysis of characterized genes corroborated previous findings that CLF proteins group by chain length, in both the large dataset of 544 putative type II PKS clusters (Fig. 1) and the smaller set of 78 characterized gene clusters (Fig. 2). We computationally tested the long-standing hypothesis that the volume of the KS-CLF amphipathic cavity, which houses the growing polyketide chain during biosynthesis, is a main determinant of polyketide chain length (5, 8–15). Using the solved actinorhodin KS-CLF structure as a template for in silico mutagenesis, we calculated the predicted volume of the KS-CLF cavity for five PKS clades (Fig. 2): C16–C18, C20, C24, C26, and C28–C30 (SI Appendix, Fig. S6 and Table S1). Larger KS-CLF cavities are correlated with longer polyketides (SI Appendix, Table S2). Interestingly, the predicted change in cavity volume is more pronounced between C16–C24 (347 A3) than C24–C30 (81 A3). This observation could reflect the limitation of relying on homology models built from a single, short-chain template structure, or it may suggest that the cavity size of the KS-CLFs encoding the largest polyketides does not expand enough to accommodate the entire nascent polyketide chain. It is possible that in the case of the longest polyketides (C28–C30), auxiliary enzyme(s) create an expanded solvent-excluded cage, which serves to protect reactive polyketide intermediates (5, 6, 12). Our cluster-wide analysis reveals that CLF mutations are correlated with changes to the gene roster (discussed below).
Fig. 2.
Phylogeny of 78 CLF protein sequences from the reference set and selected orphan genes. Accessory genes identified in the same gene cluster (within 30 kb) as the CLF are shown at each leaf. Leaf colors represent phylum of origin. Node support for the CLF phylogeny is shown as Bayesian posterior probabilities.
Evolution of Type II PKS Accessory Enzymes.
The origin and diversification of type II PKS enzymes outside of the KS and CLF are not well understood. To study cluster-wide evolution, we first identified classes of accessory genes frequently clustered within 30 kb of the KS-CLF gene pair (Fig. 2, Table 1, and SI Appendix, Fig. S4). We developed a method to detect gene swap events, building upon existing approaches (19, 20) to quantify gene pair coevolution by comparing protein similarity scores between pairs of homologs. Correlated similarity scores suggest that gene types coevolved (Fig. 3 A and B). Our results confirmed that the core KS and CLF coevolved with little to no gene swaps: when two KSs from different genomes have high similarity, the neighboring CLFs also have high similarity, and when the KSs have low similarity, the neighboring CLFs also have low similarity (Fig. 3C and SI Appendix, Fig. S7A). We applied this framework to detect coevolution of tailoring genes with the core KS, and extended it to detect discrete homologous gene swap events as off-diagonal groups (Fig. 3B).
Table 1.
Number of gene clusters having at least one of the listed accessory genes clustered within 30 kb of KS-CLF genes
| Gene | Percentage of reference set (78 clusters), % | Percentage of 544 putative clusters, % |
| KS | 100 | 100 |
| CLF | 100 | 100 |
| ACP | 97 | 77 |
| Acyltransferase | 29 | 15 |
| KSIII | 12 | 7 |
| C9 KR | 55 | 36 |
| C15 KR | 14 | 8 |
| C17 KR | 8 | 3 |
| C19 KR | 18 | 8 |
| TcmN cyclase | 94 | 77 |
| TcmI cyclase | 55 | 46 |
| TcmJ cyclase | 28 | 32 |
| OxyN cyclase | 21 | 13 |
Fig. 3.
Coevolution of type II PKS KS with partner genes. (A) Schematic illustrating two nucleotide records and the clustered (within 30 kb) KS + partner on each record. (B) KS1-KS2 pairwise amino acid identities are plotted vs. pairwise identities of a clustered partner. (C–I) Correlation of evolutionary histories of the KS with partner genes.
Polyketide backbone.
