Significance
Energy from the sun is captured by the biosphere through photosynthetic antenna proteins, which function by integration of a set of pigments into a protein scaffold. Nature uses relatively similar pigment building blocks to achieve diverse functions, and the organizing principles behind this process are not fully understood. Using a single-molecule microfluidic trap and multiparameter fluorescence spectroscopy, we explore how the photophysical properties of individual pigment sites are modulated by the protein’s oligomerization states, using single proteins of a model antenna from cyanobacteria. Our measurements achieve a bottom-up dissection of the structural-function relation of the model pigment−protein complex with important implications for photosynthetic antenna biophysics.
Keywords: photosynthetic protein, allophycocyanin, ABEL trap, single-molecule spectroscopy
Abstract
Oligomerization plays a critical role in shaping the light-harvesting properties of many photosynthetic pigment−protein complexes, but a detailed understanding of this process at the level of individual pigments is still lacking. To study the effects of oligomerization, we designed a single-molecule approach to probe the photophysical properties of individual pigment sites as a function of protein assembly state. Our method, based on the principles of anti-Brownian electrokinetic trapping of single fluorescent proteins, step-wise photobleaching, and multiparameter spectroscopy, allows pigment-specific spectroscopic information on single multipigment antennae to be recorded in a nonperturbative aqueous environment with unprecedented detail. We focus on the monomer-to-trimer transformation of allophycocyanin (APC), an important antenna protein in cyanobacteria. Our data reveal that the two chemically identical pigments in APC have different roles. One (α) is the functional pigment that red-shifts its spectral properties upon trimer formation, whereas the other (β) is a “protective” pigment that persistently quenches the excited state of α in the prefunctional, monomer state of the protein. These results show how subtleties in pigment organization give rise to functionally important aspects of energy transfer and photoprotection in antenna complexes. The method developed here should find immediate application in understanding the emergent properties of other natural and artificial light-harvesting systems.
The first step in photosynthesis is the capture of solar radiation by light-harvesting antenna complexes (1). In these complexes, pigments are three-dimensionally organized in a protein scaffold with specific positions, orientations, densities, and chemistry. The organizing principles (2) of these pigments are evolutionarily refined to achieve wide spectral coverage, efficient energy transport over long distances (3) and even a degree of quantum coherence (4). For example, the phycobiliproteins (5) fulfill their roles as light harvesters and energy transfer chains in cyanobacteria, by covalently binding multiple open-chain tetrapyrrole molecules (bilins), shaping the photophysical and photochemical properties of these otherwise flexible and photolabile pigments via pigment−protein interactions and forming quaternary structures that provide an extra level of pigment distance and orientation control (6).
A deep understanding of the physicochemical principles of pigment organization in antenna proteins not only sheds light on the fundamentals of the light-harvesting process but can also provide guidance to the design and optimization of artificial systems (7). Critical to elucidation of these principles are the properties of the individual pigments in their native biological environment (“site properties”). However, characterization of site properties in a multipigment antenna has been difficult because the contribution of a single pigment is often obscured by signals from other pigments on the same protein. Although single-pigment variants can be prepared by denaturation (8) or mutagenesis (9), the perturbation imposed by these methods might be undesirable. Single-molecule fluorescence (10–15) can provide subcomplex information, but the common practice of immobilization might perturb the delicate integrity of these highly optimized antenna complexes. Moreover, previous studies lack sufficient spectroscopic resolution to resolve individual pigments.
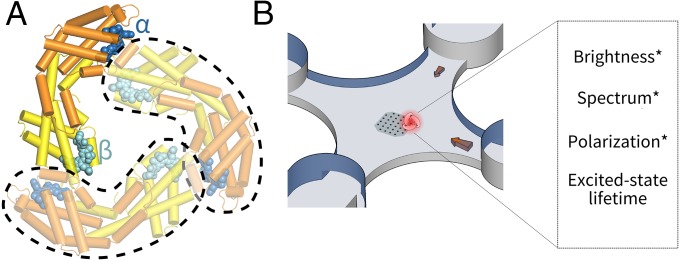
Here, to overcome these limitations, we conduct nonperturbative measurements on fully rotationally mobile single pigment−protein complexes in aqueous solution using an Anti-Brownian ELectrokinetic (ABEL) trap. Furthermore, we use step-wise photobleaching and polarization-resolved multiparameter fluorescence spectroscopy to identify and characterize individual pigment sites. We focus on the pigment organization principles of a key antenna protein allophycocyanin (APC), which is important in light harvesting, energy transfer (16), and photoprotection (17) in cyanobacteria. Its monomer form (Fig. 1A), which binds two (α and β) phycocyanobilin (PCB) pigments in similar but nonidentical protein environments, self-associates to form a trimer (Fig. 1A). This trimerization process reorganizes the pigments to generate a ∼30-nm red shift in absorption, which is critical for APC’s function. To trace this reorganization process to its molecular origin, we first present a detailed optical study of the individual α- and β-pigments on the monomer and characterize their photophysical properties. A previously unknown photochemical form of the β-pigment is discovered. We then use the monomer information to unravel the photodynamics of the trimers with single-pigment resolution, and we discuss a model of pigment organization based on these measurements.
Fig. 1.
Experimental scheme. (A) Crystal structure of the model antenna protein used in this study: APC (PDB ID code 1ALL) is a trimer with C3 symmetry. Each constituent monomer (circled in dashed lines) binds two heterogeneous PCB pigments (α, blue; β, cyan). In the trimer, a β-pigment is brought to close proximity to the α-pigment of a neighboring monomer. (B) A single-molecule fluorescence assay to probe the photodynamics of individual APCs in solution. Molecules are held in an anti-Brownian ELectrokinetic trap and interrogated by polarization-resolved multiparameter fluorescence spectroscopy.
Results
Measurement Strategy.
To interrogate the photophysical states of APC in a nonperturbative solution environment, we use the ABEL trap (18, 19), which integrates high-speed fluorescence tracking, feedback control, and microfluidics to counteract Brownian motion in aqueous solution. The ABEL trap enables >1,000 times longer observation time (approximately seconds) of single biomolecules in solution (20) compared with diffusion-based modalities, during which multiparameter interrogation of both spectroscopic and transport properties (21) can be performed.