The nascent polyketide chain is constructed through the collaboration of the KS-CLF with the ACP (SI Appendix, Fig. S1). All characterized gene clusters for which there is sufficient coverage (30 kb flanking the KS-CLF) contain an ACP (Table 1). Large-scale phylogenetic analysis of diverse ACP homologs revealed that type II PKS ACPs form a clade distinct from primary FAS and other secondary ACPs (SI Appendix, Fig. S8). The KS and clustered ACP genes share a correlated evolutionary history, suggesting they coevolved (Fig. 3D and SI Appendix, Fig. S7 B and C). Interestingly, several anciently diverged orphan clusters (top of Figs. 1 and 2), do not harbor an ACP homolog, suggesting that this gene was either absent from the initial ancestor or lost from multiple extant clusters. The ACP-less clusters must use either an alternative mechanism of biosynthesis or an ACP encoded outside the gene cluster.
Besides the ACP, other genes involved in backbone biosynthesis include acyltransferase (AT) and priming KS (KSIII). Most characterized type II systems are primed with acetyl units and “borrow” these enzymes from the FAS pathway (8–15, 21–23). Systems using nonacetate starting units rely on secondary AT and KSIII enzymes (Fig. 2, Table 1, and SI Appendix, Fig. S1) and produce polyketides with longer alkyl and alkenyl substituents. We found KSIIIs clustered primarily with long-chain (C28–C30) systems and ATs clustered with C21 systems (Fig. 2), and both underwent gene swaps (SI Appendix, Fig. S9).
Oxidation state of the polyketide.
Ketoreductase (KR) domains have a profound effect on the final product, because the oxidation state of the nascent polyketide chain can direct the regiochemistry of subsequent cyclizations (6). We found that most gene clusters contain at least one KR gene (Table 1). Previous phylogenetic analysis suggested there are four main classes of type II PKS KRs, which correlate with the regiospecificity of the reduction event: C9, C15, C17, and C19 (24). The four main classes of KRs are distinct at the sequence level, evolved with their clustered KS (SI Appendix, Figs. S9 and S10), and underwent clear swapping events (Fig. 3E).
Cyclization of the polyketide.
Cyclases function in a chaperone-like manner to direct regio- and stereoselective intramolecular cyclization of the polyketide chain (6, 25, 26). We identified four commonly occurring, nonhomologous categories of cyclases in type II PKS gene clusters: TcmN-like, OxyN-like, TcmI-like, and TcmJ-like (Fig. 3 and Table 1). We found no recognizable sequence or structural similarity between the four categories, suggesting that they evolved from four distinct ancestors. The striking finding that a TcmN cyclase homolog is present in 94% of characterized type II PKS gene clusters (Table 1) prompted us to focus on its role in the origin and evolution of type II PKS gene clusters. Remarkably, genes encoding TcmN and OxyN cyclase genes are present in even the most anciently diverged type II PKS clusters, suggesting that the introduction of these genes into the ancestral gene cluster coincided in time with the ancestral KS-CLF duplication (Fig. 2). The TcmN-like cyclase participates in first- and second-ring cyclizations (27–29), whereas TcmI-, OxyN-, and TcmJ-like cyclases are thought to direct subsequent ring closures (5, 30). All four of these categories share an evolutionary history with their clustered KS (Fig. 3 F–I), and the TcmN and OxyN genes display evidence of homologous gene swapping (Fig. 3 F and G).
Inferring the Ancestral Type II PKS Gene Cluster.
We found that gene cluster architecture is remarkably predictable based only on CLF protein sequence, with accessory gene architectures consistent within each CLF clade (Fig. 2). This finding suggests that CLF sequence mutations are correlated with the presence or absence of surrounding genes, and there have been a finite number of major evolutionary events, each represented by a different CLF clade. Each clade in the phylogeny represents a discrete evolutionary “solution” developed by the clade’s single ancestor, and the architecture of each ancestor is generally revealed by the shared makeup of its descendants. Fig. 4 summarizes the observed evolution of these key ancestors (represented as red numbers in Figs. 1, 2, and 4), in terms of both their encoded chain length and gene cluster architectures (accessory enzymes).
Fig. 4.
Evolution of type II PKS gene clusters by coordinated gene swaps. The tree traces the key ancestors from the most anciently diverged type II PKS gene clusters (Top) to the more recently diverged C20 systems, such as oxytetracycline and landomycin (Bottom). Highlighted in red are key ancestors, whose gene cluster architecture is inferred on the left. Representative polyketide structures from each clade are shown, and activity sites for KR (gold), TcmN cyclase (green), and KSIII (light blue) are shown on the chemical structures.
To elucidate the architecture of the ancestral type II PKS gene cluster existing immediately after the KS-CLF pairing (ancestor 1 in Figs. 1, 2, and 4), we studied the most anciently diverged clusters that descended directly from this single ancestor (Fig. 4 and SI Appendix, Fig. S11). These gene clusters are orphans encoding unknown molecules, but their cluster architectures are available from our analysis and exhibit remarkable conservation in gene makeup: Nearly all harbor a KS, CLF, TcmN cyclase, and OxyN cyclase, and many harbor an ACP, suggesting that the ancestor of all PKS gene clusters (ancestor 1) harbored these five genes. It is possible that some of these genes were clustered with the KS before the KS-CLF pairing. To investigate this possibility, we performed a detailed analysis of the origin of the tandem KS-CLF. These two genes are homologous, and previous suggestions that they arose from a gene duplication implies that this duplication occurred in a single species, at a single genetic locus (8). However, an alternative hypothesis, that the ancestral KS diverged by evolution in two different species followed by a later reunion of the two genes in a single species, cannot be ruled out from the existing sequence data. We identified a clade of gene clusters comprising a single KS clustered with an ACP and TcmN homolog, which could represent either (i) descendants of an ancestral cluster before a “swapping in” of the CLF or (ii) descendants of an ancestor that harbored the KS, CLF, ACP, and TcmN but subsequently lost the CLF. Very few gene clusters have been sequenced that descended from these key intermediate ancestors, unfortunately obscuring the order of events in which these five key genes became clustered. Additional sequencing data will better elucidate these early evolutionary paths.
The apparent ancientness of the TcmN and OxyN cyclase genes prompted us to investigate their evolutionary origins. It has been shown that the 3D structure of TcmN cyclase bears similarity to the “hot dog” fold of dehydratase proteins, which are often clustered with PKS/FAS systems (27). Our own homology searches found that OxyN-like cyclases share similarity and active site motifs with formamidases and metal-dependent hydrolases (31). Both of these protein classes are ancient, with homologs performing diverse functions in diverse species. The type II PKS homologs comprise only a small, recently evolved subset of each class (SI Appendix, Figs. S12 and S13), suggesting that these cyclases were swapped into the gene clusters from other systems and subsequently evolved PKS-specific functions.
Functional Consequences of Gene Swaps in Type II PKS Gene Clusters.
Having established the likely architecture of the ancestral type II PKS gene cluster as KS, CLF, ACP, and TcmN and OxyN cyclases, we traced the evolution of this ancestral cluster into characterized extant clusters surviving today. We inferred key ancestors from Fig. 2 and deduced functional consequences of the observed gene swaps; the results are summarized in Fig. 4.
Resistomycin represents an important type II polyketide, because its cluster is the most anciently diverged of the characterized set of 78. We studied all sequenced homologs of resistomycin to infer the common ancestor (ancestor 2) of all 78 characterized systems (Fig. 4 and SI Appendix, Fig. S11). Few systems related to resistomycin have been sequenced; many of the nearest relatives are from metagenomic sequencing projects, including uncultivable bacteria. Of the sequences that are available, the resistomycin-like architecture of KS-CLF, ACP, TcmN, OxyN, and TcmJ is conserved, suggesting that ancestor 2 gained a TcmJ cyclase. Interestingly, some distant relatives of resistomycin harbor no ACP gene but retain the three cyclases. Such anciently diverged gene clusters may represent interesting targets for bioprospecting, given their unique sequence and nonactinomycete origin.