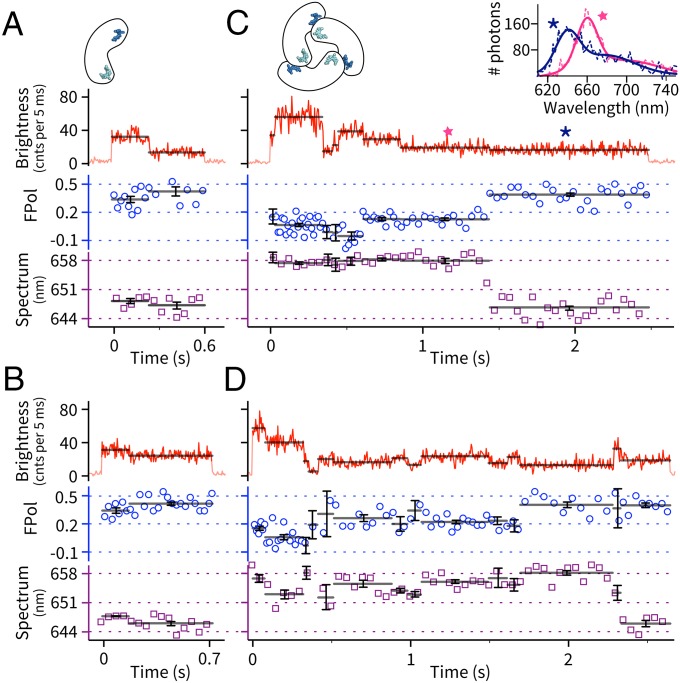
We investigated both the monomers and trimers (SI Appendix, SI Materials and Methods) of APC. Single proteins were probed in the ABEL trap for durations limited by complete photobleaching of the whole complex (Fig. 2). Monomers were typically observed for ∼0.5 s with clear step-wise digital switching in brightness (Fig. 2 A and B). This observation contrasts with previous unsuccessful attempts to observe single monomers with surface immobilization (11), highlighting the importance of using a nonperturbative ABEL trap to study APC photophysics. Trimers (crosslinked to prevent dissociation, SI Appendix, Fig. S3), on the other hand, were observed to show longer trapping times and more complex stepping behavior in fluorescence brightness, similar to previous work (11, 13, 22).
Fig. 2.
Simultaneously recorded brightness−FPol−spectrum dynamics of individual APC proteins in solution. (A and B) Two example molecules of the monomer. (C and D) Two example molecules of the trimer. Solid black lines indicate the mean parameter value of an identified brightness level. Error bars indicate 68% confidence intervals and are defined in SI Appendix, SI Materials and Methods. In the FPol channel, blue circles indicate FPol values calculated from every 250 total photons. In the spectral channel, purple squares indicate spectral CM of each 50-ms frame. (Inset) Full emission spectra (dashed, raw data; solid, double-Gaussian fits) of molecule C's last two emission levels (stars).
To better characterize the photophysics of these brightness levels, we measured multiple fluorescence parameters from trapped single proteins (Fig. 1B). In particular, we monitored fluorescence emission spectra (23), which report the electronic and vibronic energy landscape of the pigments in their protein environments, and excited-state lifetime, which provides signatures of fluorescence quenching and energy transfer. In this work, we also introduced single-molecule fluorescence polarization (FPol) measurements in the ABEL trap, which is the single-molecule analog of fluorescence anisotropy (24). Fundamentally, the emission FPol is determined by the degree of dipole reorientation between absorption and fluorescence emission. In the case of a single solution-phase antenna complex containing multiple pigments, FPol is most sensitive to the directions of energy transfer pathways within the complex (SI Appendix, SI Materials and Methods). Although single-molecule FPol measurements in solution have been performed in the past (25, 26), they have been limited by molecular diffusion to provide only snapshots of FPol states with large uncertainties. Here, combining the long observation times enabled by the ABEL trap with FPol, we observed, for the first time, to our knowledge, dynamic transitions between distinct FPol levels of the same protein, as shown in Fig. 2 C and D. Moreover, brightness, FPol and emission spectra of a single APC were recorded simultaneously, as correlated dynamics across multiple parameters offer additional insights in defining and characterizing the emission states of the system (23, 27, 28).
Emission States of the Monomer.
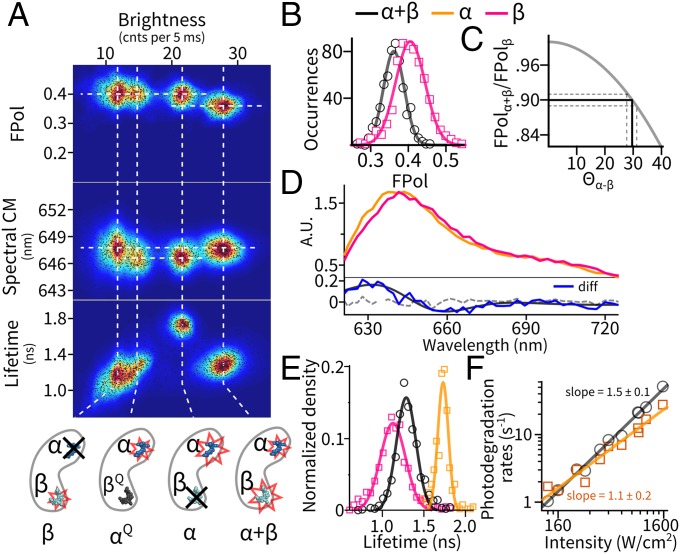
In monomers (Fig. 2 A and B), step-wise decreases in fluorescence brightness are accompanied by small changes in FPol and spectra. We first mapped each level’s average brightness, FPol, and spectral center of mass (CM) as a single point in a 3D parameter space (Fig. 3A) and then aggregated data acquired from 1941 monomers to create a scatter plot. Further, we measured brightness−lifetime distributions in a separate experiment using pulsed excitation (Fig. 3A, Bottom).
Fig. 3.