The next key ancestor, which gave rise to the spore pigments and all other systems (ancestor 3), underwent a swap of TcmI with OxyN (Figs. 2 and 4), both of which are thought to catalyze late ring cyclization events. The TcmI and OxyN cyclase genes are observed together in none of the 544 putative clusters (SI Appendix, Fig. S14), so these genes may be mutually exclusive. Interestingly, OxyN reappears in one of the C20 subclades (Fig. 2), corresponding to a loss in TcmI (ancestors 7 and 8). This gene swap appears correlated with changes to polyketide ring topologies, because those C20 PKSs that use OxyN produce molecules with linear topology, whereas those C20 PKSs that use TcmI display a kink in the polyaromatic backbone (Fig. 4 and SI Appendix, Table S3).
Further diversification of type II PKS gene clusters occurred upon the introduction of KR genes (Fig. 4). We observe that the C19 KR was introduced into the ancestor of frankiamycin (32) and all other characterized clusters (ancestor 4).
One of the most striking findings of the cluster-wide phylogenetic analysis is that all C16- to C21-encoding gene clusters arose from a single common ancestor (ancestor 7). This ancestor was likely C20, because there are nonmonophyletic clades of C20 systems descending from it (Fig. 2). This C20 ancestor proliferated rapidly and gave rise to the majority of characterized type II polyketide antibiotics known today (e.g., tetracyclines) (Figs. 1, 2, and 4). Ancestor 7 underwent a major, coordinated set of mutations and gene swaps. We established above that the cavity volume of these systems is significantly smaller than the cavity volume of the C24–C30 systems (SI Appendix, Table S2). The gene architectures are also significantly different. The more anciently diverged, longer-chain systems harbor a monomeric TcmN gene, whereas the more recently diverged CLFs encoding shorter chain lengths (C16–C21) harbor a dimeric form of the gene (e.g., oxytetracycline otcD1). We hypothesized that the ancestral TcmN monomer had duplicated to become the OtcD1-like dimer, but, surprisingly, phylogenetic analysis of TcmN homologs refuted this hypothesis; rather, the dimer and the monomer diverged before type II PKSs proliferated in Actinobacteria, and the sequence-divergent dimeric homolog (OtcD1-like) was swapped into the C16–C21 common ancestor (ancestor 7), replacing the monomeric TcmN-like form (Figs. 3F and 4 and SI Appendix, Fig. S12).
An additional major change in key ancestor 7 was the replacement of a C19 KR with a C9 KR. Extensive biochemical analysis and docking studies suggest that KR interactions with the minimal PKS are essential during biosynthesis (33, 34), which could explain why most clusters harbor either a C9 or C19 KR (although there are exceptions, such as resistomycin, the spore pigments, and anciently diverged orphans that harbor no KR). The C19- to C9-KR gene swap events can be seen at the bottom of Fig. 3E, where KSs of high sequence identity are clustered with KRs with low sequence identity. Finally, ancestor 7 lost a TcmJ homolog. Taken together, these cluster-wide observations suggest that the transition from ancestor 6 to ancestor 7 involved the coordinated swaps of cyclases (TcmN monomer with TcmN dimer and loss of TcmJ) and KRs (C19 with C9). This ancestor further diversified via the introduction of additional KRs (C15 and C17) and cyclase gene swaps (TcmI to OxyN) to yield the diverse extant clusters seen today (Figs. 2 and 4).
The functional consequences of key evolutionary events (Fig. 4) can be predicted based on the known biochemistries of PKS enzymes (SI Appendix, Fig. S1). The product of the original, ancestral type II PKS gene cluster (ancestor 1 on Fig. 4) was likely an acetyl-primed C20–C24 polyketide cyclized at the C9–C14 position, because TcmN-like cyclases almost always facilitate C9–C14 cyclization events in the absence of a C9 KR (95% of pathways; SI Appendix, Table S4). The introduction of the C19 KR in ancestor 4 likely changed the oxidation state of the polyaromatic core. The gene swap of C9 for C19 likely changed both the oxidation state and the cyclization pattern, because in the presence of a C9 KR, the TcmN-like cyclase catalyzes C7–C12 ring closure (5, 10, 25, 35). The swap of TcmI for OxyN in ancestor 8 is correlated with a change in the polyketide structure from bent to linear. The global chemical innovation was likely driven by a combination of these key gene swap events with mutations to the KS-CLF core.