Pigment-specific dissection of the emission states of the monomer. (A) Multiparameter mapping of all emissive levels observed in APC monomers. Dashed lines are guides to the eye for alignment. State identifications are illustrated at the bottom; see Emission States of the Monomer for details. (B–F) Emissive properties of the states. The legends apply to all panels. (B) Distribution of FPol in the intact monomer (α+β, black) and the β-only state (red). (C) Estimation of the angular separation (Θα-β) between the α- and β-pigment dipoles on the monomer by a FRET model. Solid gray, expected FPol of the α+β state as a function of Θα-β; solid black, experimentally observed FPol and its corresponding Θα-β. Dashed lines indicate 95% confidence interval. (D, Top) Emission spectra of the individual α- and β-pigment sites. (Bottom) Difference spectrum (blue) between the two pigments. Dashed line denotes difference spectrum between two randomly picked subsets of the β-only state. (E) Decomposed excited-state lifetime distributions of the individual pigments and the intact monomer. (F) Power dependence of the photodegradation rates with fits to a power law.
This analysis revealed four states in the multidimensional parameter space. These states separate mainly by brightness and lifetime, and show small but distinctive differences in emission spectra and FPol. We interpret these states as follows. First, we assign the most luminous state (brightness, 28 counts per 5 ms; FPol, 0.36; spectrum, 647.6 nm; lifetime, 1.3 ns) to be the “pristine” monomer containing intact α- and β-pigments (α+β). Indeed, the majority (∼60%) of the single molecules entered the trap with this state. Interestingly, this state has a slightly lower FPol (∼0.36) compared with the other three (Fig. 3B), which we interpret as resulting from energy transfer between the two nonparallel pigment dipoles (to be analyzed in Coupling Between the Two Pigments on the Monomer). The three less-luminous states all have FPol values corresponding to the theoretical maximum of a single, rotationally mobile dipole emitter in our setup (∼0.40, SI Appendix, SI Materials and Methods and Fig. S2), which suggests that each represents a state with only one remaining emissive pigment, as a result of light-driven dynamics.
We next assign the second brightest state (∼22 counts per 5 ms) to be the monomer with only the α-pigment emitting and the dimmest state (∼12 counts per 5 ms) to be the monomer with only the β-pigment emitting. Previous spectroscopic data on biochemically purified α- and β-subunits (29, 30) established that the α-pigment is twice as bright as the β-pigment in APC from a variety of strains. Interestingly, between the α-only and β-only populations, there exists another state with near-identical FPol and spectral CM compared with the α-pigment, but reduced excited-state lifetime (∼1.3 ns). We identify this to be the α-pigment in a quenched form (αQ). Moreover, this αQ-state is a single-step photoproduct of the intact monomer, as 72% of this state results from direct transitions originating from the α+β state. We subsequently attribute the source of quenching to be the photoproduct(s) of the β-pigment acting as a nonemissive Fӧrster resonance energy transfer (FRET) acceptor.
Multiparameter Fluorescence Dynamics of Trimers.
Compared with the monomers, trimers showed more complicated dynamics in the simultaneously recorded single-molecule brightness, FPol, and spectrum traces. This is exemplified by multiple intermediate states and frequent digital jumps in all three parameters (Fig. 2 C and D and SI Appendix, Fig. S8). Both correlated and uncorrelated transitions between the different parameters were frequently observed. To understand this complex data set, we first extract some common features of the data. Trimers typically start with a homogeneous state with 3 times the monomer brightness and a mean FPol centered around 0.025, a value much lower than the monomer FPol of 0.36 and consistent with ensemble measurements (31). The mean spectral CM of this state (658 nm) is ∼10 nm red-shifted from the monomer and agrees with the bulk spectra. We thus assign this homogenous state to be the pristine APC trimer with six intact pigments.
In the spectral channel, about 25% (215/847) of the molecules showed a prominent (∼10 nm) spectral blue shift near the end of the trapping event. Most of these shifts (>90%) were permanent before trap exit (Fig. 2 C and D), but reversible shifts were also observed (SI Appendix, Fig. S8 C and D). Blue-shifting of the last unbleached step was previously reported on surface-immobilized APC trimers (11, 13). In addition to the major blue shift, small spectral fluctuations (∼1–5 nm) were also observed in many single-molecule traces (Fig. 2D). In the polarization channel, FPol generally increased as the complex bleached down, but there was a high degree of molecule-to-molecule heterogeneity. In some cases, switches in brightness were accompanied by discrete steps in FPol (Fig. 2C, ∼1.4 s). In others, FPol remained the same regardless of the brightness changes (Fig. 2D, ∼2.3 s). Spectral and FPol dynamics within a single brightness level were rare, but were clearly identified in ∼3% of the observed traces (SI Appendix, Fig. S8 F and G).
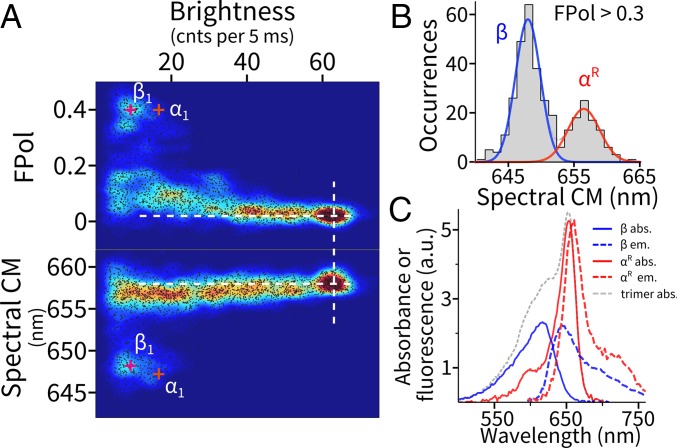
When projecting all brightness levels measured from 847 trimers onto a multiparameter map (Fig. 4A), we observed a distribution that can be best described as a few well-defined clusters plus continua with reproducible density variations. Clearly, after the homogeneous high-brightness initial state, there exists a significant amount of heterogeneity in the emission states of the trimers. We examine the origin of the intermediate states below.
Fig. 4.