Conclusions
Type II PKSs are responsible for the biosynthesis of pharmacologically relevant natural products, and discovery and engineering of new polyketides are of great interest. Investigating the evolution of entire microbial gene clusters, as opposed to individual genes, is a nascent field that has become possible only recently with the large number of microbial genomes being sequenced. We tackled the question of how type II PKS gene clusters evolved by studying the evolution of the KS-CLF and its coevolution with accessory genes. Remarkably, divergence of the PKS KS (and CLF) from the FabF KS apparently occurred before bacterial speciation. Chemical innovation was driven by mutations in the KS-CLF gene pair to accommodate polyketides of variable length and changes to the accessory gene roster to diversify the oxidation state, priming unit, and cyclization patterns of the natural product.
The methods presented here for studying the evolution of type II PKSs are generalizable and can be applied to other biosynthetic systems, where gene swaps may also be a source of chemical diversity. In addition to reconstructing the evolutionary history of an important class of natural products, these results represent a launching point for bioprospecting in three ways. First, the lessons learned about nature’s strategies to generate chemical diversity may guide rational design and engineering of hybrid synthases (36). Our study reveals patterns in gene compatibility that can be used as ground rules for engineering. Second, we identified orphan gene clusters from diverse phyla that may encode novel chemical structures that could potentially be expressed in tractable nonactinobacterial hosts (18, 37, 38). Finally, ancestral sequence reconstruction, which has been applied to single proteins and whole genomes (39, 40), could be applied to ancestral gene clusters, leading to a potentially novel source of chemical diversity: the resurrection of extinct gene clusters and chemicals.
Methods
Materials and methods, along with supporting figures and a complete reference set of the 78 characterized type II polyketides, are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Colin Harvey, Yi Tang, Marnix Medema, and Raeka Aiyar for helpful discussions. We acknowledge support from Haverford College (L.K.C. and E.E.B.), the Research Corporation for Science Advancement Cottrell College Scholars Award (to L.K.C.), the Burroughs Wellcome Fund Career Award at the Scientific Interface and NIH Grant U01 GM110706 (to M.E.H.), and the Stanford School of Medicine Dean’s Office (G.A.V.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: Data are available for download and visualization at sequence.stanford.edu/TypeIIPKS/.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1511688112/-/DCSupplemental.
References
- 1.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: How natural selection drives chemical innovation. Proc Natl Acad Sci USA. 2008;105(12):4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Medema MH, et al. antiSMASH: Rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011;39(Web Server issue):W339–W346. doi: 10.1093/nar/gkr466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jenke-Kodama H, Börner T, Dittmann E. Natural biocombinatorics in the polyketide synthase genes of the actinobacterium Streptomyces avermitilis. PLOS Comput Biol. 2006;2(10):e132. doi: 10.1371/journal.pcbi.0020132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jenke-Kodama H, Sandmann A, Müller R, Dittmann E. Evolutionary implications of bacterial polyketide synthases. Mol Biol Evol. 2005;22(10):2027–2039. doi: 10.1093/molbev/msi193. [DOI] [PubMed] [Google Scholar]
- 5.Hertweck C. The biosynthetic logic of polyketide diversity. Angew Chem Int Ed Engl. 2009;48(26):4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 6.Hertweck C, Luzhetskyy A, Rebets Y, Bechthold A. Type II polyketide synthases: Gaining a deeper insight into enzymatic teamwork. Nat Prod Rep. 2007;24(1):162–190. doi: 10.1039/b507395m. [DOI] [PubMed] [Google Scholar]