Dissecting pigment organization in APC trimers. (A) Multiparameter (brightness, FPol, and emission spectrum) mapping of all emissive levels observed in APC trimers. Dashed lines are guides to the eye for alignment. (B) Distribution of the spectral CM of levels showing a high degree of FPol. (C) Decomposed pigment-specific optical properties of the trimer. The two emission spectra were estimated by pooling the full spectra from each population in B separately. The absorption spectrum of β was approximated by one-half of that of the monomer. The absorption spectrum of αR was estimated by subtracting the three β-pigments’ contribution from the absorption spectrum of the trimer (SI Appendix, Fig. S4).
Discussion
Our single-molecule method uses step-wise photobleaching to sequentially eliminate the spectroscopic contribution from individual pigments within a complex until a single emitting site is exposed. This unique aspect, together with information along multiple fluorescence dimensions in a nonperturbative ABEL trap environment, enables us to carefully examine the emissive properties and photophysics of the individual sites and subsequently provides a “bottom-up” route to understanding the collective and synergistic behaviors of the protein complex at different aggregation stages.
Pigment-Specific Spectroscopic Properties of the Monomer.
The ability to separate, localize and isolate the single-pigment (α, β, and αQ) states allows us to examine the site properties of the monomer in great detail. By pooling the spectra from their respective clusters, we obtained the full emission spectra of the individual α- and β-pigments (Fig. 3D). Although the overall shapes of the two spectra are similar, β is red-shifted by ∼4 nm in peak position and serves as the lower-energy site on the monomer. Moreover, the β-pigment features shortened excited-state lifetime (∼1.1 ns, Fig. 3E) and more heterogeneity in the brightness−lifetime map (i.e., slight correlation in the β-only and α+β clusters) compared with the α-pigment (∼1.8 ns). It is well established that photoexcited PCB pigments in solution undergo fast internal relaxation and photochemistry (<40 ps) that outcompetes fluorescence (32). A protein environment generally rigidifies the pigment, leading to suppressed nonradiative decay and restricted photochemical pathways. In cyanobacteria, this process is tuned by pigment−protein interactions to favor either excited-state longevity (as in biliproteins) or photochemistry [as in phytochromes (33)]. Here, our measurements reveal the different degrees of flexibility of the two pigments: Although both pigments are optimized for radiative decay, the β-pigment is less so and retains more conformational and photochemical flexibility. One of the β-photoproducts is directly visualized as a dark quencher to the remaining α-pigment (the αQ-form).
To investigate the photodegradation mechanism of the individual sites, we measured photodegradation rates (from fits to survival time distributions, SI Appendix, Fig. S5) of the α+β and the α-only states as a function of excitation intensity (Fig. 3F). Interestingly, the photodegradation rate of the intact monomer shows a nonlinear dependence (an exponent of ∼1.5) on excitation power, whereas the α-only state shows a much more linear dependence (an exponent of ∼1.1). Pulsed excitation sped up photodegradation (11, 22), but more so for the α+β state (2.3-fold increase) than the α-only state (1.5-fold increase, SI Appendix, Fig. S5). Based on those observations and the fact that pump−probe experiments (34, 35) revealed large excited-state (S1) absorption cross-sections in the 580- to 650-nm spectral window, we propose that absorption from S1 (or an S1-mediated photoproduct) is involved in photodegradation. On an intact monomer, this process happens more often at the β-site. Further evidence of S1 absorption is provided by fluorescence antibunching experiments (36), which revealed efficient singlet−singlet annihilation processes between the two pigments on the monomer (SI Appendix, SI Results and Discussion and Fig. S7).
Coupling Between the Two Pigments on the Monomer.
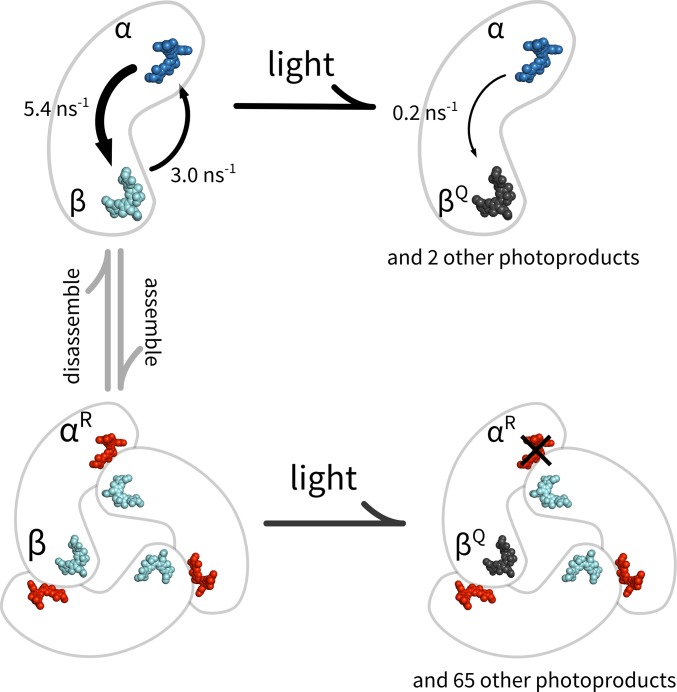
The “state-purified” pigment-specific information allows us to construct a detailed model of the coupling between the α- and β-pigments on the monomer, which we build up in Fig. 5. Given the ∼5-nm spatial separation between the two pigments, FRET is expected to be the dominant coupling mechanism. Although our multiparameter characterization directly provides emission parameters of the individual pigments as needed for a Fӧrster equation analysis, direct calculation of the energy transfer rates is hampered by insufficient knowledge regarding the absorption profiles of the two pigments. On the other hand, we can take advantage of the measured properties of the intact monomer. For example, the brightness of the intact monomer is only 83% of the sum of its constituent pigments (α-state + β-state), which indicates energy redistribution toward the dimmer β-pigment site. We thus sought a FRET model that is most consistent with the collective behavior of the α+β state. Toward this end, we treated the ratio of energy transfer rates (kα→β/kβ→α) and absorption probability (Aβ/Aα) as free parameters and calculated the expected brightness and excited-state lifetime of the α+β state as the two parameters were varied (SI Appendix, Fig. S6). We found that best agreement with experimental data occurred when the α-to-β transfer rate is about 2 times faster than the β-to-α rate on the APC monomer (kα→β/kβ→α ≈ 2) and the β-pigment has a slightly lower absorption probability (Aβ/Aα ≈ 0.9). The large asymmetry in the bidirectional energy transfer rates is consistent with a direct Fӧrster equation calculation (kα→β = 5.4 ns−1, kβ→α = 3.0 ns−1, and a ratio of 1.8), in which we assumed the absorption lineshapes of the individual pigments to be identical to that of the monomer (SI Appendix, Fig. S4) with a peak absorbance ratio given by Aβ/Aα. We subsequently used these rates for further modeling.
Fig. 5.
A model of pigment organization in APC.
Using the measured FPol and the quantitative FRET model, we estimated the angular separation between the α- and β-pigments on the monomer. This is based on the interpretation that the modest depolarization of the α+β state (FPol ∼0.36) originates from bidirectional energy exchange between the nonparallel α- and β-pigment dipoles. In Fig. 3C, we calculated the expected (relative) FPol of the monomer as a function of the angular separation between the two pigment dipoles. Using this approach, our measured FPol corresponds to an angle of 29.5 ± 1.5° between the α- and β-pigment dipoles. We note that our optical estimate for the monomer is different from previous estimates from the crystal structure of the trimer (ref. 37, ∼0°), perhaps due to changes in the protein nanoenvironment and the precise electronic conformation of the chromophore during trimer formation.
The α-Pigment Is Actively Quenched in the Monomer.
Because the α-to-β energy transfer pathway is more efficient, the α-pigment in the intact monomer is effectively quenched. Using the quantitative FRET model, we calculated that the emission rate of the α-site on the monomer is reduced by ∼43% in the presence of the β-pigment. Intriguingly, our study revealed a previously unknown state (αQ) of the APC monomer, in which the α-pigment is observed in a quenched form. We hypothesize that this quenched α-state is produced by the β-pigment entering into a nonemissive but still absorptive state, likely a photoproduct of the PCB pigment (Fig. 5). The αQ-state is fairly common, as quenching was observed in ∼55% of the cases where β emission terminates. In this quenched state, the brightness of the α-pigment is reduced by ∼25%. From the reduction of the excited-state lifetime (αQ compared with α), we extracted a quenching rate of 0.2 ns−1. The finding that the α-pigment is actively quenched in an intact monomer, even after partial photodegradation of the β-pigment, is interesting and could have functional implications (discussed in Pigment Organization Principles in APC). On the other hand, our data do not show a quenched β-state, suggesting that the photoproduct of the α-pigment does not act as a quencher.
The α-Pigment Is the Site of 657-nm Emission in the Trimer.
When three monomers form a trimer, the pigments reorganize to generate a new absorption band at 650 nm. To trace the molecular origin of this spectral red shift, we first overlaid the emission characteristics of the α- and β-pigments in the monomer (α1 and β1), on top of the trimer map (Fig. 4A, “+” signs). Intriguingly, although the β features (β1) are preserved in the trimer map, the α-features (α1) mostly disappeared. This observation suggests that the β-pigment retains its emission characteristics after trimer assembly and is a common intermediate along the photodegradation pathway of the trimer while the α-pigment, on the other hand, either could not be observed emitting by itself (e.g., quenched) or has altered emission characteristics in the trimer following assembly. To test these hypotheses, we selected levels with a high emission FPol (>0.3) and analyzed their associated emission spectra (Fig. 4 B and C). These levels most likely represent trimers with only one (or two on the same constituent monomer) remaining emitter, because all other configurations activate energy transfer pathways across neighboring monomers, which, due to the geometry of the pigments in the trimer (37), would significantly lower FPol. Strikingly, we found two spectral populations in these levels. One population retains the emission spectrum of the β-pigment on the monomer (648-nm spectral CM) whereas the second population has a red-shifted spectrum centered at ∼657 nm that we label αR. Averaged full spectra of the two populations are shown in Fig. 4C (with absorption spectra explained in the legend). This red-shifted high FPol population is found only in the trimer. Taken together, we assign the origin of the 657-nm population to the spectrally altered α-pigment in the trimer, which shifted to the red upon trimer formation (Fig. 5).
Our model that assigns α as the site of the 657-nm emission is supported by resonance-enhanced coherent anti-Stokes Raman spectroscopy (38) and is consistent with spectral−structural analysis of APC trimers with a linker peptide (39), where structural perturbation of the β-pigment caused suppression of only the 620-nm bands but not the 650-nm band or the emission spectrum. Moreover, a recent structural study of a red-shifted APC variant (APC-B) (40) is also consistent with our model. There, the red-shifted, trimer-only band at ∼670 nm correlated well with an altered and more planer geometry of the α-pigment.
Our data disfavor the model that the bathochromic shift is the result of exciton band formation by the strongly coupled, near-degenerate α/β-pigment dimers at the monomer−monomer interface (16), because, if this were the case, the spectral content of the high FPol levels would have been identical to that of the monomer (Fig. 3). Recently, Womick and Moran (41, 42) proposed a vibronic-enhanced exciton model that interprets the strong ultrafast exciton signature of APC trimers (41, 43) on the basis of a strongly coupled dimer consisting of two highly nondegenerate (∼750 cm−1 gap in site energy) α- and β-pigments. Our model is fully consistent with their view. Still, the physicochemical nature of this trimerization-induced spectral red shift of the α-pigment remains to be fully elucidated. In addition to an increased coplanarity of the α-pigment, other factors, including hydrophobicity (44) or an altered hydrogen-bonding network of the D ring (40) induced by the contact of the neighboring monomer, have also been suggested to play a role. Interestingly, the spectral characteristics of ApcE (45), which is similar to the red-shifted α-pigment here, have been elucidated to originate from pigment conformation rather than excitonic splitting (46). A comparative structural analysis of ApcE with APC could potentially provide key molecular insights into the spectral shift of the α-pigment.
Emission Model of the Trimer.