- 7.Fischbach MA, Walsh CT. Antibiotics for emerging pathogens. Science. 2009;325(5944):1089–1093. doi: 10.1126/science.1176667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridley CP, Lee HY, Khosla C. Evolution of polyketide synthases in bacteria. Proc Natl Acad Sci USA. 2008;105(12):4595–4600. doi: 10.1073/pnas.0710107105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tang Y, Tsai S-C, Khosla C. Polyketide chain length control by chain length factor. J Am Chem Soc. 2003;125(42):12708–12709. doi: 10.1021/ja0378759. [DOI] [PubMed] [Google Scholar]
- 10.Keatinge-Clay AT, Maltby DA, Medzihradszky KF, Khosla C, Stroud RM. An antibiotic factory caught in action. Nat Struct Mol Biol. 2004;11(9):888–893. doi: 10.1038/nsmb808. [DOI] [PubMed] [Google Scholar]
- 11.Nicholson TP, et al. First in vitro directed biosynthesis of new compounds by a minimal type II polyketide synthase: Evidence for the mechanism of chain length determination. Chem Commun (Camb) 2003;(6):686–687. doi: 10.1039/b300847a. [DOI] [PubMed] [Google Scholar]
- 12.Szu P-H, et al. Analysis of the ketosynthase-chain length factor heterodimer from the fredericamycin polyketide synthase. Chem Biol. 2011;18(8):1021–1031. doi: 10.1016/j.chembiol.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav G, Gokhale RS, Mohanty D. Towards prediction of metabolic products of polyketide synthases: An in silico analysis. PLOS Comput Biol. 2009;5(4):e1000351. doi: 10.1371/journal.pcbi.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dreier J, Khosla C. Mechanistic analysis of a type II polyketide synthase. Role of conserved residues in the β-ketoacyl synthase-chain length factor heterodimer. Biochemistry. 2000;39(8):2088–2095. doi: 10.1021/bi992121l. [DOI] [PubMed] [Google Scholar]
- 15.Burson KK, Khosla C. Dissecting the chain length specificity in bacterial aromatic polyketide synthases using chimeric genes. Tetrahedron. 2000;56(48):9401–9408. [Google Scholar]
- 16.Höfle G, Kunze B. Biosynthesis of aurachins A-L in Stigmatella aurantiaca: A feeding study. J Nat Prod. 2008;71(11):1843–1849. doi: 10.1021/np8003084. [DOI] [PubMed] [Google Scholar]
- 17.Pistorius D, Li Y, Sandmann A, Müller R. Completing the puzzle of aurachin biosynthesis in Stigmatella aurantiaca Sg a15. Mol Biosyst. 2011;7(12):3308–3315. doi: 10.1039/c1mb05328k. [DOI] [PubMed] [Google Scholar]
- 18.Cimermancic P, et al. Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell. 2014;158(2):412–421. doi: 10.1016/j.cell.2014.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cohen O, Ashkenazy H, Burstein D, Pupko T. Uncovering the co-evolutionary network among prokaryotic genes. Bioinformatics. 2012;28(18):i389–i394. doi: 10.1093/bioinformatics/bts396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scornavacca C, Zickmann F, Huson DH. Tanglegrams for rooted phylogenetic trees and networks. Bioinformatics. 2011;27(13):i248–i256. doi: 10.1093/bioinformatics/btr210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Summers RG, Ali A, Shen B, Wessel WA, Hutchinson CR. Malonyl-coenzyme A:acyl carrier protein acyltransferase of Streptomyces glaucescens: A possible link between fatty acid and polyketide biosynthesis. Biochemistry. 1995;34(29):9389–9402. doi: 10.1021/bi00029a015. [DOI] [PubMed] [Google Scholar]
- 22.Arthur CJ, et al. The malonyl transferase activity of type II polyketide synthase acyl carrier proteins. Chem Biol. 2006;13(6):587–596. doi: 10.1016/j.chembiol.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 23.Moore BS, Hertweck C. Biosynthesis and attachment of novel bacterial polyketide synthase starter units. Nat Prod Rep. 2002;19(1):70–99. doi: 10.1039/b003939j. [DOI] [PubMed] [Google Scholar]
- 24.Lackner G, et al. Biosynthesis of pentangular polyphenols: Deductions from the benastatin and griseorhodin pathways. J Am Chem Soc. 2007;129(30):9306–9312. doi: 10.1021/ja0718624. [DOI] [PubMed] [Google Scholar]