Next, we examine the molecular origin behind the numerous intermediate states observed in the trimer photodynamics (Fig. 4A). We start by constructing a spectroscopic model based on the single-molecule data, bulk absorption profiles, and the crystal structure, with the following assumptions: (i) The β-pigment maintains the same spectroscopic properties after trimerization, including the propensity to photoconvert into a nonemissive quencher. (ii) The α-pigment is red-shifted in the trimer (subsequently referred to as αR) and acts as the major emissive species on a pristine trimer. We estimated the absorption profile of αR by subtracting one-half of the monomer absorption profile (SI Appendix, Fig. S4) (normalized to absorbance at 280 nm) from that of the trimer (Fig. 4C). (iii) Pigment dipoles are oriented according to the crystal structure (37). (iv) Pigments are coupled thorough a FRET mechanism. We then enumerated all possible photophysical configurations, taking into account that β can be either “on”, “bleached” or “a quencher” and αR can be either “on” or “bleached.” Specifically, there are 23, 31, and 13 unique configurations for zero, one, and two β-quenchers (a total of 67 configurations). For each configuration, we calculated the expected brightness, FPol, emission spectrum, and excited-state lifetime by solving a multipigment FRET network model using the site properties (SI Appendix, SI Materials and Methods and Fig. S9). Repeated interaction with the excitation laser gradually transitions the protein from one configuration to another.
This simple model captures many essential features of the trimer data. First, the FPol value of the intact trimer is correctly predicted. Our model interprets the emission from the intact APC (SI Appendix, Table S1) as originating from direct excitation of αR, efficient β-to-αR energy transfer within the closely spaced αR/β-pairs and energy equilibration between the three identical αR/β-pairs. Second, the model successfully predicts the overall shape of the multiparameter map. Initially, β efficiently transfers to αR of the αR/β-pair due to proximity and large spectral overlap. Once the acceptor photobleaches, β turns to αR on the same monomer as the FRET acceptor and subsequently to αR at the distal end. As a result, the emission spectrum of the trimer is resistant to photodamage until all three αR-pigments on the complex photobleach. Meanwhile, the switching of FRET pathways is sensed as changes in FPol. The large heterogeneity in FPol (at low brightness) reflects the multitude of energy flow pathways on the partially bleached complexes, sampled by stochastic photobleaching. Third, some trimers display large, sudden decreases in fluorescent brightness similar to that previously observed (11, 13, 22) (SI Appendix, Fig. S8F). This behavior is understood in our model as the generation of nonemissive quenchers localized at the β-sites, analogous to the αQ-state in the monomers. We calculated that a single β-quencher can reduce the brightness of the whole complex by 80% (SI Appendix, Fig. S9, configuration A′), which is consistent with experimental observations. Nevertheless, our simple model cannot explain the slight blue shift (∼1 nm) of spectral CM observed at intermediate brightness levels. It is possible that light-induced conformational changes (22) play a role in these cases.
Pigment Organization Principles in APC.
In cyanobacteria and red algae, APC resides in the core of a multiprotein antenna complex called the phycobilisome. It has two major functions: harvesting sunlight in the red spectral window and funneling excitation energy to the chlorophylls in the reaction center (47). The ∼30-nm red shift in absorption (∼20-nm red shift in emission) upon trimer formation gives APC its unique optical properties and is thus of critical importance to the protein’s function. Our data corroborate the view that this emergent property originates from a site-specific conformational alteration of the PCB pigment. Moreover, we pinpoint the spectral change to the three α-sites. These results might also be important in understanding the orange carotenoid protein-mediated nonphotochemical quenching pathway (48) in cyanobacteria, in which APC was recently found to be the quenching site of the phycobilisome (17) (SI Appendix, SI Results and Discussion).
Given that the α-pigment defines the spectral signature as well as the function of APC, it is intriguing to discover here, for the first time, that the β-pigment, either itself or in a photoactivated form, acts as a quencher to the α-pigment in the preassembly, monomer form of the protein. We propose here that β serves a protective role that reduces α’s exposure in the excited state, from which premature photodamage could occur. This mechanism is likely more important in the early stages of the APC maturation process (49, 50): i.e., after monomer formation but before full integration of the trimeric APC into the phycobilisome.
Conclusion
A fascinating feature of antenna complexes is that nature uses seemingly uniform building blocks and reorganizes them to produce synergistic and diverse functions. Unraveling these principles requires nonperturbative investigation of the individual components and tracking of their properties during reorganization. We have shown here that a single-molecule, multiparameter fluorescence spectroscopy approach combined with anti-Brownian trapping achieves a bottom-up dissection of the previously unknown (to our knowledge) functional roles of the individual pigments in APC. This methodology should be broadly applicable for interrogating other naturally existing or artificial multichromophore systems.
Materials and Methods
A detailed description of the materials and methods is given in SI Appendix, SI Materials and Methods. Briefly, the ABEL trap was implemented as previously described (23), with the addition of polarization sensitive optics to measure FPol (SI Appendix, Fig. S1). APC from Spirulina sp. was purchased from Prozyme. A pure monomer population was prepared by the spontaneous dissociation of the wild-type protein at 1 nM concentration for >15 h (SI Appendix, Fig. S3). Measurements on a pure trimer population utilized cross-linked APCs to prevent dissociation at single-molecule concentrations.
Supplementary Material
Acknowledgments
The authors thank Randall Goldsmith for early contributions to this project, Gabriela Schlau-Cohen for helpful discussion, and Nick Bertone from PicoQuant for the loan of two τ-SPAD detectors. This work is supported in part by the Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through Grant DE-FG02-07ER15892.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 13751.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1514027112/-/DCSupplemental.