- 25.Fritzsche K, Ishida K, Hertweck C. Orchestration of discoid polyketide cyclization in the resistomycin pathway. J Am Chem Soc. 2008;130(26):8307–8316. doi: 10.1021/ja800251m. [DOI] [PubMed] [Google Scholar]
- 26.Jakobi K, Hertweck C. A gene cluster encoding resistomycin biosynthesis in Streptomyces resistomycificus; exploring polyketide cyclization beyond linear and angucyclic patterns. J Am Chem Soc. 2004;126(8):2298–2299. doi: 10.1021/ja0390698. [DOI] [PubMed] [Google Scholar]
- 27.Ames BD, et al. Crystal structure and functional analysis of tetracenomycin ARO/CYC: Implications for cyclization specificity of aromatic polyketides. Proc Natl Acad Sci USA. 2008;105(14):5349–5354. doi: 10.1073/pnas.0709223105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shen B, Hutchinson CR. Deciphering the mechanism for the assembly of aromatic polyketides by a bacterial polyketide synthase. Proc Natl Acad Sci USA. 1996;93(13):6600–6604. doi: 10.1073/pnas.93.13.6600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson TB, Katayama K, Watanabe K, Hutchinson CR, Rayment I. Structural and functional analysis of tetracenomycin F2 cyclase from Streptomyces glaucescens. A type II polyketide cyclase. J Biol Chem. 2004;279(36):37956–37963. doi: 10.1074/jbc.M406144200. [DOI] [PubMed] [Google Scholar]
- 30.Pickens LB, Tang Y. Oxytetracycline biosynthesis. J Biol Chem. 2010;285(36):27509–27515. doi: 10.1074/jbc.R110.130419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Díaz-Sáez L, Srikannathasan V, Zoltner M, Hunter WN. Structures of bacterial kynurenine formamidase reveal a crowded binuclear zinc catalytic site primed to generate a potent nucleophile. Biochem J. 2014;462(3):581–589. doi: 10.1042/BJ20140511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogasawara Y, Yackley BJ, Greenberg JA, Rogelj S, Melançon CE., 3rd Expanding our understanding of sequence-function relationships of type II polyketide biosynthetic gene clusters: Bioinformatics-guided identification of Frankiamicin A from Frankia sp. EAN1pec. PLOS One. 2015;10(4):e0121505. doi: 10.1371/journal.pone.0121505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hertweck C, et al. Context-dependent behavior of the enterocin iterative polyketide synthase; a new model for ketoreduction. Chem Biol. 2004;11(4):461–468. doi: 10.1016/j.chembiol.2004.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Javidpour P, et al. The determinants of activity and specificity in actinorhodin type II polyketide ketoreductase. Chem Biol. 2013;20(10):1225–1234. doi: 10.1016/j.chembiol.2013.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korman TP, Tan Y-H, Wong J, Luo R, Tsai S-C. Inhibition kinetics and emodin cocrystal structure of a type II polyketide ketoreductase. Biochemistry. 2008;47(7):1837–1847. doi: 10.1021/bi7016427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Poust S, Hagen A, Katz L, Keasling JD. Narrowing the gap between the promise and reality of polyketide synthases as a synthetic biology platform. Curr Opin Biotechnol. 2014;30:32–39. doi: 10.1016/j.copbio.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 37.O’Brien RV, Davis RW, Khosla C, Hillenmeyer ME. Computational identification and analysis of orphan assembly-line polyketide synthases. J Antibiot (Tokyo) 2014;67(1):89–97. doi: 10.1038/ja.2013.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bachmann BO, Van Lanen SG, Baltz RH. Microbial genome mining for accelerated natural products discovery: Is a renaissance in the making? J Ind Microbiol Biotechnol. 2014;41(2):175–184. doi: 10.1007/s10295-013-1389-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma J, et al. Reconstructing contiguous regions of an ancestral genome. Genome Res. 2006;16(12):1557–1565. doi: 10.1101/gr.5383506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gaucher EA, Govindarajan S, Ganesh OK. Palaeotemperature trend for Precambrian life inferred from resurrected proteins. Nature. 2008;451(7179):704–707. doi: 10.1038/nature06510. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.