References
- 1.Blankenship RE. Molecular Mechanisms of Photosynthesis. 2nd Ed Wiley-Blackwell; Oxford: 2014. [Google Scholar]
- 2.Croce R, van Amerongen H. Natural strategies for photosynthetic light harvesting. Nat Chem Biol. 2014;10(7):492–501. doi: 10.1038/nchembio.1555. [DOI] [PubMed] [Google Scholar]
- 3.van Grondelle R, Dekker JP, Gillbro T, Sundstrom V. Energy transfer and trapping in photosynthesis. Biochim Biophys Acta. 1994;1187(1):1–65. [Google Scholar]
- 4.Anna JM, Scholes GD, van Grondelle R. A little coherence in photosynthetic light harvesting. Bioscience. 2014;64(1):14–25. [Google Scholar]
- 5.Scheer H. Biliproteins. Angew Chem Int Ed Engl. 1981;20(3):241–261. [Google Scholar]
- 6.Adir N. Elucidation of the molecular structures of components of the phycobilisome: reconstructing a giant. Photosynth Res. 2005;85(1):15–32. doi: 10.1007/s11120-004-2143-y. [DOI] [PubMed] [Google Scholar]
- 7.Miller RA, Presley AD, Francis MB. Self-assembling light-harvesting systems from synthetically modified tobacco mosaic virus coat proteins. J Am Chem Soc. 2007;129(11):3104–3109. doi: 10.1021/ja063887t. [DOI] [PubMed] [Google Scholar]
- 8.MacColl R, Berns DS, Koven NL. Effect of salts on C-phycocyanin. Arch Biochem Biophys. 1971;146(2):477–482. doi: 10.1016/0003-9861(71)90151-2. [DOI] [PubMed] [Google Scholar]
- 9.Debreczeny MP, Sauer K, Zhou J, Bryant DA. Monomeric C-phycocyanin at room temperature and 77 K: Resolution of the absorption and fluorescence spectra of the individual chromophores and the energy-transfer rate constants. J Phys Chem. 1993;97(38):9852–9862. [Google Scholar]
- 10.Bopp MA, Jia Y, Li L, Cogdell RJ, Hochstrasser RM. Fluorescence and photobleaching dynamics of single light-harvesting complexes. Proc Natl Acad Sci USA. 1997;94(20):10630–10635. doi: 10.1073/pnas.94.20.10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ying L, Xie XS. Fluorescence spectroscopy, exciton dynamics, and photochemistry of single allophycocyanin trimers. J Phys Chem B. 1998;102(50):10399–10409. [Google Scholar]
- 12.van Oijen AM, Ketelaars M, Kohler J, Aartsma TJ, Schmidt J. Unraveling the electronic structure of individual photosynthetic pigment−protein complexes. Science. 1999;285(5426):400–402. doi: 10.1126/science.285.5426.400. [DOI] [PubMed] [Google Scholar]
- 13.Loos D, Cotlet M, De Schryver F, Habuchi S, Hofkens J. Single-molecule spectroscopy selectively probes donor and acceptor chromophores in the phycobiliprotein allophycocyanin. Biophys J. 2004;87(4):2598–2608. doi: 10.1529/biophysj.104.046219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rutkauskas D, Novoderezkhin V, Cogdell RJ, van Grondelle R. Fluorescence spectral fluctuations of single LH2 complexes from Rhodopseudomonas acidophila strain 10050. Biochemistry. 2004;43(15):4431–4438. doi: 10.1021/bi0497648. [DOI] [PubMed] [Google Scholar]
- 15.Krüger TP, Novoderezhkin VI, Ilioaia C, van Grondelle R. Fluorescence spectral dynamics of single LHCII trimers. Biophys J. 2010;98(12):3093–3101. doi: 10.1016/j.bpj.2010.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.MacColl R. Allophycocyanin and energy transfer. Biochim Biophys Acta. 2004;1657(2-3):73–81. doi: 10.1016/j.bbabio.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 17.Tian L, et al. Site, rate, and mechanism of photoprotective quenching in cyanobacteria. J Am Chem Soc. 2011;133(45):18304–18311. doi: 10.1021/ja206414m. [DOI] [PubMed] [Google Scholar]
- 18.Cohen AE, Moerner WE. Suppressing Brownian motion of individual biomolecules in solution. Proc Natl Acad Sci USA. 2006;103(12):4362–4365. doi: 10.1073/pnas.0509976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schlau-Cohen GS, Bockenhauer S, Wang Q, Moerner WE. Single-molecule spectroscopy of photosynthetic proteins in solution: Exploration of structure–function relationships. Chem Sci. 2014;5(8):2933–2939. [Google Scholar]
- 20.Wang Q, Goldsmith RH, Jiang Y, Bockenhauer SD, Moerner WE. Probing single biomolecules in solution using the anti-Brownian electrokinetic (ABEL) trap. Acc Chem Res. 2012;45(11):1955–1964. doi: 10.1021/ar200304t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Q, Moerner WE. Single-molecule motions enable direct visualization of biomolecular interactions in solution. Nat Methods. 2014;11(5):555–558. doi: 10.1038/nmeth.2882. [DOI] [PubMed] [Google Scholar]
- 22.Goldsmith RH, Moerner WE. Watching conformational- and photo-dynamics of single fluorescent proteins in solution. Nat Chem. 2010;2(3):179–186. doi: 10.1038/nchem.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Q, Moerner WE. Lifetime and spectrally resolved characterization of the photodynamics of single fluorophores in solution using the anti-Brownian electrokinetic trap. J Phys Chem B. 2013;117(16):4641–4648. doi: 10.1021/jp308949d. [DOI] [PubMed] [Google Scholar]
- 24.Lakowicz JR. Principles of Fluorescence Spectroscopy. Springer; New York: 2006. [Google Scholar]
- 25.Ha T, Laurence TA, Chemla DS, Weiss S. Polarization spectroscopy of single fluorescent molecules. J Phys Chem B. 1999;103(33):6839–6850. [Google Scholar]
- 26.Schaffer J, et al. Identification of single molecules in aqueous solution by time-resolved fluorescence anisotropy. J Phys Chem A. 1999;103(3):331–336. [Google Scholar]
- 27.Tinnefeld P, Sauer M. Branching out of single-molecule fluorescence spectroscopy: Challenges for chemistry and influence on biology. Angew Chem Int Ed Engl. 2005;44(18):2642–2671. doi: 10.1002/anie.200300647. [DOI] [PubMed] [Google Scholar]
- 28.Börner R, Kowerko D, Krause S, von Borczyskowski C, Hübner CG. Efficient simultaneous fluorescence orientation, spectrum, and lifetime detection for single molecule dynamics. J Chem Phys. 2012;137(16):164202. doi: 10.1063/1.4759108. [DOI] [PubMed] [Google Scholar]
- 29.Cohen-Bazire G, Béguin S, Rimon S, Glazer AN, Brown DM. Physico-chemical and immunological properties of allophycocyanins. Arch Microbiol. 1977;111(3):225–238. doi: 10.1007/BF00549359. [DOI] [PubMed] [Google Scholar]
- 30.Füglistaller P, Mimuro M, Suter F, Zuber H. Allophycocyanin complexes of the phycobilisome from Mastigocladus laminosus. Influence of the linker polypeptide L8.9C on the spectral properties of the phycobiliprotein subunits. Biol Chem Hoppe Seyler. 1987;368(4):353–367. doi: 10.1515/bchm3.1987.368.1.353. [DOI] [PubMed] [Google Scholar]
- 31.Canaani OD, Gantt E. Circular dichroism and polarized fluorescence characteristics of blue-green algal allophycocyanins. Biochemistry. 1980;19(13):2950–2956. doi: 10.1021/bi00554a021. [DOI] [PubMed] [Google Scholar]
- 32.Bischoff M, et al. Excited-state processes in phycocyanobilin studied by femtosecond spectroscopy. J Phys Chem B. 2000;104(8):1810–1816. [Google Scholar]
- 33.Kim PW, Rockwell NC, Martin SS, Lagarias JC, Larsen DS. Dynamic inhomogeneity in the photodynamics of cyanobacterial phytochrome Cph1. Biochemistry. 2014;53(17):2818–2826. doi: 10.1021/bi500108s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riter RR, Edington MD, Beck WF. Protein-matrix solvation dynamics in the α subunit of C-phycocyanin. J Phys Chem. 1996;100(33):14198–14205. [Google Scholar]
- 35.Shiu YJ, et al. A transient absorption study of allophycocyanin. Proc Indian Acad Sci (Chem Sci) 2002;114(6):611–621. [Google Scholar]
- 36.Tinnefeld P, et al. Higher-excited-state photophysical pathways in multichromophoric systems revealed by single-molecule fluorescence spectroscopy. ChemPhysChem. 2004;5(11):1786–1790. doi: 10.1002/cphc.200400325. [DOI] [PubMed] [Google Scholar]
- 37.Brejc K, Ficner R, Huber R, Steinbacher S. Isolation, crystallization, crystal structure analysis and refinement of allophycocyanin from the cyanobacterium Spirulina platensis at 2.3 Å resolution. J Mol Biol. 1995;249(2):424–440. doi: 10.1006/jmbi.1995.0307. [DOI] [PubMed] [Google Scholar]
- 38.Schneider S, et al. Resonance-enhanced CARS spectroscopy of biliproteins. Influence of aggregation and linker proteins on chromophore structure in allophycocyanin (Mastigocladus laminosus) Photochem Photobiol. 1995;62(5):847–854. [Google Scholar]
- 39.Reuter W, Wiegand G, Huber R, Than ME. Structural analysis at 2.2 Å of orthorhombic crystals presents the asymmetry of the allophycocyanin-linker complex, AP·LC7.8, from phycobilisomes of Mastigocladus laminosus. Proc Natl Acad Sci USA. 1999;96(4):1363–1368. doi: 10.1073/pnas.96.4.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peng PP, et al. The structure of allophycocyanin B from Synechocystis PCC 6803 reveals the structural basis for the extreme redshift of the terminal emitter in phycobilisomes. Acta Crystallogr D Biol Crystallogr. 2014;70(Pt 10):2558–2569. doi: 10.1107/S1399004714015776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Womick JM, Moran AM. Exciton coherence and energy transport in the light-harvesting dimers of allophycocyanin. J Phys Chem B. 2009;113(48):15747–15759. doi: 10.1021/jp907644h. [DOI] [PubMed] [Google Scholar]
- 42.Womick JM, Moran AM. Vibronic enhancement of exciton sizes and energy transport in photosynthetic complexes. J Phys Chem B. 2011;115(6):1347–1356. doi: 10.1021/jp106713q. [DOI] [PubMed] [Google Scholar]
- 43.Edington MD, Riter RE, Beck WF. Evidence for coherent energy-transfer in allophycocyanin trimers. J Phys Chem. 1995;99(43):15699–15704. [Google Scholar]
- 44.McGregor A, Klartag M, David L, Adir N. Allophycocyanin trimer stability and functionality are primarily due to polar enhanced hydrophobicity of the phycocyanobilin binding pocket. J Mol Biol. 2008;384(2):406–421. doi: 10.1016/j.jmb.2008.09.018. [DOI] [PubMed] [Google Scholar]
- 45.Zhao K-H, et al. Reconstitution of phycobilisome core-membrane linker, LCM, by autocatalytic chromophore binding to ApcE. Biochim Biophys Acta. 2005;1706(1-2):81–87. doi: 10.1016/j.bbabio.2004.09.008. [DOI] [PubMed] [Google Scholar]
- 46.Long S, et al. Single-molecule spectroscopy and femtosecond transient absorption studies on the excitation energy transfer process in ApcE(1-240) dimers. Phys Chem Chem Phys. 2015;17(20):13387–13396. doi: 10.1039/c5cp01687h. [DOI] [PubMed] [Google Scholar]
- 47.Liu H, et al. Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science. 2013;342(6162):1104–1107. doi: 10.1126/science.1242321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kirilovsky D. Modulating energy arriving at photochemical reaction centers: Orange carotenoid protein-related photoprotection and state transitions. Photosynth Res. 2014;126(1):3–17. doi: 10.1007/s11120-014-0031-7. [DOI] [PubMed] [Google Scholar]
- 49.Anderson LK, Toole CM. A model for early events in the assembly pathway of cyanobacterial phycobilisomes. Mol Microbiol. 1998;30(3):467–474. doi: 10.1046/j.1365-2958.1998.01081.x. [DOI] [PubMed] [Google Scholar]
- 50.Scheer H, Zhao K-H. Biliprotein maturation: The chromophore attachment. Mol Microbiol. 2008;68(2):263–276. doi: 10.1111/j.1365-2958.2008.06160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.